Abstract
We report new substrates for quantitative enzyme activity measurements of human palmitoyl protein thioesterase (PPT1) and tripeptidyl peptidase (TPP1) in dried blood spots from newborns using tandem mass spectrometry. Deficiencies in these enzyme activities due to inborn errors of metabolism cause neuronal ceroid lipofuscinoses. The assays use synthetic compounds that were designed to mimic the natural substrates. Incubation produces nanomole quantities of enzymatic products per a blood spot that are quantified by tandem mass spectrometry using synthetic internal standards and selected reaction monitoring. The assays utilize a minimum steps for sample workup and can be run in a duplex format for the detection of neuronal ceroid lipofuscinoses or potentially multiplexed with other mass spectrometry-based assays for newborn screening of lysosomal storage disorders.
The neuronal ceroid lipofuscinoses (NCLs) are a group of lysosomal storage disorders primarily affecting children and adolescents. NCLs are inherited as autosomal recessive disorders that cause neurodegenerative diseases manifesting similar clinical features, including seizures, mental regression, visual loss, behavior changes, movement disorders, and shortened life expectancy in affected individuals.1 Of the two most prevalent forms, infantile neuronal ceroid lipofuscinosis (INCL), also called the Santavuori-Haltia disease,2 is caused by mutations in the CLN1 gene located on chromosome 1p32, which encodes the lysosomal enzyme palmitoyl protein thioesterase I (PPT1, EC3.1.2.22). Over 40 mutations of the CLN1 gene are known.3 PPT1 cleaves thioester-linked fatty acid groups from C-terminal cysteine residues in lipoproteins. PPT1 is structurally similar to lipases and has a peptide binding site as well as a well-defined fatty acid binding pocket.4 The other NCL form, classic late infantile neuronal ceroid lipofuscinosis (NCL II, also called the Jansky-Bielschowsky disease5) results from mutations in the TPPI gene (previously named CLN2)6 which is located on chromosome 11p15 and encodes the lysosomal enzyme tripeptidylpeptidase 1 (TPP1).1,2 TPP1 is a serine protease that cleaves three amino acid residues from unsubstituted protein N-termini. The human form of TPP1 shows preferential cleavage of the Ala-Ala-Phe peptide triad.7
Collectively, NCLs are estimated to constitute the most common hereditary neurodegenerative disorder in childhood2 with an estimated prevalence of 1:12 500 newborns in the U.S.2,3 A particularly high incidence of INCL is found in Finland where it is due to a missense mutation (W122R).2 NCLs are incurable disorders, and treatment of affected children has been mainly supportive. However, recent reports of enzyme replacement therapies8−10 and neuronal stem transplantation11,12 indicated that animals treated with recombinant enzymes showed significantly decreased levels of lysosomal storage material. This could possibly open an avenue for the development of a therapy for human NCLs.12 Because INCL is characterized by early onset (6 months to 1 year) and rapid progression, the success of any potential therapy strongly depends on early diagnosis. A detection strategy for such rare metabolic disorders, which is currently being explored on a large scale,13 is by screening the enzyme defects in dried blood spots (DBSs) collected from entire newborn populations.
The most direct and specific diagnosis of NCLs relies on enzymatic assays in biological samples such as leukocytes or cultured skin fibroblasts, using radiometric or fluorescence detection. The radiometric method for PPT1 measures the release of tritium-labeled palmitate from a palmitate-labeled H-Ras protein.14 The fluorometric assay for PPT1, developed by van Diggelen et al.,15 uses an S-palmitoyl-6-thiogalactosylcoumarin conjugate that requires a coupling enzyme to release the fluorescent 7-hydroxy-4-methylcoumarin for detection. The fluorometric assay for TPP1 uses Ala-Ala-Pro-7-amido-4-methylcoumarin15 that releases the fluorescent 7-amino-4-methylcoumarin upon enzymatic hydrolysis.16 These assays have been applied to analysis of dried blood spots from newborns.17
We have previously reported several assays of lysosomal enzyme activities in DBSs using tandem mass spectrometry and selected reaction monitoring (SRM).18 Our strategy has been to assay the enzymes with synthetic compounds that are structurally similar to the natural enzyme substrates and at the same time allow highly sensitive and specific detection of products by SRM-tandem mass spectrometry. The assays are designed to allow for simple sample handling and purification to be compatible with the work flow in newborn screening laboratories. The substrates are designed for multiplexed detection of several enzyme products and their quantitation by SRM in a single analytical run. Recent results of a pilot triplex study of over 100 000 DBS samples from newborns in Washington state showed a very low rate (<0.005%) of false positives, which indicated that multiplex analysis by tandem mass spectrometry is a robust and practical method of newborn screening for lysosomal storage disorders.13
Here, we report two new assays of TPP1 and PPT1 that utilize synthetic substrates closely mimicking the natural ones. The synthetic substrates are designed such as to make the new assays potentially multiplexable for tandem mass spectrometry analysis with assays developed so far for several other lysosomal enzymes.19,20
Experimental Section
Materials
All water used was purified by a Millipore Milli-Q 18 MΩ System. N-acetylated heptapeptides ALLPFGC and AAAPFGC, and Fmoc-protected AAF were purchased from Lifetein (Hillsborough, NJ). Palmitoyl chloride (98% pure), Fmoc-4-aminomethylbenzoic acid, N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride, and tris(2-carboxyethyl)phosphine hydrochloride were purchased from Sigma-Aldrich (St Louis, MO). All solvents used were technical grade as supplied by Sigma-Aldrich. Triton X-100 was purchased from ACROS (NJ, USA). Solid phase extraction C18 Omix pipette tips were supplied by Agilent (Santa Clara, CA). All experiments and sample handling were conducted in compliance with Institutional Review Board guidelines. All infantile CLN1 and CLN2 affected patients had been diagnosed previously with established clinical and biochemical procedures. DBSs were stored at −20 °C in zip-lock plastic bags (one bag sealed inside a second bag). Zip-lock bags were kept in a sealed plastic box containing desiccant (anhydrous CaSO4 granules). Details of the substrate synthesis are given in the Supporting Information.
