Significance
p63, the sister homolog of p53, is a master regulator of epithelial stem cell (SC) biology. p63 is indeed intimately implicated in the maintenance of the self-renewal capacity of stratified epithelia and their derivatives, including the mammary gland. Although the physiological role of p63 in normal mammary SCs is now acknowledged, proof of its implications in breast cancer SCs remains elusive. Here, we find that mammary cancer stem cells (CSCs) possess increased levels of p63 expression compared with normal progenitors. p63 promotes self-renewal and expansion of mammary CSCs and breast tumor growth in vivo. Additionally, this study provides a link between p63 and the Sonic Hedgehog signaling pathway in the regulation of breast cancer stemness.
Keywords: mammary stem cells, p53 family, breast cancer
Abstract
The predominant p63 isoform, ΔNp63, is a master regulator of normal epithelial stem cell (SC) maintenance. However, in vivo evidence of the regulation of cancer stem cell (CSC) properties by p63 is still limited. Here, we exploit the transgenic MMTV-ErbB2 (v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2) mouse model of carcinogenesis to dissect the role of p63 in the regulation of mammary CSC self-renewal and breast tumorigenesis. ErbB2 tumor cells enriched for SC-like properties display increased levels of ΔNp63 expression compared with normal mammary progenitors. Down-regulation of p63 in ErbB2 mammospheres markedly restricts self-renewal and expansion of CSCs, and this action is fully independent of p53. Furthermore, transplantation of ErbB2 progenitors expressing shRNAs against p63 into the mammary fat pads of syngeneic mice delays tumor growth in vivo. p63 knockdown in ErbB2 progenitors diminishes the expression of genes encoding components of the Sonic Hedgehog (Hh) signaling pathway, a driver of mammary SC self-renewal. Remarkably, p63 regulates the expression of Sonic Hedgehog (Shh), GLI family zinc finger 2 (Gli2), and Patched1 (Ptch1) genes by directly binding to their gene regulatory regions, and eventually contributes to pathway activation. Collectively, these studies highlight the importance of p63 in maintaining the self-renewal potential of mammary CSCs via a positive modulation of the Hh signaling pathway.
Deregulation of self-renewal may be a crucial event underlying tumorigenesis. Cancer stem cells (CSCs) are thought to use numerous signaling pathways underlying normal SC biology aberrantly (1). As a result, CSCs acquire unlimited self-renewal potential and drive tumor growth and metastases. Thus, unveiling the molecular basis of stem cell (SC) self-renewal may be extremely relevant in understanding how this process contributes to cancer development.
Emerging evidence intimately links the p53 sister homolog tumor protein p63 (p63) to SC biology (2–4). The TP63 gene is expressed as multiple protein isoforms (5). The use of alternative transcription start sites (TSSs) produces transactivating (TA) isoforms that contain an N-terminal exon encoding a p53-like transactivation domain (TAD) and ΔN isoforms partially lacking this domain. ΔNp63 proteins can directly regulate transcription due to the presence of alternative TADs (6). Both the TAp63 and ΔNp63 transcripts are differentially spliced at their 3′ ends to generate proteins with unique C termini designated α, β, and γ (5), whose biology has not yet been deeply studied.
ΔNp63, the predominant p63 isoform in epithelial tissues, is indispensable for preserving the self-renewal capacity of SCs in adult stratified epithelia and glandular structures (2–4). In normal breast tissue, the expression of ΔNp63 is strictly restricted to the basal/myoepithelial compartment (7), where SCs are expected to reside. The requirement for p63 in the morphogenesis of the mammary gland was originally suggested by the observation that complete abrogation of TP63 gene function in animal models causes loss of the mammary epithelium (8, 9). Emerging evidence is pointing toward a role for p63 in regulating stemness in the adult mammary gland (3, 10, 11), although the clinical relevance of p63 as a SC marker in breast cancer remains poorly understood.
Ectopic expression of ΔNp63 in breast carcinoma cell lines increases the percentage of CSC-like subpopulations and leads to augmented cancer cell clonogenicity and tumor xenograft incidence (12). Accordingly, ΔNp63 is overexpressed in the basal-like subtype of breast cancer (13, 14), in which a specific role for ΔNp63 in regulating CSC activity has been reported very recently (11). In differentiated mammary tumors, such as luminal-like breast carcinomas, ΔNp63 expression is instead reduced during cancer progression (7). In invasive ductal carcinomas, ΔNp63 positivity is generally restricted to rare tumor cells. Occasional nuclear positivity for ΔNp63 in malignant cells is consistent with a stem/progenitor identity and suggests that ΔNp63 enrichment would confer increased self-renewal potential. Here, we demonstrate that ΔNp63 promotes the stemness properties of luminal-type breast cancer progenitors and identify the Hedgehog (Hh) signaling pathway as a downstream effector of p63 activity in mammary CSCs.
Results
Mammary Cancer Progenitors Are Enriched for ΔNp63 Expression.
To dissect the role of p63 in breast CSC self-renewal, we used transgenic mice expressing mammary gland-targeted ErbB2 (MMTV-ErbB2). Expression of the activated ErbB2 oncogene in murine mammary epithelium efficiently induces multifocal luminal-like mammary carcinomas (15). The ErbB2 tumor has been shown to classify with human luminal-type carcinoma (16, 17), and represents a well-established SC model of mammary carcinogenesis (15). The expression pattern of p63 in the ErbB2-derived mammary tumors closely mirrors the expression pattern of p63 in human ductal carcinomas (7). p63 positivity indeed diminishes during progression from hyperplasia to invasive lesions that ultimately display rare immunoreactive tumor cells (Fig. S1 A–D). Interestingly, we found that a fraction of the p63-expressing cells were also positive for stem cell antigen 1 (Sca-1) (Fig. S1E), which identifies a CSC population highly enriched for tumorigenic capability in this breast cancer model (15). Thus, p63 positivity in tumor cells is consistent with a stem/progenitor identity in the ErbB2 tumors.
