Abstract
Ceramides (CER) are involved in alcohol-induced neuroinflammation. In a mouse model of chronic alcohol exposure, 16 CER and 18 sphingomyelin (SM) concentrations from whole brain lipid extracts were measured using electrospray mass spectrometry. All 18 CER concentrations in alcohol exposed adults increased significantly (range: 25–607%); in juveniles, 6 CER decreased (range: −9 to −37%). In contrast, only three SM decreased in adult and one increased significantly in juvenile. Next, regional identification at 50 μm spatial resolution from coronal sections was obtained with matrix implanted laser desorption/ionization mass spectrometry imaging (MILDI-MSI) by implanting silver nanoparticulate matrices followed by focused laser desorption. Most of the CER and SM quantified in whole brain extracts were detected in MILDI images. Coronal sections from three brain levels show qualitative regional changes in CER-SM ion intensities, as a function of group and brain region, in cortex, striatum, accumbens, habenula, and hippocampus. Highly correlated changes in certain white matter CER-SM pairs occur in regions across all groups, including the hippocampus and the lateral (but not medial) cerebellar cortex of adult mice. Our data provide the first microscale MS evidence of regional lipid intensity variations induced by alcohol.
Keywords: Ceramide, sphingomyelin, lipids, alcohol, mass spectrometry imaging, matrix-assisted laser desorption ionization, silver nanoparticles, matrix implanted laser desorption ionization
Chronic exposure to high doses of alcohol is associated with neurotoxicity that may result in various neurological dysfunctions including dementia.1 Indeed, numerous clinical studies have documented cognitive impairments in alcoholics that encompass multiple domains including perceptual, motor, visuospatial, and executive functions (working memory, attention control, and response inhibition).2 Similarly, multiple brain imaging studies have reported pathology in the brain of alcoholics when compared with controls. Specifically, structural studies have revealed cortical atrophy, ventricular enlargement, and white matter degradation in alcoholics.3−9 Recently, preclinical imaging data has shown that this may be mediated by genetic influence.10 Functional brain imaging studies to assess baseline cerebral blood flow (CBF) or regional brain glucose metabolism, which serve as markers of brain activity, have revealed significant decreases that are most pronounced in the frontal cortical regions.11,12 Studies that used fMRI to assess activation responses during task processing have reported disrupted brain activation including abnormalities in prefrontal and anterior cingulate cortices.13,14 Recently, electroencephalographic studies that track functional connectivity between brain regions and their organization into dynamic networks, have shown disrupted resting functional connectivity in the default mode network (DMN), which is involved with interoceptive awareness, and in the saliency network.15,16
Mechanisms underlying neurotoxicity from exposure to high doses of alcohol are not well understood. However, there is increased awareness that alcohol-induced neuroinflammation contributes to ethanol neuropathogenesis.17 In animal models, administration of alcohol at doses that mimic binge drinking in humans activates microglia and increase the release of proinflammatory cytokines and chemokines.18,19 Several mechanisms have been proposed by which alcohol could activate microglia and induce neuroinflammation, including increase of extracellular glutamate in the brain;20 promotion of toll-like receptor 4 (TLR4) translocation to the membrane21 and activation of NF-κB, a transcription factor involved in proinflammatory gene activation in glial cells.22,23 In the rat, chronic alcohol activates NF-κB transcription of proinflammatory genes and formation of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) and reactive oxygen species (ROS) that are associated with ethanol induced neurodegeneration.24 In humans, alcohol abuse also induces inflammation in the brain and body25 as evidenced by increases in systemic markers of inflammation such as cytokines26 and C-reactive protein.27 Similarly studies on postmortem alcoholic brains showed increases in cytokines (monocyte chemoattractant protein 1 or MCP-1) and markers of microglial activation19 as well as increases in the expression of genes involved in inflammation.28
Ceramides (CER), a diverse family of sphingolipid messenger molecules, are implicated in inflammation, neurodegeneration, senescence, and apoptosis.29−36 CER with different acyl chain lengths are regulated differently in various tissues.37,38 Differential effects on CER species have been noted in aging, after neurotoxin exposure, and in neurodegenerative disorders/models.39,40 Studies indicate that affected structures include microglia and reactive astrocytes.41,42 Hence, experimental approaches to study regional concentration changes of brain lipid species may yield insight into basic mechanisms of the neurotoxic responses to drugs of abuse such as alcohol.
Sphingolipids are sphingosine-based lipids built by attachment of different polar head groups to the primary alcohol (C1–OH) of a precursor CER backbone.43 Depending on the type of polar group, phosphosphingolipids or glycosphingolipids are synthesized. Sphingomyelin (SM) is generated by the transfer of the phosphorylcholine moiety (from phosphatidylcholine) to the C1–OH of CER. CER44 is an important second messenger that affects cellular processes such as cell growth, differentiation, and apoptosis.
We used two types of mass spectrometry to assess the changes of CER and SM in chronic alcohol exposure on brain lipid profiles. First, whole brain lipid extracts were quantified using electrospray ionization45,46 (ESI), and revealed that CER and SM concentrations in whole brain lipid profiles were altered in adult and juvenile mice after chronic exposure to alcohol. Next, qualitative MILDI-MS imaging47−49 of CER and SM at three coronal brain levels were obtained and imaged using a new technique of Silver nanoparticulate (AgNP) matrix implantation which enables simultaneous identification of most brain lipid classes, except gangliosides.50 Imaging reveals regional differences in sphingolipid ion intensities of certain CER and SM species from three coronal regions, showing pronounced regional effects from alcohol intake in both adults and juveniles.
