Summary
The structure of the I domain of integrin αLβ 2 bound to the Ig superfamily ligand ICAM-1 reveals the open ligand binding conformation and the first example of an integrin-IgSF interface. The I domain Mg2+ directly coordinates Glu-34 of ICAM-1, and a dramatic swing of I domain residue Glu-241 enables a critical salt bridge. Liganded and unliganded structures for both high- and intermediate-affinity mutant I domains reveal that ligand binding can induce conformational change in the αL I domain and that allosteric signals can convert the closed conformation to intermediate or open conformations without ligand binding. Pulling down on the C-terminal α7 helix with introduced disulfide bonds ratchets the β6-α7 loop into three different positions in the closed, intermediate, and open conformations, with a progressive increase in affinity.
Introduction
Integrins are a family of noncovalently associated, αβ heterodimeric transmembrane molecules that mediate cell-cell and cell-extracellular matrix adhesion. Lympho cyte function-associated antigen-1 (LFA-1, αLβ2) is an integrin that is critically important in antigen-specific responses and homing by lymphocytes and together with other β2 integrins in diapedesis by monocytes and neutrophils at inflammatory sites (Gahmberg et al., 1997; Larson and Springer, 1990; Shimaoka et al., 2002). αLβ2 recognizes intercellular adhesion molecules (ICAMs), members of the Ig superfamily (IgSF) of which ICAM-1 is the most biologically important (Dustin and Springer, 1999). ICAM-1 is highly inducible on antigen-presenting cells and endothelium by cytokines in inflammation.
Although the extracellular domains of αL and β2 are each large and structurally complex, the ligand binding site is contained solely within the 180 residue inserted (I) domain of αL (Shimaoka et al., 2001, 2002). The I domain is important in ligand binding in the 9 of 18 integrin α subunits in which it is present. Crystal structures of integrin I domains reveal a dinucleotide binding or Rossmann fold, with a central hydrophobic β sheet surrounded by seven amphipathic α helices (Lee et al., 1995b; Shimaoka et al., 2002). A Mg2+ ion is coordinated at the “top” of the domain in a metal ion-dependent adhesion site (MIDAS).
Integrins trigger “outside-in” signals in response to ligand binding (Giancotti and Ruoslahti, 1999; Schwartz and Ginsberg, 2002). In B and T cell responses, αLβ2 augments proliferation and protects against apoptosis (Koopman et al., 1994; Van Seventer et al., 1990). Ligand binding induces significant structural rearrangements in the I domain of the integrin α2 subunit, as seen in a complex with a collagen-like peptide (Emsley et al., 2000). Compared to the unliganded “closed” conformation of the α2 I domain, the liganded “open” conformation exhibits a large 10Å movement of the C-terminal α helix down the side of the domain, and a rearrange ment in metal coordination at the MIDAS. The metal ion is central in the binding site and directly coordinates a Glu residue in the ligand. A similar conformational change has been observed in the αM I domain; however, in this case it is induced by a ligand-like contact of the metal in the MIDAS with a Glu residue of a neighboring I domain in the crystal lattice (Lee et al., 1995b). By contrast, multiple αL I domain structures have consis tently revealed the unliganded, closed conformation (Kallen et al., 1999; Legge et al., 2000; Qu and Leahy, 1995, 1996).
In the absence of activation, αLβ2 has low affinity for ligand (Dustin and Springer, 1989; Lollo et al., 1993). In inside-out signaling by integrins, signals received by other receptors activate intracellular signaling pathways that impinge on integrin cytoplasmic domains and make the extracellular domain competent for ligand binding on a timescale of less than 1 s (Hughes and Pfaff, 1998; Lollo et al., 1993; Shimaoka et al., 2002). This unique property enables leukocytes to rapidly respond to signals in the environment, such as foreign antigen or che-moattractants, to activate adhesion and direct cell migration. A disulfide bond has been introduced into the αL I domain to stabilize the predicted open conformation and was found to increase the affinity for ICAM-1 by 10,000-fold (Shimaoka et al., 2001). This strongly supports the idea that conformation regulates affinity; however, whether the open conformation would be stable in the absence of bound ligand has not been established because all open I domain structures determined to date are ligand bound (Emsley et al., 2000; Lee et al., 1995b) Two activation states have been defined functionally for αLβ2 on cells activated by differing stimuli. Both states are competent for cell adhesion, but high-affinity soluble ligand binding is only detectable for one of these states (Constantin et al., 2000; Stewart et al., 1996; van Kooyk et al., 1999). However, the concept that I domains might exist in intermediate- as well as high-affinity states (Shimaoka et al., 2001) has not been tested by attempting to mutationally stabilize such states or define their structure.
Because of the key role of the interaction between LFA-1 and ICAM-1 in immune responses, defining its structural basis is of great interest. Furthermore, development of pharmaceutical antagonists of this interac tion is of great importance for treatment of autoimmune disease (Giblin and Kelly, 2001; Gottlieb et al., 2000). Crystal structures have been determined for IgSF molecules recognized by integrins, i.e., ICAM-1 (Bella et al., 1998; Casasnovas et al., 1998), ICAM-2 (Casasnovas et al., 1997), VCAM-1, and MAdCAM-1 (Wang and Springer, 1998), but not for IgSF complexes with integrins. Here, the crystal structure of the I domain:ICAM-1 complex reveals an atomic view of an integrin-IgSF ligand interface for the first time. Furthermore, multiple I domain structures reveal an intermediate state in the shape-shifting pathway and show that the I domain can be stabilized in the open conformation in the absence of bound ligand.
Results and Discussion
Designed αL I Domains
Previously, αL residues 289 and 294 or residues 287 and 294 were mutated to cysteine to stabilize the closed, or the predicted open, conformations of the I domain, respectively (Table 1; Shimaoka et al., 2001). We made four further pairs of cysteine substitutions where the Cβ-Cβ distances were predicted to be closer in the open than closed conformation and hence should form disulfides that stabilized the open conformation, and we made one pair of cysteine substitutions where the Cβ-Cβ distances were more favorable for the closed conformation (Table 1). All cysteine substitutions were distant from the MIDAS and ligand binding surface so that direct involvement of the mutated residues in ligand binding could be ruled out. All mutations included one residue in the C-terminal α7 helix. None of the new mutations had Cβ-Cβ distances that were optimal for disul-fide formation (3.3 to 4.4Å); however, one or both residues of each pair were in an α helix or loop where backbone movements should be allowable that would permit disulfide formation.
Table 1.
αL I Domain Design and Binding to ICAM-1
| I Domain | Open Cβ - Cβ | Closed Distance (Å) | kon (M–1s–1 × 10–3) | koff (s–1) | KD (μM) | Class |
|---|---|---|---|---|---|---|
| K287C/K294C | 3.8 | 9.1 | 115 ± 7 | 0.014 ± 0.001 | 0.15 ± 0.016 | High |
| E284C/E301C | 7.0 | 12.5 | 105 ± 3 | 0.045 ± 0.006 | 0.36 ± 0.04 | High |
| L161C/F299C | 8.1 | 11.4 | 133 ± 10 | 0.43 ± 0.07 | 3.0 ± 0.44 | Inter. |
| K160C/F299C | 7.8 | 8.0 | 103 ± 15 | 0.77 ± 0.07 | 8.4 ± 2.4 | Inter. |
| L161C/T300C | 13.0 | 14.9 | 89 ± 12 | 0.76 ± 0.07 | 9.4 ± 2.4 | Inter. |
| K160C/T300C | 12.8 | 10.9 | 3.4 ± 0.9 | 1.2 ± 0.08 | 450 ± 210 | Low |
| L289C/K294C | 8.0 | 3.9 | 2.3 ± 0.3 | 3.6 ± 0.34 | 1600 ± 170 | Low |
| Wild-type | N/A | N/A | 3.1 ± 0.1 | 4.6 ± 0.36 | 1500 ± 200 | Low |
The distances between wild-type residue Cβ atoms replaced with cysteine in mutants were measured in the open model (Shimaoka et al., 2001) or closed aL structure (1ZON) (Qu and Leahy, 1996).