Mass Spectrometry
Electrospray-MS/MS was carried out on a Waters Quattro Micro tandem quadrupole (quadrupole-hexapole-quadrupole) instrument using a positive ionization mode and selected reaction monitoring (SRM). Samples (10 μL) were flow-injected with an autosampler in an acetonitrile/water (v/v = 80:20) solution containing 0.1% formic acid at a flow rate of 0.2 mL/min. The mass spectrometer settings were as follows: capillary voltage, 3.5 kV; cone, 35 V; extractor, 2.0 V; RF lens, 0.2 V; source temperature, 120 °C; desolvation temperature, 250 °C; cone gas flow, 50 L/h; desolvation gas flow, 500 L/h; LM 1 resolution, 14.8; HM 1 resolution, 14.8; ion energy 1, 0.2 eV; entrance, 2 V; collision, 25 eV; exit, 15 V; LM 2 resolution, 14.8; HM 2 resolution, 14.8; ion energy 2, 1.0 eV; multiplier, 650 V; gas cell Pirani pressure, 2.21 × 10–3 mbar; dwell time, 100 ms. Shorter dwell times (10 ms) were also investigated on the Quattro Micro instrument but led to increased coefficients of variation (CV) for intra-assay measurements from 1.5% at 100 ms to 4% at 10 ms, all for triplicate injections. The precursor ion-fragment ion SRM transitions for the PPT1 assays were monitored at m/z 762.5 → m/z 423.5 and m/z 678.4 → m/z 423.4 for PPT1-P and PPTI-IS, respectively, corresponding to the formation of the abundant y4 fragment ion of the peptide (Figure 1). The SRM transitions for the TPP1 assays were monitored at m/z 308.3 → m/z 208.3 and m/z 317.3 → m/z 209.3 for TPP1-P and TPP1-IS, respectively, as shown in Figure 2.
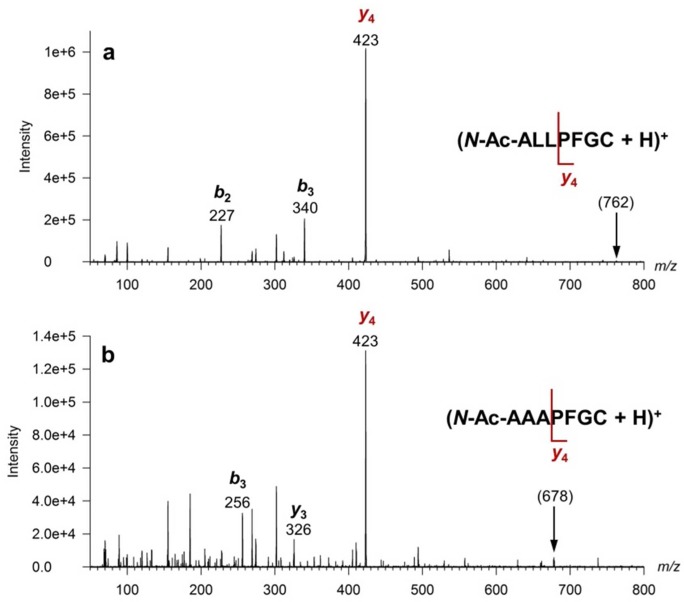
Figure 1.
Tandem mass spectra of (a) (N-Ac-ALLPFGC + H)+ ion at m/z 762, and (b) (N-Ac-AAAPFGC + H)+ ion at m/z 678, both obtained at 25 eV laboratory ion collision energy.