Finally, the quantitative ESI data is compared to the qualitative MILDI regional imaging using bivariate statistical analysis. The high correlation between the concentrations of lipids measured by ESI with their qualitative intensity in the rostral coronal sections presage the possible quantitative calibration of MILDI-MSI.
Results and Discusion
Quantitative Whole Brain Extracts
Ten whole mouse brains from each group were homogenized, and lipids extracted and fractionated. Quantitative assessment of 16 CER species and 18 SM species was performed by ESI analysis. Brain lipid extract data of SM and CER were first analyzed by multivariate analysis to visualize the sample clustering and underscore discriminating variables (Supporting Information Figure S1). Posthoc statistics were performed on both CER and SM intensities to confirm differences observed in multivariate discriminant analysis. CER and SM show significant changes in response to alcohol intake (Table 1).
Table 1. Concentrations of CER and SM Lipids in Whole Brain Extracts Analyzed by ESI-MSa.
| avg concn (×103) ± SEM |
% change EtOH/H2O |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| m/z ESI | lipid | JUV H2O | JUV EtOH | ADU H2O | ADU EtOH | JUV | ADU | ||
| 534.4891 | Cer C16:0/d18:2 | 0.41 ± 0.05 | 0.21 ± 0.04 | 0.17 ± 0.03 | 0.93 ± 0.15 | ns | –47% | *** | 436% |
| 536.5048 | Cer C16:0/d18:1 | 6.86 ± 0.36 | 5.42 ± 0.21 | 4.19 ± 0.35 | 11.37 ± 0.54 | ** | –21% | *** | 171% |
| 550.5205 | Cer C17:0/d18:1 | 0.21 ± 0.05 | 0.09 ± 0.02 | 0.08 ± 0.02 | 0.53 ± 0.09 | ns | –57% | *** | 607% |
| 562.5204 | Cer C18:1/18:1 | 124 ± 3.7 | 94.2 ± 3.6 | 73.8 ± 3.9 | 184 ± 6.6 | *** | –24% | *** | 150% |
| 564.5361 | Cer C18:0/d18:1 | 310 ± 14.3 | 264 ± 8.4 | 223 ± 14.8 | 448 ± 20.5 | * | –15% | *** | 101% |
| 566.5518 | Cer C18:0/d18:0 | 1.73 ± 0.22 | 1.44 ± 0.10 | 1.27 ± 0.18 | 2.10 ± 0.18 | ns | –17% | * | 66% |
| 578.5518 | Cer C19:0/d18:1 | 0.60 ± 0.07 | 0.38 ± 0.05 | 0.35 ± 0.08 | 0.75 ± 0.11 | ns | –37% | ** | 114% |
| 590.5518 | Cer C20:0/d18:2 | 7.26 ± 0.26 | 5.84 ± 0.23 | 4.98 ± 0.28 | 9.88 ± 0.41 | ** | –20% | *** | 98% |
| 592.5674 | Cer C20:0/d18:1 | 41.4 ± 2.30 | 39.4 ± 1.38 | 36.9 ± 2.53 | 45.9 ± 2.30 | ns | –5% | * | 25% |
| 618.5831 | Cer C22:0/d18:2 | 3.24 ± 0.23 | 2.53 ± 0.18 | 2.00 ± 0.18 | 5.29 ± 0.27 | * | –22% | *** | 164% |
| 620.5987 | Cer C22:0/d18:1 | 5.61 ± 0.26 | 4.87 ± 0.16 | 4.27 ± 0.13 | 7.30 ± 0.39 | ns | –13% | *** | 71% |
| 632.5987 | Cer C23:0/d18:2 | 0.51 ± 0.08 | 0.45 ± 0.05 | 0.27 ± 0.04 | 1.17 ± 0.16 | ns | –11% | *** | 335% |
| 634.6144 | Cer C23:0/d18:1 | 1.52 ± 0.13 | 1.37 ± 0.11 | 1.09 ± 0.10 | 2.43 ± 0.23 | ns | –10% | *** | 122% |
| 644.5987 | Cer C24:1/d18:2 | 2.40 ± 0.20 | 1.53 ± 0.16 | 1.12 ± 0.17 | 4.11 ± 0.25 | ** | –36% | *** | 268% |
| 646.6144 | Cer C24:1/d18:1 | 21.5 ± 1.36 | 17.5 ± 1.06 | 12.8 ± 1.07 | 37.1 ± 2.17 | ns | –19% | *** | 189% |
| 648.6300 | Cer C24:0/d18:1 | 2.65 ± 0.18 | 2.25 ± 0.21 | 1.71 ± 0.16 | 3.96 ± 0.28 | ns | –15% | *** | 131% |
| 701.5592 | SM C16:0/d18:2 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.00 | ns/too weak | |||
| 703.5749 | SM C16:0/d18:1 | 4.27 ± 0.09 | 4.34 ± 0.09 | 4.24 ± 0.10 | 4.18 ± 0.13 | ns | 2% | ns | –1% |
| 705.5905 | SM C16:0/d18:0 | 0.014 ± 0.005 | 0.009 ± 0.005 | 0.008 ± 0.006 | 0.006 ± 0.003 | ns/too weak | |||
| 729.5905 | SM C18:1/18:1 | 8.49 ± 0.17 | 9.44 ± 0.17 | 9.48 ± 0.24 | 6.22 ± 0.09 | *** | 11% | *** | –34% |
| 731.6062 | SM C18:0/d18:1 | 110 ± 1.81 | 113 ± 1.17 | 110 ± 1.61 | 99.9 ± 0.99 | ns | 3% | *** | –9% |
| 745.6218 | SM C19:0/d18:1 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.02 | 0.05 ± 0.01 | ns/too weak | |||
| 757.6218 | SM C20:1/d18:1 | 0.26 ± 0.02 | 0.28 ± 0.03 | 0.27 ± 0.03 | 0.17 ± 0.02 | ns | 9% | * | –37% |
| 759.6375 | SM C20:0/d18:1 | 9.33 ± 0.17 | 9.45 ± 0.09 | 9.28 ± 0.18 | 8.75 ± 0.14 | ns | 1% | ns | –6% |
| 785.6531 | SM C22:0/d18:2 | 1.71 ± 0.06 | 1.67 ± 0.02 | 1.58 ± 0.07 | 1.59 ± 0.03 | ns | –2% | ns | 0% |
| 787.6688 | SM C22:0/d18:1 | 4.95 ± 0.16 | 4.80 ± 0.07 | 4.64 ± 0.12 | 4.55 ± 0.10 | ns | –3% | ns | –2% |
| 799.6688 | SM C23:0/d18:2 | 0.18 ± 0.02 | 0.15 ± 0.01 | 0.19 ± 0.04 | 0.14 ± 0.01 | ns | –14% | ns | –26% |
| 801.6844 | SM C23:0/d18:1 | 1.17 ± 0.06 | 1.22 ± 0.03 | 1.08 ± 0.09 | 1.17 ± 0.03 | ns | 4% | ns | 8% |
| 811.6688 | SM C24:1/d18:2 | 1.20 ± 0.05 | 1.20 ± 0.03 | 1.14 ± 0.09 | 1.02 ± 0.05 | ns | 1% | ns | –10% |
| 813.6844 | SM C24:1/d18:1 | 25.5 ± 0.70 | 25.0 ± 0.33 | 25.1 ± 0.67 | 24.0 ± 0.38 | ns | –2% | ns | –4% |
| 815.7001 | SM C24:0/d18:1 | 5.84 ± 0.25 | 5.66 ± 0.13 | 5.40 ± 0.18 | 5.34 ± 0.12 | ns | –3% | ns | –1% |
| 827.7001 | SM C25:1/d18:1 | 0.04 ± 0.01 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.03 ± 0.01 | ns/too weak | |||
| 829.7157 | SM C25:0/d18:1 | 0.004 ± 0.002 | 0.001 ± 0.001 | 0.003 ± 0.002 | 0.001 ± 0.001 | ns/too weak | |||
| 841.7157 | SM C26:1/d18:1 | 0.016 ± 0.005 | 0.019 ± 0.006 | 0.018 ± 0.008 | 0.014 ± 0.007 | ns/too weak | |||
Tabulated data shows the percentage change in the CER and SM concentrations of each lipid in the two groups. Results of Bonferroni: post-test *p < 0.05, **p < 0.01, ***p < 0.001. (Further results in Table S1 in the Supporting Information.)
Table 1 shows that all CER concentrations increased significantly in chronically exposed adult (ADU EtOH) mice compared to controls (ADU H2O). In juveniles, six CER species show a significant decrease in the JUV EtOH group as compared to the JUV H2O group. By contrast, alcohol consumption significantly decreased only 3 of the 20 SM concentrations in the ADU EtOH group, while in the JUV ETOH group only one SM lipid significantly increased. The percentage changes and significance levels for the detected CER-SM pairs obtained from the ESI extractions are reported in Table 1. Data in Figure 1 provide examples from three CER and SM concentrations, which mirror the trend found for the lipids measured (Table 1).
Figure 1.
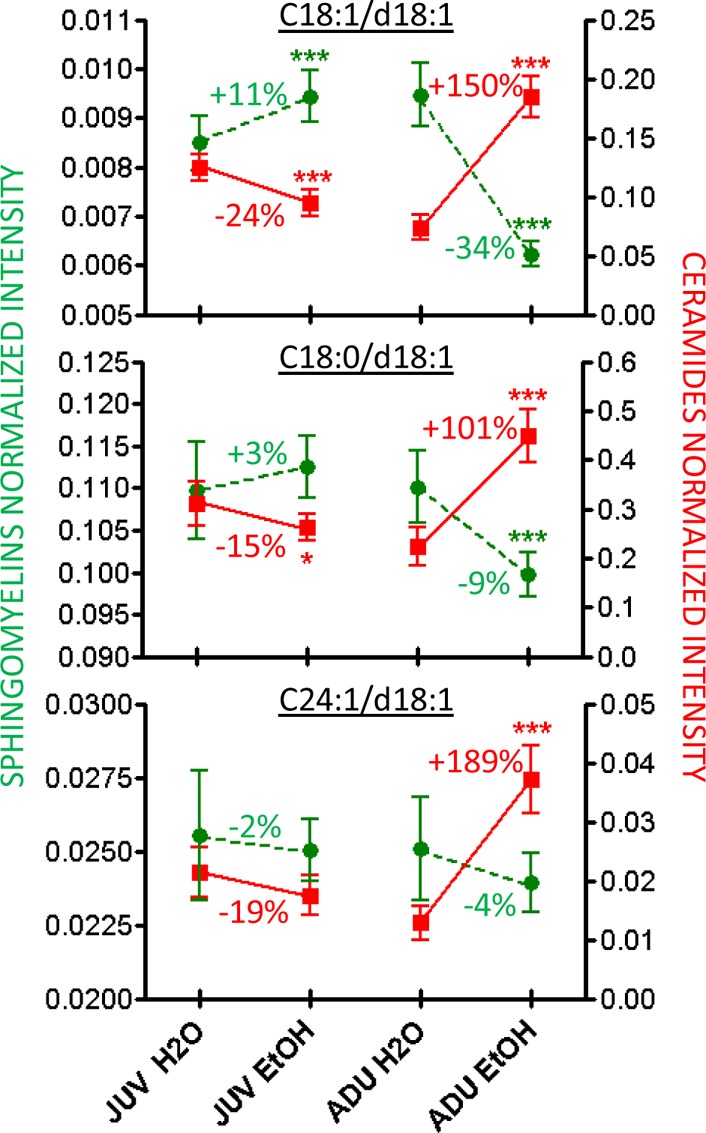
Comparison of the concentration of six brain lipids (3 SM (green) and 3 CER (red)) in chronically alcohol exposed JUV and ADU mice.
