Abstract
Oligomers that contain both α- and β-amino acid residues, or “α/β-peptides”, have emerged as promising mimics of signal-bearing polypeptides that can inhibit or augment natural protein–protein interactions. α/β-Peptides that contain a sufficient proportion of β residues evenly distributed along the sequence can be highly resistant to enzymatic degradation, which is favorable with regard to in vivo applications. Little is known, however, about recognition of α/β-peptides by the immune system. Prior studies have focused almost entirely on examples that contain a single β residue; such α/β-peptides frequently retain the immunological profile of the analogous α-peptide. We have conducted α-peptide vs α/β-peptide comparisons involving higher β residue content, focusing on molecules with αααβ and ααβαααβ backbone repeat patterns. Among analogues of an 18-mer derived from the Bim BH3 domain and an 8-mer derived from secreted phospholipase-2 (sPLA2), we find that recognition by antibodies raised against the prototype α-peptide is suppressed by periodic α → β replacements. Complementary studies reveal that antibodies raised against Bim BH3- or sPLA2-derived α/β-peptides fail to recognize prototype α-peptides displaying identical side chain repertoires. Because polypeptides containing d-α-amino acid residues are of growing interest for biomedical applications, we included the enantiomer of the sPLA2-derived α-peptide in these studies; this d-peptide is fully competent as a hapten, but the resulting antibodies do not cross react with the enantiomeric peptide. Among analogues of the 9-mer CD8+ T-cell viral epitope GP33, we observe that periodic α → β replacements suppress participation in the MHC I + peptide + T-cell receptor ternary complexes that activate cytotoxic T-lymphocytes, due in part to disruption of MHC binding.
Polypeptides are crucial for transmission of biological information, and the messages encoded in amino acid sequences are often read by multiple partners, with divergent outcomes.1 Peptide hormones, growth factors, kinases, phosphatases, glycosyl transferases, transcriptional regulators, and many other signal-bearing or signal-reading proteins bind to specific partners in order to play their designated roles in information transfer pathways.2 In addition, polypeptides interact with proteases and peptidases, sometimes in highly specific ways for targeted cleavage,3 and in more general ways for wholesale degradation.4 The adaptive immune system represents a polypeptide recognition network that features several different modes of evaluating peptidic information, including peptide presentation within major histocompatibilty class I or II (MHC I or II) complexes for interrogation by T-cell receptors (TCRs), and complexation to antibodies and B-cell receptors.5
Many specific protein–protein recognition events are attractive targets for therapeutic intervention.6 The importance of such targets is illustrated by the commercial success of agents that block interactions of vascular endothelial growth factor (VEGF) or tumor necrosis factor-α (TNFα) with their cell-surface receptors, and agents that activate receptors for glucagon-like peptide-1 (GLP-1) or parathyroid hormone (PTH).7 Such drugs are usually themselves polypeptides; in addition to binding to their intended targets (e.g., VEGF, TNFα, or the receptor for GLP-1 or PTH), these polypeptides are subject to recognition and processing by proteases and various immune system components. These latter forms of recognition can be deleterious in terms of clinical applications: proteolysis can lead to poor drug pharmacokinetics, and immunological neutralization can result in a loss of drug efficacy over time.8
The high specificity of macromolecular recognition involving polypeptides has inspired efforts to identify unnatural oligomers that mimic the target specificity of prototype peptides or proteins but avoid enzymatic degradation mechanisms. Examples include oligomers of d-α-amino acids (“d-peptides”),9N-alkyl-glycines (“peptoids”),10 aromatic subunits,11 β-amino acids (“β-peptides”)12 and combinations of α- and β-amino acids (“α/β-peptides”).13−15 Many such studies have focused on mimicry of a specific α-helix that mediates a particular protein–protein interaction. α/β-Peptides appear to be especially well-suited for recapitulating the information encoded by an α-helix, particularly for helices comprising five or more turns.15,16 Functional α/β-peptide analogues can be generated from an α-helix-forming sequence via periodic replacement of α residues with appropriately selected β residues.17 The most straightforward replacement for a given α residue is the β3 homologue, that is, the β residue that bears the original side chain on the backbone carbon adjacent to nitrogen. Three patterns of α → β replacement, ααβ, αααβ or ααβαααβ, have been shown to support helical conformations that are very similar to the α-helix formed by conventional peptides and proteins.18,19 Oligomers based on these patterns contain 25–33% β residues that are evenly distributed along the sequence; such backbones are often highly resistant to proteolytic degradation.13−19