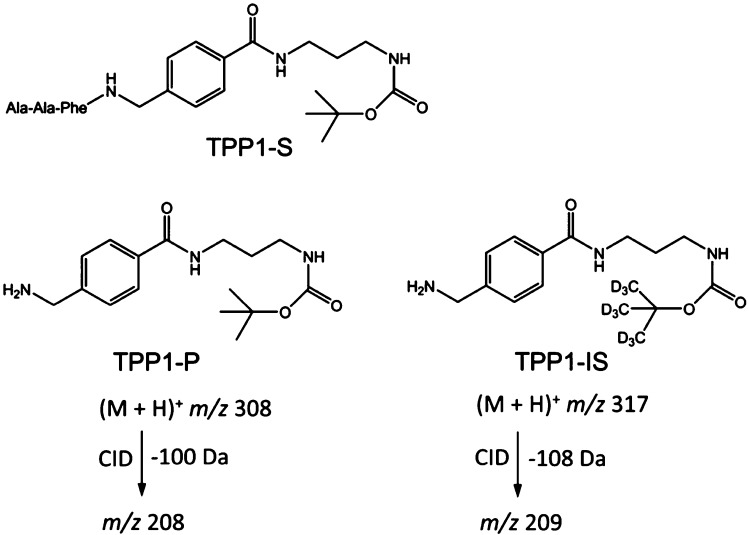
Figure 2.
Structures of TPP1 substrate (TPP1-S), enzyme product (TPP1-P), internal standard (TPP1-IS), and the pertinent SRM transitions.
Assay Protocols
PPT1 Assay
A 3 mm punch of a dried blood spot was placed in a 1.5 mL polypropylene tube (Eppendorf) and 100 μL of 100 mM phosphate buffer solution (pH 7) containing 46 nmol PPT1 substrate (PPT1-S) and 2.6 nmol PPT1 internal standard (PPT1-IS) and 0.08% v/v Triton X-100(ACROS, Cat No. 21568) was added. The solution was then vortexed briefly and incubated for 10 h at 37 °C in a thermostated air shaker at 250 rpm. After the incubation period was over, the sample was placed in an ice bath and the reaction was quenched by addition of 300 μL of ethyl acetate and 100 μL of deionized water. The tubes were vortexed and centrifuged, and the ethyl acetate layer containing Triton X-100 was separated and discarded. The assay product (PPT1-P) and internal standard (PPT1-IS) were isolated and desalted by solid phase extraction on C18 pipette tips (Omix Tips, Agilent Cat No. A57003100) followed by elution with 200 μL of a 50:50 acetonitrile:water with 1% acetic acid. Prior to injection into the mass spectrometer, the samples were treated with 800 μM tris(2-carboxyethyl)phosphine hydrochloride, TCEP (Sigma-Aldrich, Cat. No. C4706) and incubated for 30 min at 37 °C to reduce disulfide bonds formed by cysteine oxidation during the assay.
TPP1 Assay
TPP1 substrate solution was prepared by dissolving 5 mg of TPP1 substrate in 1.5 mL of dimethyl sulfoxide (DMSO). One milligram of TPP1 IS was dissolved in 1.5 mL of assay buffer, which contained 0.15 M NaCl and 0.1 M sodium acetate buffer at pH 4.0. This concentrated TPP1 IS solution was further diluted with stock assay buffer to make a 300 μM IS solution. The assay cocktail was a 100 μL mixture composed of 7.2 μL of substrate solution (40 nmol), 5 μL of 300 μM IS solution (1.5 nmol), and an additional 87.8 μL of stock assay buffer. A 3 mm punch of a dried blood spot (DBS) was placed in a 1.5 mL polypropylene Eppendorf tube containing 100 μL of assay cocktail. The solution was then vortexed briefly and incubated for 10 h at 37 °C in a thermostated air shaker at 250 rpm. After incubation, 10 μL of 15% aqueous trifluoroacetic acid was added to quench the assay, followed by 200 μL of 1 M NaOH, and then 800 μL of ethyl acetate. The mixture was vortexed, centrifuged, and 750 μL of the top ethyl acetate layer was collected to dry. Then the dried residue was reconstituted in 200 μL of acetonitrile/water (v/v = 50:50) with 1% acetic acid for analysis by MS.