The interaction of I domains with immobilized ICAM-1 in the presence of Mg2+ was monitored with surface plasmon resonance. kon and koff were obtained by curve fitting using a 1:1 binding model. KD was calculated by Scatchard plot using binding at steady state.
Structures of the underlined mutants K287C/K294C and L161C/F299C are determined here. N/A, not applicable.
Affinity and kinetic measurements using surface plasmon resonance (Shimaoka et al., 2001) showed that the mutant αL I domains fell into three classes (Table 1). Two mutants with the closest Cβ-Cβ distances in the predicted open conformation, and the greatest increase in Cβ-Cβ distances in the closed conformation, bound to ICAM-1 with high affinity (KD of 150 to 360 nM). Three mutants, which also had Cβ-Cβ atoms that were closer in the predicted open than the closed conformation, bound ICAM-1 with intermediate affinity (KD of 3 to 9 μM). Two mutants which had Cβ-Cβ atoms that were closer in the closed than the predicted open conforma tion had low affinity (KD of 0.5 to 1.6 mM), similar to the KD of 1.5 mM of the wild-type I domain. DTT reduction of the disulfides of the mutants of the high- and intermediate-affinity classes reduced their affinity to wild-type levels, and EDTA abolished binding, showing that the interactions were Mg2+ dependent (data not shown). All of the αL I domain disulfide bond mutants we have made are described here. It is interesting that the affinities of the mutants cluster into three groups of high, intermediate, and low affinity, because this clustering is consistent with the existence of three distinct conformational states, as shown below. The kinetics of the mutants also cluster. Interestingly, the kon of the intermediate- and high-affinity mutants are increased by a similar amount (Table 1).
Four Intermediate- and High-Affinity αL I Domain Crystal Structures
Crystal structures were determined for representative intermediate-affinity (L161C/F299C) and high-affinity (K287C/K294C) αL I domain mutants (Table 2). Each was determined in two forms. Intermediate-affinity I domain structures were determined in the unliganded state (1.3Å resolution) or in complex with ICAM-1 (3.3Å). High-affinity I domain structures were determined in the unliganded state (2.5Å) or in the “pseudo-liganded” state (2.0Å),i.e., with a ligand-mimetic lattice contact at the MIDAS. All structures were solved using molecular replacement (Table 2).
Table 2.
Statistics of X-Ray Diffraction Data Collection and Structure Refinement
| Intermediate-Affinity I Domain |
High-Affinity I Domain |
|||
|---|---|---|---|---|
| Unliganded | + ICAM-1 | Unliganded | Pseudo-liganded | |
| Space group | P212121 | P1 | C2221 | P41212 |
| Unit cell | ||||
| a, b, c (Å) | 40.4, 57.4, 66.9 | 46.6, 62.9, 81.5 | 61.9, 121.3, 54.1 | 35.6, 35.6, 269.6 |
| α, β, γ (°) | 90, 90, 90 | 95.4, 106.7, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50-1.3 | 50-3.3 | 50-2.5 | 50-2.0 |
| Number of reflections (total/unique) | 197,616/35,329 | 23,330/12,338 | 41,352/7,271 | 57,584/10,569 |
| αL residues | 128-306 | 128-306 | 128-300 | 128-300 |
| Number of protein/ hetero-atoms | 1427/329 | 5680/172 | 1388/31 | 1380/91 |
| Completeness (%) | 90.2/47.0a | 92.3/70.7a | 94.2/60.4a | 82.1/67.5a |
| I/σ(I) | 17.8/2.4a | 7.7/1.5a | 8.4/2.0a | 12.6/3.5a |
| Rmerge (%)b | 8.8/35.2a | 9.4/41.2a | 18.4/41.3a | 9.4/38.1a |
| Rmsd bond lengths | 0.012 Å | 0.016 Å | 0.005 Å | 0.006 Å |
| Rmsd bond angles | 2.4° | 1.7° | 1.3° | 1.3° |
| Rworkc | 15.6% | 26.4% | 25.0% | 21.3% |
| Rfreed | 20.0% | 31.3% | 28.7% | 26.5% |
| Average B factor | 15.9 Å2 | 67.3 Å2 | 34.7 Å2 | 23.7 Å2 |
| Ramachandran plot (core/disallowed)e | 91.6%/0% | 72.7%/0% | 80.5%/0% | 90.6%/0% |
| PDB code | 1MJN | 1MQ8 | 1MQA | 1MQ9 |
The second of each pair of numbers corresponds to the last resolution shell.
ΣhΣi|Ii(h) - <I(h)>|/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
Σh∥Fobs (h)| - |Fcalc (h)∥/Σh|Fobs (h)|, where Fobs (h) and Fcalc (h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 10% (for the unliganded intermediate-affinity, unliganded and pseudo-liganded high-affinity structures) or 5% (the I domain-ICAM-1 complex structure) subset of the data set excluded from refinement.
Residues in core (most favorable) and disallowed regions of the Ramachandran plot as reported by Procheck (Laskowski et al., 1993).
Structure of an Integrin—IgSF Complex
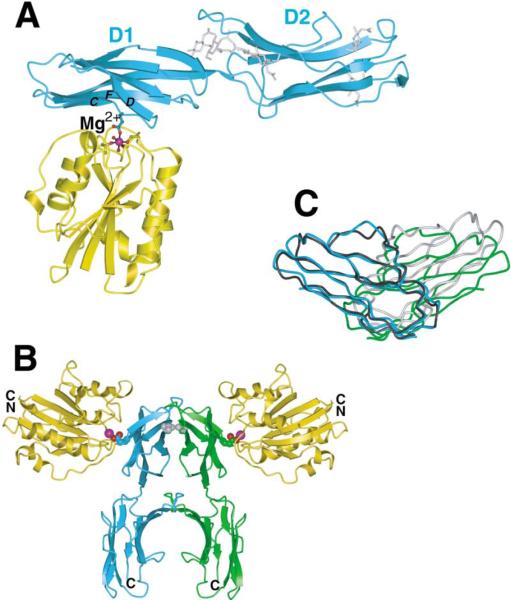
A fragment of ICAM-1 containing IgSF domains 1–3 (residues 1–291) crystallized in complex with the intermediate-affinity αL I domain (Figure 1A). There is no meaningful density for IgSF domain 3 of ICAM-1, which appears to be mobile within the crystal lattice. Domains 1 and 2 of ICAM-1 have an extended orientation with an angle between domains 1 and 2 within the range previously observed (Bella et al., 1998; Casasnovas et al., 1998). There are no significant rearrangements in ICAM-1 asso ciated with binding to the αL I domain.
Figure 1. Two αL I Domains Bound to an ICAM-1 Dimer.
(A) Ribbon diagram of one monomeric unit of the intermediate-affinity αL I domain (gold) complex with ICAM-1 domains 1-2 (cyan). The Mg2+ ion is shown as a magenta sphere. I domain MIDAS and ICAM-1 Glu-34 side chains are shown as ball-and-stick with red oxygen atoms. The interacting β strands C, D, and F of ICAM-1 are labeled. N-acetyl glucosamine residues of ICAM-1 are shown with silver bonds.
(B) The two ICAM-1-I domain complexes in the crystallographic asymmetric unit are shown with the I domains colored gold and the two ICAM-1 molecules colored cyan and green. The 2-fold axis between the ICAM-1 molecules is in the vertical direction, normal to the predicted membrane plane. Positions of the magnesium ions (magenta spheres) and Glu-34 of ICAM-1 (CPK) are shown for reference. The ICAM-1 Val-51 residues at the center of the ICAM-1 dimer interface are shown as gray CPK models.
(C) A view of domains 1 of the ICAM-1 dimer (cyan and green), rotated about 90° from the view in (B), with domain 1 of the uncomplexed ICAM-1 molecule A dimer (Casasnovas et al., 1998) superimposed using domain 1 of one of the ICAM-1 molecules (black), whereas the other one is colored gray. Figures 1–5 are prepared with programs GLR (provided by L. Esser), Molscript (Kraulis, 1991), Bobscript (Esnouf, 1997), Raster3D (Merritt and Murphy, 1994), GRASP (Nicholls et al., 1991), and Povray (The Povray Team, http://www.povray.org).