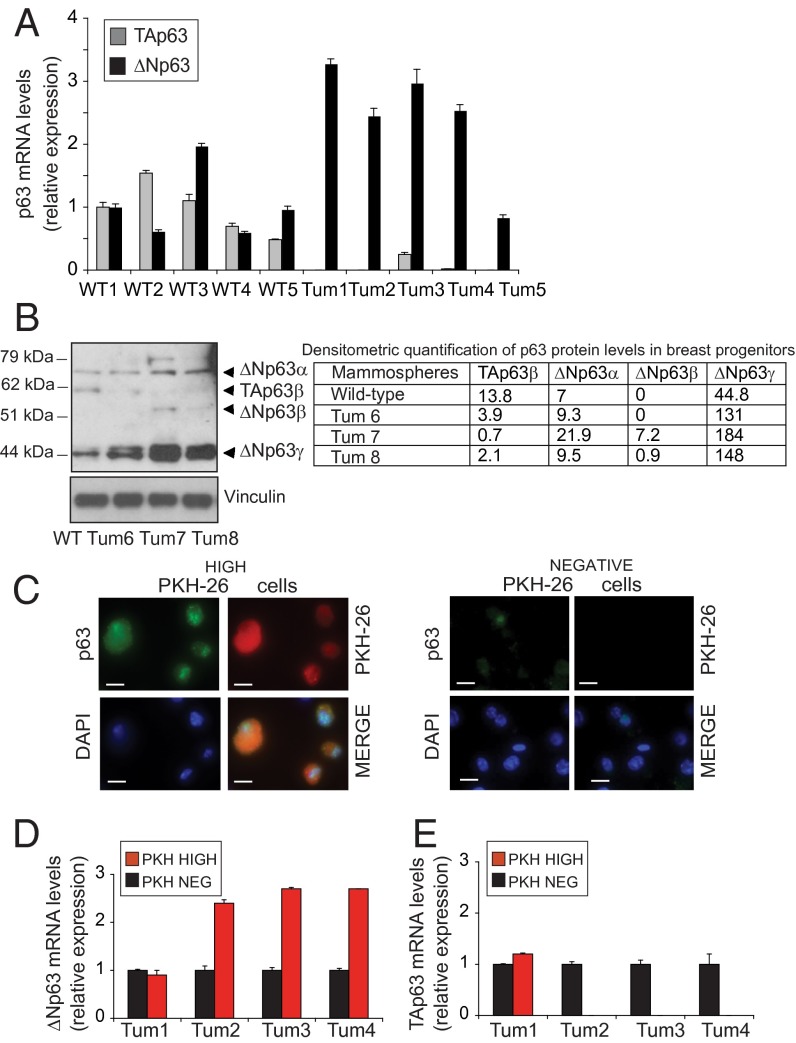
To examine the expression pattern of p63 in mammary CSCs, we took advantage of the mammosphere culture system, which enriches for self-renewing tumorigenic progenitors and mimics the original tumor upon transplantation into syngeneic mice (18). The ΔNp63 transcript (Fig. 1A) and protein (Fig. 1B) levels were consistently increased in ErbB2 mammospheres compared with their normal counterparts. By contrast, TAp63 expression was very low or absent in the ErbB2 tumor spheres (Fig. 1 A and B and Fig. S1 F and H), indicating that ΔNp63 is the predominant p63 isoform. As shown by Western blot analyses (Fig. 1B and Fig. S1 F and H), the expression pattern of the ΔNp63 isoforms was very heterogeneous in distinct tumor sphere preparations, with the ΔNp63γ variant being the most abundant isoform. To assess whether ErbB2 CSCs are enriched for p63 expression, we isolated quiescent progenitors on the basis of their ability to retain the lipophilic fluorescent dye PKH-26 (15, 19). Immunostaining of PKH-26negative (progenitors) and PKH-26high (stem) epithelial cells revealed that p63 was preferentially expressed in the quiescent population (Fig. 1C). As shown in Fig. 1D, the PKH-26high subpopulations displayed augmented levels of ΔNp63 transcripts compared with the PKH-26negative cells. TAp63 expression was barely detectable in the PKH-26high cells (Fig. 1E). These findings reveal that ΔNp63 is the primary p63 isoform expressed in ErbB2 CSCs.
Fig. 1.
Expression pattern of p63 in mammary cancer progenitors. (A) ΔNp63 and TAp63 mRNA expression levels in wild-type (WT) and ErbB2 (Tum) secondary mammospheres measured by quantitative real-time PCR (qPCR). TATA box-binding protein (TBP) mRNA levels were used as an internal normalization control. Data are presented as mean ± SD. Five independent experiments were performed for each mouse genotype. (B, Left) Immunoblot (IB) analysis of p63 protein levels in total cellular extracts of control (WT) and ErbB2 (Tum) mammospheres obtained from three independent tumors. (B, Right) Densitometric quantification of p63 protein levels in WT and ErbB2 spheres. Band intensity was normalized against vinculin. Values are expressed as the ratio of p63/vinculin amount × 100. (C) Representative photographs of p63 immunofluorescence staining of PKH-26high (Left) and PKH-26negative (Right) cells isolated from ErbB2 spheres. Nuclei were counterstained with DAPI. Green indicates antibody staining, red indicates PKH fluorescence, and blue indicates DAPI. (Scale bars: 10 μm.) Transcript levels of ΔNp63 (D) and TAp63 (E) in PKH-26high and PKH-26negative populations that were FACS-sorted from four independent labeled ErbB2 mammosphere preparations. TBP mRNA levels were used as an internal normalization control. Data are presented as mean ± SD.