The promising α-helix-mimetic properties of α/β-peptides lead to a question: will such oligomers be subject to recognition by the immune system? The literature offers only limited insight on this possibility. Three groups have reported “β3 scan” studies based on epitopes known to form MHC I complexes; each member of a β3 scan set contains only one α → β3 replacement. In the first such study, Guichard et al. evaluated the impact of single α → β3 replacements in the peptide ALGIGILTV on binding to the human HLA-A2 MHC I.20 Moderate to strong binding was observed for nearly all members of the β3 scan set. Similarly, a β3 scan of the epitope RRFFPYYV by Reinelt et al. revealed that most single replacements did not impair binding to the human HLA-B*2705 MHC I molecule.21 This group went on to provide evidence that MHC I complexes formed by all members of the β3 scan set could engage the TCR on a cognate CD8+ T-cell, as indicated by induction of target cell lysis; however, the α/β analogues were 102- to 105-fold less effective than the α-peptide RRFFPYYV in this regard. Webb et al. conducted a β3 scan of the epitope SIINFEKL, which binds to the mouse H2-Kb MHC I.22 Four of the eight α → β3 replacement analogues matched the prototype α-peptide in terms of MHC I affinity, and most of the remaining analogues retained measurable affinity. The susceptibility of these α/β-peptide+MHC I complexes to recognition by two cognate TCRs was assessed by monitoring induction of IL-2 secretion upon exposure of T-cells to antigen-presenting cells that express H2-Kb after the latter had been incubated with the α/β-peptide. All peptides containing single α → β3 replacements were able to elicit a response, although with varying efficacies; some members of the β3 scan set matched the original α-peptide. Two of the β3-containing analogues of SIINFEKL were cocrystallized with H2-Kb, providing atomic-level insight on the accommodation of single unnatural residues by an MHC I partner. Collectively, these three studies indicate that MHC I binding is relatively forgiving toward many single α → β3 replacements in peptide epitopes, and that many of the resulting MHC I complexes can engage cognate TCRs productively.
Dali et al. evaluated aza-β3-containing analogues of YALKRQGRTLYG in terms of binding to I-Ad or I-Ed MHC II (mouse).23 These studies included most of the single α → aza-β3 replacements and a few analogues that contained two or three consecutive α → aza-β3 replacements. In general, single replacements were well tolerated in terms of MHC II binding, but replacements in only the C-terminal region supported TCR recognition. These workers explored another aspect of immunological recognition by immunizing rabbits with the peptide YALKRQGRTLYG and determining whether analogues containing α → aza-β3 replacements could compete with the original peptide for binding to the resulting antisera. Binding was quite strong for most replacements, but α → aza-β3 replacement in the final three residues substantially impaired binding to the antisera.
Based on the few available precedents, which are summarized above, it is difficult to predict the immunological consequences of α-helix-mimetic α/β patterns that contain 20–40% α → β replacement. The degree to which single α → β substitutions have been tolerated in prior studies raises the possibility that the higher β residue density of the α-helix mimics would support some form of recognition by the immune system. Understanding whether immune recognition mechanisms tolerate higher α → β replacement densities is important because β residue proportions >20%, with even β distribution along the sequence, are likely to be required for robust resistance to proteolysis, as suggested by our own observations16 and by the relatively rapid degradation reported for peptides containing single α → β3 or α → aza-β3 replacements.23
The immunological consequences of structural modifications that modulate peptide susceptibility to proteolysis are of interest not only for designing therapeutic agents that target protein–protein recognition events but also for developing more effective peptide-based vaccines.24 Recent studies of the effect of introducing noncanonical amino acid residues or amide bond mimics on ligand presentation in MHC complexes have provided insight into the importance of hydrogen bonding25 and secondary structure formation26 in T-cell-mediated recognition, which provides additional context for our efforts to evaluate the impact of α → β backbone modification on immunological responses. Here we report experimental evaluation of the effects of multiple α → β substitutions, in either the αααβ or ααβαααβ pattern, on several types of immunological recognition. A commercially available polyclonal antibody preparation that recognizes an 18-mer peptide corresponding to the human Bim BH3 domain was used to interrogate α/β analogues. These studies were complemented by evaluating the recognition of human BimBH3 18-mer analogues by custom-produced chicken polyclonal antibodies raised against either the prototype α-peptide or an α/β analogue. The generality of these findings was probed by characterizing the properties of custom-produced chicken polyclonal antibodies raised against an 8-mer peptide or α/β analogues derived from murine secreted phospholipase-2 (sPLA2). In addition, a nine-residue viral epitope and α/β analogues were evaluated for their ability bind H2-Db MHC I, induce interferon γ responses in CD8+ T-cells, and stimulate CD8+ peptide-specific T-cell expansion in mice.