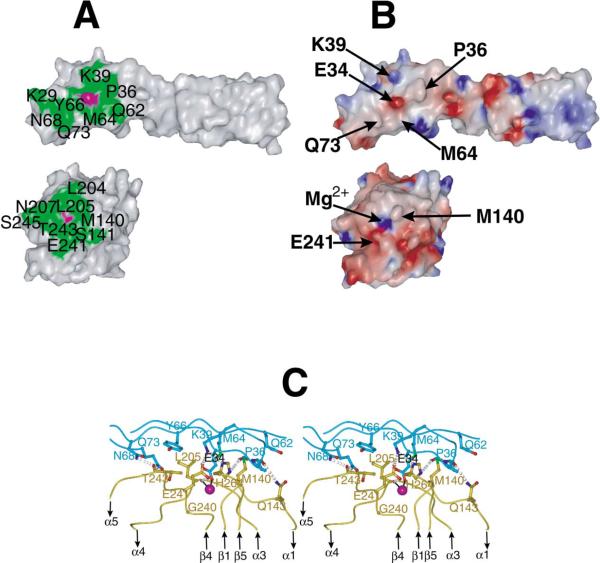
A total of 1250Å2 of solvent-accessible surface is buried in the I domain:ICAM-1 contact. This is a small interface for a protein:protein interaction (1600 ± 400Å2 ) (Conte et al., 1999). A shallow groove bearing the MIDAS on the “top” face of the I domain binds to the side of domain 1 of ICAM-1, making no contacts with the flexible loops at the N-terminal end of domain 1 (Casasnovas et al., 1998) or with domain 2 (Figure 1A). The contacting ICAM-1 β strands lie parallel to the groove (Figure 1A). These are the F and C β strands on the edge of one sheet and the D β strand on the edge of the other sheet. Glu-34 at the end of β strand C of ICAM-1 forms a direct coordination through its acidic side chain to the Mg2+ ion held in the I domain MIDAS, which is in the open conformation. Glu-34 sits precisely in the middle of the contact surface on ICAM-1 (magenta surface, Figure 2A, top), as does the Mg2+ ion on the I domain (magenta, Figure 2A, bottom). The contact surface is relatively flat, with the exception that Met-140 on the I domain (Figure 2B, bottom) protrudes and inserts its side chain into a shallow cleft between Met-64 and Pro-36 on ICAM-1 (Figure 2B, top).
Figure 2. The Interface between the αL I Domain and ICAM-1.
(A) Residues in the I domain:ICAM-1 interface in an open-book representation of the protein surface with the book folded open horizontally, with ICAM-1 above and the I domain below. Interacting residues are colored green and labeled, except for Glu-34 in ICAM-1 and the MIDAS Mg2+ ion in ICAM-1, which are magenta and unlabeled.
(B) The electrostatic surface of the complex in the same open-book representation. The color scale for the charge distribution extends from −10 kT/e− (red) to +10 kT/e− (blue) calculated using GRASP (Nicholls et al., 1991). The position of bulges in the interface are marked by arrows, as are the salt-bridged residues Lys-39 in ICAM-1 and Glu-241 in the I domain and Glu-34 in ICAM-1 and the Mg2+ ion in the I domain.
(C) Stereo view of interface details with ICAM-1 in cyan and I domain in gold. The view is about the same as in Figure 1A. The metal ion, oxygens, and nitrogens are represented as magenta, red, and blue spheres, respectively. Metal coordination and hydrogen bonds are represented by solid black lines and gray dotted lines, respectively. Contacting residues are shown as ball-and-stick models. For clarity, MIDAS residues are omitted.
The metal coordination bond between the Mg2+ and Glu-34 is surrounded on the I domain by a ring of hy drophobic residues (Leu-204, Leu-205, and Met-140) and the aliphatic portion of Thr-243 (Figure 2A, bottom). All interacting residues are contributed by the β1-α1, α3-α4, and β4-α5 loops, which also bear metal-coordi nating residues and form the MIDAS (Figures 2C and 3A). The ring of hydrophobic residues on the I domain is in contact with a similar ring of hydrophobic residues on ICAM-1 that surround Glu-34: Pro-36, Tyr-66, Met- 64, and the aliphatic portions of Gln-62 and Gln-73 (Figure 2A, top). The surrounding nonpolar environment will strengthen the Coulombic interaction between the Glu- 34 of ICAM-1 and Mg2+ of the MIDAS.
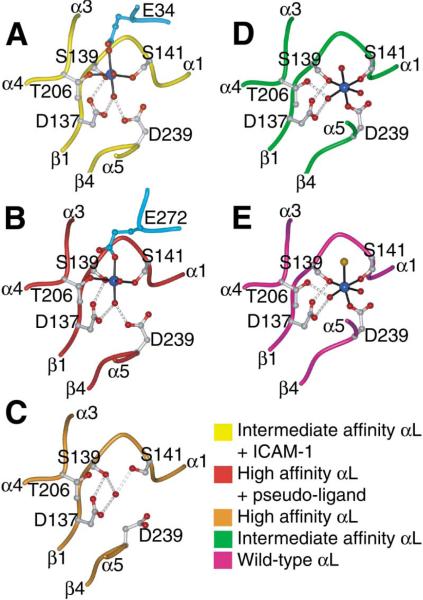
Figure 3. MIDAS Structures.
Structures are from the I domain:ICAM-1 complex (A), the pseudo-liganded high-affinity I domain (B), the unliganded high-affinity I domain (C), the unliganded intermediate-affinity I domain (D), and the wild-type I domain (E) (1LFA) (Qu and Leahy, 1996). The keys to the color scheme are shown below, with the ICAM-1 or ligand mimetic molecule colored cyan in (A) and (B). The metal ions are colored blue, water molecule and ligating side chain oxygen atoms are colored red, and the chloride ion from the wild-type I domain structure is colored orange. The MIDAS residues and Glu-34 from ICAM-1 in (A) and Glu-272 from a lattice mate I domain in (B) are shown as ball-and-stick models. Metal coordination and hydrogen bonds are represented by solid black lines and gray dotted lines, respectively.
Surrounding the hydrophobic ring are polar interactions involving hydrogen bonds and salt bridges that appear to orient ICAM-1 for optimal contact with the I domain and further strengthen their interaction. Many of the hydrogen bonds are backbone to side chain. Hydrogen bonding residues include Thr-35, Pro-36, Asn- 68 in ICAM-1, and Gln-143, Thr-243, and His-264 in the I domain (Figure 2C). The overall electrostatic surface of the contact area displays good charge complementarity (Figure 2B). A salt bridge between Glu-241 of the I domain and Lys-39 of ICAM-1, both of which are crucial for ligand binding (Edwards et al., 1998; Fisher et al., 1997), also facilitates positioning of ICAM-1 and the I domain for optimal interaction (Figures 2B and 2C). This contact is only possible because of a dramatic reorienta tion of the Glu-241 side chain upon conversion of the αL I domain to the open conformation (Figure 4A), as described below.
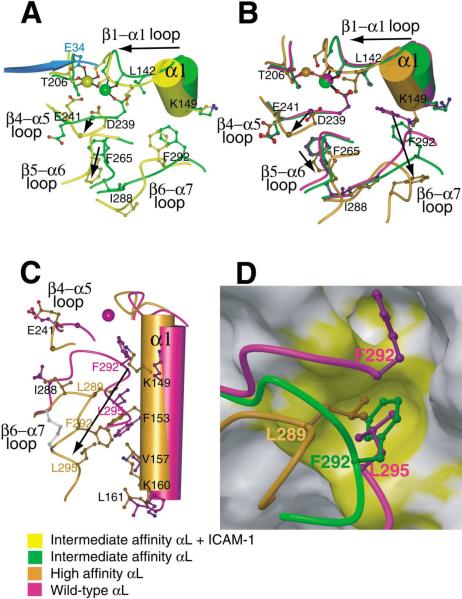
Figure 4. Propagation of Conformational Change in the I Domain.