Loss of ΔNp63 Impairs Self-Renewal and Expansion of Mammary CSCs.
Serial sphere-forming capacity represents a valid surrogate for assessing CSC self-renewal (18). To test whether ΔNp63 affects the self-renewal potential of mammary CSCs, we transduced ErbB2 secondary mammospheres with lentiviral shRNA targeting p63 mRNA. Assessment of gene knockdown revealed that ΔNp63 mRNA (Fig. 2A, Right and Fig. S1G) and protein (Fig. S1H) levels were down-regulated by ∼80% upon short hairpin p63 (shp63) delivery. Functional inactivation of ΔNp63 in shp63-transduced ErbB2 progenitors was confirmed by reduced transcript levels of its direct target genes (Fig. S1I).
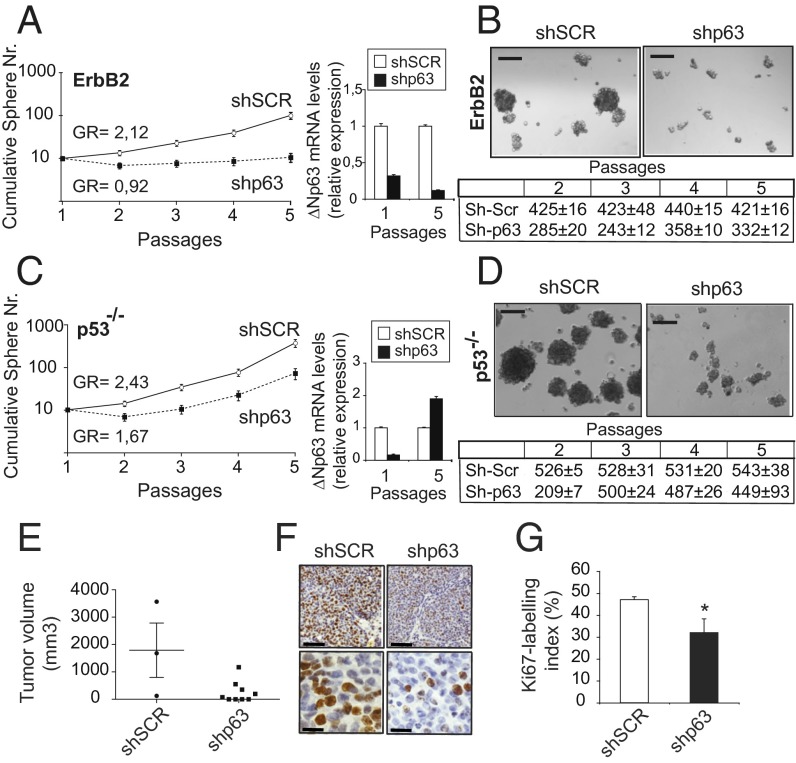
Fig. 2.
p63 supports mammary CSC self-renewal and mammary tumor growth. (A, Left) Semilogarithmic plotting of cumulative sphere number (±SD of quadruplicates) during in vitro passaging of control scrambled (shSCR) and p63-depleted (shp63) ErbB2 spheres. Data are representative of four individual experiments. GR, growth rate. (A, Right) ΔNp63 mRNA levels were measured at passages 1 and 5. TBP expression was used as an internal control. (B, Upper) Representative phase-contrast photographs of shSCR and shp63 ErbB2 spheres. (Scale bars: 100 μm.) (B, Lower) Average mammosphere size during serial replatings of shSCR and shp63 ErbB2 progenitors, calculated as cell number per sphere ± SD (P < 0.01). (C, Left) Cumulative sphere number (±SD of quadruplicates) of shSCR and shp63 mammospheres obtained from the p53 null (−/−) mammary epithelium. (C, Right) Data are representative of three individual experiments. Relative ΔNp63 mRNA levels were measured at passages 1 and 5. (D, Upper) Representative photographs of control and p63-depleted p53−/− spheres. (Scale bars: 100 μm.) (D, Lower) Average size (±SD) of p53−/− mammospheres expressing shRNA-targeting p63 relative to scrambled controls (P < 0.01). (E) Growth rate of secondary breast tumors arising from transplantation of shSCR or shp63 ErbB2 epithelial cells into the mammary fat pad of female syngeneic recipients. Ten weeks postinjection, all animals were killed and tumor volume was quantified. Data represent mean ± SEM (P < 0.05). (F) Representative Ki-67 staining of shSCR and shp63 transplanted tumors. (Scale bars: Upper, 100 μm; Lower, 50 μm). (G) Proliferative activity of shSCR and shp63 transplanted tumors assessed as the Ki-67 labeling index. Error bars represent mean ± SD. Data were generated from three animals from each experimental group. *P < 0.01.
Depletion of p63 caused a significant decrease in the number of sphere-forming cells during serial passages (Fig. 2A, Left), demonstrating that the self-renewal capacity of tumor progenitors was impaired in the absence of p63. Furthermore, the size of shp63 mammospheres was reduced compared with the controls (Fig. 2B), suggesting that p63 down-regulation may negatively affect either the frequency of CSCs or their proliferation potential.
To corroborate further the role of ΔNp63 in the regulation of breast cancer stemness, we performed isoform-specific inactivation of TAp63 or ΔNp63 in the human HCC1937 breast cancer cell line that expresses both isoforms at the endogenous level (Fig. S2A). Specific loss of ΔNp63, but not TAp63 (Fig. S2B), decreased the ability of HCC1937 cells to form mammospheres (Fig. S2C). Similarly, transduction of MCF-7 cells, lacking detectable endogenous TAp63 mRNA (Fig. S2A), with a pan-p63 shRNA-harboring lentivirus significantly reduced their mammosphere-forming efficiency (Fig. S2 D and E). Taken together, these findings indicate that ΔNp63, but not TAp63, contributes to sustain mammary CSC expansion by promoting CSC self-renewal and proliferation.
