Abstract
Ribonucleoprotein complexes involved in pre-mRNA splicing and mRNA decay are often regulated by phosphorylation of RNA-binding proteins. Cells use phosphorylation-dependent signaling pathways to turn on and off gene expression. Not much is known about how phosphorylation-dependent signals transmitted by exogenous factors or cell cycle checkpoints regulate RNA-mediated gene expression at the atomic level. Several human diseases are linked to an altered phosphorylation state of an RNA binding protein. Understanding the structural response to the phosphorylation “signal” and its effect on ribonucleoprotein assembly provides mechanistic understanding, as well as new information for the design of novel drugs. In this review, I highlight recent structural studies that reveal the mechanisms by which phosphorylation can regulate protein–protein and protein–RNA interactions in ribonucleoprotein complexes.
Protein phosphorylation is a ubiquitous regulatory mechanism in eukaryotes and is essential for the control of gene expression.1,2 Phosphorylation of RNA-binding proteins by protein kinases and their dephosphorylation by protein phosphatases is an important on/off switch to control RNA processing and mRNA decay in response to extracellular signals or cell cycle checkpoints.3 Structural studies on phosphorylated and nonphosphorylated RNA binding proteins can provide important insights as to how an RNA binding protein executes the phosphorylation signal to control gene expression. The structural consequence of phosphorylating an RNA-binding protein, and the effect of phosphorylation on RNA–protein and protein–protein interactions in ribonucleoprotein complexes is just beginning to be understood.
Protein phosphorylation usually occurs in dynamic or disordered regions in RNA binding proteins and there are diverse mechanisms by which the phosphate moiety may participate in executing a structural response to the signal. The dianionic nature of a phosphoryl group (pKa ∼ 6.9) at physiological pH, its tetrahedral geometry, presence of electron-rich oxygens, and its steric bulk allows it to participate in a network of hydrogen bonding interactions. A phosphate–arginine salt-bridge is the most common interaction observed. The arginine guanidinium group is unique in that it has a pKa > 12, a rigid planar structure, and a charge distribution that allows it to form a tight interaction with the doubly charged phosphate at physiological pH. Arginine–phosphate bonds have been proposed to have “covalent-like” stability.4 Other residues involved in interaction with a phosphate are lysine, tyrosine, serine, threonine, asparagine, histidine, and metal ions. Phosphorylation may induce a global conformational change in a protein that may allosterically either promote or inhibit protein–protein or protein–RNA interactions. Silent phosphorylation effects can be observed where there is no conformational change induced in the RNA-binding protein, and the negative charge and steric bulk of the phosphate directly alters its association with an effector protein or RNA. Disorder-to-order or order-to-disorder transitions are most frequently reported, where the structural and dynamic changes are localized to the site of phosphorylation. In analogy to protein kinases, that have an “activation segment” that harbors a phosphorylated threonine or tyrosine residue that is required for kinase activation,5 many RNA binding proteins have a “signal response segment” (SRS). The SRS usually comprises a disordered region of the protein that may become ordered upon phosphorylation. Structural studies on RNA binding proteins to date have shown that the structural consequences of phosphorylation tend to be localized in tertiary structure. Phosphorylation may organize the SRS to be a “hub” or epitope that serves as a recognition site for another protein in the ribonucleoprotein complex.
In this review, I describe the structural outcome to phosphorylation observed in RNA binding proteins where such information is available for intact phosphorylated and nonphosphorylated proteins, or their subdomains, from X-ray crystallography or NMR spectroscopy. In particular, RNA binding proteins that play a role in splicing and mRNA decay have been highlighted.
Structural Control of Alternative Splicing by Phosphorylation
The functional consequences of reversible phosphorylation in regulating alternative splicing and splice-site recognition are well established.6 Proteomics studies have identified >300 proteins as being associated with splicing complexes, and many of these factors are either phosphorylated or glycosylated.7 A rapid change in alternative splice site selection has been observed in response to cellular stimuli that correlates well with a change in the phosphorylation state of splicing factors.8−10 The effects of phosphorylation can be mediated either by remodeling ribonucleoprotein complexes (mRNPs) via altering protein–protein or protein–RNA interactions11−13 or by changing the subcellular localization of the splicing factor.14,15 Phosphorylation of splicing regulatory proteins or SR proteins can occur in arginine and serine rich (RS) domains that are between 50 and 300 residues. These RS domains can be hypophosphorylated at 8–10 residues or hyperphosphorylated by multiple kinases at >20 serine residues.16 Phosphorylation of SR proteins can alter subcellular localization and macromolecular interactions in spliceosomes and hence affect splice site selection. Several diseases17−19 show variation in splicing activity due to either altered kinase activity or a change the phosphorylation state of splicing factors. Therefore, understanding the molecular basis as to how phosphorylation affects the structure of splicing factors in the context of larger spliceosomal assemblies may be a useful strategy for drug development.
RS Domains In SR Splicing Factor 1 (SFRS1)
The SR protein SFRS1 (previously known as Alternative Splicing Factor (ASF)/Splicing Factor 2 (SF2))20 is the best-studied member of the SR protein family. SFRS1 has two RNA recognition motifs (RRMs) in the N-terminus and the RS domain in the C-terminus with 20 serines (Figure 1A, B). SFRS1 is phosphorylated on 12 of these serines in the N-terminal portion of the RS domain (RS1) (Figure 1B) by the kinase SR protein kinase-1 (SRPK1).21,22 Hyperphosphorylation of RS1 in the cytoplasm by SRPK1 and cdc2-like (Clk/Sty) kinases facilitates dissociation of SFRS1 from mRNP complexes, promoting its re-entry into the nucleus13 by associating with the nuclear import factor Transportin 3 (or Transportin-SR2).23 Once in the nucleus, phosphorylated SFRS1 associates with U1–70K and U2AF proteins during spliceosome assembly.24,25 SFRS1 may also undergo additional phosphorylation by the cdc2-like (Clk/Sty) kinases in the nucleus resulting in its dissociation from speckles.26
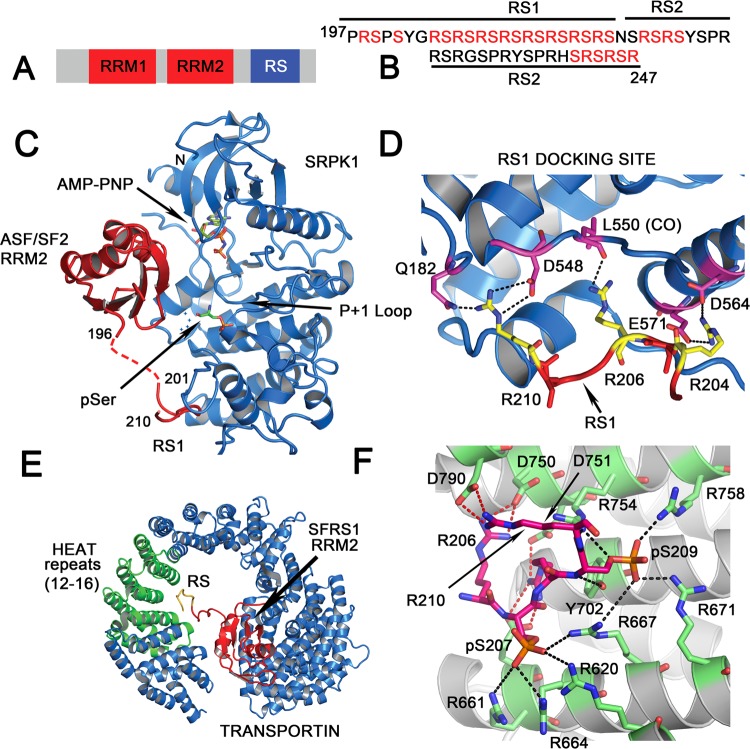
Figure 1.
Crystal structures of phosphorylated SFRS1 bound to Transporting 3 and SRPK1. (A) Domain structure of SFRS1 is depicted. (B) The sequence of the RS domain consisting of the N-terminal RS1, which is phosphorylated by SRPK1, is shown. The C-terminal RS2 region is phosphorylated by Clk/Sty kinases. (C) The 2.9 Å crystal structure of the SRPK1- SFRS1 complex (PDB code 3BEG) is shown. The kinase is in blue ribbon and SFRS1 in red. (D) Specific interactions between protein side chains of SRPK1 and the RS1 domain of SFRS1 in the docking site are depicted. (E) Cartoon representation of the 2.6 Å crystal structure of Transportin 3 (in blue) bound to the SFRS1 RRM2-RS1 region (in red) (PDB code 4C0O). Heat repeats 12–16 (shown in green) primarily interact with RS1 (in gold). (F) The specific interactions between protein side chains of Transportin 3 and the RS1 domain of SFRS1 is depicted. An extensive arginine-zipper-like interface is observed. The Transportin 3 helices are in gray and the RS1 domain in magenta. Arginines (R206, R208, and R210) from SFRS1 are shown in magenta/blue stick, the phosphoserines (S209 and S207) are in orange. Residues from Transportin 3 that interact with the RS1 peptide are in green stick. Black dashes denote salt bridge interactions between arginines from Transportin 3 and red dashes denote salt bridge interactions between arginines from SFRS1.
The SFRS1 RS domain is unstructured in the absence of bound targets by circular dichroism (CD)27 and NMR.28 In contrast to experimental evidence from CD and NMR that support the unstructured state of the RS domain, molecular dynamics (MD) simulations previously suggested that the unphosphorylated RS domain adopts a α-helical structure.30,31 Upon phosphorylation, the RS domain was predicted to undergo a conformational change to form either an extended structure or an “arginine claw”.30,31 A recent quantitative NMR and MD study28 shows unequivocally that the unphosphorylated RS domain is highly disordered, and it switches to a compact, partly ordered arched shape upon phosphorylation. NMR spin relaxation measurements show small (<0.1) 15N–1H heteronuclear nuclear Overhauser effects (hetNOEs), an indicator of fast motions in the pico-to-nanosecond time scale. The hetNOE values coupled with large NMR line-widths suggest that the unphosphorylated RS1 experiences both fast and slow motions in the micro-to-millisecond time scale. Upon phosphorylation, the hetNOE values increase to 0.6 ± 0.1 and the vicinal three-bond couplings, which are correlated to the backbone dihedral angles, indicate the presence of helical or turn-like structure. A structural ensemble was calculated based on NMR data that includes heteronuclear chemical shifts, one-bond and three-bond scalar couplings, and residual dipolar couplings that were combined with MD simulations for the unphosphorylated RS1, and the monoionic (PO4–) and dianionic (PO42–) protonation states of the phosphoserines. The ensemble provides no evidence for an “arginine claw”, but the RS1 domain is partly ordered assuming an arch-like structure upon phosphorylation, with the arginine and phosphoserine side-chains adopting preferred rotamer conformations. This decrease in conformational entropy upon phosphorylation of the RS domain may promote association of SFRS1 with a wide range of protein targets. Therefore, RS domains are an example where the signal response segment of the RNA binding protein undergoes a disorder to partly ordered state upon state upon hypo- or hyperphosphorylation.