Results
Design and Synthesis of Substrates and Internal Standards
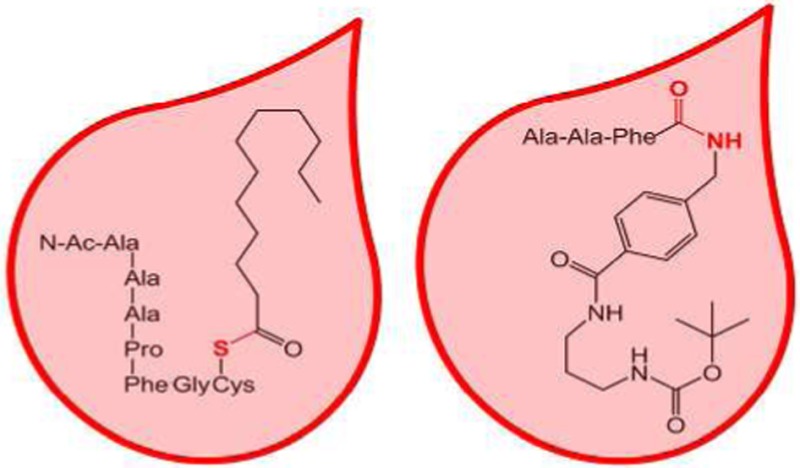
The PPT1 substrate is a cysteine terminated heptapeptide carrying an S-palmitoyl group, N-Ac-Ala-Leu-Leu-Pro-Phe-Gly-Cys-S-COC11H23. Acetylation of the N-terminus protects the substrate and the PPT1 product from being attacked by serum exopeptidases that are present and may be active in rehydrated DBSs. The internal standard, N-Ac-Ala-Ala-Ala-Pro-Phe-Gly-Cys, is homologous with the depalmitylated enzyme product, N-Ac-Ala-Leu-Leu-Pro-Phe-Gly-Cys. The amino acid residues in the substrate, product, and internal standard were designed with two features in mind. One was to produce peptides that would be sufficiently lipophilic to be readily extracted on the C18 solid-phase support. The other feature was to promote residue-selective ion fragmentation upon collisional activation that would focus most of the fragment ion signal into a single dominant dissociation channel. This was accomplished by incorporating in the peptides the proline residue that directs the backbone fragmentation to form a dominant y4 ion at m/z 423, which is common for the enzyme product and the internal standard (Figure 1). The PPT1 substrate was prepared by the single step palmitoylation of a readily available peptide precursor according to Rijkers et al.21 (Scheme S1, Supporting Information).
The substrate for TPP1 (TPP1-S, Figure 2) is a conjugate containing the Ala-Ala-Phe triad, which is preferentially cleaved by the enzyme,7 that is linked by an amide bond to a nonpeptidic moiety. The latter consists of an 4-aminobenzylcarboxamide, a 1,3-propanediamine, and a t-butyloxycarbamate group (t-BOC). The product (TPP1-P) and internal standard (TPP1-IS) retain the hydrophobic benzyl, propylene, and t-BOC linkers to be readily extracted in ethyl acetate or on C18 solid-phase support. The t-BOC group allows for the introduction of a deuterium label in the IS.20 Upon collisional activation, ions containing the t-BOC group readily dissociate by consecutive loss of C4H8 and CO2,22 which provides suitable channels for selected reaction monitoring. The ion transitions of the TPP1 product and IS, m/z 308 → m/z 208 and m/z 317 → m/z 209, respectively (Figure S1, Supporting Information), overlap with none of the other products and internal standards in the LSD assay cassette,19,20,23 thus allowing multiplex quantitation by SRM. The TPP1 substrate was synthesized from the components by standard coupling reactions in five steps (Scheme S2), as described in the Supporting Information. The product and d9-internal standard were prepared in four steps (Scheme S3, Supporting Information).
Assay Evaluation and Optimization
Both assays were evaluated and optimized regarding the incubation conditions affecting the enzyme activity, workup procedures affecting the product and IS recovery, and mass spectrometric analysis affecting the ionization efficiency and collision induced dissociation. Results of the relevant measurements are summarized in Figures S2–S8 in the Supporting Information.
The PPT1 activity is known to have an optimum activity at pH 7 when acting on a natural substrate.14 The PPT1 activity in the DBS with respect to the synthetic PPT1 substrate showed a similar pH dependence, increasing from pH 4 to 8 (Figure S2, top, Supporting Information). However, nonenzymatic hydrolysis of the thioester bond in the PPT1 substrate substantially increased between pH 7 and 8. Therefore, the assay pH was adjusted to pH 7, which showed a maximum activity after a blank correction (Figure S2, bottom, Supporting Information). The TPP1 activity toward TPP1-S showed a pH dependence profile that was more typical of a lysosomal enzyme, peaking at pH 4 (Figure S3, Supporting Information). The production of TPP-P by nonenzymatic hydrolysis in the blank sample at pH 4 was ca. 15-fold lower than that due to TPP1 activity in the DBS.
The PPT1 activity as a function of the amount of enzyme was established by conducting the assays with an increasing number of DBS punches from a single donor. Figure S4 (Supporting Information) shows that the PPT1 activity increased approximately linearly with the number of DBSs used for the assay. The amount of product obtained from a single DBS punch (2 nmol) was sufficient for activity enzyme measurements in the assays.