Qualitative MILDI-MSI Distributions of CER and SM within Each of Three Coronal Levels
Coronal sections at brain levels (+1.54 mm, −1.70 mm, and −5.88 mm re: bregma, Franklin & Paxinos mouse atlas) were selected for mass spectrometric imaging (MSI), as they contain regions that are known to be affected by chronic alcohol consumption. For each of these three levels, an image is shown in Figure S2 (Supporting Information) upon which are drawn the regions of interest (ROIs) defining the following subregions: (1) The most rostral level (+1.54 mm re: bregma) contains three structures involved in addiction, the striatum, nucleus accumbens, and frontal cortex. (2) The intermediate section (−1.70 mm re: bregma) contains the hippocampus, which plays a role in memory impairments, the hypothalamus, and the habenula. (3) The caudal level (−5.88 mm re: bregma) includes the cerebellum which is subject to alcoholic cerebellar degeneration51 and plays an important role in motor control (and may be involved in some cognitive functions such as attention and language).52−57
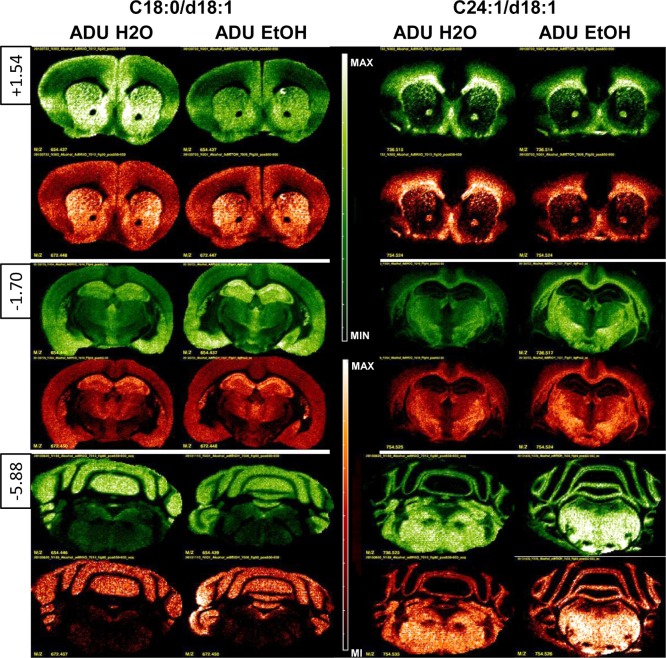
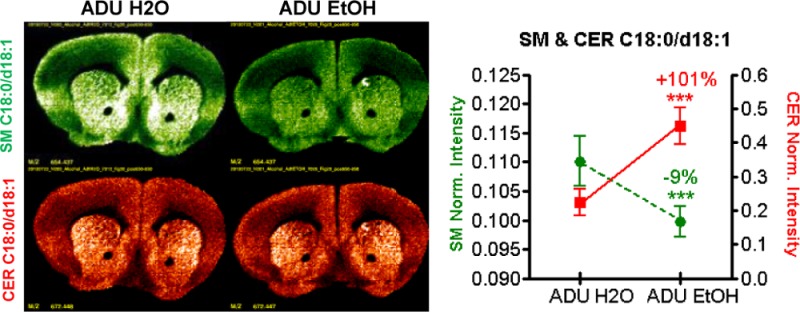
Figure 2 shows images from two of the more intense lipid signals measured in each of the three levels taken from ADU water and ethanol groups. The left column shows the distribution of CER (in red) and SM (in green) C18:0/d18:1, which are predominant gray matter species. The right column illustrates CER (in red) and SM (in green) C24:1/d18:1, which are predominant white matter species. The spatial distribution of CER and SM intensities as measured by MILDI-MSI revealed patterns of regional changes at different levels of the neuraxis some of which are at odds with the overall concentration changes measured from adult whole brain extract. For example, at level +1.54 mm re: bregma, CER C18:0/d18:1 MILDI intensity was unchanged and CER C24:1/d18.1 decreased in chronically exposed ADU. Also in the ADU alcohol group at level −1.70 mm re: bregma, a correlated intensity increase of both C18 and C24 backbone CER–SM lipid pairs is apparent within the hippocampus and other regions.
Figure 2.
Comparison of MS images in the adult groups (ADU H2O and ADU EtOH) for SM (green) and CER (red) C18:0/d18:1 and C24:1/d18:1 for coronal sections at levels +1.54 mm, −1.70 mm, and −5.88 mm re: bregma. Note that SM and CER C18:0/d18:1 are located predominantly in gray matter, while SM and CER C24:1/d18:1 are associated with white matter (i.e., regions containing myelinated axon tracts).
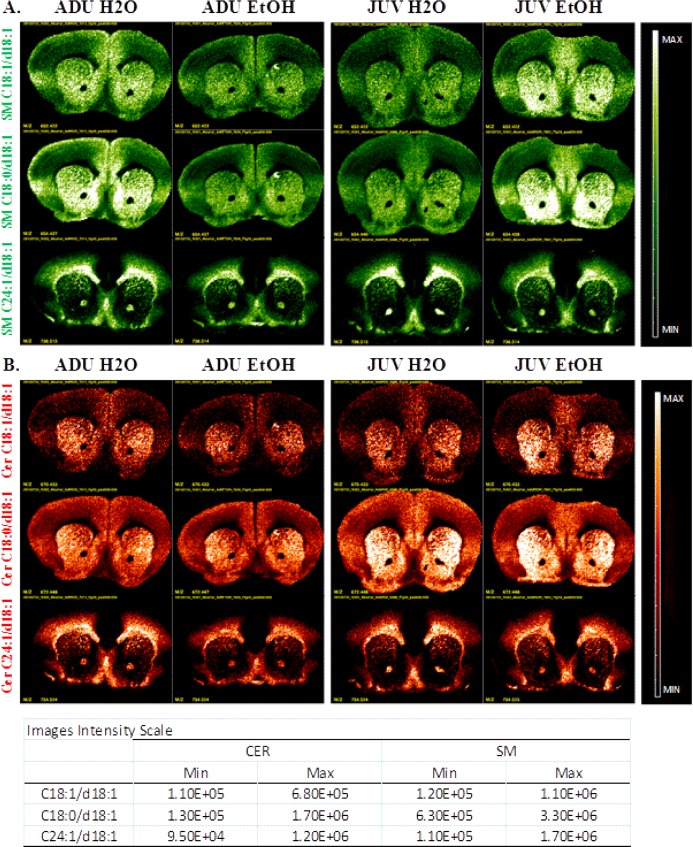
Figure 3 shows MS images from the most rostral section (+1.54 mm re: bregma) for the ADU and JUV groups (Analogous images for ADU and JUV mice at levels −1.70 and −5.88 appear in the Supporting Information as Figure S4A and B, respectively). The +1.54 mm level includes the striatum, nucleus accumbens, prelimbic cortex, piriform cortex, and to a lesser extent the primary and secondary motor cortex. Note the association of the C18 backbone species with gray matter and of the C24 species with white matter. In the ADU group, SM (C18:1/d18:1 and C18:0/d18:1) ion counts decreased with alcohol consumption (Figure 3A, left panels), without a commensurate change in the corresponding CER species (Figure 3B, left panels). As observed in the whole brain extracts analyzed by ESI, changes in the JUV group differed from those in ADU, as SM (C18:1/d18:1 and C18:0/d18:1) ion counts increased with alcohol consumption, while SM (C24:1/d18:1) did not show obvious changes in JUV exposed to alcohol (Figure 3A, right panels).
Figure 3.
MS Images of coronal sections at level bregma +1.54 mm. (A) Images of the three detected SM (green) (B) Images of the three detected CER (red).
Figure 3B shows no obvious changes in CER intensities with alcohol consumption in either ADU (left panels) or JUV (right panels) group. However, basal levels of CER (C18:1/d18:1 and C18:0/d18:1) seem to be higher in JUV than in ADU. Because CER intensities at this brain level do not show the same increase detected in the whole brain extract results, the CER changes observed by ESI may be either diffuse or localized in another bregma. (MSI of bregmas −1.70 mm and −5.88 mm are available in the Supporting Information.)
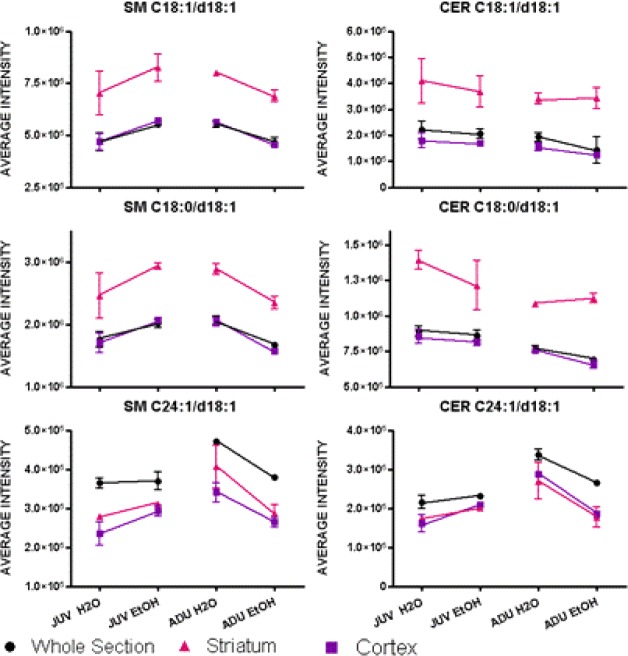
Figure 4 shows regional changes for CER and SM at rostral level bregma +1.54 using the average ion counts in the whole section, the striatum (including the nucleus accumbens, the dorsomedial and the ventral lateral striatum), and the cerebral cortex (including the insula, the piriform, parietal and cingulate cortex). The sample size of 2–3 animals per group limits opportunities for statistical inferences beyond displaying the median and range of the intensities as shown in Figure 4. The trends and consistency of the imaged SM and CER in the JUV with the ESI results are clear. In contrast, the imaged CER in the ADU ethanol group shows minimal changes in the whole section, the striatum, and the cortex, whereas ESI showed significant increases in the whole brain extract. Figure 4 summarizes the findings from rostral sections (+1.54 mm re: bregma), which differ in ADU and JUV mice. SM graphs in Figure 4 show that, as observed in the images in Figure 3A, the predominantly gray matter SM (C18:1/d18:1 and C18:0/d18:1) intensity decreased with alcohol intake in the ADU group, and increased with alcohol consumption in the JUV group for measurements from the whole section, striatum, and cortex as did the predominantly white matter SM (C24:1/d18:1) (even though no obvious changes were observed in the images of CER in Figure 3B). The CER (C18:1/d18:1 and C18:0/d18:1) intensities that are predominant in gray matter decreased with alcohol consumption in the JUV group and trended toward a slight increase in the striatum of the ADU group, while remaining unchanged in the cortex or the overall section. The distribution of white matter CER (C24:1/d18:1) intensities shows the same trend as the corresponding SM, which is consistent with Figure 3A.