The mechanism by which the RS domain of SFRS1 is hypophosphorylated in a sequential and processive manner from the C- to the N-terminal end of RS1 is unique and has been well studied by X-ray crystallography and kinetic measurements.33−36In vitro, SRPK1 binds SFRS1 with high affinity, with a Kd between 50 and 100 nM and can stay bound to the RS substrate for eight cycles of phosphorylation before releasing the substrate. SR protein kinases have unusual specificity in that they only phosphorylate serines but not threonines that lie adjacent to arginines.
The 2.9 Å crystal structure of the SRPK1–SFRS1 complex (PDB code 3BEG) illustrates the mechanism of hypophosphorylation of RS137 (Figure 1C). There are three regions on SFRS1 that make extensive contacts with SRPK1. First, the second RRM (RRM2) contacts both lobes of the kinase domain via three loops (the α1/β1 loop, the β2/β3 loop, and the βN/β4 loop) burying 1079 Å2 of accessible surface area between RRM2 and the kinase domain. Intriguingly the mode of recognition of SRPK1 by SFRS1 via α-helix-1 is very similar to how RRM2 recognizes RNA.38 The SFRS1 RRM2 (also called a pseudo-RRM) has a canonical α/β fold observed in most RRM domains; however, β4 is longer, and it has an additional β-strand (βN) to form a five-stranded β–sheet, which extends the interaction surface with the kinase. Solution NMR studies38,39 show that the region corresponding to the βN/β4 hairpin observed in the crystal is highly mobile, and in solution, the shorter β4-strand is followed by a break in the strand and a β3′-β3″ hairpin. From the kinase perspective, the glycine-rich loop, the α-helices D and F, and the nucleotide-binding pocket make contacts with RRM2. The interactions with the glycine-rich loop appear to orient the nucleotide for phosphoryl transfer and catalysis. This structure, along with the structure of RRM2/RS1 bound to Transportin 3, demonstrate the pivotal role played by RRM2 in facilitating protein–protein interactions mediated by RS domains by acting as an accessory domain for high-affinity binding. RRM2 also binds RNA using the same surface in α-helix-1,38 indicating it plays an essential role in regulating both protein–protein and protein–RNA interactions.
Second, the N-terminal portion of RS1 sits in a docking groove in the kinase domain (Figure 1D). The interaction between the RS region and the kinase are electrostatic. Alternate arginines on the RS domain form salt-bridge interactions with acidic residues in the docking site. The network of electrostatic interactions likely facilitates movement of the substrate RS from the docking site to the catalytic site without energetic penalty. Only a portion of the RS1 peptide was observed in the crystal structure. No electron density is observed for the linker connecting the RRM2 to residue 201 of the RS1 domain, indicating the RS1 remains flexible when bound to the kinase.
Finally, the C-terminal phosphoserine in RS1 contacts a highly basic region near the P+1 loop of the kinase by interacting with Arg515, Arg518, and Arg561 of SRPK1. The structure explains how SRPK1 binds SFRS1 with high affinity and how it phosphorylates processively in a directional manner from the C- to the N-terminus. The C-terminal end of the RS1 domain sits in the active site and RS1 is threaded through the basic groove until it dissociates from the kinase. Dissociation is facilitated by unfolding of the RRM2 β4 strand when the last serine approaches the active site.
A 2.6 Å crystal structure (PDB code 4C0O) of Transportin-3 complexed to the phosphorylated RRM2-RS1 domain of SFRS1 (Figure 1E) reveals an extensive arginine-zipper-like interaction between two serine phosphates (out of three phosphoserines in the crystallized molecule) and arginine residues in HEAT repeats 14–17 of Transportin 3.32 The arginines of the RS domain in turn form salt bridges with Asp and Glu residues in Transportin 3 (Figure 1F). Most of the interactions are electrostatic, with 11 salt-bridges in the complex between an extended phosphorylated RS domain and the inner face of Transportin 3. Phospho-Ser207 of the RS domain interacts with the guanidinium groups of Arg620, Arg661, Arg664, and Arg667 of Transportin 3, and phospho-Ser209 of the RS domain interacts with the guanidinium groups of Arg667, Arg671, Arg754, and Arg758 of Transportin 3. The RRM2 domain is also involved in protein–protein contacts and is sequestered in the inner concave face of Transportin 3 between HEAT repeats 4–7 and 19–20. The total interface between the cargo (RRM2-RS1 domain of SFRS1) and Transportin 3 is ∼4300 Å2. Hypophosphorylation of RS1 is essential for the interaction between SFRS1 and Transportin 3, and is required for its import into the nucleus in vivo.23
How does hypo- or hyperphosphorylation of the SFRS1 RS domain affect its interactions with RNA and U1–70K during spliceosome assembly? Cho et al.40 examined the interaction of RRMs 1 and 2 in the SFRS1 N-terminus with a 13 nt exonic splicing enhancer (ESE) sequence present in the receptor tyrosine kinase (Ron) mRNA in the presence and absence of unphosphorylated, hypophosphorylated (p-SFRS1), or hyperphosphorylated (pp-SFRS1) RS1 domains using filter binding experiments as well as EMSA. The hypophosphorylated RS1 domain was phosphorylated at 8–10 serines whereas the hyperphosphorylated RS1 domain was phosphorylated at 20–22 serines in RS1 as confirmed by mass spectrometry.41 Unphosphorylated SFRS1 bound the Ron ESE in a specific manner, with a Kd of 172 ± 34 nM by filter binding and 50 ± 5 nM by EMSA. p-SFRS1 bound the Ron ESE with affinity comparable to the unphosphorylated SFRS1 protein (Kd of 180 ± 38 nM by filter binding and 26 ± 11 nM by EMSA). Hyperphosphorylation of SFRS1 at 20–22 serines in its RS domain weakened affinity for the RNA by ∼10-fold (Kd of 540 ± 16 nM by filter binding). Furthermore, the unphosphorylated RS1 domain was found to interact directly with RRMs 1–2 of SFRS1, thereby stabilizing the SFRS1–RNA complex. In contrast, hyperphosphorylated RS1 did not interact with RRMs 1–2 of SFRS1 but did promote interaction of SFRS1 with U1–70K. Therefore, the main role of RS1 hyperphosphorylation appears to be to promote protein–protein interactions with U1–70K during spliceosome assembly. Only a small decrease in affinity (of ∼10-fold) of pp-SFRS1 toward Ron ESE mRNA is observed.40
These biochemical, structural, and dynamics studies (summarized in Table 1) reveal that the RS domain acts as a phosphorylation-dependent switch during spliceosome assembly. RS domains usually undergo two transitions in their phospho-dependent interactions with other proteins. The highly disordered unphosphorylated RS state interacts nonspecifically with its RRMs to stabilize the SFRS1–RNA complex by ∼5–10 fold.40 Upon hyperphosphorylation, the RS1 domain dissociates from its RRMs and likely forms a more compact structure, which is “primed” to form a “fuzzy”42 extended charged platform that docks into the binding partner via a network of electrostatic interactions. Hyperphosphorylation of RS1 weakens affinity of SFRS1 for the RNA by ∼10 fold.40 The RS1 domain also appears to require the neighboring RRM to modulate its affinity toward protein targets by extending the buried surface area between SFRS1 and its interacting partner. Therefore, phosphorylation of RS domains modulates protein–protein and protein–RNA interactions of SFRS1 during splicing by undergoing a disorder-to-order transition that is coupled to protein (such as U1–70K or Transportin 3) binding.
Table 1. Summary of Structural and Biochemical Data on Phosphorylated RNA Binding Proteins.