The PPT1 activity showed a pseudolinear increase with incubation times from 1 to 10 h and then it leveled off at longer incubation times (Figure S5, top, Supporting Information). The leveling-off effect can be in part due to enzymatic digestion of PPT1-P by proteolytic enzymes in rehydrated DBSs. This was checked by incubating synthetic PPT1-P in the assay buffer with a DBS for 18 h. The incubation resulted in 25–27% decrease of PPT1-P compared to a control sample. Because PPT1-P and PPT1-IS are similar hydrophobic peptides, it is safe to presume that their depletion will occur at similar rates and will not affect their molar ratio. The TPP1 activity showed a gradual increase with incubation time with a pseudolinear portion of the curve between 3 and 13 h (Figure S5, bottom, Supporting Information). On the basis of these measurements, the incubation time for both PPT1 and TPP1 was set at 10 h. The average PPT1 and TPP1 substrate conversions at 10 h of incubation were 5% and 1%, respectively.
The enzyme kinetics with respect to the synthetic substrates was established through Michaelis–Menten plots, which were fitted to obtain the Km and Vmax values (Figure S6, Supporting Information). Each Michaelis–Menten curve was obtained from triplicate activity measurements over an appropriate substrate concentration range (0–500 and 0–300 μM). The PPT1 substrate showed Km = 0.23 mM and Vmax = 217.0 μmol L–1 h–1. The TPP1 substrate showed Km = 53.3 μM and Vmax = 46.0 μmol L–1 h–1. The measured Km were used to adjust the initial substrate concentrations in the assays to ≥2 Km value, e.g., 0.46 and 0.40 mM for PPT1-S and TPP1-S, respectively.
Assay and Sample Workup Conditions
ESI-MS/MS analysis of the assays requires that the samples be dissolved in a compatible solvent free of nonvolatile salts and detergents. The presence of a surfactant in the PPT1 assay buffer further accentuates the need for matrix removal. Liquid–liquid extraction is a well-established method of assay sample purification, and it was investigated for PPT1-P and IS using different organic solvents. However, we found that ethyl acetate, n-butanol, and 8:1:1 ethyl acetate-n-butanol-n-hexanol mixture were ineffective in extracting the product and internal standard. In addition, the nonionic detergent (Triton X-100) was also extracted into the organic layer and interfered with mass spectrometric analysis. Therefore, liquid–liquid extraction with ethyl acetate was utilized to first remove Triton X-100 from the samples, whereas recovery of PPT1-P and IS was achieved by subsequent solid phase extraction (SPE). Standard C18 SPE pipette tips (Omix Tips) were found to selectively extract PPT1-P and IS from the aqueous phase of the assay whereas buffer salts can be washed away. The peptides were released by elution with a 50:50 mixture of acetonitrile–water that achieved recoveries of 52 and 50% (both ±2%) for PPT1-P and IS, respectively. This acetonitrile–water mixture does not elute the substantially more lipophilic PPT1-S from the solid phase, so the substrate does not interfere with mass spectrometric analysis. Prior to injecting the samples in the mass spectrometer, disulfide bonds spuriously formed by cysteine oxidation under the assay conditions must be reduced. TCEP was selected as the reducing agent, and the optimal concentration required for quantitative reduction was determined to be 0.8 mM (Figure S7, Supporting Information).
Liquid–liquid extraction into ethyl acetate of TPP1-P and IS performed with 76 ± 2% recovery in a single step. These compounds are chemically identical except for the presence of deuterium isotopes in TPP1-IS and can be expected to have similar partition coefficients for extraction into ethyl acetate. The different incubation and workup conditions for PPT1 and TPP1 assays precluded coincubation in a single duplex assay or sample combination prior to workup. However, duplex mass spectrometric quantitation was possible, as described below.
Mass Spectrometric Response
The workup procedures for TPP1 and PPT1 products and internal standards resulted in very similar recoveries, indicating practically no bias in the concentrations of the P and IS in the samples subjected for mass spectrometric analysis. The responses in SRM for TPP1 and PPT1 products and internal standards were determined to ensure accurate quantitation of the product formation in enzyme assays. Figure S8 (Supporting Information) shows the observed relative responses (P/IS reporter ion intensity ratios) plotted against the calculated concentration ratios. Both response curves show a satisfactory linearity (r2 ≈ 0.99) and slopes close to 1. SRM of PPT1-P shows about a 10% higher response than that of PPT1-IS. The nature of this difference has not been determined, although slightly different ionization efficiencies of the N-Ac-Ala-Ala-Ala-Pro-Phe-Gly-Cys (PPT1-P) and N-Ac-Ala-Leu-Leu-Pro-Phe-Gly-Cys (PPT1-IS) peptides in electrospray, as well as different ion fragmentation efficiencies for the reporter y4ion formation, are not unexpected. The slope for the TPP1-P/IS response, 0.9995 ± 0.003, r2 = 0.998, indicated a nearly identical response to the product and internal standard.