Results and Discussion
Cross-recognition of α/β-peptide analogues of the Bim BH3 domain by polyclonal antibodies?
The Bcl-2 homology domain 3 (BH3 domain) mediates interactions among pro- and antiapoptotic members of the Bcl-2 protein family.27 Pro-apoptotic family members such as Bim, Puma, Bak, and Bax are bound by antiapoptotic family members such as Bcl-2, Bcl-xL, and Mcl-1 via association of the BH3 domain on the former with a complementary cleft on the latter. In the bound state, BH3 domains adopt an α-helical conformation.28 Four key hydrophobic side chains are aligned along one side of this helix, and these side chains are accommodated by pockets in the floor of each BH3-recognition cleft. We previously examined α/β analogues of the 18-residue Bim BH3 peptide 1 for the ability to bind to Bcl-xL and Mcl-1.19 Comprehensive evaluation of all possible analogues based on the ααβ3, αααβ3, or ααβ3αααβ3 backbone pattern (14 α/β-peptides in total) revealed two that retained affinity for both partner proteins. One of these dual-binding α/β-peptides had the ααβαααβ pattern (2), and the other had the αααβ pattern (3a). Co-crystal structures of 2+Bcl-xL and 3a+Bcl-xL revealed in atomic detail the close match between the α/β-peptide helices and the α-helix formed by the Bim BH3 domain itself. This conformational mimicry leads to similar three-dimensional presentations of key side chains from the original α-helix and the helices formed by the α/β-peptides.
We used a commercially available polyclonal rabbit antibody raised against the 36-mer AMASMRQAEPADMRPEIWIAQELRRIGDEFNAYYAR to probe for cross-reactivity between an α-peptide epitope and analogous α/β-peptides containing multiple, regularly spaced α → β3 replacements. The sequence of Bim BH3 α-peptide 1 is contained within the 36-mer (underlined). Polyclonal antibody preparations were used for these studies instead of monoclonal antibody because polyclonal antibodies provide a greater opportunity to observe cross-reactivity between antibodies and closely related peptide species. Control enzyme-linked immunosorbent assays (ELISA) showed that at least a subset of the antibodies in the polyclonal commercial preparation could bind to immobilized 18-mer 1 (indirect ELISA). Moreover, this binding could be competitively disrupted if soluble 1 was introduced along with the polyclonal antibodies (competition ELISA).
The ELISA format was used to ask whether α/β-peptide 2 or any of the four possible 18-mers with the αααβ3 pattern, 3a–d (Figure 1), could be recognized by polyclonal anti-Bim BH3 antibodies. For the commercial BimBH3 antibody preparation, no antibody binding could be detected to immobilized samples of any of these five α/β-peptides (Figure 2a). In addition, none of these α/β-peptides could competitively disrupt commercial antibody binding to immobilized 1 (Figure 2b). Thus, although Dali et al. found that most analogues of 12-mer YALKRQGRTLYG containing a single α → aza-β3 replacement could be recognized by polyclonal antibodies resulting from immunization with the α-peptide,23 we observe that making four or five evenly spaced α → β3 replacements in an 18-mer sequence appears to eliminate polyclonal antibody recognition. Since two of the α/β-peptides examined are known to mimic the α-peptide 18-mer in terms of recognition by partner proteins Bcl-xL and Mcl-1, our failure to detect antibody binding to these α/β-peptides suggests that the antibodies mostly or entirely recognize a molecular surface different from that required for binding to Bcl-2-family proteins such as Bcl-xL or Mcl-1. Such a difference could arise if the conformation recognized by the antibodies is not α-helical.
Figure 1.