| RNA-binding protein (PDB codes available) | structure determination method | phosphorylated site/s | mechanism | kinase | change in Kd toward RNA due to phosphorylation | change in Kd toward protein target due to phosphorylation |
|---|---|---|---|---|---|---|
| SFRS1 | NMR (1), MD (1), X-ray (2) | ∼8–10 serines in RS1 by SRPK1; 20 serines by both SRPK1 and Clk | localized disorder to partial order transition upon phosphorylation | SRPK1 Clk/Sty | 5–10 fold decrease in Ron ESE mRNA affinity due to hyperphosphorylation at 20 sites by EMSA and filter binding40 | not determined quantitatively |
| (1) RS1 domain only (no PDB code available) | ||||||
| interaction of SFRS1 with Transportin 3 is salt dependent with significant reduction in binding in 500 mM NaCl in GST pull-down experiments consistent with an electrostatic interaction32 | ||||||
| (2) RRM2-RS domain (3BEG bound to SRPK1; 4C0O bound to Transportin 3) | ||||||
| localized disorder to order transition upon SRPK1 and Transportin 3 binding | ||||||
| SF1 | X-ray and NMR | Ser80, Ser82 | localized disorder to order transition upon phosphorylation | KIS kinase | not quantified; small increase in efficiency of RNA binding by SAXS and EMSA.43,44 | Small increase (<2-fold) in association by ITC43 |
| 2M09 (free HH- NMR) | ||||||
| 2M0G (unphosphorylated SF1 NYD bound to U2AF UHM – NMR) | ||||||
| Kd values: pSF1 + U2AF65RRM123 = 96 ± 32 nM; SF1 + U2AF65RRM123 = 114 ± 23 nM | ||||||
| 4FXW | ||||||
| (phosphorylated SF1 NTD bound to the U2AF UHM – X-ray) 4FXX (unphosphorylated SF1 NTD (residues 26–132)) | ||||||
| SLBPL-motif | X-ray | Thr171(hSLBP)/ Thr230 (dSLBP) | localized disorder to order transition upon phosphorylation | unknown | 7–11 fold increase in affinity toward histone mRNA stem-loop75,76 | >32-fold toward Pin1 using fluorescence anisotropy80 |
| unknown possibly CK2 | ||||||
| unknown | ∼27 fold increase in affinity toward histone mRNA stem-loop by dSLBP75 | |||||
| 4QOZ (T171 phos) 4L8R (T171 nonphos) | ||||||
| Ser221, Ser222, and Thr226 (hSLBP) | ||||||
| Ser269, Ser271, Ser273, Ser275 (dSLBP) | ||||||
| KSRP KH1 | NMR, X-ray | Ser193 | order to disorder, i.e., global unfolding of KH1 domain | AKT | no detectable difference by EMSA and ITC89 | >1000-fold increase in affinity toward 14–3–3ζ upon phosphorylation by ITC89 |
| 2OPU (nonphos KH domain) 1QJB (phospho-Ser peptide bound to 14–3–3ζ) | ||||||
| Upf1/SMG proteins | X-ray | Thr28, Ser1078, Ser1096, and Ser1116 in SQ motifs in Upf1 | localized disorder to order transition upon phosphorylation | SMG-1; possibly other SQ kinases such as ATR, ATM, DNA-PK | unknown | >100-fold increase in affinity toward SMG7 upon phosphorylation by ITC105 Binding to SMG5–SMG7, SMG6, and SMG7 is phosphorylation dependent in GST-pull down and coimmunoprecipitation experiments103,104 |
| 1YA0 (N-terminal domain of SMG7) | ||||||
| 4UM2 (TPR domain of SMG6) | ||||||
| 3ZHE (C. elegans SMG5–SMG7) | ||||||
| TiaS | X-ray | Thr18 | silent phosphorylation | unknown | no significant difference toward tRNA is predicted111,112 | N/A |
| (phosphorylated) | ||||||
| 3AMT 3AMU 3AU7 | ||||||
| Ire1 | X-ray | Ser724 in activation segment (in human Ire1) | global conformational change involving dimerization and domain reorientation | Ire1 autophosphorylation | 3–5 fold decrease in Km for the RNA substrate due to phosphorylation at 1–4 sites; 2–40 fold effect on Kcat/Km118 | N/A |
| 3P23 2RIO/3LJ0 3FBV |
SPSP motif in Splicing Factor 1 (SF1)
Insights into the role of phosphorylation in regulating alternative splicing have also come from structural studies of phosphorylated and nonphosphorylated splicing factor 1 (SF1).43,44 SF1 participates in the assembly of complex E of the spliceosome45 and is important for 3′-splice site recognition by binding to the branch point sequence (BPS) of the RNA.46 SF1 also interacts with the essential U2 snRNP auxiliary factor large subunit (U2AF65) that in turn binds the poly pyrimidine tract located at the 3′ splice site.47,48
The SF1 protein consists of five structural domains (Figure 2A): (1) N-terminal U2AF ligand motif (ULM) that binds the U2AF65 homology motif (UHM), (2) a helix hairpin (HH) motif containing the phosphorylated SPSP containing domain, (3) the K homology (KH) and Quaking homology 2 (QUA2) KH-QUA2 domain that binds RNA, (4) a zinc knuckle motif that binds RNA nonspecifically, and (5) a proline rich region at the C-terminus that may interact with SH2 containing proteins in signaling pathways. U2AF65 has RS and ULM domains at the N-terminus followed by RRM1, RRM2 that bind RNA, and the UHM domain that mediates protein–protein interactions with SF1 (Figure 2A). Structures of the SF1 KH-QUA2 domain bound to BPS RNA49 and the U2AF65 RRM1-RRM2 domains bound to the poly-pyrimidine tract RNA50,51 have revealed how these proteins contact the RNA at the 3′-splice site. The structure of the UHM domain of U2AF65 complexed to the ULM domain of SF1 shows how protein–protein interactions occur via the ULM motif.52
Figure 2.
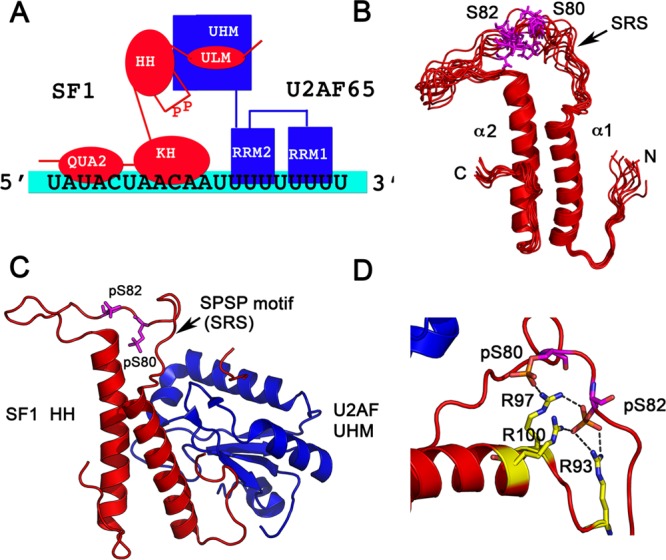
X-ray crystallographic and NMR structures of phosphorylated SF1. (A) Schematic showing the domain organization and interactions between SF1, U2AF, and the RNA. (B) Solution NMR structural ensemble (PDB code 2M09) of the free helix hairpin (HH) motif consisting of two α-helices in an antiparallel arrangement connected by a flexible linker is shown. The serines that are phosphorylated are in magenta in a dynamic SPSP loop. (C) The 2.29 Å crystal structure of phosphorylated SF1 HH bound to the U2AF UHM (PDB code 4FXW) is shown. SF1 is in red, and U2AF in blue. The phosphorylated serines are depicted in magenta. (D) Interactions of the phosphates with neighboring arginines to form an “arginine claw” are shown.
SF1 is constitutively phosphorylated at two conserved serines in the SPSP motif (Ser80–Pro81–Ser82–Pro83) in HEK cells,53 and serine phosphorylation may be important for cancer progression.54,55 Phosphorylation of SF1 at Ser80 and Ser82 by KIS kinase56 has been suggested to be important for the stability of the SF1–U2AF65–RNA ternary complex.53,57 It is also required for cell proliferation in NIH 3T3 cells, and contributes to the SF1-U2AF65 interaction in vivo.44 Intriguingly, this SPSP motif lies between the KH-QUA2 domain and the ULM that binds the UHM of U2AF65 (Figure 2A). How does phosphorylation at the SPSP sequence that lies between the ULM and the KH-QUA2 motifs of SF1, affect its interaction with RNA or U2AF65? Solution NMR and X-ray crystallographic studies from the Kielkopf44 and Sattler43 laboratories have revealed the structural and dynamic changes that occur in the binary SF1–U2AF65 complex in response to phosphorylation at the SPSP motif, providing new insight into how phosphorylation may promote assembly of complex E of the spliceosome.
Zhang et al.43 determined the solution NMR structure of the helix hairpin (HH) motif (residues 27–145) of SF1 and the entire SF1 N-terminus (residues 1–145) consisting of the ULM and HH domains bound to U2AF65. The HH region consists of two α-helices in an antiparallel arrangement connected by a flexible linker (PDB code 2M09) that has the site of SPSP phosphorylation (Figure 2B). The helices are stabilized by hydrophobic contacts between aliphatic Leu, Ile, and Met residues that interdigitate to form a hydrophobic core. The helix hairpin structure is further stabilized by the C-terminal extension that folds back and packs against helix α2 of the hairpin fold. The SPSP motif is solvent exposed in a flexible linker between the two helices (Figure 2B). The solution NMR structure of the unphosphorylated SF1ULM+HH −U2AF65-UHM complex shows a burial of an additional ∼500 Å2 of a hydrophobic interface between the two proteins due to the HH domain. This interaction therefore forms a secondary binding interface between SF1-U2AF65 proteins, extending the interaction interface by ∼500 Å2. NMR 15N-relaxation measurements demonstrate that the SRS “linker” region, which comprises the site of phosphorylation, becomes rigid via interaction of the phosphates with the guanidinium groups of Arg93, Arg97, and Arg100.43 These solution NMR studies indicate that the effects of phosphorylation are localized to the SPSP loop and do not drastically affect the overall structure or dynamics of the SF1ULM+HH −U2AF65-UHM complex.
How does phosphorylation of the SPSP motif of SF1 affect binding affinity toward U2AF65? The unphosphorylated helix hairpin motif only slightly increases the affinity of SF1 for the U2AF65-UHM by isothermal titration calorimetry (ITC), when measured as isolated domains. The Kd for the interaction of the SF1ULM and U2AF65-UHM is reported to be 127 ± 48 nM and that of SF1ULM+HH and U2AF65-UHM is reported to be 84 ± 24 nM.43 Phosphorylation of the SPSP motif did not alter the Kd for the interaction of the SF1ULM+HH and U2AF65-UHM by ITC. In contrast to the ITC measurements performed by Zhang et al.43 on the individual domains, GST pull-down experiments performed by Wang et al.44 on almost full-length proteins shows that the SASA double mutant is less efficient than the phosphorylated protein in its interaction with U2AF65, and phosphorylation of SF1 at the SPSP motif contributes to interaction with U2AF65. The small increase in binding observed in GST pull-down experiments upon phosphorylation in the Wang et al.44 study could be due to inclusion of the SF1 KH-QUA2 domain, a difference in methodology, or buffers used. Although both studies show that phosphorylation of the SPSP motif is not necessary for the interaction of SF1 and U2AF65in vitro, the apparently small but significant effects of phosphorylation on binding of full-length proteins in vitro and in vivo in the presence of RNA should be investigated in greater detail. The mechanism by which phosphorylation may affect the affinity, albeit to a small extent, between full-length SF1 and U2AF65 is not fully resolved.
In general, there is agreement between the solution NMR studies of Zhang et al.43 and the X-ray crystallographic studies of Wang et al.,44 in that the effects of phosphorylation on the SF-U2AF65 complex are localized to the SRS and similar hydrophobic interfaces are observed between unphosphorylated SF1 and U2AF65 in the NMR and crystal structures. Calorimetric studies performed by Wang et al.44 on nearly full length SF1 (residues 1–255; comprising the ULM, HH/SPSP, and KH-QUA2 domains) and the U2AF65-UHM indicate that there is a large change in the difference in heat capacity (ΔCp) that corresponds to burial of an additional ∼1900 Å2 of surface area in the unphosphorylated SF1-U2AF heterodimer. The 2.29 Å crystal structure of phosphorylated SF1 (residues 14–132)-U2AF65-UHM (PDB code 4FXW) confirms that ∼1400 Å2 of hydrophobic surface area is buried in the complex due to the presence of the HH/SPSP domain, corroborating their calorimetry studies (Figure 2C). Phosphorylation at the SPSP motif induces an “arginine claw” in the crystal structure (Figure 2D). Three arginines of the HH/SPSP domain of SF1 (Arg93, Arg97, and Arg100) form strong intramolecular interactions with the SF1 phosphoserines so as to stabilize the loop conformation. The side chains of Lys104 and Arg79 are also in proximity to the phosphates, thereby providing a favorable positively charged electrostatic environment to surround the phosphoryl groups. In contrast, the 2.48 Å structure of the unphosphorylated SF1 domain bound to U2AF65-UHM shows little electron density for loop residues 74–81 indicating structural disorder for these residues. The structural data is also corroborated by thermostability measurements that show significant stabilization of the phosphorylated SF1 (residues 14–132)–U2AF65-UHM complex compared to the unphosphorylated complex. The backbone Cα atoms of SF1 HH domain (residues 46–66/97–115; without the loop) have an rmsd of 0.50–0.68 Å between the phosphorylated and nonphosphorylated structures, indicating that with the exception of the SPSP loop, the HH domain structure is similar in the two phosphorylated and nonphosphorylated structures, consistent with NMR studies.