Clinical Sample Analysis
PPT1 assays were performed with 62 random newborn samples that were obtained from the Washington State Newborn Screening Laboratory under Institutional Review Board guidelines, and five previously diagnosed infantile NCL patients (Figure 3). The amount of product formed was calculated using the SRM intensity ratios of the product to the internal standard, the known concentration of the internal standard, and the response ratio (P/IS = 1.104, Figure S8, top, Supporting Information). The data are compiled in Table S1 (Supporting Information). A small amount of PPT1-P is formed by nonenzymatic hydrolysis of the substrate at pH 7.0, giving a mean assay/blank ratio of 16.3. Therefore, all assay data were subjected to blank correction. Enzymatic activity was calculated as μmol h–1 (L of blood)−1 from the amount of product formed, incubation time, and volume of blood. The blood volume in the DBS was estimated at 3.2 μL, based on the estimated volume of a blood spot (10 μL) and the punch/DBS area ratio.
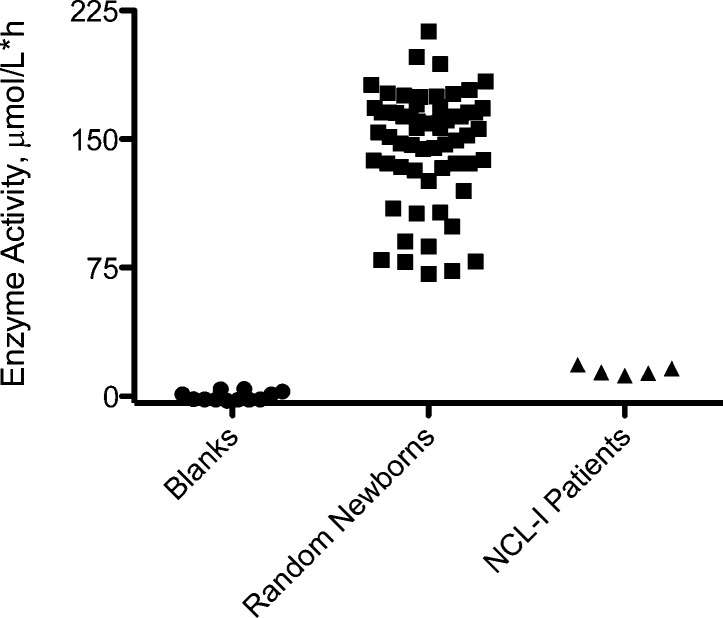
Figure 3.
Graphical representation of PPT1 activities in DBS from simplex assays.
Unaffected newborns showed a range of PPT1 activities from 71 to 213 μmol h–1 L–1 with a mean at 147 μmol h–1 L–1. Patients affected with infantile NCL (PPT1 deficiency) displayed a range of activities between 12 and 18 μ mol h–1 L–1 with a mean value of 15 μmol h–1 L–1. Blanks combining all the components of the assay but replacing the DBS punch with a filter paper punch were analyzed, and the activities were in the range of 6.5 to 13.4 μmol h–1 L–1 with a mean value of 9.0 μmol h–1 L–1. Assay precision was calculated using DBSs from a healthy adult control sample. The intra-assay coefficient of variation (CV) was 3.2% (n = 5), calculated from five injections of the sample from the incubation of a single DBS. The interassay CV was 15% that involved 10 injections from different DBS punches while avoiding the blood spot perimeter.
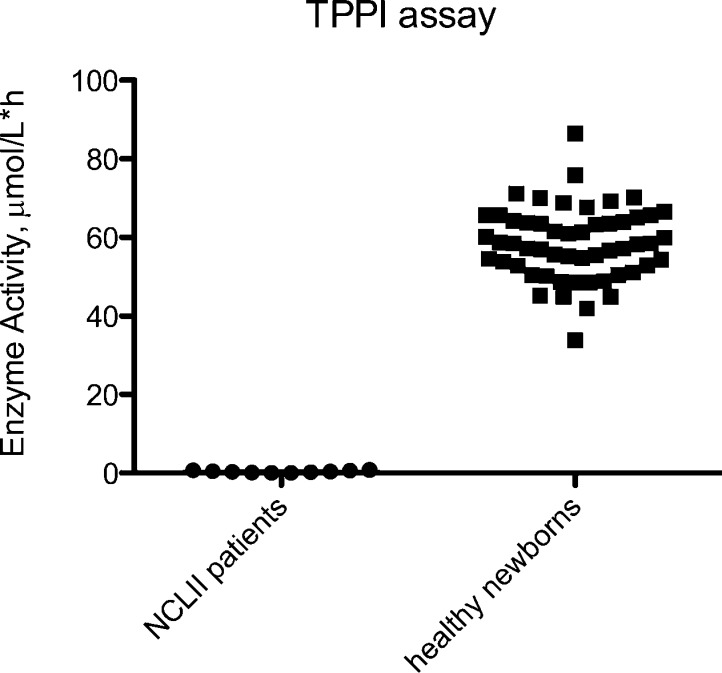
TPP1 assays were performed with DBS samples from 54 random newborns, obtained with IRB approval from the Washington State Newborn Screening Laboratory, and 10 previously diagnosed NCL II patients (Figure 4). Unaffected newborns showed a range of activities from 34 to 87 μmol h–1 L–1 with an average of 58 μmol h–1 L–1. NCL II patients (TPP1 deficiency) displayed a range between 0.02 to 0.8 μmol h–1 L–1 and an average of 0.3 μmol h–1 L–1, all for blank corrected data. Blanks were carried out by combining all the components of the assay but replacing the DBS with a filter paper punch. The sample mean/blank ratios were >60 and the blanks showed 14% CV. The assays showed a clear distinction between NCL II affected patients and healthy newborns. TPP1 assay precision was calculated using a DBS from a control sample. The intra-assay CV was 1.4% (n = 5) involving five injections of the sample from the incubation of a single blood spot. The interassay CV was 8.7% when based on 15 injections from different blood spots from the same individual.