(A–C) The effect of ligand binding (A) and remodeling of the β6-α7 loop in absence of ligand binding (B and C) are compared on I domain structures near the MIDAS (A and B) and the α7-α1 interface (C). Views of (A) and (B) are similar and (C) is approximately orthogonal to (B). Key residues in conformational change are shown in ball-and-stick and labeled, except Gly-240 is unlabeled and shown as a large Cα atom sphere and MIDAS residues Ser-139 and Ser-141 are shown but not labeled. Metal ions are shown as large spheres with the same color as the backbone of the corresponding I domain. The position of the missing metal ion in the unliganded high-affinity structure is simulated by superimposing the metal ion from the pseudo-liganded high-affinity structure and is shown smaller than the other metals. Side chain oxygen atoms are shown as red; water oxygens are omitted for clarity. The directions of major shifts from closed to open conformations are shown with arrows. A portion of ICAM-1 domain-1 containing the metal-coordinating residue E34 is shown in cyan. In (C), the side chain bonds in the designed disulfide bridge (Cys-287-Cys-294) are shown in gray.
(D) The hydrophobic pocket that acts as a detent for the rachet-like movement of the β6-α7 loop. The backbone of the β6-α7 loop and the three residues that occupy the same hydrophobic pocket in the three different conformational states are shown in the same color key as in (A)–(C). The pocket is shown as a GRASP van der Waals surface using the wild-type 1LFA structure with the residues from 287 to the C terminus deleted. The upper hydrophobic pocket is also shown, which is occupied only in the closed conformation (by F292 which is shown in the wild-type structure along with L295). For the closed conformation, the β6-α7 main chain trace is broken between F292 and L295 for clarity. The view is about the same as in (C). On the otherwise gray GRASP surface, residues Ile-150, Phe-153, Ile-237, Ile-259, and Ile-261 are colored yellow to show the hydrophobic pockets.
Previous mutational studies define the relative importance of the interactions revealed in the interface (Edwards et al., 1998; Fisher et al., 1997; Huang and Springer, 1995; Kamata et al., 1995; Staunton et al., 1990). Glu-34 of ICAM-1 and the residues that coordinate with the Mg2+ in the MIDAS of the I domain have long been identified as crucial. The ICAM-1 contact residues Lys-39, Met-64, Tyr-66, Asn-68, and Gln-73 were also previously recognized as important in mutational studies, as were the I domain contact residues Met-140, Glu-146, Leu-205, Glu-241, Thr-243, and Ser-243.
A Dimeric Integrin-Ligand Complex
The crystal asymmetric unit contains two ICAM-1:I domain complexes that are related by 2-fold noncrystallo-graphic symmetry (Figure 1B). The two ICAM-1 molecules dimerize at a hydrophobic interface on the BED sheet of domain 1. A total of 1100Å2 of solvent-accessible surface is buried. Dimerization at the same hy drophobic interface centered on Val-51 was seen in crystals of ICAM-1 domains 1-2 (Casasnovas et al., 1998) and was suggested to mediate physiologic dimerization on the cell surface. Comparison of ICAM-1 di mers shows some flexibility at the dimer interface (Figure 1C).
The spatial arrangement of the dimeric ICAM-1:I domain complex appears ideal for physiologic binding of two αLβ2 heterodimers to a cell surface ICAM-1 dimer. The bottoms of the two I domains bearing the connections to other integrin domains point in opposite directions and away from the connection of ICAM-1 domain 2 to domains 3–5 (Figure 1B). Orienting each half of the dimer similarly in the interface between two adherent cells places the dimer symmetry axis normal to the two cell membranes (Figure 1B). Thus, our structure provides the first glimpse of the orientation between two cells of LFA-1 and ICAM-1 in the immunological synapse (Grakoui et al., 1999). Our structure also provides the first glimpse of a dimeric integrin-ligand interaction. Since essentially all integrin ligands are either dimeric (in solution) or multimeric (on cell surfaces and the extracellular matrix), dimeric interactions of integrins with ligands are likely to be of broad significance.
Ligand-Induced Conformational Change in the I Domain: Outside-In Signaling
Comparison between the unliganded and liganded intermediate affinity I domain structures reveals the critical rearrangements in the ligand binding site that are induced by binding to ICAM-1 (see Supplemental Movie S1 at http://www.cell.com/cgi/content/full/112/1/99/DC1). The MIDAS of the unliganded intermediate-affinity I domain is in the closed conformation (Figure 3D), with a disposition of coordinating residues and loops identical to that of the wild-type αL I domain (Figure 3E). In the open conformation of the MIDAS stabilized by ICAM-1, the direct coordination of I domain residue Asp-239 to the metal is replaced by an indirect coordination mediated by a water molecule, and the metal moves 2Å toward Thr-206, forming a direct coordination to it (Figure 3A). The change in coordination is linked to a movement in the portion of the β1-αl loop bearing MIDAS residue Ser-141 and the β4-α5 loop bearing MIDAS residue Asp-239 (Figure 4A). The loss of direct metal coordination to Asp-239 not only increases the electro-philicity of the metal for Glu-34 of ICAM-1, but the metal also moves markedly closer to Glu-34 (Figure 4A). The 2Å movement of Ser-141 enables it to make many contacts with the Glu-34 side chain of ICAM-1 and is associated with a 2.5Å backbone movement at I domain residue Gln-143 in the β1-αl loop, which brings its side chain into position to hydrogen bond with the backbone of Pro-36 of ICAM-1 (Figure 2C). The change in coordina tion of Asp-239 is associated with a dramatic rearrangement in the I domain β4-α5 loop bearing this residue (Figure 4A). The loop now bends at Gly-240 away from instead of toward the Mg2+, with a 4Å change in Cα position and a backbone flip of Gly-240. The resulting 100° rotation in the side chain of Glu-241 (Figure 4A) brings it into position to form the crucial salt bridge to the side chain of Lys-39 in ICAM-1 (Figure 2C).
The movement of key MIDAS and ligand binding residues in the β1-α1 loop “pulls” the entire α1 helix toward the center of the domain and is linked to key rearrangements in the hydrophobic core of the I domain (Figure 4A). The side chain of Leu-142 in the β1-α1 loop is in contact with Asp-239 and helps “push” it out of direct coordination with Mg2+ and induce the movements in the β4-α5 loop at Gly-240 and Glu-241. The side chain of Phe-265 in the β5-α6 loop swings away from the β4-α5 loop and toward the β6-α7 loop to make way for these changes. Previous NMR chemical shift perturbation studies have shown that binding to ICAM-1 induced significant changes at the MIDAS and the C-terminal region of the αL I domain (Huth et al., 2000). The changes in the β1-α1 and β4-α5 MIDAS loops and the inward rigid body movement of the α1 helix, which “squeezes” the hydrophobic core of the domain, are essentially identical to those observed in the transition from the closed to open conformation of the α2 and αM I domains (Emsley et al., 2000; Lee et al., 1995a). However, in the absence of ligand binding, the β6-α7 loop in the intermediate-affinity I domain is already in a conformation that is intermediate between its open and closed conformations (see below). Upon ligand binding, some readjustments occur in this loop and in β6 and α7, but the β6-α7 loop does not assume the open conformation. This appears to be a consequence of constraints imposed by the Cys-161-Cys-299 disulfide bond. The electron density is poorest in the complex structure nearby this disulfide and in β6 and α7, suggesting structural strain, whereas the density is excellent in all regions of the uncomplexed intermediate αL I domain.
The interaction between LFA-1 and ICAM-1, described here in atomic detail, is paradigmatic of many other cell adhesive interactions between integrins and IgSF members (Wang and Springer, 1998). The binding site has several remarkable features. Both the shift in coordination at the MIDAS and the remarkable swing of the Glu-241 side chain in the β4-α5 MIDAS loop appear to be important in switching the I domain into a high-affinity state. Our finding that Glu-241 forms a salt bridge to Lys-39 of ICAM-1 demonstrates a novel mechanism for regulating LFA-1 binding to ICAM-1. This Glu residue is highly conserved, and its swing may be a general mechanism for regulating ligand binding by I domains. Furthermore, the binding interface described here provides an atomic framework for the development of an important class of pharmaceutical antagonists.