Because p63 and p53 physically interact (20), ΔNp63 can act in a dominant negative fashion to inhibit p53 tumor-suppressive properties. Hence, one potential mechanism by which ΔNp63 may preserve self-renewal is by functionally counteracting p53, which crucially controls the replicative properties of mammary CSCs (15). To test this possibility, we down-regulated p63 in secondary mammospheres obtained from the mammary gland of p53 KO mice. We observed that p63 depletion impaired the self-renewing ability of mammary progenitors (Fig. 2C, Left) and reduced sphere size (Fig. 2D) even in a p53 null background. These findings demonstrate that p63 sustains mammary CSC replicative potential through a p53-independent mechanism.
Notably, down-regulation of p63 expression was maintained throughout serial replatings in both ErbB2 and p53 null progenitors. However, at later passages, we always observed re-expression of p63 correlating with increased self-renewing potential (e.g., Fig. 2C, Right), indicating that a positive selection reactivated the self-renewal of CSCs.
Depletion of p63 in ErbB2 Progenitors Attenuates Mammary Tumor Growth.
To assess whether loss of p63 in mammary cancer progenitors may halt ErbB2-driven tumorigenesis, we used p63-depleted spheres for in vivo transplantation assays. We found that depletion of p63 markedly suppressed breast cancer growth in vivo relative to control mice (Fig. 2E). Tumor growth inhibition was paralleled by decreased frequency of Ki-67–positive nuclei in shp63 compared with short hairpin scrambled (shSCR) tumors (Fig. 2 F and G).
To determine whether reactivation of p63 might also occur in vivo in the shp63-transplanted tumors, we evaluated p63 immunoreactivity in tumor sections. Interestingly, we observed that the percentage of p63-positive cells was comparable in shp63 (0.025% ± 0.02) and control (0.044% ± 0.017) tumors (P = 0.3). Thus, given the similarity to the mammosphere culture system, the proliferation of residual p63 proficient cells within the implanted shp63 cell population might have overtaken the p63 KO during tumor progression.
p63 Regulates the Hh Signaling Pathway in Mammary CSCs.
The Hh signaling pathway is a relevant driver of epithelial SC self-renewal (21). The Hh signaling cascade is initiated when one of the three secreted ligands, Sonic Hedgehog (SHH), Desert Hedgehog, or Indian Hedgehog (IHH), binds and inactivates the Patched1 (PTCH1) receptor. Upon binding, PTCH1 relieves its inhibition on Smoothened (SMO), which then activates the Glioma-associated oncogene homologues 1, 2, and 3 (GLI 1,2,3) family of transcription factors. The expression levels of Hh target genes, such as Gli1 and Ptch1, are used as markers of active canonical Hh signaling (22). The effects of Hh signaling on mammary SC self-renewal have been ascribed to the transcriptional activation of the SC marker B-cell–specific Moloney murine leukemia virus Integration site1 (BMI1) (21). Aberrant regulation of the Hh pathway promotes self-renewal and expansion of CSCs in several human cancers (21–24).
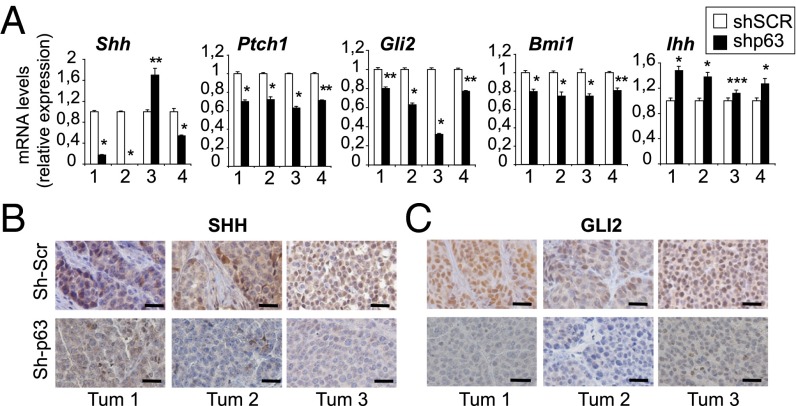
Several reports demonstrated that in tumor cell lines, the p63 isoforms regulate the transcription of some key players of the Hh signaling (25, 26). However, the potential contribution of the p63-Hh cross-talk to breast cancer stemness remains unexplored. Therefore, we examined whether p63 may sustain mammary CSC self-renewal through the activation of the Hh pathway. Interestingly, p63 knockdown in ErbB2 progenitors resulted in altered expression of various Hh signaling components (Fig. 3A). Shh, Ptch1, Gli2, and Bmi1 transcript levels were reduced in shp63 compared with control mammospheres. Conversely, the expression of Ihh was up-regulated in shp63 compared with shSCR spheres. This finding is in accordance with the observation that Ihh is targeted by ΔNp63 for transcriptional repression in quiescent normal mammary SCs (3). The expression of other Hh molecules, such GLI1 and SMO, was not significantly affected by p63 loss. In addition, histological examination of p63 shRNA-expressing tumor transplants revealed in vivo down-regulation of SHH (Fig. 3B) and GLI2 (Fig. 3C) compared with control tumors.
Fig. 3.
p63 activates the Hh signaling pathway in mammary CSCs. (A) Shh, Ptch1, Gli2, Bmi1, and Ihh gene expression was quantified by qPCR using four independent preparations of RNA obtained from shSCR and shp63 ErbB2 mammospheres. Data are expressed as mean ± SD and are normalized to TBP expression. *P < 0.001; **P < 0.003; ***P < 0.005. Tumors from shSCR- and shp63-transplanted mice were analyzed using SHH (B) and GLI2 (C) immunohistochemistry. Representative images from three independent tumors for each experimental group are presented. (Scale bars: 100 μm.)