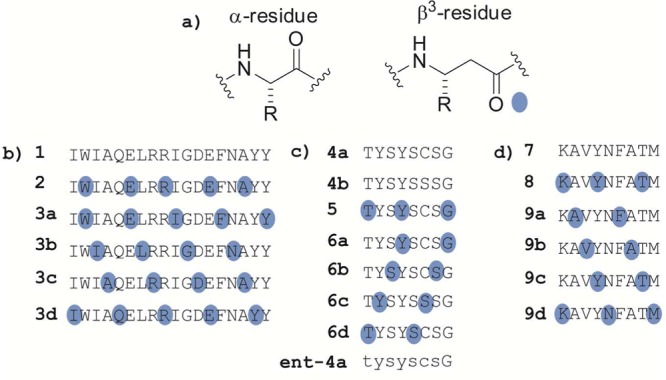
α/β-Peptide analogues of fragments derived from Bim BH3, sPLA2, and GP33 used in this study. (a) Structures of an α-amino acid residue and the homologous β3-amino acid residue. (b) Sequences of Bim BH3-derived peptides. The conventional single-letter code is used to indicate α residue identity. Blue dots indicate sites of α → β3 replacement; each β3 residue bears the side chain of the α-residue indicated by the letter. (c) Sequences sPLA2-derived peptides. Peptide ent-4a is the enantiomer of 4a. (d) Sequences GP33-derived peptides.
Figure 2.
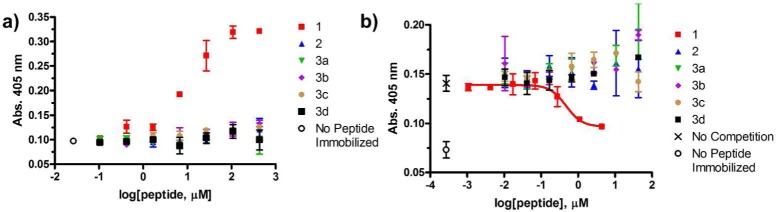
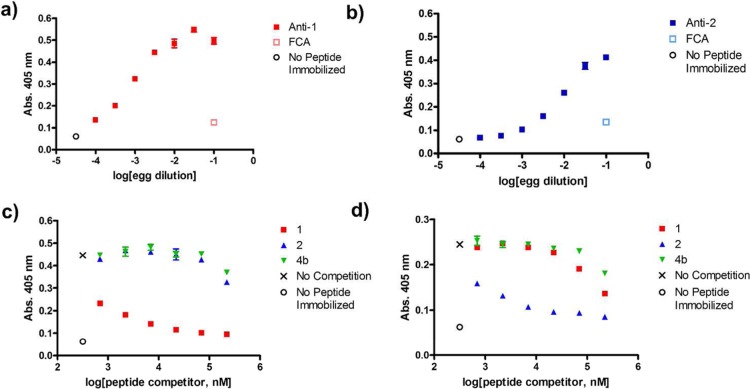
Commercial polyclonal anti-BimBH3 antibody preparation binds to an α-peptide derived from BimBH3 (1) but not to α/β homologues (2,3a-d). Data points represent mean ± SD, with each condition run in duplicate. (a) Indirect ELISA analysis of anti-BimBH3 commercial antibody binding to immobilized synthetic BimBH3 analogues. The x-axis represents the concentration of peptide in solutions used during the plate-coating step. “No peptide immobilized” is a negative control. (b) Competitive ELISA analysis of peptides in solution inhibiting the binding of commercial anti-BimBH3 antibody to immobilized peptide 1. The x-axis represents the concentration of peptide incubated with polyclonal antibody before adding this solution to wells containing immobilized α-peptide 1. “No competition” denotes the absorbance observed when no competitor peptide was incubated with polyclonal antibody prior to addition. The red curve represents the fitting of a sigmoidal dose response model to the data for α-peptide 1 (IC50 = 3 μM). None of the α/β-peptide analogues inhibits binding to immobilized 1 at the highest α/β-peptide concentration tested (43 μM).
Circular dichroism measurements indicated that α-peptide 1 and α/β-peptides 2 and 3a–d do not adopt a regular secondary structure in aqueous solution (Supporting Information Figure S1). These results suggest that the failure of the polyclonal Bim BH3 antibody preparation to recognize α/β-peptides 2 and 3a–d is not due to changes in the extent or type of secondary structure sampled by these Bim BH3 α/β analogues relative to the prototype α-peptide.