Figure 4.
Graphical representation of TPP1 activities in DBS from simplex assays.
Duplex Analysis
The PPT1 and TPP1 assays were combined in a single injection for tandem mass spectrometry SRM analysis with the goal of speeding up data acquisition and increasing sample throughput. Due to different assay pH conditions and workup procedures, the assays could not be run as single DBS incubations, but rather samples from each assay were combined after workup. The results of this duplex assay of 40 random samples are presented in Figure S9 (Supporting Information), and the data are compiled in Table S3 (Supporting Information) In the duplex format, random samples showed PPT1 activities in the range of 81 to 268 μmol h–1 L–1 with a mean activity of 156 μmol h–1 L–1. These results were comparable to the data from the above-described simplex PPT1 assay. The five infantile NCL-affected newborns had a range of activity of 14–40 μmol h–1 L–1, with a mean at 29 μmol h–1 L–1. The PPT1 activity of the five late infantile NCL II affected newborns (TPP1 deficiency) ranged between 30 and 82 μmol h–1 L–1, with an average activity of 61 mmol h–1 L–1. This was lower than the mean activity for random TPP1 samples, but within the normal range. Blanks were evaluated for the duplex assay to produce PPT1 activities of 13 to 15 mmol h–1 L–1 with a mean of 14 mmol h–1 L–1.
Duplex data from incubation of TPP1 activity in 40 random samples gave a range of activities of 40 to 106 μmol h–1 L–1 with a mean activity of 63 μmol h–1 L–1 (Figure S10 and Table S4, Supporting Information) These values are comparable to those from simplex assay measurements with a different set of 54 random samples (see above). TPP1 activity of five NCL II (TPP1 deficiency) affected newborns ranged between 2.6 and 2.8 μmol h–1 L–1 with an average activity of 2.7 μmol h–1 L–1, which dropped to 0.2 μmol h–1 L–1 after blank correction. The TPP1 activity for the affected individuals from the duplex assay were very similar to those from the simplex assay. The TPP1 activities of the five infantile NCL1 affected newborns with diagnosed PPT1 deficiency had a mean of 27 μmol h–1 L–1, which was at the low end but still within the normal range for random samples.
Discussion
The new substrates showed robust performance in PPT1 and TPP1 assays based on tandem mass spectrometric activity measurements. Compared to fluorometric assays,17 the new protocols do not require enzyme extraction from the DBS nor chloroform extraction of the PPT1 substrate, which may be problematic in a clinical laboratory. Enzymatic product formation from the new substrates was several times higher than that reported for the fluorometric assays.17 For example, a 46 h incubation of the fluorometric PPT1 substrate was reported to produce, on average, 0.82 nmol of product per DBS.17 Our new PPT1 substrate was shown to produce on average 4.7 nmol of product per DBS after 10 h of incubation. The fluorometric TPP1 substrate was reported to produce, on average, 0.27 nmol of product per DBS after 46 h of incubation.17 This is to be compared with 1.7 nmol/DBS produced from our new substrate after 10 h of incubation. These product quantities, when combined with efficient liquid–liquid or solid-phase extraction procedures, provide high ion counts in the mass spectrometric analysis by SRM.
The lower activities measured in samples from the cross-affected children are most likely due to partial deterioration of the enzymes in the DBSs.25 Whereas the DBSs from random newborns were less than 6 months old and were stored at low temperatures;24 the much rarer samples from the affected children had been collected over several years and were stored at room temperature. We observed a 15% decrease of TPP1 activity in a 6-month old DBS that was stored for an additional 12 months at 4 °C. A more systematic study of DBS aging carried out with multiple samples would be necessary to address this issue before the PPT1 and TPP1 activity assays are used for large-scale screening.13,25
Conclusions
The results reported here illustrate the power of tandem mass spectrometry in performing enzyme assays in dried blood spots from human subjects using synthetic compounds that closely mimic natural substrates. Using this approach, it is possible to increase enzymatic product formation in the DBS to facilitate accurate quantitation of enzyme activity. Through careful design of enzyme substrates and internal standards for human palmitoyl protein thioesterase and tripeptidyl peptidase, the analytical procedures for the detection of neuronal ceroid lipofuscinoses can be multiplexed with each other and potentially also with the previously developed methods of detecting lysosomal enzyme deficiencies.