Could the wild-type I domain bind in the closed or intermediate conformation to ICAM-1? Superposition on the cocrystal structure shows no significant clashes. The β6-α7 loop in the closed and intermediate structures is too far from the bound ICAM-1 to block ligand binding. Thus, the downward movement of this loop upon I domain activation has no direct role in exposing the ICAM-1 binding site. In the closed conformation, the Mg2+ ion would be too far away for direct coordination to Glu-34 of ICAM-1, but would be at an appropriate distance for indirect coordination. Therefore, subsequent to ligand binding, reshaping to the open conformation could occur, including lateral movement of the β1-α1 backbone with readjustments of its side chains that contact ICAM-1. The shifts in MIDAS coordination and the swing of the Glu-241 side chain toward Lys-39 of ICAM-1 could be accommodated subsequent to ICAM-1 binding. Thus, ICAM-1 could bind to an αL I domain in the closed or intermediate conformation, and then favor a shift to the open conformation.
Allosteric Regulation of Ligand Binding by theβ6-α7 Loop: Inside-Out Signaling
The high-affinity αL I domain crystallized in unliganded and pseudo-liganded forms. The two structures have essentially identical open conformations, with a root mean square deviation of 0.57Å for all Cα atoms. The MIDAS residues of both structures are in the open conformation (Figures 3B and 3C); however, the unliganded high-affinity structure lacks a metal ion at the MIDAS (Figure 3C). In the absence of a metal and a ligand, a water molecule mediates a hydrogen bond network among Asp-137, Ser-139, and Ser-141. The absence of metal coordination and the presence of electrostatic repulsion by Asp-137 cause the side chain of Asp-239 to swing away from other MIDAS residues (Figure 3C). In the pseudo-liganded high-affinity I domain, the metal coordination is completed by an exogenous Glu-272 from a crystallographic symmetry mate (Figure 3B). This lattice contact is similar to that in the open conformation of the αM I domain (Lee et al., 1995b), except that the Glu is donated by α helix 6 instead of α helix 7. The Glu in the ligand-mimetic lattice contact binds in a similar orientation to the Glu in ICAM-1 (Figures 3A and 3B).
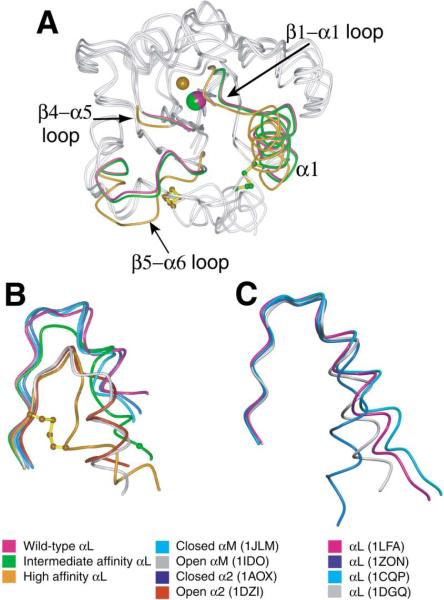
The effect of inside-out signaling on I domain confor mation is mimicked by pulling down the C-terminal α7 helix with disulfide bonds in the absence of ligand bind ing (Figure 5A). In comparison to the wild-type αL I do main, which assumes the closed conformation (Qu and Leahy, 1995), the β6-α7 loop of the unliganded intermediate-affinity I domain is partially pulled down, whereas that of the unliganded high-affinity I domain is markedly displaced downward (Figures 4C and 5B). The struc tures of the αM and α2 I domains in the open and closed conformations are shown for comparison (Figure 5B; Emsley et al., 1997, 2000; Lee et al., 1995a, 1995b). The closed αL, αM, and α2 I domains share an identical conformation of the β6-α7 loop. The open α2 and αM I domain structures also have an identical conformation of the β6-α7 loop, which differs markedly from the closed conformation of this loop. Importantly, the unli ganded, high-affinity αL I domain mutant has an identical open conformation in the β6 strand and the β6-α7 loop (Figure 5B). It is these regions that have the important interactions with the hydrophobic core that regulate the conformation of the MIDAS. The conformation of α7 in the high-affinity αL mutant differs markedly, partially due to the constraint from the engineered disulfide bond; however, the conformation of α7 is highly variable, as shown by comparison of different αL structures in the closed conformation (Figure 5C). Because the remainder of the high-affinity mutant I domain including the MIDAS is in the open, ligand binding configuration, the structural comparison suggests that the conformation of β strand 6 and the β6-α7 loop, but not the structural details of α7 per se, are critical for the packing interactions in the hydrophobic core that open the ligand binding site. In intact integrins, α7 helix movement would therefore appear to be linked to ligand binding only through its effect on β6-α7 loop conformation.
Figure 5. The Structures of the Unliganded Wild-Type, Intermediate-Affinity, and High-Affinity αL I Domains.
(A) The three unliganded αL I domain backbones are shown superimposed and viewed centered on the MIDAS. Regions of the backbones that differ structurally are labeled and color keyed; other backbone regions are gray. The metal ions at the MIDAS and the atoms in disulfide-linked cysteines are shown in the same colors as the backbone differences; the cysteine side chain bonds are yellow. As in Figure 4B, the position of the missing metal ion in the high-affinity structure is simulated with a smaller sphere. β6-α7 loop and α7 are shown in gray for clarity; differences in these regions are shown in (B).
(B) The C-terminal fragments encompassing the β6-α7 loop for the three unligated αL conformations and open and closed α2 and αM I domain structures (color keys on bottom left and middle). The side chain bonds of Cys-287 and Cys-294 in the designed disulfide bridge in the high-affinity mutant are shown in yellow; the Cα atom of Cys-299 in the designed disulfide of the intermediate mutant is shown as a green sphere.
(C) The C-terminal fragments encompassing the β6-α7 loop for four different wild-type αL structures in the closed conformation (color keys on bottom right).
The β6-α7 loop of the intermediate-affinity I domain is in a position intermediate between that exhibited in the open and closed conformations of the αL, αM, and α2 I domains (Figure 5B). In the low-affinity, closed conformation, Phe-292 is in the top of the β6-α7 loop, buried in a hydrophobic pocket (Figure 4D). The removal of Phe-292 from this pocket is key to enabling rearrangement at the MIDAS (Figures 4B and 4C). Furthermore, another hydrophobic pocket ~5.5Å lower down the side of the domain is occupied successively by Leu-295, Phe-292, and Leu-289 in the low-, intermediate-, and high-affinity structures, respectively (Figure 4D). Although the α7 he lix is disrupted in the high-affinity αL I domain by the disulfide bond, in the α2 and αM I domains the α7 helix is displaced by two turns of helix down the side of the domain between the open and closed conformations (Emsley et al., 2000; Lee et al., 1995a). In the closed conformation of the αL I domain, residues 292–295 in the upper part of α7 form a 310 helix (Qu and Leahy, 1996). In the intermediate conformation of the αL I domain, this turn of 310 helix beginning with Phe-292 is displaced downward a distance corresponding to one turn of helix, and Phe-292 is deeply buried in the hydrophobic pocket (Figure 4D). The pocket thus acts as a detent that holds the β6-α7 loop in three different ratchet positions, and each movement displaces the α7 helix a distance down ward corresponding to one turn of helix (see Supplemen tal Movie S1 at http://www.cell.com/cgi/content/full/112/1/99/DC1).