Mammary SC self-renewal is regulated by a number of additional signaling pathways, including Notch, TGF-β, and Wnt/β-catenin, which are aberrantly activated in CSCs (27). Interestingly, some of the components of these pathways are under the transcriptional control of p63. For instance, p63 regulates the expression of several molecules of the Notch pathway (28, 29). Moreover, in basal-like breast tumors, ΔNp63 regulates CSC properties through the induction of Fzd7 expression and subsequent activation of the Wnt/β-catenin pathway (11). To assess whether p63 also promotes SC activity through the modulation of these two pathways, we measured the expression levels of Notch receptors and Notch target genes, as well as the expression levels of Fzd7, in p63-depleted ErbB2 mammospheres. We found that silencing of p63 does not significantly affect the expression of any of these genes (Fig. S3), demonstrating that neither Notch nor Wnt/β-catenin is a relevant signaling mediator of p63 stemness activity in our model system. This observation may be in line with a divergent cluster-of-cluster-assignment evolution of basal vs. luminal breast cancers (14).
p63 Directly Regulates the Transcription of Hh Signaling Components.
We then asked whether p63 directly binds to the regulatory regions of some of the Hh components. Genome-wide profiling of p63-binding sites by ChIP-sequencing analysis of human cells (30) (Fig. S4 A–C) and mouse cells (31) (Fig. S4G) revealed peaks of p63 binding to regions encompassing or within the Shh, Gli2, and Ptch1 loci.
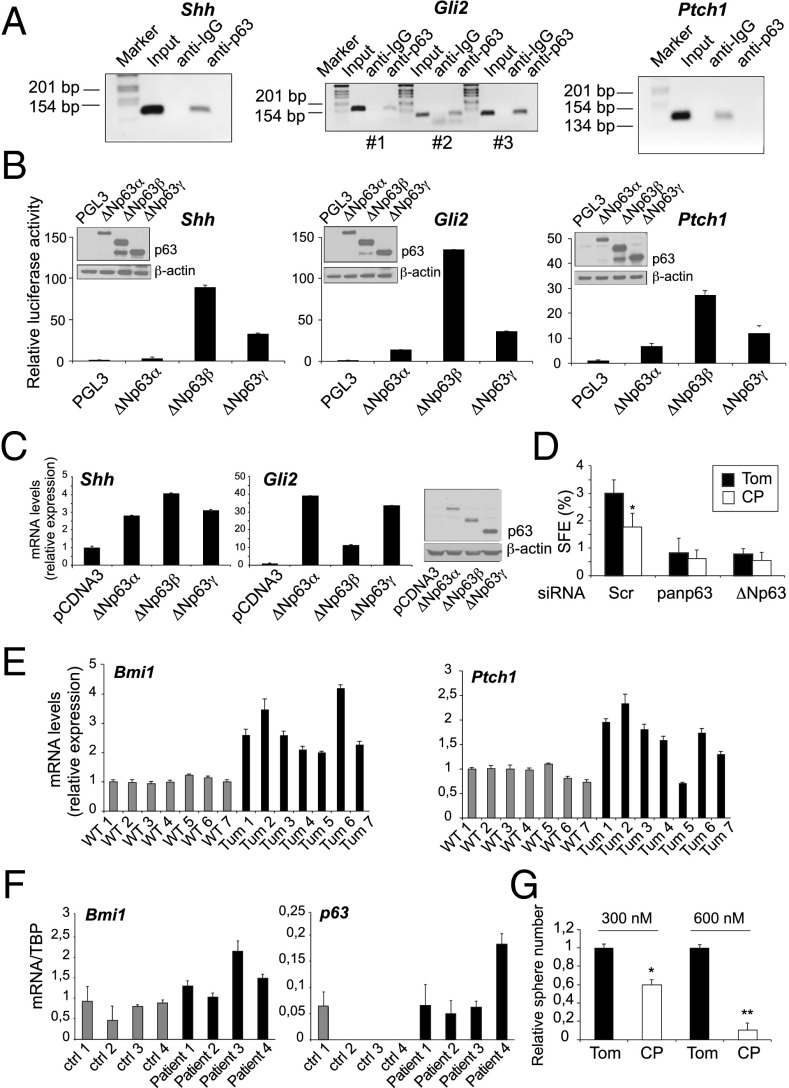
To validate the direct interaction of ΔNp63 with the regulatory regions of these genes, we performed ChIP experiments in breast cancer cells and demonstrated binding of endogenous ΔNp63 to genomic sites located in proximity to or within the Shh, Gli2, and Ptch1 loci (Fig. 4A). To characterize the binding capabilities of p63 to the regulatory regions of these genes further, we performed ChIP assays in MCF-7 cells overexpressing TAp63 or ΔNp63. We found that both isoforms are able to occupy binding sites encompassing or within the Shh (Fig. S4D), Gli2 (Fig. S4E), and Ptch1 (Fig. S4F) genes.
Fig. 4.