Figure 4.
Average SM and CER intensities in sections at level bregma +1.54 mm for the whole section, the striatum, and the cortex.
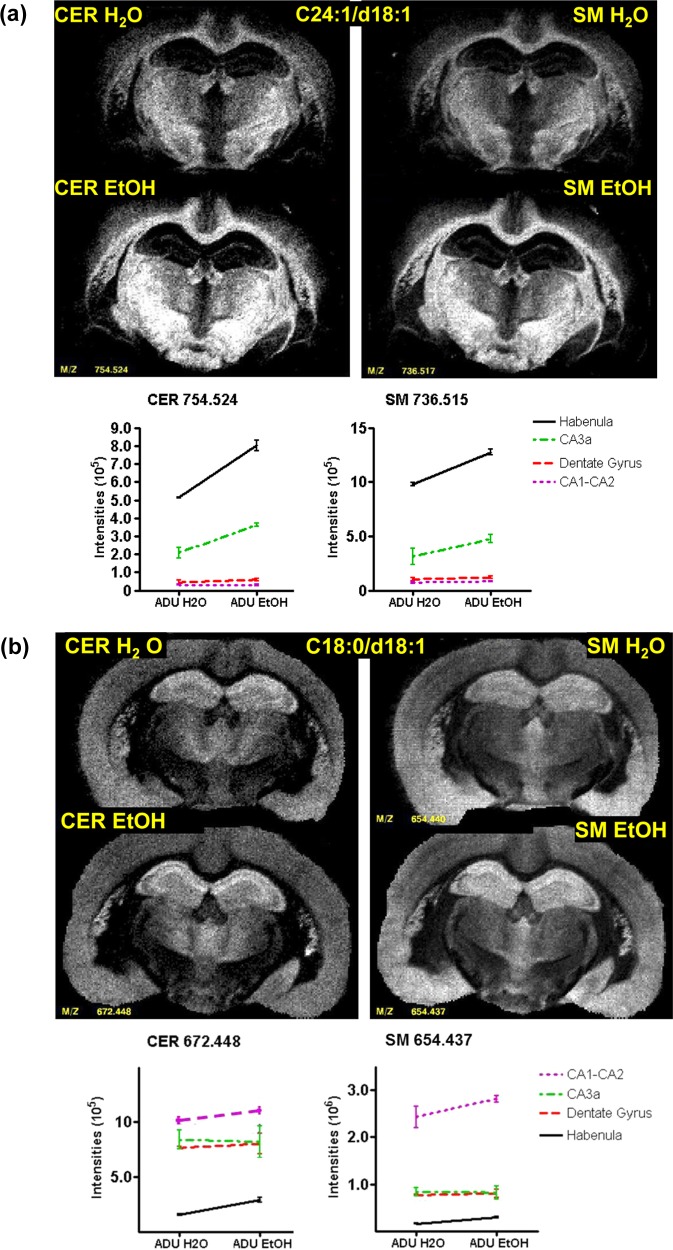
Figure 5a shows MS images of the CER-SM (C24:1/d18:1) pair in sections through the hippocampus (+1.70 mm re: bregma). The intensities of this pair show striking regional variations in the hippocampus and habenula responses to EtOH consumption. Although the dentate gyrus intensities were unaffected, there was a marked increase in the habenula in the EtOH group. Hippocampal regions CA3a showed a smaller increase in these CER–SM pair. A different EtOH response pattern appeared for the gray matter CER–SM (C18:1/d18:0) pair (Figure 5b); the dentate gyrus and CA3a showed no effects, but CA1-2 and the habenula showed increases with EtOH consumption.
Figure 5.
(a) Comparison of lipid pair SM–CER C24:1/d18:0 in adult alcohol at level −1.70 re:bregma. (b) Comparison of lipid pair SM–CER C18:0/d18:1 in adult alcohol at level −1.70 re:bregma.
Bivariate Analysis of Correlated Regional Lipid Intensities for CER-SM Pairs
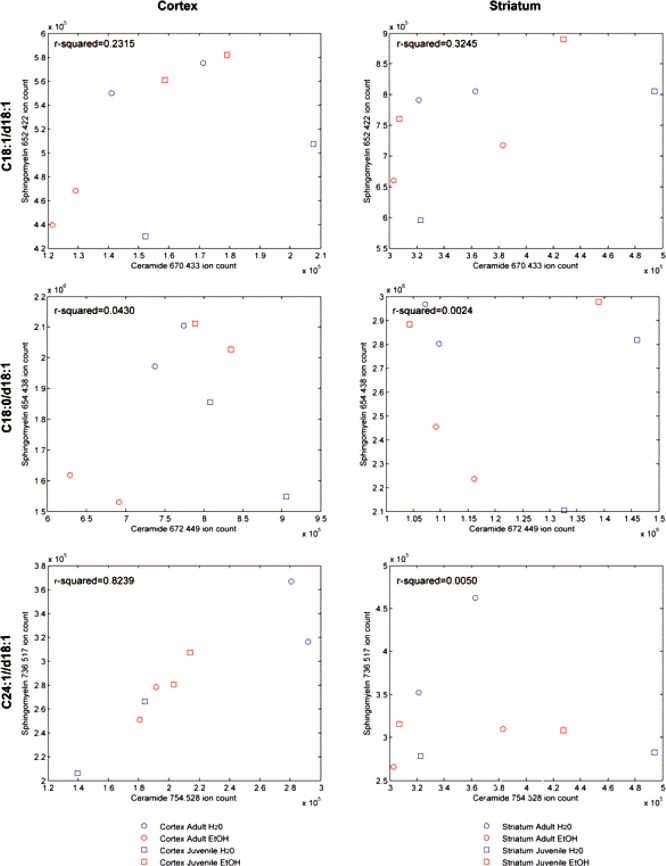
As the CER species are the precursors of the corresponding SM species, we included bivariate plots and calculation of coefficients of determination (R-squared) to examine the statistical independence of counts of the respective CER precursors and SM ion species in the cerebral cortex, striatum (bregma +1.54 mm, Figure 6), and lateral and medial cerebellar cortex (bregma −5.88 mm, Figure 7).
Figure 6.
CER versus SM plots: cerebral cortex and striatum.
Figure 7.

Cerebellar cortex SM and CER plots.
Figure 6 shows a positive correlation between C18:1/d18:1 CER and SM species in level +1.54 across ages and alcohol consumption. In ADU animals, alcohol treatment reduced both CER and SM C18:1/d18:1 ion counts in the cerebral cortex, but only SM counts in the striatum. In JUV, though, ethanol treatment produced a strong increase in SM counts in cortex and a moderate increase in the striatum. Although the CER and SM C18:0/d18:1 ion counts were uncorrelated in either cerebral cortex or striatum, the response pattern to alcohol treatment was the same. Alcohol intake of ADU animals reduced both CER and SM C18:1/d18:1 ion counts in cerebral cortex, but only the SM counts in the striatum. JUV animals showed alcohol-related increases in SM counts in both regions. The CER and SM C24:1/d18:1 ion species were also affected by ethanol consumption in both structures. There was a very strong linear positive correlation between these species in the cerebral cortex, but no correlation in their ion counts in the striatum. The cortex of ADU rats showed a strong alcohol induced decrease in both CER and SM, while JUV rats showed the opposite effect in alcohol consumption. The SM ion counts also decreased in ADU striatum after alcohol treatment, but the JUV striatum was unaffected (Figure 6).
The ion counts from the medial and lateral cerebellar cortex (Figure 7) showed strong correlations between CER and SM for each species (C18:1/d18:1, C18:0/d18:1, and C24:1/d18:1) across ADU and JUV groups, with coefficients of variation ranging from 0.48 for the C18:1/d18:1 forms to 0.95 for the C24:1/d18:1 species. The data suggest a very high degree of coregulation.
Figure 8 excludes the JUV groups of Figure 7 and shows only the ADU groups which are most affected by alcohol in this study. The ADU groups exhibit a high correlation in which the most prominent alcohol effect was a coordinated increase in SM and CER C24:1/d18:1 intensity in the lateral cerebellar cortex of ADU animals in the alcohol consumption group (MANOVA, F(1,4) = 83.7, p < 0.01), but no corresponding effect in the medial cerebellar cortex.
Figure 8.
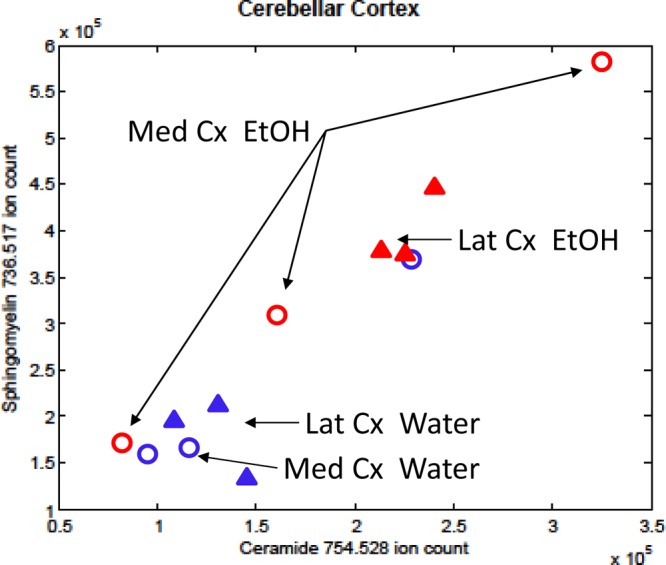
Correlated change of MILDI-MSI regional intensities of CER and SM C24:1/d18:1 in ADU cerebellar cortex.