Comparison of the unliganded, high-affinity open structure and the wild-type closed structure shows that the conformational changes induced by pulling down the β6-α7 loop with a disulfide bridge are first propagated to the hydrophobic core of the I domain and then to the MIDAS loops. Pulling down the β6-α7 loop would have exposed hydrophobic core residues on α1, β4, and β5, had there not been nearby structural rearrange ments. To shield the hydrophobic core from solvent, α1 moves ~2 Å inward (Figures 4B, 4C, and 5A). In the β1-α1 loop, the third of three residues that form MIDAS coordinations, Ser-141, shifts 2Å in backbone position, which in effect changes the metal coordination from the closed to the open conformation. The movement of Leu- 142 in the β1-α1 loop pushes Asp-239 in the β4-α5 loop from the primary to the secondary metal coordination sphere (Figure 4B). Second, there are concerted downward movements of the β5-α6 and β4-α5 loops in order to fill the space left open by the β6-α7 loop (Figures 4B and 5A). The downward movement of Phe-265 in the β5-α6 loop allows the flip of the Gly-240 main chain in the β4-α5 loop. This flip positions the Glu-241 side chain for contact with Lys-39 of ICAM-1 as discussed above. The β5-α6 loop, which bears Phe-265, possesses high temperature factors and differs significantly among the four structures, largely due to its prominent role in dif fering crystal lattice contacts. However, residue Phe-265 on this loop is well ordered, and its position appears to be related to the structural transition from the closed to open conformation (Figure 4A compared to 4B). The conformational changes induced in unliganded struc tures by downward movement of the β6-α7 loop (Figure 4B) are identical in all important respects to those induced by ligand binding to the intermediate conformation (Figure 4A), except that after ligation the β6-α7 loop remains in the intermediate conformation as already discussed.
The unliganded intermediate-affinity and wild-type structures are well superimposable in the regions mentioned above (Figures 4B and 5A). Therefore, pulling down the β6-α7 loop half-way between the closed and open conformations does not cause significant main chain conformational change elsewhere in the domain and leaves the ligand binding site in the closed conformation (see Supplemental Movie S1 at http://www.cell.com/cgi/content/full/112/1/99/DC1). However, this intermediate movement produces an I domain that is ener getically primed for the above mentioned structural rearrangements. The key Phe-292 residue in the β6-α7 loop is already removed from its hydrophobic pocket between β strands 4 and 5 and α helix 1, and the back bone of the β6-α7 loop has moved outward, away from the hydrophobic core. Therefore, the lateral movement of the α1 helix, the flip of the β4-α5 loop, and the down ward shift of the β5-α6 loop are achievable with a lower energetic cost, accounting for the increased affinity for ligand.
Our results show that in intact integrins, downward movement of the I domain C-terminal α helix would be sufficient to convey conformational movements from other domains to the I domain, providing a mechanism for I domain activation in inside-out signaling. The down ward movement of the β6-α7 loop induced here by muta tionally introduced disulfide bonds mimics a downward pull thought to be induced in intact integrins by interac tion with the neighboring β subunit I-like domain. The idea that the C-terminal α7 helix and the following linker to the β-propeller domain act as a “bell rope” is sup ported by findings that mutations in these regions can either activate or inhibit ICAM-1 binding (Alonso et al., 2002; Huth et al., 2000; Lupher et al., 2001; Oxvig et al., 1999). The β2 I-like domain regulates ligand binding by the I domain instead of directly participating in ligand binding (Lu et al., 2001), and its MIDAS has been pro posed to interact with an acidic residue in the I domain linker to exert the downward pull on the bell rope, which activates ligand binding by the I domain (Alonso et al., 2002; Huth et al., 2000; Lu et al., 2001; Shimaoka et al., 2002).
Our structure of the unliganded high-affinity αL I do main shows for the first time that the open conformation is stable in the absence of binding to a ligand or pseudo-ligand. Since the disulfide bonds introduced here into isolated I domains mimic a downward pull exerted on the C-terminal α helix by neighboring domains in intact integrins, our findings suggest that conformational change in I domains could precede ligand binding in intact integrins. This is of considerable biological impor tance. The on rate for ICAM-1 binding by the closed conformation of the I domain is slow, and the 30-fold increase in kon observed for the intermediate and open conformations (Table 1) would facilitate the rapid kinetics of integrin-mediated adhesion by leukocytes in the bloodstream, where cellular activation can lead to integrin activation and ligand binding in less than 1 s. Our finding that the open conformation is stable in the ab sence of ligand binding is supported by studies with activation-sensitive antibodies to the αL and αM I domains, which show that changes in I domain conformation occur in intact cell surface integrins upon activation in the absence of ligand binding (Diamond and Springer, 1993; Li et al., 2002; Ma et al., 2002).
Our complex structure and previous crystal studies on αM and α2 I domains (Emsley et al., 2000; Lee et al., 1995b) show that ligand binding can also stabilize the open conformation of the I domain. Thus, although it has often been disputed whether conformational change precedes or follows ligand binding, the findings actually support both concepts. Furthermore, the ligand binding site of the unliganded, high-affinity αL I domain has the same open conformation as the liganded or pseudo-liganded I domains. This demonstrates that signal transmission through the hydrophobic core of the I domain works the same in both directions, i.e., in both inside-out and outside-in signaling.
An Intermediate Conformation for I Domains
The finding of an intermediate conformational state of the I domain is novel. It is intriguing that there is excellent evidence for two different activation states for αLβ2 on cell surfaces (Constantin et al., 2000; Stewart et al., 1996; van Kooyk et al., 1999). Both states are active in binding to ICAM-1 in adhesion assays, whereas only one is sufficiently high affinity to detectably bind soluble ICAM-1. The intermediate conformation of the αL I domain is very well folded, and indeed the 1.3Å crystal structure is the highest resolution solved for an I domain to date. The β6-α7 loop and upper portion of α7 are very well packed, with an average B factor of 24Å2 for all nonhydrogen atoms of residues 288–296. These findings suggest that the intermediate conformation of the β6-α7 loop represents a discrete conformational state that could exist in intact integrins, and that the intermediate conformation could represent the second active state of αLβ2 that is active in cell adhesion but does not bind ligand with high affinity. However, we cannot rule out the possibility that the intermediate conformation could represent a transition state during equilibration of I domains between the open and closed conformations (Shimaoka et al., 2001). A recent study with a fluorescent ligand-mimetic peptide has shown that the α4β1 integrin exhibits multiple affinity states on the cell surface depending on the activation condition (Chigaev et al., 2001). The presence of multiple affinity states allows more precise regulation of ligand binding and could be essential for the physiological functions of integrins.
The structures of the unliganded intermediate-affinity and high-affinity αL I domains suggest two discrete steps in a shape-shifting pathway by which inside-out signals can activate integrins for ligand binding. The finding that the affinities of other mutant I domains cluster near those of the I domains with structurally defined open, intermediate, and closed conformations also suggests that these are discrete conformational states in the shape-shifting pathway. Recent averaging of electron microscopic negatively stained images of the extracellular domain of integrin αVβ3 suggest that integrins adopt at least three overall conformations: highly bent, extended with a closed headpiece, and extended with an open headpiece (Takagi et al., 2002). It will be very interesting to determine whether integrins that contain I domains also exist in multiple overall shapes and how conformational change in I domains is linked to specific structural alterations elsewhere in integrins.
Experimental Procedures
Protein Expression and Purification
Wild-type and mutant I domains were expressed in E. coli BL21(DE3) (Novagen, Madison, WI), refolded, and purified as described (Legge et al., 2000; Shimaoka et al., 2001), except 0.2 mM CuSO4 and 1 mM o-phenanthroline were used to catalyze disulfide bond formation and purification was on a monoQ HR5/5 ion-exchange column (Pharmacia) and a Superdex 200 column (Pharmacia). Intramolecular disulfide bonds were confirmed in all mutants by decreased mobility in SDS-PAGE after reduction (Shimaoka et al., 2001).
A cDNA encoding the ICAM-1 signal sequence, residues Q1 to T291, and an additional C-terminal alanine was subcloned into pMT/BiP/V5-His insect cell expression vector (Invitrogen). S2 insect cells (Invitrogen) grown in Drosophila serum-free medium (Invitrogen) were cotransfected with 19 μg pMT/BiP/V5-His containing the ICAM-1 fragment and 1 μg pCoHYGRO (Invitrogen) with the calcium phosphate method and selected with 300 μg/ml hygromycin. Induction with 500 μM CuSO4 for more than 16 hr yielded 1 mg/l ICAM-1. ICAM-1 was purified as described (Dustin et al., 1992) and further with monoQ ion-exchange chromatography in 20 mM Tris HCl (pH 8) with a gradient of 0 to 1 M NaCl.