An important question that remains unanswered is how phosphorylation at the SPSP motif affects spliceosome assembly by the two proteins? Are there global changes observed in the RNA bound functionally relevant ternary complexes? A comparison of the pairwise distribution functions of the nonphosphorylated and phosphorylated SF1ULM+HH −U2AF65-UHM complexes by low-resolution small-angle X-ray scattering (SAXS) shows only minor differences in the derived radii of gyration for these complexes in both studies.43,44 Whereas the addition of RNA does induce a large change in the radii of gyration (Rg) of both phosphorylated and nonphosphorylated complexes by SAXS (δRg of 0.52 nm corresponding to Dmax of 3 nm), phosphorylation at the SPSP motif has a smaller effect on Rg values (δRg of 0.06 nm, no significant difference in Dmax).43 When the effect of phosphorylation at the SPSP motif on RNA binding was tested using electrophoretic mobility shift assays (EMSA), a small but significant change was noted in that the phosphorylated protein complex bound RNA with slightly higher efficiency,43 as previously reported.53 Both the SAXS and EMSA studies indicate that the effects of phosphorylation on RNA association are likely to be minor. Future high-resolution structures on the ternary nonphosphorylated and phosphorylated SF1ULM+HH −U2AF65-UHM–RNA bound complexes should reveal the precise role of phosphorylation in the assembly of the E complex of the spliceosome.
These studies by NMR spectroscopy, X-ray crystallography, and SAXS, combined with biochemical and biophysical data on phosphorylated and nonphosphorylated SF1, both free and complexed to U2AF65 have provided detailed insight into the structural effects of SF1 phosphorylation (see Table 1 for a summary of structural and biochemical data). The data indicate that phosphorylation of the SPSP motif in SF1 results in localized changes in dynamics surrounding the site of phosphorylation. Phosphorylation of SF1 does not appear to have a large effect on RNA binding for either SF1 or U2AF65 in the RNA bound ternary complex. Surprisingly, phosphorylation also does not affect the affinity or structure of the SF1-U2AF65 binary complex. How does SPSP phosphorylation affect spliceosome assembly? The structural studies suggest that SPSP phosphorylation of SF1 is required to structure the loop so as to “signal” the recruitment of a third protein in complex E of the spliceosome. Future studies should test this hypothesis.
Phosphorylation-Dependent Control of mRNA Decay
It has long been known that mRNA decay rates can change dramatically in the presence of extracellular stimuli such as hormones, stress, viral infections, cytokines, etc. Several RNA binding proteins such as proteins that bind AU-rich elements (AREs) are directly under the control of signaling pathways58,59 and play central roles in regulating the immune response and human diseases. We are only beginning to understand how kinase activated pathways regulate the phosphorylation state of RNA-binding proteins and mRNA decay factors downstream.3 In this section, I discuss three decay factors that are regulated by phosphorylation: SLBP, KSRP, and Upf1.
The L-Motif of Stem-Loop Binding Protein (SLBP)
Histone proteins are required for packaging DNA into chromatin as well as for epigenetic regulation via the histone code.60,61 In mammals, five major classes of the canonical histone proteins (H1, H2A, H2B, H3, and H4) are encoded by ∼65 genes, their biogenesis being tightly coupled to DNA replication.62 These replication-dependent histone mRNAs are cell cycle regulated, and their expression is coordinated with DNA replication during the “synthesis-phase” or S-phase of the cell cycle.63 The major cis-element responsible for cell cycle regulation of the histone mRNAs is a highly conserved 16-nucleotide stem-loop present in the unique 3′ untranslated region (3′ UTR) of histone mRNAs that binds Stem Loop Binding Protein (SLBP).64,65 The hairpin sequence includes a six base pair stem that is capped by a UYUM tetraloop. SLBP66 and CstF64200 are the only two cell cycle regulated trans-acting factors whose expression during S-phase is correlated to expression of histone mRNAs. SLBP is an architectural protein that is important for efficient pre-mRNA processing of histone mRNAs,67,68 histone mRNA export,69 translation,70 and degradation71 of these mRNAs by its ability to interact with the RNA as well as a plethora of proteins in ribonucleoprotein complexes.
All SLBP proteins form a highly stable complex with histone stem-loop RNA.72,73 An apparent equilibrium dissociation constant (Kd) of between 1.5 nM-47.5 nM has been measured for the interaction of human, Drosophila, and Xenopus SLBP RNA Binding Domains (RBDs) with the stem-loop by several laboratories.72,74−76 The off-rate for dissociation of the complex is very slow (koff = 5.4 × 10–4 min–1; t1/2 ∼ 21.4 h)75 indicating that once formed, the SLBP–RNA complex functions as an integral unit. The minimal protein sequence required for specific and high affinity RNA binding is a ∼73 residue segment that bears no sequence homology to any other protein in the eukaryotic genome75 (Figure 3A). In the absence of RNA, the SLBP RBD is intrinsically disordered in solution73,75 and folds in the presence of RNA.73 The crystal structure of human SLBP in a ternary complex with the histone mRNA stem-loop and the exonuclease ERI-1/3′hExo (PDB codes 4L8R and 4QOZ)77 reveals that the SLBP RBD forms a characteristic inverted L-shaped structure consisting of three α-helices that are connected by loops. Helix-2 lies orthogonal to helices 1 and 3 forming a novel RNA binding fold called the SLBP L-motif (Figure 3B). Recognition of the RNA stem-loop occurs by the outer face of the SLBP L-motif by unfolding the tetraloop of the RNA.74,78 SLBP is anchored by sequence specific side-chain-to-base contacts between the guanidinium group of an arginine (Arg181) to the O6 of the first and second guanines, G6 and G7, in G-C base pairs at the bottom of the RNA helix, and a third hydrogen-bond to the G6 phosphate at the base of the stem.
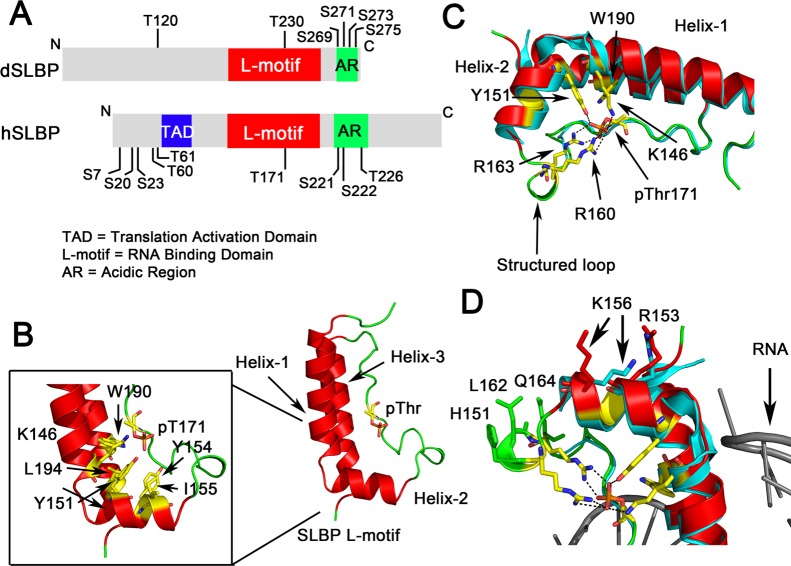
Figure 3.
X-ray crystal structure of phosphorylated human SLBP. (A) Schematic showing the domain organization of human SLBP (hSLBP) and Drosophila SLBP (dSLBP). The RNA binding domain is designated the “L-motif” and is followed by an acidic region, rich in Asp and Glu residues, that is also phosphorylated. The N-terminal domain is involved in translation activation (TAD). Phosphorylation sites that have been mapped in vivo(76,80,124) are indicated. (B) The T171 phosphorylated SLBP L-motif is shown with a characteristic L-shape as seen in the crystal structure of the hSLBP/histone mRNA/3′hExo ternary complex (PDB code 4QOZ). The fold consists of three α-helices connected by a 20-residue flexible loop that has the site of phosphorylation (shown in stick). Hydrophobic residues at the junction of the helices are shown in yellow (inset). (C) Hydrogen bonding interactions mediated by the phosphothreonine with R163, R169, K146, Y151, and W190 (via a water molecule) are shown. The structured loop that is disordered in the unphosphorylated SLBP structure is fully ordered in phosphorylated SLBP. The unphosphorylated structure is shown in blue and the phosphorylated structure in red ribbon. (D) Residues in helix-2 and the structured loop that undergo a conformational change upon SLBP phosphorylation and have been implicated in RNA processing are highlighted.
A unique feature of SLBP proteins is that they are constitutively phosphorylated at an invariant threonine (Thr171 in hSLBP and Thr230 in dSLBP) in vivo(76) (Figure 3A). Phosphorylation of SLBP at Thr230 is essential for nuclear localization of dSLBP, is essential for viability of Drosophila embryos and for histone pre-mRNA processing in flies in vivo.79 In mammalian HEK293 cells, the T171A mutant SLBP is more cytoplasmic and is present in significant number of non-S-phase cells.80 In contrast, the T171D and T171E phosphomimic mutants are almost exclusively nuclear throughout the cell cycle.80 Therefore, phosphorylation at Thr171 is required for efficient import into the nucleus and proper cell cycle regulation of SLBP.80 Phosphorylation at Thr171 (in a TPNK sequence) also makes SLBP a substrate for the prolyl isomerase Pin1.80 The SLBP L-motif undergoes cis–trans proline isomerization in solution.75 Pin1 acts with protein phosphatase PP2A to dephosphorylate SLBP at the end of S-phase, dissociating SLBP from the histone mRNA, thereby facilitating mRNA decay.80 Pin1 knockdown promotes nuclear localization of SLBP and increases histone mRNA abundance and stability.80,81 Affinity measurements using nitrocellulose filter binding76 and kinetic measurements using SPR75 reveal that dephosphorylation of Thr171/Thr230 increases the on-rate of the more basic SLBP protein by 10-fold and also increases the off-rate by 100-fold, thereby decreasing the overall binding affinity (Kd) for the histone stem-loop by ∼7–11 fold.