Bivariate Correlation between Quantitative ESI and Qualitative MILDI
The remaining question in the study is how ESI quantitation correlates to MILDI-MSI lipid ion intensities. Table 2 shows that all of the CER and SM lipids detected quantitatively by ESI in brain extracts are also detectable in tissue sections with MILDI-MSI. Bivariate correlations are then calculated from the data in Table 2 between the quantitative SM and CER concentrations from ESI versus the qualitative MILDI-MSI lipid ion regional intensities found in each of the three sections. For the SM species, the ESI values are highly correlated with the MILDI ion species’ counts from the two most rostral sections (r = 0.975 and 0.934, respectively). For the CER species, only the most rostral section shows a high correlation (r = 0.938) between ESI and MILDI ion species counts; however, the others do not (for the thalamus/hippocampus level section, r = 0.572 (or r2 = 0.327, NS by Bonferroni). For the cerebellum level section, r = 0.094 (or r2 = 0.009, NS). These correlations between ESI and MILDI data highlight the potential for future experiments in which regional quantitative ESI from thick (150 μm) brain sections will be used to calibrate MILDI-MSI lipid ion intensities from adjacent sections on either side.
Table 2. Qualitative MILDI Intensity from Three Different Brain Levels from One ADU (EtOH) Animal Compared to Quantitative ESI Extractsa.
| |
MILDI-MSI |
|||||
|---|---|---|---|---|---|---|
| quantitative ESI extraction |
intensity |
|||||
| lipid | m/z | concn, I/Istd × 10–3 | adu etoh +1.5 (18) | aud etoh –1.7 (45) | adu etoh –5.88 (80) | m/z |
| Cer C17:0/d18:1 | 550.5205 | 0.5 | 2.00 × 101 | 1.00 × 101 | 2.80 × 101 | 658.4335 |
| Cer C18:1/18:1 | 562.52 | 200 | 1.95 × 105 | 1.08 × 105 | 6.67 × 103 | 670.4335 |
| Cer C18:0/d18:1 | 564.5361 | 500 | 7.38 × 105 | 5.72 × 105 | 1.42 × 105 | 672.449 |
| Cer C18:0/d18:0 | 566.5518 | 2 | 1.70 × 101 | 1.80 × 101 | 2.10 × 101 | 674.4645 |
| Cer C19:0/d18:1 | 578.5518 | 0.75 | 1.80 × 101 | 1.30 × 101 | 1.70 × 101 | 686.4646 |
| Cer C20:0/d18:2 | 590.5518 | 10 | 5.80 × 101 | 3.15 × 102 | 1.27 × 102 | 698.4652 |
| Cer C20:0/d18:1 | 592.5674 | 45 | 1.10 × 104 | 3.15 × 104 | 2.46 × 104 | 700.4807 |
| Cer C22:0/d18:2 | 618.5831 | 5 | 9.95 × 104 | 2.51 × 105 | 1.79 × 105 | 726.4969 |
| Cer C22:0/d18:1 | 620.5987 | 5 | 9.50 × 101 | 1.06 × 103 | 7.10 × 102 | 728.5132 |
| Cer C23:0/d18:2 | 632.5987 | 1.25 | 2.04 × 104 | 7.47 × 104 | 3.97 × 104 | 740.5124 |
| Cer C23:0/d18:1 | 634.6144 | 2.5 | 8.80 × 101 | 5.40 × 101 | 7.50 × 101 | 742.5288 |
| Cer C24:1/d18:2 | 644.5987 | 4 | 6.00 × 104 | 2.23 × 105 | 1.79 × 105 | 752.5125 |
| Cer C24:1/d18:1 | 646.6144 | 40 | 2.67 × 105 | 6.61 × 105 | 6.50 × 105 | 754.528 |
| Cer C24:0/d18:1 | 648.6303 | 4 | 7.40 × 101 | 1.30 × 102 | 1.91 × 102 | 756.5443 |
| SM C18:1/18:1 | 729.5905 | 9.5 | 4.89 × 105 | 4.02 × 105 | 1.04 × 105 | 652.422 |
| SM C18:0/d18:1 | 731.6062 | 115 | 1.72 × 106 | 1.60 × 106 | 7.21 × 105 | 654.438 |
| SM C19:0/d18:1 | 745.6218 | 0.07 | 2.70 × 101 | 2.83 × 102 | 2.20 × 101 | 668.4549 |
| SM C20:1/d18:1 | 757.6218 | 0.3 | 1.51 × 103 | 1.75 × 104 | 1.13 × 104 | 680.4547 |
| SM C20:0/d18:1 | 759.6375 | 9.5 | 4.01 × 104 | 1.09 × 105 | 9.21 × 104 | 682.471 |
| SM C22:0/d18:2 | 785.6531 | 1.75 | 1.16 × 104 | 7.39 × 104 | 4.75 × 104 | 708.486 |
| SM C22:0/d18:1 | 787.6688 | 7 | 7.97 × 104 | 9.36 × 104 | 1.04 × 105 | 710.5016 |
| SM C23:0/d18:2 | 799.6688 | 0.2 | 7.20 × 103 | 3.82 × 104 | 2.19 × 104 | 722.5016 |
| SM C23:0/d18:1 | 801.6844 | 1.25 | 5.18 × 102 | 4.64 × 103 | 2.48 × 103 | 724.5172 |
| SM C24:1/d18:2 | 811.6688 | 1.25 | 3.82 × 104 | 1.38 × 105 | 1.51 × 105 | 734.5018 |
| SM C24:1/d18:1 | 813.6844 | 25 | 3.88 × 105 | 9.16 × 105 | 1.06 × 106 | 736.5175 |
| SM C24:0/d18:1 | 815.701 | 6 | 1.02 × 103 | 3.05 × 103 | 2.54 × 103 | 738.533 |
| SM C25:1/d18:1 | 827.701 | 0.05 | 2.12 × 102 | 2.97 × 103 | 8.39 × 102 | 750.5336 |
| SM C25:0/d18:1 | 829.7157 | 0.005 | 2.30 × 101 | 2.80 × 101 | 1.70 × 101 | 752.5491 |
| SM C26:1/d18:1 | 841.7157 | 0.025 | 1.60 × 101 | 3.56 × 102 | 4.00 × 101 | 764.5497 |
The mass resolved lipid intensity is compared to the quantitative lipid concentration of the ADU alcohol group obtained by ESI from extracts.
We have shown that alcohol consumption effects on brain lipid content vary with age and brain regions. Quantitative analysis of whole brain extracts showed that chronic alcohol consumption decreases SM and substantially increases CER concentrations in ADU. In JUV mice, though, chronic alcohol consumption produced mild increases in SM and decreases in CER concentrations. In parallel, qualitative MILDI-MS imaging using AgNP matrix implantation showed that CER and SM spatial distributions varied regionally in the cerebellar cortex, hippocampus, striatum, and cerebral cortex, structures which have been previously implicated in structural and functional studies on acute and chronic effects of alcohol.58 The magnitudes of the effects of alcohol intake on lipid profiles were greater in adult than in juvenile animals both from the brain extracts and the MILDI-MSI studies.
Pairs of CER and SM ion species showed a more complicated dependence in specific brain regions with MILDI-MSI. Specifically, highly coregulated intensity increases for some CER and SM pairs occur in restricted regions within the hippocampus, habenula, and cerebellum. These effects were particularly prominent for C24:1/d18:1 CER and SM species in the lateral cerebellar cortex, cerebral cortex, hippocampus, and striatum, suggesting a very high degree of local coregulation. Regional lipid profiling by MILDI-MS imaging is a promising technique for probing cellular consequences of ethanol consumption, as it allows localization of molecular changes that could not be surmised from quantitative brain extracts.
There is increased recognition that alcohol-induced neuroinflammation contributes to ethanol neuropathogenesis.17 In animal models administration of alcohol at doses that mimic binge drinking in humans was shown to activate microglia and increase the release of proinflammatory cytokines and chemokines.18,19 CER, a diverse family of sphingolipid intracellular and extracellular signaling molecules, have been implicated in a broad range of cellular processes associated with inflammation, neurodegeneration, angiogenesis, metabolic syndromes and cancer biology.29−36 CER with different acyl chain lengths are regulated differentially in different tissues.37,38 Differential effects on CER species have been noted in aging, neurotoxin exposure, and neurodegenerative disorders/models39,40 including ethanol induced neonatal neurodegeneration.59 Interestingly, interventions indicate that affected sites include microglia and reactive astrocytes.41,42 Hence, future use of the experimental approaches for determining local lipid intensity changes demonstrated in this study may yield insight into basic mechanisms of cellular response in normal function, disease and substance abuse.