Experiments using commercial polyclonal Bim BH3 antibody indicate that antibodies capable of recognizing α-peptide 1 do not cross react with α/β-peptides 2 or 3a–d; however, these experiments do not test whether antibodies can be produced that recognize α/β-peptides. We undertook a new set of experiments to address this question, and to determine whether antibodies specific for an α/β-peptide would cross-react with a homologous α-peptide. Injecting female, laying chickens with short peptides conjugated to carrier proteins, which provide T-helper cell epitopes, along with Freund’s adjuvant induces large quantities of antigen-specific IgY antibodies that can be harvested from eggs laid by these hens.29 Minimally processed yolks from these eggs can be used in the ELISA format to detect production of peptide-specific polyclonal antibodies and to determine whether those peptide-specific antibodies recognize related molecules. We evaluated the abilities of α-peptide 1 or α/β-peptide 2 to induce peptide-specific antibody responses using this approach. Control indirect ELISAs indicated that inoculation with α-peptide 1 or α/β-peptide 2 conjugated to bovine γ-globulin carrier protein induces the production of IgY antibodies capable of binding to the peptide antigen when that peptide has been conjugated to ovalbumin and immobilized. Egg yolks from control chickens injected with adjuvant but no peptide contained negligible amounts of antibodies with these recognition capabilities (“FCA” data points, Figure 3a, b).
Figure 3.
Inoculation of chickens with α-peptide 1 or α/β-peptide 2 conjugated to bovine γ-globulin with adjuvant stimulates production of peptide-specific responses (“anti-peptide” IgY antibodies). Association of antibody with immobilized peptide was evaluated only between an antibody and the peptide injected to stimulate production of that antibody. Data points in panels a–d represent mean ± SD, with each condition run in duplicate. In all panels, the indicated peptides were conjugated to ovalbumin and were immobilized using a 1:100 dilution of the solution produced from the conjugation reaction. (a, b) Indirect ELISA analysis of IgY antibodies induced by injection of conjugated α-peptide 1 or α/β-peptide 2, respectively. The x-axis represents the dilution of egg yolk used for each condition. Anti-1 and anti-2 data are from egg yolks of chickens injected with conjugated peptides 1 or 2, respectively. “FCA” data points are generated using egg yolks from chickens injected only with Freund’s complete adjuvant (FCA), which serve as negative controls. (c, d) Competitive ELISA analysis of peptides in solution inhibiting the binding of egg yolk-derived antibodies to immobilized α-peptide 1 or α/β-peptide 2, respectively. The x-axis represents the concentration of peptide incubated with egg yolk preparations before adding this solution to wells containing immobilized peptides. Peptide 4b is included as a negative control to account for nonspecific inhibition of antibody-peptide complex formation by a solution of unrelated peptide in DMSO.
Since identical chemistry was used to conjugate peptides to bovine gamma globulin for injection and to conjugate peptides to ovalbumin for ELISA it is possible that some of the response observed in these ELISA studies results from antibody recognition of cross-linker structure. This possibility was addressed with competition ELISA studies, in which interactions between antibodies and an immobilized peptide are assessed in the presence of a soluble peptide. If the soluble peptide binds to the antibodies, then antibody binding to immobilized peptide, which is detected in the ELISA, will be diminished. Since the soluble peptides we used did not contain the linker substructure, the competitive effects we observed must reflect binding to the antibodies mediated by the peptides themselves rather than linkers.
Competitive ELISA using antibodies from a chicken injected with α-peptide 1 conjugated to the carrier show that free peptide 1 disrupts binding of these polyclonal antibodies (anti-1) to immobilized 1 more effectively than α/β-peptide 2 disrupts this binding (Figure 3c). Conversely, α/β-peptide 2 is more effective than α-peptide 1 at disrupting interaction of anti-2 with immobilized 2 (Figure 3d). These results indicate that the polyclonal IgY antibody preparations contain significant capacity for recognition of the peptide used for immunization rather than the linker. In addition, the competitive ELISA data are consistent with the lack of cross-reactivity observed for commercial Bim BH3 polyclonal antibody with α/β-peptide 2. The data show that an α/β-peptide hapten can induce a B-cell-mediated immune response, and that at least in this case the resulting antibodies show little cross-reactivity with the homologous α-peptide, which displays an identical side chain sequence.
Cross-Recognition of α/β-Peptide Analogues or the Enantiomer of an Antigenic Fragment of sPLA2 by Polyclonal Antibodies?