The crystal structure of the phosphorylated L-motif (PDB code 4QOZ) shows that Thr171 lies in a long flexible loop between helix-2 and helix-3 (residues Val158-Ser179) on the concave face of the L-motif (Figure 3B) and is shielded away from the RNA. The fold is stabilized by a major hydrophobic core at the junction of the two α-helices (Figure 3B). In the unphosphorylated structure,77 no electron density is observed for residues Pro159-Gln164; however, this segment is ordered in the structure of the phosphorylated Thr171 SLBP–RNA complex. The phosphate has a depressed pKa (<4.5),82 exhibits torsional strain in solution,82 and is involved in a network of hydrogen bonding interactions with Arg163, Tyr151, Lys146, and Arg160 in the crystal structure (Figure 3C). The phosphoryl oxygen also makes a hydrogen bond to a water molecule that, in turn, is coordinated to the Trp190 indole nitrogen. A stabilizing γ-methyl-π interaction is observed between the Thr171 γ-methyl group and the indole ring of Trp190, suggesting why threonine, and not serine, may be conserved at this position in all SLBPs. No global conformational changes are observed in the L-motif and the effects of phosphorylation appear to be localized to secondary structure around the site of phosphoryation. The backbone Cα atoms show an r.m.s.d. of 0.2 Å between the unphosphorylated and phosphorylated SLBP L-motif (with the exception of residues 165–169).
The structure reveals how phosphorylation at Thr171 may create a binding epitope for another protein in the RNA processing complex (Figure 3D). Previous mutagenesis studies have shown that when Asp152, Arg153, Ile155, Lys156 in helix-2, and Arg160, His161, Leu162, Gln164, and Pro165 in the disordered loop are mutated together, it abolishes histone pre-mRNA processing in vitro.83 Mutation of Arg160 would remove a stabilizing contact with the phosphoryl oxygens, likely making the loop more flexible. However, the side-chains of Arg153, Lys156 in helix-2 and His161, Leu162, and Gln164 are solvent exposed (Figure 3D), lie on the same face, and are ordered in the SLBP–RNA complex in response to phosphorylation. The likely role of Thr171 phosphorylation is to orient these five residues so their side-chains are in a binding competent mode for interaction with an unidentified RNA processing factor.
Therefore, constitutive phosphorylation at Thr171 in the SLBP L-motif is an important “hub” for regulation of the SLBP–histone mRNA complex. The phosphate forms a network of hydrogen-bonding interactions that structures the Pro159-Gln164 loop that is the likely binding site for another protein in the RNA processing complex. At the end of S-phase, Pin1 and PP2A act in concert to dephosphorylate SLBP at Thr171. This would make the loop disordered, likely destabilizing protein–protein interactions in the RNA processing complex. Dephosphorylation at this site facilitates dissociation of the L-motif from the RNA, thereby facilitating histone mRNA degradation.
Besides Thr171, human and Drosophila SLBP proteins80,84 are also phosphorylated at 3–4 Ser/Thr residues in an acidic region that lies C-terminal to the L-motif (Figure 3A). The functional consequence/s of phosphorylating human SLBP at Ser221, Ser222, and Thr226 in the acidic C-terminal region is currently unknown, although these sites are phosphorylated in vivo by mass spectrometry.80 Inhibition of Pin1 by PiB in HEK cells in vivo enriched phosphates at these sites in human SLBP by mass spectrometry, indicating that phosphorylation at these sites may be regulated.80 In Drosophila SLBP, phosphorylation of the C-terminal tail at four serines is required for RNA processing in vivo, likely via association with an unknown processing factor.84 Phosphorylation of the Drosophila SLBP C-terminal tail increases binding affinity for histone mRNA stem-loop ∼27-fold by Surface Plasmon Resonance (SPR),75 although the mechanism for increased affinity is not clear. No structures of SLBP proteins that are phosphorylated at the C-terminus are currently available. Conformational stability measurements using circular dichroism show that C-terminal phosphorylation of dSLBP forms a more stable SLBP–RNA complex,73 and the effect of phosphorylation is not on the on-rate but on the off-rate of histone mRNA binding.75 One interpretation of this data is that the phosphorylated C-terminus may interact transiently and nonspecifically with the L-motif in the RNA-bound complex, thereby stabilizing the SLBP–RNA complex. Regardless of the smaller effects of phosphorylation on histone mRNA binding (summarized in Table 1), phosphorylation at these sites is likely to be more important for recruitment of another protein in the histone pre-mRNA processing complex.
Phosphorylated RNA Decay Factors that Bind 14–3–3 and 14–3–3-like Domains
14–3–3 proteins are a family of dimeric, all α-helical domains that bind phosphorylated Ser/Thr residues in eukaryotes.85 Two optimal binding motifs recognized by 14–3–3 proteins have been identified: Arg–Ser–Xaa–[pSer/pThr]–Xaa–Pro or Arg–Xaa–Xaa–Xaa–[pSer/pThr]–Xaa–Pro where Xaa denotes any amino acid. Several phosphorylated RNA decay factors interact with 14–3–3 isoforms or with proteins that have 14–3–3-like domains. K-homology splicing regulator protein (KSRP) is an mRNA decay promoting factor that binds AU-rich elements (ARE) in the 3′ UTRs of a number of cytokine mRNAs that play important roles in the immune response, initiating ARE-mediated decay (AMD) of these mRNAs.86,87 Phosphorylation of KSRP at a serine residue (Ser193) in its N-terminal RNA binding domain (KH1) impairs its ability to recruit the exosome and trigger AMD88 (Figure 4A). Phosphorylation at Ser193 makes KSRP a substrate for 14–3–3ζ and the phosphorylated KSRP-14–3–3ζ complex translocates from the cytoplasm to the nucleus, thereby decreasing its ability to participate in mRNA decay.89 The solution NMR structure of KH1 (PDB code 2OPU) shows a typical KH α/β fold with three α-helices packed against a three-stranded antiparallel β-sheet89 (Figure 4B). Ser193 lies in a structured part of the β-sheet and unphosphorylated KSRP is not capable of associating with 14–3–3ζ (Figure 4B). Intriguingly, phosphorylation at Ser193 destabilizes and unfolds the KH1 domain as determined by NMR spectroscopy and circular dichroism (CD).89 This order-to-disorder transition is reversible, and dephosphorylation of KH1 at Ser193 results in refolding of the KH domain. Unfolding of KH1 creates a binding site for 14–3–3ζ, which is capable of recognizing the phosphoserine in an extended conformation (Figure 4C). Consistent with this, a 15-residue peptide comprising the KSRP sequence around Ser193 (residues Gly186-Ser200) can effectively compete with KSRP for 14–3–3ζ. The peptide segment Gly186-Ser200 can therefore be considered as the KSRP SRS, which forms a β-strand in the unphosphorylated KH domain. Unfolding of KH1 does not impair the ability of KSRP to bind the mRNA, since the KH3 and KH4 domains of KSRP mediate high affinity RNA binding to the ARE in TNF-α.90 The role of the SRS in the KH1 domain appears to be to induce a phosphorylation-dependent order-to-disorder switch in KSRP so as to change its subcellular localization via interaction with 14–3–3ζ.
Figure 4.
Structures of phosphorylated mRNA decay factors that bind 14–3–3 or 14–3–3-like domains. (A) Schematic showing domain organization of KSRP. The four KH domains are shown in red and the nuclear localization signals are in blue. (B) Solution NMR structure of the first KH domain (PDB code 2OPU) of KSRP when unphosphorylated. The site of phosphorylation, Ser193, is shown in stick. (C) Comparison of the folding topologies of 14–3–3ζ with 14–3–3-like domains from SMG6, SMG7, and the SMG5-SMG7 complex is shown in purple. The phosphoserine binding site is indicated. (D) Interaction of a phosphoserine peptide with 14–3–3ζ as seen in the crystal structure of the cocomplex (PDB code 1QJB) is depicted. The phosphoserine binds in an extended conformation to 14–3–3 proteins.
The zinc finger protein Tristetraprolin (TTP) is a well characterized ARE binding protein and is phosphorylated by the kinase MK2 at two serines (Ser60 and Ser186), and is also recognized by 14–3–3. Phosphorylation of TTP by MK2 has no effect on the binding affinity of TTP for the TNF-α ARE in EMSA assays, but promotes its interaction with 14–3–3 proteins thereby altering its subcellular distribution and inhibiting its mRNA decay activity.91 However, in contrast to KSRP, the TTP SRS that binds 14–3–3 is located in an unfolded region of the protein. No structural information is currently available for phosphorylated TTP, either free or bound to 14–3–3.