Surface Plasmon Resonance
Soluble monomeric ICAM-1, or BSA as a negative control, was immobilized on a CM-5 sensor chip by the amine coupling method. I domains were perfused onto the chip in Tris-buffered saline solution plus 1 mM MgCl2, at a flow rate of 10 μl/min, at 25°C (Shimaoka et al., 2001).
Crystallization and Data Collection
Crystals were grown by the hanging drop vapor diffusion method at room temperature. One to two microliters of protein solution (10 to 20 mg/ml) was mixed with an equal volume of well solution on a siliconized glass coverslide and equilibrated against 1 ml of the well solution. The intermediate-affinity I domain was crystallized with 30% PEG 4000, 0.05 M MgCl2, and 0.1 M Tris·Cl (pH 8.5). The inter mediate affinity I domain in complex with ICAM-1 domains 1 to 3 was crystallized with 25% PEG-4000 and 0.1 M sodium acetate (pH 4.6). The high-affinity I domain crystallized in two forms. A well solution of 20% PEG 2000 monomethyl ether, 0.025 M MnCl2, and 0.1 M HEPES·Na (pH 7.0) yielded crystals of the pseudo-liganded form. A well solution of 1.2 M ammonium phosphate and 0.1 M Tris·Cl (pH 8.0) yielded crystals of the unliganded form. Crystals were harvested in their mother liquor supplemented with 12%–15% glycerol as cryoprotectant, then flash frozen in liquid nitrogen. Diffraction data were collected at the 19-ID station of the Advanced Photon Source at the Argonne National Laboratory with an SBC2 3k × 3k CCD detector and processed with program suite HKL2000 (Otwinowski and Minor, 1997).
Structure Determination and Model Refinement
All four structures were solved using Amore (Navaza, 1994) for molecular replacement and O for visual inspection and model rebuilding (Jones and Kjeldgaard, 1993).
The 1.3Å structure of the intermediate-affinity I domain was solved using the closed αL I domain (Qu and Leahy, 1995) as search model. A complete model was built with ARP_WARP 5.0 (Perrakis et al., 2001). The addition of hydrogen atoms, refinement of multiple side chain conformations, and sulfur atom anisotropy was carried out with SHELX 97_2 (Sheldrick and Schneider, 1997).
The other three structures at 2.0 to 3.3Å resolution were refined using CNS version 1.0 protocols for rigid body refinement, positional refinement by Powell minimization, group B factor refinement, and slow-cool simulated annealing molecular dynamics (Brunger et al., 1998). Anisotropic temperature factor correction, bulk solvent correction, and maximum likelihood refinement target were applied throughout.
The structure of the intermediate-affinity I domain ICAM-1 complex was solved using ICAM-1 domains 1 and 2 (Casasnovas et al., 1998) as a search model. The location of the I domain was readily discernible in the electron density map calculated using phases from the ICAM-1 model, and the above intermediate-affinity I domain structure was manually positioned in the map. There are two essentially identical I domain:ICAM-1 complexes in the crystallographic asymmetric unit. Stringent noncrystallographic symmetry (NCS) restraints applied throughout the refinement protocol were monitored by the decrease of Rfree. Specifically, NCS restraints were applied for ICAM-1 domain 1 (except for residues 43–47, which display significant differences in conformation), domain 2, and I domain residues 130–264. The C-terminal part of the I domain was excluded from the NCS restraints due to its flexibility, and this exclusion was monitored by the lowered free R factor. Sigma A weighted 2Fo − Fc and Fo − Fc maps were computed for visual inspection during rebuilding of the model with O. Analysis of the packing of the molecules in the crystal lattice indicated that domain 3 of ICAM-1 could be accommodated in the lattice. However, the diffuse electron density following domain 2 prohibited modeling domain 3. The final model includes residues 1 to 184 of ICAM-1 and one or two N-acetylglucosamine residues at each of the four N-linked glycosylation sites. The electron density for I domain residues 285–298 is relatively poor. However, a continuous density that connects the main chain is clearly visible at 0.7δ contour level, with the exception of residues 286 and 287 for both I domains. It appears that these two residues could adopt multiple conformations. Therefore, the Val-286 and Lys-287 side chains are modeled with an occupancy of 0.50 and probably represent the most stable conformation.
The pseudo-liganded high-affinity I domain with a Mn2+ ion at the MIDAS was solved using the uncomplexed intermediate-affinity I domain structure as a search model. Water molecules with at least one hydrogen bond to protein atoms and a temperature factor below 80Å2 were included.
The structure of the unliganded high-affinity I domain was solved using the liganded high-affinity structure as a search model. To minimize model bias, regions of the search model including the three MIDAS loops, the β5-α6 loop, and the C-terminal fragment starting from β6 were removed during initial cycles of refinement. Simulated annealing omit maps (Hodel et al., 1992) were computed and inspected during model rebuilding. The identity of a water molecule instead of a metal ion at the MIDAS was confirmed by the hydrogen bonding distance (>2.6Å) as opposed to the magnesium coordinating distance of about 2.0Å, and the absence of a bipyramidal coordination sphere.
Supplementary Material
Acknowledgments
We would like to thank Rob Meijers for help with ARP_WARP. This project was supported by NIH grants CA31798 (T.A.S.) and HL48675 (J.-H.W.). T.X. is supported by a postdoctoral fellowship from the Irvington Institute. The calculation of the relative domain orientation of ICAM-1 utilized program HINGE developed by Peter Sun at the NIAID.