The combined analysis of tissue extracts and MILDI MS tissue imaging using silver nanoparticle matrix is an evolving tool for elucidating basic mechanisms of systemic interactions. This work demonstrates the first regionally specific lipid ion intensities from multiple SM species and their corresponding CER in murine models of alcoholism. These data suggest that CER–SM pathways are altered differentially in brain regions by chronic alcohol consumption. As CER is well-known for its involvement in inflammation this finding motivates future method development for detecting other inflammatory lipid species such as arachidonic acid, prostaglandins, and leukotrienes. Moreover, future studies with larger rat models will give a number of advantages over mice. For example, regions such as the hypothalamus and habenula can be more reliably sampled from a larger brain. The larger volume of rat brains should allow sub-millimeter (100–200 μm) thick sections to be lipid extracted and analyzed quantitatively by ESI, and a corresponding analysis of qualitative MILDI-MS images of thinner sections surrounding each thick section will allow even stronger regional correlation between quantitative and qualitative data. This approach could play a major role in further defining the pathogenesis of alcohol abuse as well as other neurodegenerative or addictive diseases.
Methods
Animals
Male C57BL/6 mice (n = 60) were obtained from Taconic at 3 weeks (juvenile) or 7 weeks old (adult) and allowed to habituate to the animal facility for 2 weeks. All mice were treated in accordance with IACUC guidelines. Five weeks old juveniles (JUV) and 9 weeks old adult (ADU) mice were placed on a drinking-in-the-dark paradigm (DID).60 Briefly, all mice were split into either control (water) or ethanol (EtOH) group (n = 15/group). The EtOH group had a daily 4 h access to a 12% ethanol bottle during their dark cycle for 52 days. Control mice were given water. Food and water were given ad libitum to all mice at all times. Body weights at the start of the DID paradigm were 20.88 ± 1.42 g for JUV water, 21.07 ± 1.55 g for JUV EtOH, 22.34 ± 1.56 g for ADU water, and 22.58 ± 1.62 g for ADU EtOH. Drinking intake and body weight were measured daily for 52 days. After 52 days, mice were euthanized 1 h after ethanol access, and then brain and liver were collected. Wet liver and brain were weighed and flash frozen in 2-methylbutane on dry ice and stored at −80 °C until further processing. Body weights for each group on the final day were 29.68 ± 3.16 g for JUV water, and 29.78 ± 4.01 g for JUV EtOH, 28.82 ± 2.54 g for ADU water, and 29.94 ± 3.44 g for ADU EtOH. Alcohol consumption was measured for 52 days; averages were 108 ± 19 g/kg for JUV EtOH and 85 ± 16 g/kg for ADU EtOH. Statistics were performed on body weight, liver wet weight, and cumulative alcohol consumption (at time of sacrifice). Body and liver weight did not show statistical differences between the four groups; however, cumulative alcohol consumption was 21% higher (unpaired t test, p-value = 0.0060) in the JUV group compared to the ADU group
Quantitative Analysis
Lipid Extraction and Fractionation
Total lipids were extracted from brain homogenate using a modified Folch extraction method.61 Brains were weighed and ground in a mixture of chloroform/methanol (2:1 v/v) (20 μL for each 1 mg tissue). Lipid standards were added to this total volume (the volume of lipid standards was subtracted from the total volume of chloroform/methanol) which depends on the weight of each brain (Table 3). Tissues were homogenized, sonicated, and vortexed. Then 4 μL water/1 mg tissue was added. The mixture was again vortexed and centrifuged. The extraction results in an upper aqueous phase rich in gangliosides and a lower organic phase containing phospholipids, CERs, fatty acids, triglycerides, and cholesterol. The aqueous phase was removed and stored at −20 °C. The organic phase was then evaporated to dryness using nitrogen, resuspended in a volume of chloroform equal to the remaining volume of extraction liquid after lipid standards were added, and lipid fractionation done. The fractionation was performed in 1 mL SuperClean LC-NH2 SPE tubes (Sigma # 504483). The columns were conditioned with 2 mL of hexane, and 200 μL of the samples chloroform solution loaded onto them. The fractions were retrieved using different solvents (Table 3). The fractions were retrieved using different solvents, and lipids contained in the resulting fractions are described in Table 3. Only fraction 2 (CERs) and fraction 5 data (SMs) are discussed in this paper.
Table 3. Different Phases/Fractions for Lipid Extraction/Fractionation and Their Parameters.
| organic/lower
phase |
|||||||
|---|---|---|---|---|---|---|---|
| aqueous/upper phase | fraction 1 | fraction 2 | fraction 3 | fraction 4 | fraction 5 | fraction 6 | |
| lipids | gangliosides | DAG, TAG, cholesterol | Cer, MAG | free FA, modified FA | cerebrosides, sphingoid bases | SM, PC, PE | ST, PS, PI, PG, CL, PA |
| fractionation | na | 2 mL diethyl ether | 1.6 mL CHL/MeOH (23/1) | 1.8 mL diisopropyl ether/acetic acid (98/4) | 2 mL acetone/Me OH (9/1.2) | 2 mL CHL/MeOH (2/1) | 2 mL 0.2 M C2H4O2·NH, 3 MeOH |
| internal standards | na | na | Cer C12:0/d18:1 | stearic acid d3 | GalCer C12:0/d18:1 | PC 14:0/14:0 | PS 12:0/12:0 |
| dilution for MS analysis | na | na | 1/10 in MeOH | 1/50 in MeOH | 1/10 in MeOH | 1/100 in C2H4O2·NH, 3 MeOH | 1/10 in MeOH |
| ion mode | na | na | NEG | NEG | NEG | POS | NEG |
| mass range | na | na | 400–800 | 200–800 | 400–2000 | 400–1000 | 400–2000 |
ESI Mass Spectrometric Analysis
Samples were diluted and analyzed in an Oribtrap Velos (Thermo Fisher) with a static nanospray source with 4 μm spray tips and a capillary temperature of 200 °C. The FTMS mode with a mass resolution of 100 K was used for all samples. Fraction 2 samples were diluted 1/10 in MeOH, and fraction 5 samples 1/100 in 10 mM ammonium acetate in MeOH. Both fractions were analyzed in negative ion mode using the following parameters: spray = 1.5 kV, Rf = 69%, and 1 scan = 500 ms. The m/z range for fraction 2 was 400–800, and 400–1000 for fraction 5. Adducts were observed in fraction 2 and were removed/reduced using the source fragmentation parameter of the mass spectrometer. MS/MS analysis was done to confirm the structure of the annotated lipids.62,63 The same mass spectrometry parameters were used and the CID energy ranged from 15 to 40%.
Data Processing
We used our lipid database for peak annotation. Intensities of all annotated lipids in each sample were automatically recorded in an Excel sheet using R.64 Data were normalized with the intensity of the internal standard of each fraction.
Statistical Analysis
Data were processed by multivariate analysis using the PLS toolbox and MatLab (MATLAB and Statistics Toolbox Release 2012b, The MathWorks, Inc., Natick, MA). The analysis allows visualization of the multivariate clusters within the samples and highlights the variables of interest (i.e., the increased or decreased variables in each group of sample). Then, each variable of interest was investigated by univariate analysis (two-way ANOVA and Bonferroni post-tests) using GraphPad Prism (GraphPad Prism 5.00, GraphPad Software Inc., San Diego, CA). The two parameters used were the age (JUV/ADU) and alcohol consumption (H2O/EtOH)].
Instrumentation for Laser Desorption/Ionization Mass Spectrometry Imaging
A Thermo Scientific MALDI LTQ- XL-Orbitrap instrument, using Xcalibur software, was used for MILDI-MS imaging data acquisition. Images were collected in positive ion mode by mechanical translation of the tissue section underneath a focused laser and acquisition of mass spectra at each position, with three laser shots/pixel at a fluency of 14 uJ. The laser focus is around 50 μm, so the target plate stepping distance was set to 50 μm for both the X and Y directions. The mass range was from 650 to 950 Da with the mass resolution set to 60 000 (@m/z 400).
Tissue Imaging Using Silver Nanoparticle Matrix Implantation
MALDI-MSI using organic matrices was first demonstrated for imaging peptides and proteins in tissue,67 and the technique was subsequently used to profile and image lipids in brain tissue.68,69 At the time, three technical issues were recognized as preventing quantitative MALDI: (1) reproducible and uniform dispersion of matrix solution across the tissue sample, (2) reliable incorporation of molecular analyte into the matrix, and (3) variability of the ionization probabilities of a desorbed molecule from one histological region of the tissue to other regions. The introduction of MILDI-MSI43−46 solves the first two issues.
The implantation of AgNP– and analysis by microfocused laser desorption proceeds by first affixing 18 μm thick coronal mice brain sections (+1.54 mm, −1.70 mm, and −5.88 mm re: bregma) onto glass microscope slides. Implantation of 6 nm diameter AgNP uses an NPlanter (Ionwerks Inc., Houston, TX) which produces Ag vapor (by magnetron sputtering). The Ag vapor condenses into small, pure, singly charged AgNP– ions within a gas filled condensation zone. The AgNP diameters range from 1 to 15 nm, and a quadrupole mass filter selects a nominal diameter of 6 nm (2.5 nm fwhm). These ions are accelerated to energies of 500 eV, formed into a beam, focused to 1 mm diameter onto the tissue sections, and electrically rastered over an area of 400 mm2 (20 × 20), thus covering the mouse brain coronal tissue surface. A portion of the ion beam current is continually measured with an electrometer and used to control the ion beam fluctuations to less than 5% maximum variation during deposition. Implantation of a whole tissue section requires 18 min.