Shh, Gli2, and Ptch1 are direct p63 target genes. (A) Specific binding of p63 to the regulatory regions of Shh (Left), Gli2 (Middle), and Ptch1 (Right) genes. ChIP assays were performed in human MCF-7 breast carcinoma cells using H129 anti-p63 antibody and control IgG. p63 binds to one regulatory region within the Shh (+22 kb from the TSS) and Ptch1 (39 kb downstream of the TSS) genes, and to three regulatory regions (+47, +151 and +191 kb from the TSS, denoted as #1, #2, and #3) of the Gli2 locus. (B) Luciferase reporter assays of Shh (Left), Gli2 (Middle), and Ptch1 (Right) regulatory regions. The reporter vectors were cotransfected with ΔNp63-expressing plasmids into the p53 null human H1299 cell line. Cellular lysates were either assayed for luciferase activity or analyzed by IB (Insets). Data are presented as mean ± SD and are representative of three independent experiments. (C) qPCR was performed to analyze the transcript levels of Shh (Left) and Gli2 (Middle) quantitatively in MCF-7 cells transfected with constructs expressing the ΔNp63 isoforms. (Right) Protein extracts were subjected to IB for p63 and β-actin detection. (D) Effect of CP on the sphere-forming efficiency (SFE) of control and p63-depleted HCC1937 cells. Cells were transfected with control, pan-p63, and ΔNp63 siRNA oligos and then plated for the mammosphere-forming assay in the presence of 300 nM CP or the inactive CP analog tomatidine (Tom). *P < 0.05. (E) RT-qPCR analysis of Hh transcriptional targets in normal (WT) and ErbB2 (Tum) mammospheres. Seven independent preparations of RNA for each genotype were analyzed. (F) qPCR analysis of Bmi1 and p63 mRNA in human mammospheres obtained from normal breast tissue [control (ctrl)] and primary mammary tumors (Patient). (G) Effect of Hh antagonists on the self-renewal of mammary CSCs. Primary spheres were dissociated and cultured with CP or Tom for 7 d. Data are expressed as mean ± SD and are representative of four independent experiments. *P < 0.05; **P < 0.01.
Remarkably, ChIP assays in a mouse mammary epithelial cell line revealed that ΔNp63 specifically binds to a predicted p63 responsive element within a genomic region encompassing the murine Shh locus (−30 kb from the TSS; Fig. S4H). These findings highlight the notion that downstream effectors of p63 in mammary CSC regulation are conserved between humans and mice.
To study the transcriptional regulation of Shh, Gli2, and Ptch1 by p63, we generated luciferase reporter constructs encompassing the p63-binding regions within these genes. Cotransfection of the Shh, Gli2, and Ptch1 reporters with the ΔNp63 isoforms increased their basal transactivation levels (Fig. 4B and Fig. S4I). In accordance with the report by Caserta et al. (25), we found that the TAp63 isoforms were also able to transactivate the Shh promoter (Fig. S4J). The ΔNp63 isoforms activated the reporter constructs to different extents, reflecting their distinct transactivating properties (32). The p63β- and p63γ-isoforms are generally more transcriptionally active in vitro than the α-proteins, which possess a C-terminal autoinhibitory domain reducing their activity. Similarly, we found that overexpression of ΔNp63 isoforms in the MCF-7 breast cancer cells determined different levels of induction of Shh and Gli2 mRNA (Fig. 4C).
To validate the Hh signaling as a downstream effector of p63 in mammary CSC regulation, we assessed the effect of the SMO inhibitor cyclopamine (CP) on the sphere-forming efficiency of p63-depleted breast cancer cells. Fig. 4D shows that although the addition of CP reduced the ability of control cells to form spheres, it failed to inhibit the sphere formation potential of tumor cells significantly further upon p63 down-regulation (Fig. S4K). Taken together, these findings demonstrate that Shh, Gli2, and Ptch1 are direct transcriptional target genes of p63 in mammary cells, and that p63 loss diminished the self-renewal potential of breast CSCs by attenuating Hh activation.
To validate the role of Hh signaling in breast cancer stemness, we compared the expression levels of Hh transcriptional targets between WT and ErbB2 mammospheres. ErbB2 progenitors displayed increased Ptch1 and Bmi1 transcript levels (Fig. 4E), indicating augmented activation of the Hh pathway in tumors compared with normal progenitors. Similarly, CSCs isolated from primary human breast tumors expressed higher levels of Bmi1 mRNA relative to normal progenitors obtained from reductive mammoplasties (Fig. 4F, Left). Consistent with the data obtained for the ErbB2 spheres (Fig. 1A), we found that the expression of p63 was more elevated in cancer than in normal mammary progenitors (Fig. 4F, Right).
We next demonstrated that pharmacological blockade of the Hh pathway by treatment of tumor spheres with CP led to a dose-dependent reduction of ErbB2 mammosphere-forming efficiency (Fig. 4G). Similar to other tumor types, collectively, these findings demonstrate that Hh function contributes to the maintenance of the self-renewing capacity of mammary CSCs.
To validate the relationship between the expression of p63 and the Hh signaling pathway in the clinical setting, we analyzed the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset (33), a collection of 2,000 clinically annotated primary breast cancer specimens. We found a strong positive correlation between p63 and Gli2 mRNA levels (Fig. S5A). The values of Pearson correlation between p63 and Gli2 are presented in Fig. S5D. Comparable results for p63 and Gli2 expression were obtained using the cBioPortal for Cancer Genomics database (www.cbioportal.org/). As previously reported (11), our analysis of the METABRIC dataset confirmed that expression of p63 is also significantly correlated with expression of Fzd7 in patients with breast cancer, at least when using one of two distinct probes (Fig. S5 B–D).
Discussion
p63, a major epithelial transcription factor, is crucial for maintenance of the regenerative capacity of several ectoderm-derived tissues, including the mammary gland (2–4). Our data highlight a role for ΔNp63 in promoting self-renewal and expansion of ErbB2 progenitors. Notably, we observed that the ability of ΔNp63 to sustain the self-replicative potential of mammary CSCs does not require p53. The observation that ΔΝp63 does not act through the inhibition of p53 suggests that p63 directly mediates transcriptional responses that confer SC properties.