References
- Alonso JL, Essafi M, Xiong JP, Stehle T, Arnaout MA. Does the integrin αA domain act as a ligand for its βA domain? Curr. Biol. 2002;12:R340–R342. doi: 10.1016/s0960-9822(02)00852-7. [DOI] [PubMed] [Google Scholar]
- Bella J, Kolatkar PR, Marlor C, Greve JM, Rossmann MG. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Casasnovas JM, Springer TA, Liu J.-h., Harrison SC, Wang J.-h. The crystal structure of ICAM-2 reveals a distinc tive integrin recognition surface. Nature. 1997;387:312–315. doi: 10.1038/387312a0. [DOI] [PubMed] [Google Scholar]
- Casasnovas JM, Stehle T, Liu J.-h., Wang J.-h., Springer TA. A dimeric crystal structure for the N-terminal two do mains of ICAM-1. Proc. Natl. Acad. Sci. USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A, Blenc AM, Braaten JV, Kumaraswamy N, Kepley CL, Andrews RP, Oliver JM, Edwards BS, Prossnitz ER, Larson RS, Sklar LA. Real-time analysis of the affinity regulation of α4-integrin: the physiologically activated receptor is intermediate in affinity between resting and Mn2+ or antibody activation. J. Biol. Chem. 2001;276:48670–48678. doi: 10.1074/jbc.M103194200. [DOI] [PubMed] [Google Scholar]
- Constantin G, Majeed M, Giagulli C, Piccib L, Kim JY, Butcher EC, Laudanna C. Chemokines trigger immediate β2 integrin affinity and mobility changes: differential regulation and roles in lymphocyte arrest under flow. Immunity. 2000;13:759–769. doi: 10.1016/s1074-7613(00)00074-1. [DOI] [PubMed] [Google Scholar]
- Conte LL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Springer TA. A subpopulation of Mac-1 (CD11b/CD18) molecules mediates neutrophil adhesion to ICAM-1 and fibrinogen. J. Cell Biol. 1993;120:545–556. doi: 10.1083/jcb.120.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Springer TA. T cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Springer TA. Intercellular adhesion mole cules (ICAMs). In: Kreis T, Vale R, editors. Guidebook to the Extracellular Matrix and Adhesion Proteins. Sambrook and Tooze; New York: 1999. [Google Scholar]
- Dustin ML, Carpen O, Springer TA. Regulation of locomotion and cell-cell contact area by the LFA-1 and ICAM-1 adhesion receptors. J. Immunol. 1992;148:2654–2663. [PubMed] [Google Scholar]
- Edwards CP, Fisher KL, Presta LG, Bodary SC. Mapping the intercellular adhesion molecule-1 and -2 binding site on the inserted domain of leukocyte function-associated antigen-1. J. Biol. Chem. 1998;273:28937–28944. doi: 10.1074/jbc.273.44.28937. [DOI] [PubMed] [Google Scholar]
- Emsley J, King SL, Bergelson JM, Liddington RC. Crystal structure of the I domain from integrin α2β1. J. Biol. Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Lidding ton RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 1997;15:132–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- Fisher KL, Lu J, Riddle L, Kim KJ, Presta LG, Bodary SC. Identification of the binding site in intercellular adhesion molecule 1 for its receptor, leukocyte function-associated antigen 1. Mol. Biol. Cell. 1997;8:501–515. doi: 10.1091/mbc.8.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahmberg CG, Tolvanen M, Kotovuori P. Leukocyte adhesion: structure and function of human leukocyte β2-integrins and their cellular ligands. Eur. J. Biochem. 1997;245:215–232. doi: 10.1111/j.1432-1033.1997.00215.x. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Giblin PA, Kelly TA. Antagonists of β2 integrin-medi ated cell adhesion. Annu. Rep. Med. Chem. 2001;36:181–190. [Google Scholar]
- Gottlieb A, Krueger JG, Bright R, Ling M, Lebwohl M, Kang S, Feldman S, Spellman M, Wittkowski K, Ochs HD, et al. Effects of administration of a single dose of a humanized monoclonal antibody to CD11a on the immunobiology and clinical activity of psoriasis. J. Am. Acad. Dermatol. 2000;42:428–435. doi: 10.1016/s0190-9622(00)90214-7. [DOI] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Hodel A, Kim S-H, Brünger AT. Model bias in macro-molecular crystal structures. Acta Crystallogr. 1992;A48:851–858. [Google Scholar]
- Huang C, Springer TA. A binding interface on the I domain of lymphocyte function associated antigen-1 (LFA-1) re quired for specific interaction with intercellular adhesion molecule 1 (ICAM-1). J. Biol. Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- Huth JR, Olejniczak ET, Mendoza R, Liang H, Harris EA, Lupher ML, Jr., Wilson AE, Fesik SW, Staunton DE. NMR and mutagenesis evidence for an I domain allosteric site that regulates lymphocyte function-associated antigen 1 ligand binding. Proc. Natl. Acad. Sci. USA. 2000;97:5231–5236. doi: 10.1073/pnas.97.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M. O Version 5.9. Uppsala University; Uppsala, Sweden: 1993. [Google Scholar]
- Kallen J, Welzenbach K, Ramage P, Geyl D, Kriwacki R, Legge G, Cottens S, Weitz-Schmidt G, Hommel U. Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain. J. Mol. Biol. 1999;292:1–9. doi: 10.1006/jmbi.1999.3047. [DOI] [PubMed] [Google Scholar]
- Kamata T, Wright R, Takada Y. Critical threonine and aspartic acid residues within the I domains of β2 integrins for interactions with intercellular adhesion molecule 1 (ICAM-1) and C3bi. J. Biol. Chem. 1995;270:12531–12535. doi: 10.1074/jbc.270.21.12531. [DOI] [PubMed] [Google Scholar]
- Koopman G, Keehnen RMJ, Lindhout E, Newman W, Shimizu Y, Van Seventer GA, deGroot C, Pals ST. Adhesion through the LFA-1 (CD11a/CD18)-ICAM-1 (CD54) and the VLA-4 (CD49d)-VCAM-1 (CD106) pathways prevents apoptosis of germinal center B cells. J. Immunol. 1994;152:3760–3767. [PubMed] [Google Scholar]
- Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Larson RS, Springer TA. Structure and function of leukocyte integrins. Immunol. Rev. 1990;114:181–217. doi: 10.1111/j.1600-065x.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee J-O, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995a;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Lee J-O, Rieu P, Arnaout MA, Liddington R. Crys tal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell. 1995b;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- Legge GB, Kriwacki RW, Chung J, Hommel U, Ramage P, Case DA, Dyson HJ, Wright PE. NMR solution struc ture of the inserted domain of human leukocyte function associated antigen-1. J. Mol. Biol. 2000;295:1251–1264. doi: 10.1006/jmbi.1999.3409. [DOI] [PubMed] [Google Scholar]
- Li R, Haruta I, Rieu P, Sugimori T, Xiong JP, Arnaout MA. Characterization of a conformationally sensitive murine monoclonal antibody directed to the metal ion-dependent adhesion site face of integrin CD11b. J. Immunol. 2002;168:1219–1225. doi: 10.4049/jimmunol.168.3.1219. [DOI] [PubMed] [Google Scholar]
- Lollo BA, Chan KWH, Hanson EM, Moy VT, Brian AA. Direct evidence for two affinity states for lymphocyte func tion-associated antigen 1 on activated T cells. J. Biol. Chem. 1993;268:21693–21700. [PubMed] [Google Scholar]
- Lu C, Shimaoka M, Zang Q, Takagi J, Springer TA. Locking in alternate conformations of the integrin αLβ2 I domain with disulfide bonds reveals functional relationships among integrin domains. Proc. Natl. Acad. Sci. USA. 2001;98:2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupher ML, Jr., Harris EA, Beals CR, Sui L, Liddington RC, Staunton DE. Cellular activation of leukocyte function-associated antigen-1 and its affinity are regulated at the I domain allosteric site. J. Immunol. 2001;167:1431–1439. doi: 10.4049/jimmunol.167.3.1431. [DOI] [PubMed] [Google Scholar]
- Ma Q, Shimaoka M, Lu C, Jing H, Carman CV, Springer TA. Activation induced conformational changes in the I do main region of LFA-1. J. Biol. Chem. 2002;277:10638–10641. doi: 10.1074/jbc.M112417200. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP. Raster 3D version 2.0: a program for photorealistic graphics. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Navaza J. Amore: an automated package for molecular re placement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association; insights from the interfacial and thermodynamic prop erties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oxvig C, Lu C, Springer TA. Conformational changes in tertiary structure near the ligand binding site of an integrin I domain. Proc. Natl. Acad. Sci. USA. 1999;96:2215–2220. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/warp and molecular replacement. Acta Crystallogr. D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Qu A, Leahy DJ. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, αLβ2) integrin. Proc. Natl. Acad. Sci. USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu A, Leahy DJ. The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin. Structure. 1996;4:931–942. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM, Schneider TR. SHELXL: high resolution refinement. In: Sweet RM, Carter CW Jr., editors. Methods in Enzymology. Academic Press; Orlando, FL: 1997. pp. 319–343. [PubMed] [Google Scholar]
- Shimaoka M, Lu C, Palframan R, von Andrian UH, Takagi J, Springer TA. Reversibly locking a protein fold in an active conformation with a disulfide bond: integrin α L I domains with high affinity and antagonist activity in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- Staunton DE, Dustin ML, Erickson HP, Springer TA. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- Stewart MP, Cabanas C, Hogg N. T cell adhesion to intercellular adhesion molecule-1 (ICAM-1) is controlled by cell spreading and the activation of integrin LFA-1. J. Immunol. 1996;156:1810–1817. [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- van Kooyk Y, van Vliet SJ, Figdor CG. The actin cytoskeleton regulates LFA-1 ligand binding through avidity rather than affinity changes. J. Biol. Chem. 1999;274:26869–26877. doi: 10.1074/jbc.274.38.26869. [DOI] [PubMed] [Google Scholar]
- Van Seventer GA, Shimizu Y, Horgan KJ, Shaw S. The LFA-1 ligand ICAM-1 provides an important costimulatory signal for T cell receptor-mediated activation of resting T cells. J. Immunol. 1990;144:4579–4586. [PubMed] [Google Scholar]
- Wang J.-h., Springer TA. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.