The reproducibility of MILDI-MSI is demonstrated by implantation of AgNP– into four tissue sections from a single normal rat which were serially cut starting at 1.80 mm re: bregma. Each of the tissues was implanted on different days to obtain an estimate of the intensity variance of each analyzed lipid due to instrumental instabilities during either the AgNP– implantation or the subsequent mass spectral image acquisition. Examples of the typical variances of lipid ion intensities from these four serial tissues sections are SM 18:0/d18:1 (5.7%), SM 24:1/d18:1(7.4%), Cer 18:0/d18:1 (10.9%), and Cer 24:1/d18:1(7.0%). The similarity of mass resolved CER images from the four serial sections is shown in the Supporting Information (Figure S5).
Though highly reproducible, the MILDI-MS lipid intensities do not represent molecular concentrations. Precise and quantitative addition of NP matrix makes the desorbed lipid signal reproducible, but as with any matrix there is no guarantee that the lipid ionization efficiency will be the same from a control and experimentally manipulated tissue. Thus, even if the molecular concentration of the lipid analyte is equal in both samples, different local chemical environments may exist in the diseased tissue which change the probability of ion formation from a local experimental (or pathological) region relative to the same region of the control.
Imaging Data Processing
Imaging data were subsequently analyzed with ImageWerks software (Ionwerks, Inc.). Average intensity/pixel of mass resolved lipid ions within user defined regions of interest (ROIs) are computed by integrating the total intensity within the ROI and dividing by the number of pixels (these intensities are not normalized by the total intensity of all ions in the spectrum as is usually done in MALDI-MSI). The ROI average intensities are compared between animal groups. Images for a specific lipid are displayed using the same linear color scale to allow a visual comparison of intensities between samples; however, the low number of samples (2–3) used for MS Imaging did not allow us to perform statistical analysis on those intensities.
Acknowledgments
The authors acknowledge Dr. Mari Prieto, Glen Gregory and Thermo Fisher Corporation for technical and instrumentation advice.
Supporting Information Available
Partial least square discriminant analysis; two-way ANOVA and Bonferroni post-test results; regions of interest drawn for each level; Figure 45 and Figure 80 are the denominations used for bregmas 1.70 and 5.88 mm by Franklin & Paxinos in their mouse brain atlas; reproducibility of two CER in four serial sections of rat brain tissue. This material is available free of charge via the Internet at http://pubs.acs.org.
This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, NIH. Ionwerks and the University of Pittsburgh gratefully acknowledge ARRA support through NIDA phase II SBIR Grant 1RC3DA031431-01.
The authors declare no competing financial interest.
Notes
∇ A.R. and L.M.: Co-first authors.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gupta S.; Warner J. (2008) Alcohol-related dementia: A 21st-century silent epidemic?. Br. J. Psychiatry 193(5), 351–353. [DOI] [PubMed] [Google Scholar]
- Kopera M.; Wojnar M.; Brower K.; Glass J.; Nowosad I.; Gmaj B.; Szelenberger W. (2012) Cognitive functions in abstinent alcohol-dependent patients. Alcohol 46(7), 665–671. [DOI] [PubMed] [Google Scholar]
- Harper C. (2009) The neuropathology of alcohol-related brain damage. Alcohol Alcohol. 44(2), 136–140. [DOI] [PubMed] [Google Scholar]
- Harris G. J.; Jaffin S. K.; Hodge S. M.; Kennedy D.; Caviness V. S.; Marinkovic K.; Papadimitriou G. M.; Makris N.; Oscar-Berman M. (2008) Frontal white matter and cingulum diffusion tensor imaging deficits in alcoholism. Alcohol.: Clin. Exp. Res. 32(6), 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A.; Sullivan E. V.; Rosenbloom M. J.; Mathalon D. H.; Lim K. O. (1998) A controlled study of cortical gray matter and ventricular changes in alcoholic men over a 5-year interval. Arch. Gen. Psychiatry 55(10), 905–912. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A.; Rosenbloom M.; Rohlfing T.; Sullivan E. V. (2009) Degradation of association and projection white matter systems in alcoholism detected with quantitative fiber tracking. Biol. Psychiatry 65(8), 680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas V. A.; Studholme C.; Meyerhoff D. J.; Song E.; Weiner M. W. (2005) Chronic active heavy drinking and family history of problem drinking modulate regional brain tissue volumes. Psychiatry Res. 138(2), 115–130. [DOI] [PubMed] [Google Scholar]
- Jernigan T. L.; Butters N.; DiTraglia G.; Schafer K.; Smith T.; Irwin M.; Grant I.; Schuckit M.; Cermak L. S. (1991) Reduced cerebral grey matter observed in alcoholics using magnetic resonance imaging. Alcohol.: Clin. Exp. Res. 15(3), 418–427. [DOI] [PubMed] [Google Scholar]
- Ratti M. T.; Soragna D.; Sibilla L.; Giardini A.; Albergati A.; Savoldi F.; Bo P. (1999) Cognitive impairment and cerebral atrophy in “heavy drinkers”. Prog. Neuropsychopharmacol. Biol. Psychiatry 23(2), 243–258. [DOI] [PubMed] [Google Scholar]
- Delis F.; Benveniste H.; Xenos M.; Grandy D.; Wang G. J.; Volkow N. D.; Thanos P. K. (2012) Loss of dopamine D2 receptors induces atrophy in the temporal and parietal cortices and the caudal thalamus of ethanol-consuming mice. Alcohol.: Clin. Exp. Res. 36(5), 815–825. [DOI] [PubMed] [Google Scholar]
- Goldstein R. Z.; Volkow N. D. (2002) Drug addiction and its underlying neurobiological basis: Neuroimaging evidence for the involvement of the frontal cortex. Am. J. Psychiatry 159(10), 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N. D.; Hitzemann R.; Wang G. J.; Fowler J. S.; Burr G.; Pascani K.; Dewey S. L.; Wolf A. P. (1992) Decreased brain metabolism in neurologically intact healthy alcoholics. Am. J. Psychiatry 149(8), 1016–1022. [DOI] [PubMed] [Google Scholar]
- Oscar-Berman M.; Marinkovic K. (2007) Alcohol: Effects on neurobehavioral functions and the brain. Neuropsychol. Rev. 17(3), 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum J. B.; Ramchandani V. A.; Bodurka J.; Rawlings R.; Momenan R.; George D.; Hommer D. W. (2007) Blunted rostral anterior cingulate response during a simplified decoding task of negative emotional facial expressions in alcoholic patients. Alcohol.: Clin. Exp. Res. 31(9), 1490–1504. [DOI] [PubMed] [Google Scholar]
- Chanraud S.; Pitel A. L.; Pfefferbaum A.; Sullivan E. V. (2011) Disruption of functional connectivity of the default-mode network in alcoholism. Cereb. Cortex 21(10), 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E. V.; Muller-Oehring E.; Pitel A. L.; Chanraud S.; Shankaranarayanan A.; Alsop D. C.; Rohlfing T.; Pfefferbaum A. (2013) A selective insular perfusion deficit contributes to compromised salience network connectivity in recovering alcoholic men. Biol. Psychiatry 74(7), 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R. J.; Lallemand F.; de W. P. (2009) Biochemical and neurotransmitter changes implicated in alcohol-induced brain damage in chronic or “binge drinking” alcohol abuse. Alcohol Alcohol. 44(2), 128–135. [DOI] [PubMed] [Google Scholar]
- Ehrlich D.; Pirchl M.; Humpel C. (2012) Effects of long-term moderate ethanol and cholesterol on cognition, cholinergic neurons, inflammation, and vascular impairment in rats. Neuroscience 205, 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.; Crews F. T. (2008) Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp. Neurol. 210(2), 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R. J.; Colivicchi M. A.; Allen R.; Schol F.; Lallemand F.; de W. P.; Ballini C.; Corte L. D.; Dexter D. (2009) Neuro-inflammation induced in the hippocampus of “binge drinking” rats may be mediated by elevated extracellular glutamate content. J. Neurochem. 111(5), 1119–1128. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lizarbe S.; Montesinos J.; Guerri C. (2013) Ethanol induces TLR4/TLR2 association, triggering an inflammatory response in microglial cells. J. Neurochem. 126(2), 261–273. [DOI] [PubMed] [Google Scholar]
- Crews F.; Nixon K.; Kim D.; Joseph J.; Shukitt-Hale B.; Qin L.; Zou J. (2006) BHT blocks NF-kappaB activation and ethanol-induced brain damage. Alcohol.: Clin. Exp. Res. 30(11), 1938–1949. [DOI] [PubMed] [Google Scholar]
- Ward R. J.; Zhang Y.; Crichton R. R.; Piret B.; Piette J.; de W. P. (1996) Identification of the nuclear transcription factor NFkappaB in rat after in vivo ethanol administration. FEBS Lett. 389(2), 119–122. [DOI] [PubMed] [Google Scholar]
- Qin L.; Crews F. T. (2012) NADPH oxidase and reactive oxygen species contribute to alcohol-induced microglial activation and neurodegeneration. J. Neuroinflammation 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M. R.; Miller A. H. (2007) Depressive disorders and immunity: 20 years of progress and discovery. Brain Behav. Immun. 21(4), 374–383. [DOI] [PubMed] [Google Scholar]