Upf1 (also known as RENT1 or SMG-2) is a nonsense-mediated decay (NMD) factor that is an ATP-dependent 5′ to 3′ RNA helicase and the main effector of NMD.92,93 NMD is a surveillance mechanism in eukaryotes and a mechanism to detect and degrade mRNA transcripts that have a premature termination codon (PTC). Several crystal structures are available for human Upf1 helicase region (residues 295–914),94 Upf1 (residues 115–914) bound to the C-terminal region of Upf2 (residues 1105–1198),95 crystal structures of Upf1–RNA and Upf1–Upf2–RNA complexes,96 a cryo-EM structure of Upf1 bound to the exon junction complex (EJC)97 and EM images of SMG1–Upf1 complexes.98 The overall structure of the helicase core of Upf1 consists of two RecA-like domains that are found in SF1 and SF2 superfamily of helicases. In addition, Upf1 has a N-terminal cysteine-histidine-rich (CH) domain (residues 115–275) that binds Upf2, and a helical domain (residues 556–609). These structures reveal how Upf1 binds RNA and Upf2, providing mechanistic insight into the role of Upf1 in NMD. Upf1 is phosphorylated by the kinase SMG-199,100 (SMG stands for suppressors of morphogenetic defects in genitalia) at Thr28, Ser1078, Ser1096, and Ser1116 SQ motifs in vivo in the unstructured N- and C-terminus.101,102 Phosphorylation–dephosphorylation cycles at these SQ motifs are essential for NMD. Phosphorylated Upf1 is also important for histone mRNA decay via direct association with SLBP.71 Phosphorylation of Upf1 at Thr28 in the N-terminus creates a binding site for SMG-6, a monomeric 14–3–3-like protein that has endonucleolytic activity and also binds the exon–exon junction complex or EJC.103 The C-terminal SQ motifs, in particular, Ser1096 and Ser1116, bind SMG-7 and SMG-5, respectively. SMG-5 and SMG-7 form a stable 14–3–3-like heterodimer (Figure 4C) that binds the C-terminal phosphoserines and catalyzes Upf1 dephosphorylation by recruiting the protein phosphatase PP2A.104 The crystal structures of SMG-5-SMG7,104 SMG-6,103 and truncated SMG-7105 reveal 14–3–3-like domains that have adjacent helical hairpin domains (Figure 4C). Collectively, the entire unit is referred to as the tetratricopeptide (TPR) region. The phosphoserine binding pockets of the SMG-5, SMG-6, and SMG-7 TPR domain is very similar to that of 14–3–3, indicating a similar mode of interaction with an extended phosphopeptide. The canonical mode of interaction of extended phosphoserine containing peptides by 14–3–3 consists of interaction of the phosphate with two arginines, a tyrosine, and a lysine residue in 14–3–3 (Figure 4D). This mode of interaction appears to be conserved in 14–3–3-like proteins such as SMG-5, SMG-6, and SMG-7. The effect of Upf1 hyperphosphorylation on RNA association has not been quantified. The interaction of the truncated 14–3–3 domain of SMG7 (residues 1–497) with a 12 residue Upf1 phosphopeptide corresponding to the region around Ser1078 has been quantified by isothermal titration calorimetry (ITC) and was found to be 50 μM.105 The Upf1 phosphopeptide bound SMG7 with a 1:1 stoichiometry. No detectable binding was observed with the unphosphorylated peptide and given the lower limit of detection by ITC, this corresponds to a >100-fold increase in affinity for SMG7 due to Upf1 phosphorylation. GST pull-down experiments show that the SMG5-SMG7 heterodimer interacts with Ser1096 and Ser1116 of Upf1 in a phosphorylation-dependent manner.103 No structural information is available on the interaction of Upf1 phosphopeptides with SMG5, SMG6, or SMG7. An important question that remains unanswered is how phosphorylated SQ motifs bind their respective 14–3–3-like domains in a specific manner. For example, the SMG5–SMG7 complex binds phosphorylated Ser1096, but does not bind phosphorylated Thr28.102 Conversely, SMG-6 binds the sequence around phosphorylated Thr28 but will not bind the region around phosphorylated Ser1096 on Upf1. Future structural studies on the respective complexes should clarify the mode of specific interaction of phosphorylated SQ motifs of Upf1 with SMG proteins in detail.
Other Phosphorylated Proteins Involved in RNA-Mediated Gene Expression
There are several proteins in eukaryotes and prokaryotes that are involved in RNA-mediated gene expression and are also regulated by phosphorylation. However, structural and kinetic information is available for only a few other proteins, either in the phosphorylated or nonphosphorylated state. The eukaryotic initiation factor 4E (eIF4E) binds the 7-methylguanine (m7G) cap to regulate cap-dependent translation initiation.106 A family of 4E binding proteins (4E-BPs) regulates translation in eukaryotes by directly associating with eIF4E in their unphosphorylated states, thereby inhibiting translation.107,108 4E-BPs are phosphorylated at four sites (Thr37, Thr46, Ser65, and Thr70) by the mammalian target of rapamycin (mTOR), resulting in dissociation of phosphorylated 4E-BP from eIF4E.109 Although 4E-BPs do not bind RNA, multisite phosphorylation of these proteins controls protein–protein interactions at the m7G cap on the 5′ end of the mRNA and hence regulates translation initiation. NMR characterization of full-length unphosphorylated 4E-BP2110 shows that almost the entire protein is intrinsically disordered with transient helical structure.110 Interaction with eIF4E occurs via a bipartite interface that is stabilized to form a helix in the 54YXXXXL motif upon association with eIF4E.110 All four phosphorylation sites lie in proximity to the interaction surface with eIF4E. Recent NMR studies201 show that phosphorylation at Thr37 and Thr46 introduces a dramatic conformational change in 4E-BP2, folding residues Pro18-Arg62 into a four-stranded β-domain that buries the 54YXXXXL motif into a β-strand, thereby abrogating interaction with eIF4E.201 This is a fascinating example of a disorder-to-order transition of an intrinsically disordered protein that weakens binding toward eIF4E by ∼4000 fold.
The only known example in prokaryotes of a phosphorylated RNA binding protein for which crystallographic information (PDB codes 3AMT, 3AMU, 3AU7) is available is tRNAIle-agm2C synthetase (TiaS).111,112 TiaS is an enzyme that modifies the cytidine (C34) in the wobble position of tRNAIle2 to 2-agmatinylcytidine or Agm2C. This base modification is required for AUA codon decoding by tRNAIle2. TiaS catalyzes Agm2C formation in an ATP-dependent manner. A 2.9 Å crystal structure of a ternary complex of TiaS bound to ATP and tRNAIle2 reveals the mechanism of base modification by this enzyme.112 Intriguingly, Thr18 in the active site of TiaS is phosphorylated in the crystal structure and phosphorylation at this site is essential for Agm2C formation. Mutation of Thr18 to alanine abrogates enzyme activity. The role of Thr18 appears to be to coordinate Mg2+ ions that are important for ATP hydrolysis, but phosphorylation at this site does not promote a conformational change in the protein. This is the only known example of “silent” phosphorylation, that is, where phosphorylation of a Thr in the active site of an RNA binding protein does not result in a conformational change. However, in this case, phosphorylation appears to be part of the catalytic mechanism and does not play an allosteric role in modulating protein–RNA recognition.
An interesting example is Ire1, a transmembrane protein kinase and RNase that is important for triggering the unfolded protein response (UPR).113,114 Ire1 is a sensor kinase that activates the UPR signal from the lumen of the endoplasmic reticulum (ER) to the cytosol. Ire1 consists of an N-terminal luminal domain, a transmembrane region, followed by a cytosolic portion that consists of the N- and C-terminal kinase domains, which is followed by the RNase domain that is similar to RNase L. The inactive, unphosphorylated Ire1 is a monomer and is bound to the Hsp70 chaperone Bip.115 Activation of UPR results in dissociation of Bip and dimerization of the cytosolic kinase domains.116,117 Dimerization results in trans-autophosphorylation of the activation segment at Ser724 in vivo, although phosphorylation has also been observed on Ser726 and Ser729 in vitro.118 Phosphorylation at Ser724 results in a global rearrangement of the kinase and RNase domains that switch from a dephosphorylated and inactive face-to-face conformation to a phosphorylated and active back-to-back conformation.119 This reorientation of the domains activates RNase endoribonuclease/splicing activity toward Xbp1 mRNA, likely via propagating conformational changes to the RNase domain active site. While a ∼3–5 fold decrease in Km for the RNA substrate is observed due to phosphorylation at 1–4 sites, a 2–40 fold effect on Kcat/Km is observed.118 The RNase activity of Ire1 is specific for Xbp1 mRNA and is not spliceosome associated.120
Although both TiaS and Ire1 are interesting variations in their structural mechanisms, they are both enzymes and not known to be associated with larger ribonucleoprotein complexes.
Concluding Remarks
Currently, 662 structures of proteins that are phosphorylated at serine residues, and 558 structures of proteins phosphorylated at threonine residues have been deposited in the protein data bank (PDB). Phosphorylated RNA binding proteins are a very small subset of these structures and mainly correspond to the proteins discussed in this review. How do the structural mechanisms for control of RNA-binding protein function by phosphorylation discussed here compare to mechanisms observed for other proteins involved in signaling or DNA-binding? Glycogen phosphorylase was the first protein to be studied in both the phosphorylated and nonphosphorylated states,121,122 and the structures revealed global conformational changes occur in the protein upon phosphorylation at Ser-14 and Thr-10 leading to allosteric activation. In contrast, phosphorylation of isocitrate dehydrogenase123 revealed silent effects and no structural change in response to phosphorylation of the active site at Ser113. Several kinase structures are now available in phosphorylated and nonphosphorylated states. Most kinases5 require phosphorylation of a disordered activation segment to trigger conformational changes in the active site, opening up the bilobed kinase structure, allowing access to the substrate. There are several examples of disorder-to-order or order-to-disorder transitions. Therefore, all the mechanisms discussed in the introduction have been observed for phosphorylated proteins.
In contrast, RNA binding proteins discussed here reveal interesting similarities in the mechanisms by which phosphorylation propagates the signal response at the atomic level (Table 1). First, with the exception of the kinase/RNase Ire1, no global conformational changes have been observed in RNA binding proteins that function as part of ribonucleoprotein (RNP) complexes. The structural and dynamic effects of phosphorylating RNA-binding proteins that are components of RNP complexes are localized in space around the phosphate. Second, phosphorylation usually occurs in a disordered region of the RNA binding domain or the SRS and stabilizes this region. Although the RS domain is highly disordered when unphosphorylated and remains flexible in the phosphorylated state, and there is evidence for formation of a transiently populated structure upon phosphorylation. The KH domain is the only report where phosphorylation occurs in a structured β-strand, which unfolds upon phosphorylation. In all cases, phosphorylation creates a binding site or “hub” for protein–protein interactions. Intriguingly, the effects of phosphorylation on association with the RNA tend to be small and range from either no change to ∼10–30-fold altered binding affinity (Kd).
Phosphorylation of RNA binding proteins also plays regulatory roles in several other RNA-mediated processes such as translation, pre-mRNA processing, and mRNA export. It is anticipated that future structural studies may reveal additional mechanisms by which ribonucleoprotein complexes are controlled by phosphorylation-dependent signaling.
Acknowledgments
R.T. was supported by the National Institutes of Health grants 1RO1-GM076660 and 1RO1-GM076660-S1 (ARRA).