We used the chicken egg-ELISA approach described above to evaluate the properties of antibodies produced after injecting chickens with an array of peptide analogues derived from a fragment of murine secreted phospholipase-2 (sPLA2; α-peptide 4a) conjugated to carrier protein. Control experiments showed that injecting adjuvant-emulsified, unconjugated peptide does not stimulate an increase in the titer of peptide-specific antibody (Supporting Information Figure S2). We evaluated a set of sPLA2 fragment analogues in addition to α-peptide 4a, including the related peptide 4b (Cys6 → Ser), five α/β-peptide analogues (5 and 6a–d), and the enantiomer of 4a (ent-4a) (Figure 1), for their efficacy in stimulating peptide-specific IgY antibody responses. α/β-Peptides 5 and 6a–d contain ααβαααβ (5) or αααβ (6a–d) backbone patterns, which are analogous to the patterns used in Bim BH3-derived peptides 2 and 3a–d, respectively. Protected β3-homocysteine is not commercially available, so α/β-peptide 6c contains β3-homoserine rather than β3-homocysteine, and α-peptide 4b is the corresponding control compound. α/β-Peptide 6c was not soluble under peptide conjugation conditions and was therefore not evaluated.
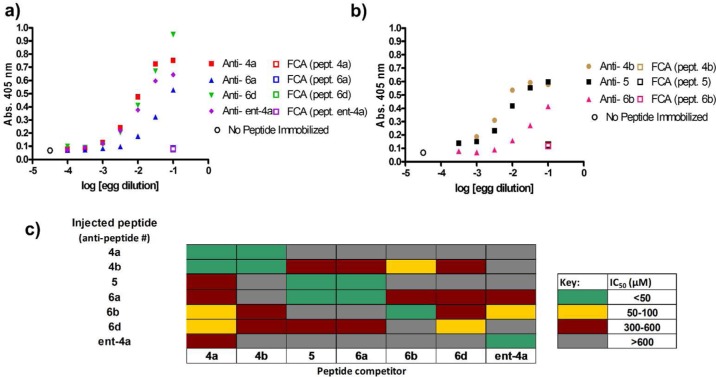
We used indirect ELISA to assess whether antibodies specific for the peptidic haptens had been induced. Using varied dilutions of egg yolk or lyophilized egg yolk powder taken either from chickens that had been injected with hapten–carrier conjugate plus adjuvant or from control chickens that had been injected only with adjuvant, we observed that all hapten-injected chickens showed increased titers of antibody capable of binding to the immobilized peptide hapten used for immunization, as compared to control chickens (Figure 4a). The egg yolk dilution curves produced were generally reproducible among multiple animals (n = 4) when the same preparation of conjugated peptide was used to inject these animals (Supporting Information Figure S3a, b). Different peptides seemed to show varying efficacies for inducing production of peptide-specific antibodies; however, these variations are difficult to interpret because the efficiencies of the gluataraldehyde cross-linking used to conjugate each peptide to carrier protein are not amenable to quantitative comparison.
Figure 4.
Inoculation of chickens with α-peptides 4a, 4b, ent-4a, or α/β-peptides 5, 6a, 6b, or 6d conjugated to bovine γ-globulin with adjuvant stimulates production of peptide-specific responses (“anti-peptide” IgY antibodies). Association of antibody with immobilized peptide was evaluated only between an antibody and the peptide injected to stimulate production of that antibody. In all panels, the indicated peptides were conjugated to ovalbumin and were immobilized using a 1:100 dilution of the solution produced from the conjugation reaction. (a, b) Indirect ELISA analysis of IgY antibodies induced from injection of conjugated peptides 4a-b, ent-4a, 5 and 6a–d. The x-axis represents the dilution of the egg yolk used. “FCA” data points represent the binding of antibodies from chickens injected only with Freund’s complete adjuvant (FCA) to the indicated immobilized peptide, which serve as negative controls. Panel a displays data generated from a single experiment, carried out separately from the single experiment performed to generate data in panel b. Data points in panels a and b represent mean ± SD, with each condition run in duplicate. (c) Competitive ELISA analysis of peptides in solution (peptide competitor) inhibiting the binding of egg yolk derived-antibodies to immobilized peptides. IC50 values were estimated manually as the concentration of peptide needed to eliminate approximately 50% of the signal (absorption at 405 nm) resulting from complex formation between IgY antibodies and immobilized peptide. All competition ELISA curves are presented in Supporting Information Figure S4.