Acknowledgments
Financial support for this research was provided by the NIH Institute for Diabetes, Digestive and Kidney Diseases (Grant R01 DK067859). Thanks are due to Dr. Martin Sadilek for technical support with mass spectrometry measurements.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ M.B. and C.X. contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hofmann S., Peltonen L.. The Neuronal Ceroid Lipofuscinoses. In The Metabolic and Molecular Basis of Inherited Disease, 8th ed.; Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Eds.; McGraw-Hill: New York, 2001; pp 3877–3894. [Google Scholar]
- Santavuori P.; Haltia M.; Rapola J. Dev. Med. Child Neurol. 1974, 16, 644. [DOI] [PubMed] [Google Scholar]
- Jalanko A.; Braulke T. Biochim. Biophys. Acta 2009, 1793, 697–709. [DOI] [PubMed] [Google Scholar]
- Bellizzi J. J. 3rd; Widom J.; Kemp C.; Lu J. Y.; Das A. K.; Hofmann S. L.; Clardy J. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 4573–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia M. Biochim. Biophys. Acta 2006, 1762, 850–856. [DOI] [PubMed] [Google Scholar]
- Williams R. E.; Mole S. Neurology 2012, 79, 183–191. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Sohar I.; Taylor J. W.; Lobel P. J. Biol. Chem. 2006, 281, 6559–6572. [DOI] [PubMed] [Google Scholar]
- Lin L.; Lobel P. Biochem. J. 2001, 357, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.; Cooper J. D.; Sleat D. E.; Cheng S. H.; Dodge J. C.; Passini M. A.; Lobel P.; Davidson B. L. Mol. Ther. 2008, 16, 649–656. [DOI] [PubMed] [Google Scholar]
- Wong A. M.; Rahim A. A.; Waddington S. N.; Cooper J. D. Biochem. Soc. Trans. 2010, 38, 1484–1488. [DOI] [PubMed] [Google Scholar]
- Selden N. R.; Al-Uzri A.; Steiner R.; Huhn S. L. 126 CNS Stem Cell Transplantation for Neuronal Ceroid Lipofuscinoses: Summary of Long-term Follow-up Study Results. Neurosurgery 2013, 60Suppl. 1161–162. [Google Scholar]
- Kohan R.; Cismondi I. A.; Oller-Ramirez A. M.; Guelbert N.; Anzolini T. V.; Alonso G.; Mole S. E.; de Kremer D. R.; de Halac N. I. Curr. Pharm. Biotechnol. 2011, 12, 867–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C. R.; Elliott S.; Buroker N.; Thomas L. I.; Keutzer J.; Glass M.; Gelb M. H.; Turecek F. J. Pediatr. 2013, 162, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp L. A.; Hofmann S. L. J. Biol. Chem. 1993, 268, 22566. [PubMed] [Google Scholar]
- van Diggelen O. P.; Keulemans J. L. M.; Winchester B.; Hofman I. L.; Vanhanen S. L.; Santavuori P.; Voznyi Y. V. Mol. Gen. Metabol. 1999, 66, 240–244. [DOI] [PubMed] [Google Scholar]
- Sohar I.; Lin L.; Lobel P. Clin. Chem. 2000, 46, 1005–1008. [PubMed] [Google Scholar]
- Lukacs Z.; Santavuori P.; Keil A.; Steinfeld R.; Kohlschütter A. Clin. Chem. 2003, 49, 509–511. [DOI] [PubMed] [Google Scholar]
- Li Y.; Scott C. R.; Chamoles N. A.; Ghavami A.; Pinto B. M.; Tureček F.; Gelb M. H. Clin. Chem. 2004, 50, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spáčil Z.; Tatipaka H.; Barcenas M.; Scott C. R.; Tureček F.; Gelb M. H. Clin. Chem. 2013, 59, 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. J.; Ghomashchi F.; Kim T.; Abam C. A.; Sadilek M.; Jack R.; Thompson J. N.; Scott C. R.; Gelb M. H.; Turecek F. Bioconjugate Chem. 2012, 28, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers D.; Kruijtzer J.; Killian J. A.; Liskamp R. M. J. Tetrahedron Lett. 2005, 46, 3341–3345. [Google Scholar]
- Spáčil Z.; Hui R.; Gelb M. H.; Tureček F. J. Mass Spectrom. 2011, 46, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennamaneni N.; Kumar A.; Barcenas M.; Spacil Z.; Scott C. R.; Tureček F.; Gelb M. H. Anal. Chem. 2014, 87, 4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffey T. A.; Bellamy G.; Elliott S.; Fox A. C.; Glass M.; Tureček F.; Gelb M. H.; Scott C. R. Clin. Chem. 2010, 56, 1854–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbin C. S.; Olivova P.; Marashio C. A.; Cooper S. K.; Cullen E.; Keutzer J. M.; Zhang X. K. Clin. Chim. Acta 2011, 412, 1207–1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.