Glossary
Keywords
- Phosphorylation
Addition of a phosphate group to a protein to alter protein function
- RNA-binding protein
A protein macromolecule that associates directly and noncovalently with RNA to regulate gene expression
- Serine/arginine-rich Splicing Factor 1 (SFRS1)
A protein that is a component of the spliceosome to regulate alternative splicing
- Splicing Factor 1 (SF1)
A protein that is involved in formation of the E complex of the spliceosome
- Stem-loop binding protein (SLBP)
A protein that binds histone mRNA stem-loop to regulate histone mRNA processing, export, translation, and decay
- K-homology splicing regulator protein (KSRP)
An RNA-binding protein that regulates the translation and stability of a subset of mRNAs that contain AU-rich elements (ARE)
- ATP-dependent RNA helicase upframeshift 1 (Upf1)
A protein that is part of the exon junction complex, that regulates degradation of nonsense mRNA transcripts
- 14–3–3 proteins
A family of conserved proteins in eukaryotes that bind other phosphorylated proteins to regulate signaling and mRNA transport processes in the cell
- pre-mRNA splicing
A biological process by which exons from primary mRNAs are joined together, with excision of introns, to produce a mature mRNA transcript
- mRNA decay
A biological process that regulates the stability of mRNAs by facilitating their degradation
- histone mRNA
Non-polyadenylated and cell-cycle regulated mRNAs that encode for histone proteins during S-phase. Histone proteins are required to package DNA into chromatin
- RNA–protein interaction
interaction between RNA and protein to regulate gene expression
The author declares no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Hunter T. (2012) Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2513–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J.; Hunter T. (2011) Protein kinase signaling networks in cancer. Curr. Opin. Genet. Dev. 21, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R.; Denmon A. P. (2013) Signaling pathways that control mRNA turnover. Cell Signal. 25, 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A. S.; Ferre S. (2005) Amazing stability of the arginine-phosphate electrostatic interaction. J. Proteome Res. 4, 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.; Taylor S.; Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675. [DOI] [PubMed] [Google Scholar]
- Stamm S. (2008) Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 283, 1223–1227. [DOI] [PubMed] [Google Scholar]
- Jurica M. S.; Moore M. J. (2003) Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12, 5–14. [DOI] [PubMed] [Google Scholar]
- Stamm S. (2002) Signals and their transduction pathways regulating alternative splicing: A new dimension of the human genome. Hum. Mol. Genet. 11, 2409–2416. [DOI] [PubMed] [Google Scholar]
- Hartmann A. M.; Rujescu D.; Giannakouros T.; Nikolakaki E.; Goedert M.; Mandelkow E. M.; Gao Q. S.; Andreadis A.; Stamm S. (2001) Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol. Cell Neurosci 18, 80–90. [DOI] [PubMed] [Google Scholar]
- Allemand E.; Hastings M. L.; Murray M. V.; Myers M. P.; Krainer A. R. (2007) Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat. Struct. Mol. Biol. 14, 630–638. [DOI] [PubMed] [Google Scholar]
- Shin C.; Feng Y.; Manley J. L. (2004) Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558. [DOI] [PubMed] [Google Scholar]
- Xiao S. H.; Manley J. L. (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Yario T. A.; Steitz J. A. (2004) A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 101, 9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Steitz J. A. (2005) SRprises along a messenger’s journey. Mol. Cell 17, 613–615. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G.; Kim V. N.; Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205. [DOI] [PubMed] [Google Scholar]
- Ghosh G.; Adams J. A. (2011) Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 278, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K.; Cooper T. A. (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 18, 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm S. M.; Tindall D. J. (2011) Alternatively spliced androgen receptor variants. Endocr. Relat. Cancer 18, R183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M. (2009) Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 74, 1–40. [DOI] [PubMed] [Google Scholar]
- Manley J. L.; Krainer A. R. (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J. F.; Tronchere H.; Chandler S. D.; Fu X. D. (1994) Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. U.S.A. 91, 10824–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J. F.; Lane W. S.; Fu X. D. (1994) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369, 678–682. [DOI] [PubMed] [Google Scholar]
- Lai M. C.; Lin R. I.; Tarn W. Y. (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Y.; Maniatis T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Kohtz J. D.; Jamison S. F.; Will C. L.; Zuo P.; Luhrmann R.; Garcia-Blanco M. A.; Manley J. L. (1994) Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Ngo J. C.; Chakrabarti S.; Ding J. H.; Velazquez-Dones A.; Nolen B.; Aubol B. E.; Adams J. A.; Fu X. D.; Ghosh G. (2005) Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell 20, 77–89. [DOI] [PubMed] [Google Scholar]
- Haynes C.; Iakoucheva L. M. (2006) Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 34, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S.; Gapsys V.; Kim H. Y.; Bessonov S.; Hsiao H. H.; Mohlmann S.; Klaukien V.; Ficner R.; Becker S.; Urlaub H.; Luhrmann R.; de Groot B.; Zweckstetter M. (2013) Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure 21, 2162–2174. [DOI] [PubMed] [Google Scholar]
- Ma C. T.; Ghosh G.; Fu X. D.; Adams J. A. (2010) Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J. Mol. Biol. 403, 386–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelberg D.; Shen T.; McCammon J. A. (2007) A proposed signaling motif for nuclear import in mRNA processing via the formation of arginine claw. Proc. Natl. Acad. Sci. U.S.A. 104, 14947–14951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D.; Drosou V.; Vlachakis D.; Voukkalis N.; Giannakouros T.; Vlassi M. (2012) Phosphorylation of the arginine/serine repeats of lamin B receptor by SRPK1-insights from molecular dynamics simulations. Biochim. Biophys. Acta 1820, 44–55. [DOI] [PubMed] [Google Scholar]
- Maertens G. N.; Cook N. J.; Wang W.; Hare S.; Gupta S. S.; Oztop I.; Lee K.; Pye V. E.; Cosnefroy O.; Snijders A. P.; KewalRamani V. N.; Fassati A.; Engelman A.; Cherepanov P. (2014) Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. U.S.A. 111, 2728–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. T.; Velazquez-Dones A.; Hagopian J. C.; Ghosh G.; Fu X. D.; Adams J. A. (2008) Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J. Mol. Biol. 376, 55–68. [DOI] [PubMed] [Google Scholar]
- Ma C. T.; Hagopian J. C.; Ghosh G.; Fu X. D.; Adams J. A. (2009) Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol. 390, 618–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian J. C.; Ma C. T.; Meade B. R.; Albuquerque C. P.; Ngo J. C.; Ghosh G.; Jennings P. A.; Fu X. D.; Adams J. A. (2008) Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol. 382, 894–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol B. E.; Jamros M. A.; McGlone M. L.; Adams J. A. (2013) Splicing kinase SRPK1 conforms to the landscape of its SR protein substrate. Biochemistry 52, 7595–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo J. C.; Giang K.; Chakrabarti S.; Ma C. T.; Huynh N.; Hagopian J. C.; Dorrestein P. C.; Fu X. D.; Adams J. A.; Ghosh G. (2008) A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell 29, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clery A.; Sinha R.; Anczukow O.; Corrionero A.; Moursy A.; Daubner G. M.; Valcarcel J.; Krainer A. R.; Allain F. H. (2013) Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc. Natl. Acad. Sci. U.S.A. 110, E2802–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintaru A. M.; Hautbergue G. M.; Hounslow A. M.; Hung M. L.; Lian L. Y.; Craven C. J.; Wilson S. A. (2007) Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 8, 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.; Hoang A.; Sinha R.; Zhong X. Y.; Fu X. D.; Krainer A. R.; Ghosh G. (2011) Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Dones A.; Hagopian J. C.; Ma C. T.; Zhong X. Y.; Zhou H.; Ghosh G.; Fu X. D.; Adams J. A. (2005) Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 280, 41761–41768. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M.; Tompa P. (2012) Fuzzy complexes: A more stochastic view of protein function. Adv. Exp. Med. Biol. 725, 1–14. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Madl T.; Bagdiul I.; Kern T.; Kang H. S.; Zou P.; Mausbacher N.; Sieber S. A.; Kramer A.; Sattler M. (2013) Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 41, 1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Maucuer A.; Gupta A.; Manceau V.; Thickman K. R.; Bauer W. J.; Kennedy S. D.; Wedekind J. E.; Green M. R.; Kielkopf C. L. (2013) Structure of phosphorylated SF1 bound to U2AF(6)(5) in an essential splicing factor complex. Structure 21, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C.; Will C. L.; Luhrmann R. (2009) The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718. [DOI] [PubMed] [Google Scholar]
- Berglund J. A.; Abovich N.; Rosbash M. (1998) A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares L. M.; Zanier K.; Mackereth C.; Sattler M.; Valcarcel J. (2006) Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312, 1961–1965. [DOI] [PubMed] [Google Scholar]
- Tavanez J. P.; Madl T.; Kooshapur H.; Sattler M.; Valcarcel J. (2012) hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol. Cell 45, 314–329. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Luyten I.; Bottomley M. J.; Messias A. C.; Houngninou-Molango S.; Sprangers R.; Zanier K.; Kramer A.; Sattler M. (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Mackereth C. D.; Madl T.; Bonnal S.; Simon B.; Zanier K.; Gasch A.; Rybin V.; Valcarcel J.; Sattler M. (2011) Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411. [DOI] [PubMed] [Google Scholar]
- Sickmier E. A.; Frato K. E.; Shen H.; Paranawithana S. R.; Green M. R.; Kielkopf C. L. (2006) Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P.; Gregorovic G.; Sprangers R.; Stier G.; Rhani Z.; Kramer A.; Sattler M. (2003) Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11, 965–976. [DOI] [PubMed] [Google Scholar]
- Manceau V.; Swenson M.; Le Caer J. P.; Sobel A.; Kielkopf C. L.; Maucuer A. (2006) Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J. 273, 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J. K.; Sadar M. D. (2012) Large scale phosphoproteome analysis of LNCaP human prostate cancer cells. Mol. Biosyst 8, 2174–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H.; Chen S.; Bi Q.; Mumby M.; Brekken D. L. (2004) Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell Proteomics 3, 279–286. [DOI] [PubMed] [Google Scholar]
- Manceau V.; Kielkopf C. L.; Sobel A.; Maucuer A. (2008) Different requirements of the kinase and UHM domains of KIS for its nuclear localization and binding to splicing factors. J. Mol. Biol. 381, 748–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Bruderer S.; Rafi Z.; Xue J.; Milburn P. J.; Kramer A.; Robinson P. J. (1999) Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 18, 4549–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y.; Shyu A. B. (2011) Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev. RNA 2, 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg D. R.; Maquat L. E. (2012) Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. E.; Allis C. D.; Strahl B. D. (2011) OPERating ON Chromatin, a Colorful Language where Context Matters. J. Mol. Biol. 9, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Reinberg D. (2011) Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 21, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F.; Gongidi P.; Woods K. R.; Jin J.; Maltais L. J. (2002) The human and mouse replication-dependent histone genes. Genomics 80, 487–498. [PubMed] [Google Scholar]