Our findings demonstrate that ΔNp63 is the predominant isoform expressed in ErbB2 progenitors, whereas TAp63 variants are detectable only in a minority of mouse mammosphere samples, and at very low levels. ΔNp63α-, β-, and γ-isoforms are all expressed in ErbB2 progenitors, although to varying extent, likely reflecting the interindividual variability across mice. The expression levels of ΔNp63 in epithelial stem/progenitor cells correlate with their proliferating capacity (2). In the normal mammary gland, the stem and progenitor cell subpopulations display segregated expression of ΔNp63 and TAp63, respectively (3). The proposed model for differential TP63 promoter use implies that the exit of SCs from the quiescent state and elaboration of mammary progenitors require the switch-off of ΔNp63 expression. In our model system, WT mammospheres express similar levels of ΔNp63 and TAp63, likely because spheres are composed of heterogeneous populations of progenitors exhibiting various degrees of maturation. Conversely, ErbB2 mammospheres and isolated CSCs are enriched for ΔNp63 expression, suggesting that aberrant levels of ΔNp63 may lead to deregulation of CSC replicative properties and expansion of the progenitor pool. Indeed, silencing the expression of ΔNp63 in ErbB2 mammospheres and in breast cancer cell lines remarkably reduced their number, size, and replating efficiency. Collectively, our data prove that ΔNp63, but not TAp63, positively regulates mammary SC activity, corroborating previous reports (11).
Consistent with a diminished self-renewing capacity, in vivo transplantation of p63-depleted mammospheres delayed breast tumor growth compared with control progenitors. In shp63 mammosphere cultures, we observed a partial reactivation of p63 over time that correlated with increased self-renewal. Similarly, re-expression of p63 in transplants during tumor growth might have attenuated the effect of p63 KO on ErbB2-driven carcinogenesis. Indeed, a delay in tumor onset was observed in mice injected with spheres transduced with p63 shRNA lentiviruses compared with their control counterparts, although the effects did not achieve statistical significance.
To date, the molecular mechanisms underlying breast CSC frequency and maintenance remain poorly understood. To gain insight into the transcriptional programs downstream of p63 in the regulation of mammary cancer stemness, we examined the endogenous mRNA expression levels of several components of the Hh signaling pathway in p63-depleted progenitors. We found that p63 down-regulation leads to reduced expression of Shh, Ptch1, and Gli2. Hence, because SHH signals GLI transcription factors via ligation of the PTCH receptors, p63 seems to affect the Hh pathway at different hierarchical levels of the signaling cascade, namely, the ligand, its receptor, and downstream transcription factors. As a result, the SC marker Bmi1, an Hh target gene, is down-regulated upon p63 depletion. Together, these findings imply that p63 activates the Hh pathway and likely influences stemness, at least in part, through the induction of Bmi1, which is indispensable for cell survival and self-renewal of SCs (21, 34). To examine further how p63 interferes with the Hh pathway, we asked whether p63 might directly control the transcription of some of the Hh molecules. We found that p63 interacts with the regulatory regions of the Shh, Gli2, and Ptch1 genes, thus inducing their expression and promoting Hh pathway activation.
Although the regulation of Shh and Gli2 expression by TAp63 and ΔNp63, respectively, has already been reported in tumor cell lines (25, 26), we now demonstrate that several components of the Hh pathway are transcriptionally controlled by ΔNp63 in mammary CSCs. Additionally, we identified Ptch1 as a previously unidentified direct target of p63 transcriptional activity.
Using p53 KO mammospheres, we showed that p63 does not require p53 to regulate mammary cancer stemness. However, the existence of interplay between p53 and the Hh pathway has been reported (35, 36). p53 was shown to repress tumorigenic Hh signals by negatively regulating the activity of GLI1 (35, 37). Interestingly, constitutively activated mutants of SMO, as well as elevated levels of GLI1 and GLI2, inhibit p53 accumulation by promoting binding of p53 to Mdm2 and its subsequent ubiquitylation (38). Accordingly, given that ΔNp63 induces Gli2 expression, it is possible that cross-talk between the p53 family members and the Hh pathway would ultimately contribute to enhance CSC self-renewal.
We also considered the possibility that p63 might interfere with additional signaling pathways implicated in SC self-renewal, such as Notch and Wnt/β-catenin. However, the expression of Notch receptors and Fzd7 was not significantly influenced by p63 silencing in our model system or by ectopic expression of ΔNp63 in the luminal-subtype MCF-7 breast cancer cell line. Fzd7-dependent enhancement of Wnt signaling by ΔNp63 may preferentially occur in basal-like mammary tumors (11). Consistently, aberrant Wnt/β-catenin pathway activation is enriched in human basal-like, rather than luminal-like, mammary tumors (39–41).
Constitutive Hh signaling activation, as a result of overexpression of Hh components, including SHH, PTCH1, GLI1, and GLI2, has been linked to breast tumorigenesis and invasiveness (42, 43). The mechanisms underlying the up-regulation of Hh components in tumors are still poorly characterized. Promoter hypomethylation and deregulation of transcription factors have been associated with the overexpression of Hh molecules. Deregulation of ΔNp63 expression in mammary CSCs might result in aberrant activation of the Hh pathway, augmented SC self-renewal, and expansion of cancer progenitors.
Overall, our findings contribute to the identification of pathways downstream of p63 involved in epithelial CSC regulation and shed light on the molecular mechanisms underlying aberrant activation of Hh signaling in breast cancer.
Materials and Methods
Mice.
FVB-Tg (MMTV-Erbb2) transgenic and p53 KO mice on a C57/BL6J background were previously described (15). Corresponding WT strains were purchased from Harlan Laboratories. Mice were bred and maintained in pathogen-free rooms under barrier conditions. All mouse experiments were performed according to local ethics committee (Ethics Committees of the University of Tor Vergata and of the European Institute of Oncology) regulation and were approved by the Italian Ministry of Health.
Mammosphere Cultures.
Mammary cells were freshly isolated from mammary glands of WT and p53 KO mice or from tumors of ErbB2 mice, as previously described (15). Each WT mammosphere preparation was obtained from pooling mammary glands from n = 20 mice. For ErbB2 and p53 KO mammosphere preparation, a single tumor or mammary gland obtained from n = 1 animal was processed. At each passage, mammospheres were mechanically dissociated and replated at 20,000 cells per milliliter. For serial passage experiments, 5,000 cells from mechanically disaggregated mammospheres were plated in quadruplicate. After 7 d, newly formed mammospheres were counted.