- Achur R. N.; Freeman W. M.; Vrana K. E. (2010) Circulating cytokines as biomarkers of alcohol abuse and alcoholism. J. Neuroimmune Pharmacol 5(1), 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alho H.; Sillanaukee P.; Kalela A.; Jaakkola O.; Laine S.; Nikkari S. T. (2004) Alcohol misuse increases serum antibodies to oxidized LDL and C-reactive protein. Alcohol Alcohol. 39(4), 312–315. [DOI] [PubMed] [Google Scholar]
- Mayfield J.; Ferguson L.; Harris R. A. (2013) Neuroimmune signaling: A key component of alcohol abuse. Curr. Opin. Neurobiol. 23(4), 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun Y. A.; Obeid L. M. (2008) Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9(2), 139–150. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A.; Obeid L. M. (2011) Many ceramides. J. Biol. Chem. 286(32), 27855–27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C.; Martinez-Martinez P. (2013) Ceramide function in the brain: When a slight tilt is enough. Cell. Mol. Life Sci. 70(2), 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleda G.; Morales L. C.; Benitez B.; Arboleda H. (2009) Regulation of ceramide-induced neuronal death: Cell metabolism meets neurodegeneration. Brain Res. Rev. 59(2), 333–346. [DOI] [PubMed] [Google Scholar]
- Bikman B. T.; Summers S. A. (2011) Ceramides as modulators of cellular and whole-body metabolism. J. Clin Invest 121(11), 4222–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Car H.; Zendzian-Piotrowska M.; Prokopiuk S.; Fiedorowicz A.; Sadowska A.; Kurek K.; Sawicka D. (2012) Ceramide profiles in the brain of rats with diabetes induced by streptozotocin. FEBS J. 279(11), 1943–1952. [DOI] [PubMed] [Google Scholar]
- Cutler R. G.; Kelly J.; Storie K.; Pedersen W. A.; Tammara A.; Hatanpaa K.; Troncoso J. C.; Mattson M. P. (2004) Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 101(7), 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana A.; Hogan E. L.; Pahan K. (2009) Ceramide and neurodegeneration: Susceptibility of neurons and oligodendrocytes to cell damage and death. J. Neurol. Sci. 278(1–2), 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviad E. L.; Albee L.; Pankova-Kholmyansky I.; Epstein S.; Park H.; Merrill A. H. Jr.; Futerman A. H. (2008) Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283(9), 5677–5684. [DOI] [PubMed] [Google Scholar]
- Levy M.; Futerman A. H. (2010) Mammalian ceramide synthases. IUBMB Life 62(5), 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann S.; Ferreiros N.; Birod K.; Eberle M.; Schreiber Y.; Pfeilschifter W.; Ziemann U.; Pierre S.; Scholich K.; Grosch S.; Geisslinger G. (2012) Ceramide synthase 6 plays a critical role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 188(11), 5723–5733. [DOI] [PubMed] [Google Scholar]
- Ben-David O.; Futerman A. H. (2010) The role of the ceramide acyl chain length in neurodegeneration: Involvement of ceramide synthases. Neuromol. Med. 12(4), 341–350. [DOI] [PubMed] [Google Scholar]
- Jung J. S.; Shin K. O.; Lee Y. M.; Shin J. A.; Park E. M.; Jeong J.; Kim D. H.; Choi J. W.; Kim H. S. (2013) Anti-inflammatory mechanism of exogenous C2 ceramide in lipopolysaccharide-stimulated microglia. Biochim. Biophys. Acta 1831(6), 1016–1026. [DOI] [PubMed] [Google Scholar]
- van Doorn R.; Nijland P. G.; Dekker N.; Witte M. E.; Lopes-Pinheiro M. A.; van het H. B.; Kooij G.; Reijerkerk A.; Dijkstra C.; van der Valk P.; van Horssen J.; de Vries H. E. (2012) Fingolimod attenuates ceramide-induced blood-brain barrier dysfunction in multiple sclerosis by targeting reactive astrocytes. Acta Neuropathol. 124(3), 397–410. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A.; Obeid L. M. (2002) The Ceramide-centric universe of lipid-mediated cell regulation: Stress encounters of the lipid kind. J. Biol. Chem. 277(29), 25847–25850. [DOI] [PubMed] [Google Scholar]
- Liu G.; Ghahremani M. H.; Banihashemi B.; Albert P. R. (2003) Diacylglycerol and ceramide formation induced by dopamine D2S receptors via Gbeta gamma-subunits in Balb/c-3T3 cells. Am. J. Physiol.: Cell Physiol. 284(3), C640–C648. [DOI] [PubMed] [Google Scholar]
- Colsch B.; Woods A. S. (2010) Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 20(6), 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Gross R. W. (2003) Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44(6), 1071–1079. [DOI] [PubMed] [Google Scholar]
- Novikov A.; Caroff M.; Della-Negra S.; Lebeyec Y.; Pautrat M.; Schultz J. A.; Tempez A.; Wang H. Y.; Jackson S. N.; Woods A. S. (2004) Matrix-implanted laser desorption/ionization mass spectrometry. Anal. Chem. 76(24), 7288–7293. [DOI] [PubMed] [Google Scholar]
- Tempez A.; Ugarov M.; Egan T.; Schultz J. A.; Novikov A.; Della-Negra S.; Lebeyec Y.; Pautrat M.; Caroff M.; Smentkowski V. S.; Wang H. Y.; Jackson S. N.; Woods A. S. (2005) Matrix implanted laser desorption ionization (MILDI) combined with ion mobility-mass spectrometry for bio-surface analysis. J. Proteome Res. 4(2), 540–545. [DOI] [PubMed] [Google Scholar]
- Jackson S. N.; Baldwin K.; Muller L.; Womack V. M.; Schultz J. A.; Balaban C.; Woods A. S. (2014) Imaging of lipids in rat heart by MALDI-MS with silver nanoparticles. Anal. Bioanal. Chem. 406(5), 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov A.; Caroff M.; Della-Negra S.; Depauw J.; Fallavier M.; Le B. Y.; Pautrat M.; Schultz J. A.; Tempez A.; Woods A. S. (2005) The Au(n) cluster probe in secondary ion mass spectrometry: Influence of the projectile size and energy on the desorption/ionization rate from biomolecular solids. Rapid Commun. Mass Spectrom. 19(13), 1851–1857. [DOI] [PubMed] [Google Scholar]
- Victor M.; Adams R. D.; MANCALL E. L. (1959) A Restricted Form of Cerebellar Cortical Degeneration Occurring in Alcoholic Patients. AMA Arch. Neurol. 1(6), 579–688. [Google Scholar]
- Abernathy K.; Chandler L. J.; Woodward J. J. (2010) Alcohol and the prefrontal cortex. Int. Rev. Neurobiol. 91, 289–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moselhy H. F.; Georgiou G.; Kahn A. (2001) Frontal lobe changes in alcoholism: A review of the literature. Alcohol Alcohol. 36(5), 357–368. [DOI] [PubMed] [Google Scholar]
- Chen G.; Cuzon Carlson V. C.; Wang J.; Beck A.; Heinz A.; Ron D.; Lovinger D. M.; Buck K. J. (2011) Striatal involvement in human alcoholism and alcohol consumption, and withdrawal in animal models. Alcohol.: Clin. Exp. Res. 35(10), 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. M.; Best P. J. (2000) Effects of ethanol on hippocampal place-cell and interneuron activity. Brain Res. 876(1–2), 154–165. [DOI] [PubMed] [Google Scholar]
- Ryabinin A. E. (1998) Role of hippocampus in alcohol-induced memory impairment: implications from behavioral and immediate early gene studies. Psychopharmacology (Berlin, Ger.) 139(1–2), 34–43. [DOI] [PubMed] [Google Scholar]
- Valenzuela C. F.; Lindquist B.; Zamudio-Bulcock P. A. (2010) A review of synaptic plasticity at Purkinje neurons with a focus on ethanol-induced cerebellar dysfunction. Int. Rev. Neurobiol. 91, 339–372. [DOI] [PubMed] [Google Scholar]
- Fadda F.; Rossetti Z. L. (1998) Chronic ethanol consumption: From neuroadaptation to neurodegeneration. Prog. Neurobiol. 56(4), 385–431. [DOI] [PubMed] [Google Scholar]
- Saito M.; Chakraborty G.; Hegde M.; Ohsie J.; Paik S. M.; Vadasz C.; Saito M. (2010) Involvement of ceramide in ethanol-induced apoptotic neurodegeneration in the neonatal mouse brain. J. Neurochem. 115(1), 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprow G. M.; Thiele T. E. (2012) The neurobiology of binge-like ethanol drinking: Evidence from rodent models. Physiol. Behav. 106(3), 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colsch B.; Woods A. S. (2010) Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 20(6), 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. (2002) Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 302(2), 199–212. [DOI] [PubMed] [Google Scholar]
- Gu M.; Kerwin J. L.; Watts J. D.; Aebersold R. (1997) Ceramide profiling of complex lipid mixtures by electrospray ionization mass spectrometry. Anal. Biochem. 244(2), 347–356. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2012) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Chaurand P.; Caprioli R. M. (2002) Direct profiling and imaging of peptides and proteins from mammalian cells and tissue sections by mass spectrometry. Electrophoresis 23(18), 3125–3135. [DOI] [PubMed] [Google Scholar]
- Jackson S. N.; Ugarov M.; Egan T.; Post J. D.; Langlais D.; Albert S. J.; Woods A. S. (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom. 42(8), 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvolve A. M.; Colsch B.; Woods A. S. (2011) Highlighting anatomical sub-structures in rat brain tissue using lipid imaging. Anal. Methods 3(8), 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.