- Marzluff W. F.; Duronio R. J. (2002) Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14, 692–699. [DOI] [PubMed] [Google Scholar]
- Birchmeier C.; Folk W.; Birnstiel M. L. (1983) The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3′ termini of sea urchin H2A mRNA. Cell 35, 433–440. [DOI] [PubMed] [Google Scholar]
- Busslinger M.; Schumperli D.; Birnstiel M. L. (1985) Regulation of histone gene expression. Cold Spring Harb Symp. Quant. Biol. 50, 665–670. [DOI] [PubMed] [Google Scholar]
- Whitfield M. L.; Zheng L. X.; Baldwin A.; Ohta T.; Hurt M. M.; Marzluff W. F. (2000) Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20, 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z.; Zheng L. X.; Sanchez R.; Marzluff W. F. (1999) Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19, 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B.; Williams A. S.; Sun J. H.; Brown V. D.; Bond U.; Marzluff W. F. (1994) Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell. Biol. 14, 1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. D.; Mullen T. E.; Marzluff W. F.; Wagner E. J. (2009) Knockdown of SLBP results in nuclear retention of histone mRNA. RNA 15, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R.; Marzluff W. F. (2002) The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22, 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J.; Ahn S. H.; Kim Y. K. (2014) The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res. 42, 9334–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle D. J.; Doudna J. A. (2001) The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA 7, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R.; Marzluff W. F.; Redinbo M. R. (2004) Electrostatic contribution of serine phosphorylation to the Drosophila SLBP–histone mRNA complex. Biochemistry 43, 9401–9412. [DOI] [PubMed] [Google Scholar]
- Zanier K.; Luyten I.; Crombie C.; Muller B.; Schumperli D.; Linge J. P.; Nilges M.; Sattler M. (2002) Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression. RNA 8, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Lam T. T.; Tonelli M.; Marzluff W. F.; Thapar R. (2012) Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP–RNA complex by phosphorylation and proline isomerization. Biochemistry 51, 3215–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers C. H.; Thapar R.; Petrotchenko E. V.; Torres M. P.; Speir J. P.; Easterling M.; Dominski Z.; Marzluff W. F. (2006) Combined top-down and bottom-up proteomics identifies a phosphorylation site in stem-loop-binding proteins that contributes to high-affinity RNA binding. Proc. Natl. Acad. Sci. U.S.A. 103, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.; Marzluff W. F.; Dominski Z.; Tong L. (2013) Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 339, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong E. S.; Marzluff W. F.; Nikonowicz E. P. (2002) NMR structure and dynamics of the RNA-binding site for the histone mRNA stem-loop binding protein. RNA 8, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti D. J.; Kupsco J. M.; Yang X. C.; Dominski Z.; Marzluff W. F.; Duronio R. J. (2004) Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell 15, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N.; Lam T. T.; Fritz A.; Rempinski D.; O’Loughlin K.; Minderman H.; Berezney R.; Marzluff W. F.; Thapar R. (2012) The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 32, 4306–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N.; Titus M. A.; Thapar R. (2014) The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS One 9, e85427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R. (2014) Contribution of protein phosphorylation to binding-induced folding of the SLBP-histone mRNA complex probed by phosphorus-31 NMR. FEBS Open Bio 4, 853–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z.; Erkmann J. A.; Greenland J. A.; Marzluff W. F. (2001) Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 21, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z.; Yang X. C.; Raska C. S.; Santiago C.; Borchers C. H.; Duronio R. J.; Marzluff W. F. (2002) 3′ end processing of Drosophila melanogaster histone pre-mRNAs: Requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 22, 6648–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt H. C.; Yaffe M. B. (2013) Phospho-Ser/Thr-binding domains: Navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14, 563–580. [DOI] [PubMed] [Google Scholar]
- Gherzi R.; Lee K. Y.; Briata P.; Wegmuller D.; Moroni C.; Karin M.; Chen C. Y. (2004) A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14, 571–583. [DOI] [PubMed] [Google Scholar]
- Chen C. Y.; Gherzi R.; Ong S. E.; Chan E. L.; Raijmakers R.; Pruijn G. J.; Stoecklin G.; Moroni C.; Mann M.; Karin M. (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Gherzi R.; Trabucchi M.; Ponassi M.; Ruggiero T.; Corte G.; Moroni C.; Chen C. Y.; Khabar K. S.; Andersen J. S.; Briata P. (2006) The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5, e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Diaz-Moreno I.; Hollingworth D.; Frenkiel T. A.; Kelly G.; Martin S.; Howell S.; Garcia-Mayoral M.; Gherzi R.; Briata P.; Ramos A. (2009) Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14–3–3 binding. Nat. Struct. Mol. Biol. 16, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mayoral M. F.; Hollingworth D.; Masino L.; Diaz-Moreno I.; Kelly G.; Gherzi R.; Chou C. F.; Chen C. Y.; Ramos A. (2007) The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure 15, 485–498. [DOI] [PubMed] [Google Scholar]
- Chrestensen C. A.; Schroeder M. J.; Shabanowitz J.; Hunt D. F.; Pelo J. W.; Worthington M. T.; Sturgill T. W. (2004) MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14–3–3 binding. J. Biol. Chem. 279, 10176–10184. [DOI] [PubMed] [Google Scholar]
- Kervestin S.; Jacobson A. (2012) NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E.; Izaurralde E. (2005) Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325. [DOI] [PubMed] [Google Scholar]
- Cheng Z.; Muhlrad D.; Lim M. K.; Parker R.; Song H. (2007) Structural and functional insights into the human Upf1 helicase core. EMBO J. 26, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M.; Mourao A.; Gutsche I.; Gehring N. H.; Hentze M. W.; Kulozik A.; Kadlec J.; Sattler M.; Cusack S. (2009) Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 28, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.; Jayachandran U.; Bonneau F.; Fiorini F.; Basquin C.; Domcke S.; Le Hir H.; Conti E. (2011) Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41, 693–703. [DOI] [PubMed] [Google Scholar]
- Melero R.; Buchwald G.; Castano R.; Raabe M.; Gil D.; Lazaro M.; Urlaub H.; Conti E.; Llorca O. (2012) The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat. Struct. Mol. Biol. 19(498–505), S491–492. [DOI] [PubMed] [Google Scholar]
- Melero R.; Uchiyama A.; Castano R.; Kataoka N.; Kurosawa H.; Ohno S.; Yamashita A.; Llorca O. (2014) Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22, 1105–1119. [DOI] [PubMed] [Google Scholar]
- Yamashita A.; Izumi N.; Kashima I.; Ohnishi T.; Saari B.; Katsuhata Y.; Muramatsu R.; Morita T.; Iwamatsu A.; Hachiya T.; Kurata R.; Hirano H.; Anderson P.; Ohno S. (2009) SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 23, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T.; Yamashita A.; Kashima I.; Schell T.; Anders K. R.; Grimson A.; Hachiya T.; Hentze M. W.; Anderson P.; Ohno S. (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200. [DOI] [PubMed] [Google Scholar]
- Yamashita A.; Ohnishi T.; Kashima I.; Taya Y.; Ohno S. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada-Katsuhata Y.; Yamashita A.; Kutsuzawa K.; Izumi N.; Hirahara F.; Ohno S. (2012) N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 40, 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.; Bonneau F.; Schussler S.; Eppinger E.; Conti E. (2014) Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 42, 9447–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S.; Weichenrieder O.; Izaurralde E. (2013) An unusual arrangement of two 14–3–3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev. 27, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N.; Ebert J.; Unterholzner L.; Lindner D.; Izaurralde E.; Conti E. (2005) SMG7 is a 14–3–3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17, 537–547. [DOI] [PubMed] [Google Scholar]
- Sonenberg N.; Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J.; Gingras A. C.; Sonenberg N.; Burley S. K. (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716. [DOI] [PubMed] [Google Scholar]
- Mader S.; Lee H.; Pause A.; Sonenberg N. (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15, 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K.; Maruki Y.; Long X.; Yoshino K.; Oshiro N.; Hidayat S.; Tokunaga C.; Avruch J.; Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. [DOI] [PubMed] [Google Scholar]
- Lukhele S.; Bah A.; Lin H.; Sonenberg N.; Forman-Kay J. D. (2013) Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure 21, 2186–2196. [DOI] [PubMed] [Google Scholar]
- Terasaka N.; Kimura S.; Osawa T.; Numata T.; Suzuki T. (2011) Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS. Nat. Struct. Mol. Biol. 18, 1268–1274. [DOI] [PubMed] [Google Scholar]
- Osawa T.; Kimura S.; Terasaka N.; Inanaga H.; Suzuki T.; Numata T. (2011) Structural basis of tRNA agmatinylation essential for AUA codon decoding. Nat. Struct. Mol. Biol. 18, 1275–1280. [DOI] [PubMed] [Google Scholar]
- Ron D.; Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Kaufman R. J. (2008) From endoplasmic reticulum stress to the inflammatory response. Nature 454, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A.; Zhang Y.; Hendershot L. M.; Harding H. P.; Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332. [DOI] [PubMed] [Google Scholar]
- Credle J. J.; Finer-Moore J. S.; Papa F. R.; Stroud R. M.; Walter P. (2005) On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 102, 18773–18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Liu C. Y.; Back S. H.; Clark R. L.; Peisach D.; Xu Z.; Kaufman R. J. (2006) The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 103, 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prischi F.; Nowak P. R.; Carrara M.; Ali M. M. (2014) Phosphoregulation of Ire1 RNase splicing activity. Nat. Commun. 5, 3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. M.; Bagratuni T.; Davenport E. L.; Nowak P. R.; Silva-Santisteban M. C.; Hardcastle A.; McAndrews C.; Rowlands M. G.; Morgan G. J.; Aherne W.; Collins I.; Davies F. E.; Pearl L. H. (2011) Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 30, 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H.; Matsui T.; Yamamoto A.; Okada T.; Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Barford D.; Hu S. H.; Johnson L. N. (1991) Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J. Mol. Biol. 218, 233–260. [DOI] [PubMed] [Google Scholar]
- Rath V. L.; Ammirati M.; LeMotte P. K.; Fennell K. F.; Mansour M. N.; Danley D. E.; Hynes T. R.; Schulte G. K.; Wasilko D. J.; Pandit J. (2000) Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core. Mol. Cell 6, 139–148. [PubMed] [Google Scholar]
- Hurley J. H.; Dean A. M.; Thorsness P. E.; Koshland D. E. Jr.; Stroud R. M. (1990) Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J. Biol. Chem. 265, 3599–3602. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Dominski Z.; Yang X. C.; Elms P.; Raska C. S.; Borchers C. H.; Marzluff W. F. (2003) Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23, 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo V.; Griesbach E.; Schumperli D. (2014) CstF64: cell cycle regulation and functional role in 3′ end processing of replication-dependent histone mRNAs. Mol Cell Biol 34, 4272–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A.; Vernon R. M.; Siddiqui Z.; Krzeminski M.; Muhandiram R.; Zhao C.; Sonenberg N.; Kay L. E.; Forman-Kay J. D. (2014) Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature E-pub Dec 22. DOI: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]