PKH-26 Assay.
Freshly isolated mammary epithelial cells were resuspended in PBS and incubated with a 2× PKH-26 dye (Sigma) solution (1:250) for 5 min, blocked with 1% BSA, washed twice, and then plated to obtain primary and secondary mammospheres. Single-cell suspensions from secondary mammospheres were FACS-sorted with a FACS Vantage SE flow cytometer (Becton Dickinson).
Statistical Analyses.
Statistical evaluation was determined using a two-tailed t test, and values were expressed as mean ± SD (or SEM). Differences were considered statistically significant at P < 0.05. Details of all other methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Daniel Aberdam for sharing his ChIP-sequencing data, Dr. Eleonora Candi for critically reading the manuscript, Dr. Giuseppina Bonizzi and Dr. Cristina Pasi for helpful discussions, Dr. Lucilla Bongiorno for technical assistance, Simona Ronzoni for technical support with flow cytometry, and Laura Tizzoni and Valentina Dallolio for quantitative PCR assays. This study was supported by Italian Association for Cancer Research Investigator Grant IG-9202 (to F.B.) and Grants IG-11955 and MCO 9979 (to G.M.), Ministero della Salute (Ricerca Oncologica and Ricerca Finalizzata), and the Medical Research Council of the United Kingdom (G.M.) and European Research Council Advanced Grant 341131 (to P.G.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500762112/-/DCSupplemental.
References
- 1.Clarke MF, Fuller M. Stem cells and cancer: Two faces of eve. Cell. 2006;124(6):1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 3.Li N, et al. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells. 2008;26(5):1253–1264. doi: 10.1634/stemcells.2007-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Candi E, et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA. 2007;104(29):11999–12004. doi: 10.1073/pnas.0703458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 6.Helton ES, Zhu J, Chen X. The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem. 2006;281(5):2533–2542. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- 7.Barbareschi M, et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol. 2001;25(8):1054–1060. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 9.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 10.Yalcin-Ozuysal O, et al. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17(10):1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti R, et al. ΔNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol. 2014;16(10):1004–1015, 1–13. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Z, et al. Overexpression of ΔNp63α induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 2010;101(11):2417–2424. doi: 10.1111/j.1349-7006.2010.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribeiro-Silva A, Zambelli Ramalho LN, Britto Garcia S, Zucoloto S. The relationship between p63 and p53 expression in normal and neoplastic breast tissue. Arch Pathol Lab Med. 2003;127(3):336–340. doi: 10.5858/2003-127-0336-TRBPAP. [DOI] [PubMed] [Google Scholar]
- 14.Hoadley KA, et al. Cancer Genome Atlas Research Network Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicalese A, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138(6):1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer. 2007;7(5):389–397. doi: 10.1038/nrc2127. [DOI] [PubMed] [Google Scholar]
- 17.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Tucci P, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109(38):15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30(6):303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dierks C, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14(3):238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Caserta TM, et al. p63 overexpression induces the expression of Sonic Hedgehog. Mol Cancer Res. 2006;4(10):759–768. doi: 10.1158/1541-7786.MCR-05-0149. [DOI] [PubMed] [Google Scholar]
- 26.Ram Kumar RM, Betz MM, Robl B, Born W, Fuchs B. ΔNp63α enhances the oncogenic phenotype of osteosarcoma cells by inducing the expression of GLI2. BMC Cancer. 2014;14:559. doi: 10.1186/1471-2407-14-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao D, et al. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Invest. 2014;124(12):5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano RA, et al. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139(4):772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terrinoni A, et al. Role of p63 and the Notch pathway in cochlea development and sensorineural deafness. Proc Natl Acad Sci USA. 2013;110(18):7300–7305. doi: 10.1073/pnas.1214498110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouwenhoven EN, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6(8):e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolchinsky Z, et al. Angiomodulin is required for cardiogenesis of embryonic stem cells and is maintained by a feedback loop network of p63 and Activin-A. Stem Cell Res (Amst) 2014;12(1):49–59. doi: 10.1016/j.scr.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Straub WE, et al. The C-terminus of p63 contains multiple regulatory elements with different functions. Cell Death Dis. 2010;1:e5. doi: 10.1038/cddis.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: Preclinical and clinical evidences. Stem Cells. 2012;30(3):372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 35.Malek R, Matta J, Taylor N, Perry ME, Mendrysa SM. The p53 inhibitor MDM2 facilitates Sonic Hedgehog-mediated tumorigenesis and influences cerebellar foliation. PLoS ONE. 2011;6(3):e17884. doi: 10.1371/journal.pone.0017884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh Y, Katoh M. Integrative genomic analyses on GLI2: Mechanism of Hedgehog priming through basal GLI2 expression, and interaction map of stem cell signaling network with P53. Int J Oncol. 2008;33(4):881–886. doi: 10.3892/ijo_00000076. [DOI] [PubMed] [Google Scholar]
- 37.Mazzà D, et al. PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell Death Differ. 2013;20(12):1688–1697. doi: 10.1038/cdd.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abe Y, et al. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci USA. 2008;105(12):4838–4843. doi: 10.1073/pnas.0712216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smid M, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 40.King TD, Zhang W, Suto MJ, Li Y. Frizzled7 as an emerging target for cancer therapy. Cell Signal. 2012;24(4):846–851. doi: 10.1016/j.cellsig.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khramtsov AI, et al. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubo M, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64(17):6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 43.O’Toole SA, et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res. 2011;71(11):4002–4014. doi: 10.1158/0008-5472.CAN-10-3738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.