It is noteworthy that hapten ent-4a, composed entirely of d-amino acid residues, stimulates production of a substantial titer of peptide-specific antibodies. Previous work indicated that an all-d protein was nonimmunogenic,30 but our observations with ent-4a show that d-polypeptides are inherently susceptible to antibody recognition, as would be expected based on physicochemical principles. A lack of T-cell help may underlie the earlier observation30 that d-rubredoxin is not immunogenic.
Since all sPLA2 fragment analogues used as haptens induced increased titers of peptide-specific antibodies, we sought to determine whether these antibodies were specific to the peptide used as hapten. We used competitive ELISA to test the ability of free peptides to disrupt binding of peptide-specific IgY antibodies in egg yolk to immobilized forms of the peptides used as haptens to induce antibody production. Cross-recognition was probed for all combinations of antibodies and sPLA2 fragment analogues. We estimated IC50 values based on the concentration of soluble peptide needed to inhibit approximately half of the signal resulting from complexation of IgY with immobilized peptide (see Supporting Information Figure S4 for curves). IgY antibody-peptide interactions are categorized as strong, medium, weak, or nonbinding based on the approximate IC50 value observed (Figure 4c).
These results show that for each antibody preparation, the soluble form of the peptide hapten is among the most potent inhibitors of complex formation with immobilized hapten, as expected. Substantial cross-reactivity is seen for α-peptides 4a and 4b and the corresponding antibodies, as might be predicted given the high degree of similarity between these two α-peptide sequences (Cys vs Ser at position 6). Additionally, cross-reactivity is observed between α/β-peptides 5 and 6a and the corresponding antibodies. These peptides vary only in the identity of their N-terminal residues (threonine or β3-homothreonine, respectively). Besides these two examples, instances of cross-reactivity are infrequent in this study, and the cross-reaction is weak when observed.
These findings with an sPLA2 fragment and analogues are consistent with the recognition profiles discussed above for antibodies that bind to Bim BH3 analogues (α-peptide 1 and α/β-peptides 2 and 3a–d). Collectively, these two data sets, based on different peptide sequences, suggest trends that may ultimately prove to be general: antibodies raised against α-peptides tend to show little cross-reactivity toward α/β-peptide homologues that contain β3-residue densities and distributions commonly used for creating α-helix mimics,13−19 and antibodies raised against such α/β-peptides tend to show little-cross reactivity toward the homologous α-peptide.
Memory CD8+ T-Cell Receptor Recognition of α/β-Peptide Analogues of the GP33 Viral Epitope within an Immunological Synapse?
C57BL/6 mice that recover from an acute infection with lymphocytic choriomeningitis virus (LCMV) contain memory CD8+ T-cells that are specific for the viral epitope designated GP33, which corresponds to the peptide KAVYNFATM (7).31 Isolation of splenocytes from such mice and ex vivo exposure to α-peptide 7 leads to activation of these memory T-cells, which can be detected by production of the cytokine interferon-γ (IFN-γ).32 This activation presumably results from binding of the exogenously added peptide to MHC I on the surfaces of antigen-presenting cells (APCs), with subsequent recognition of these peptide + MHC I complexes by cognate TCRs on the memory CD8+ T-cells.33 This cell–cell contact leads to formation of an immunological synapse and induction of T-cell receptor signaling and cytokine production.34
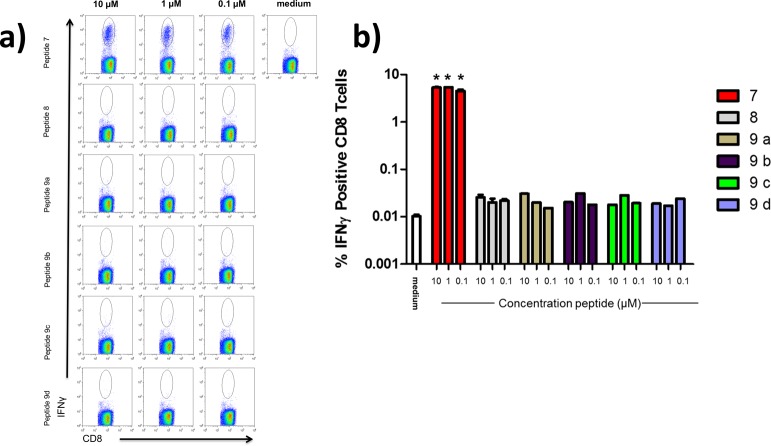
We used this system to ask whether an analogue of the 9-mer GP33 epitope containing two or three regularly spaced sites of α → β3 replacement could recapitulate the role of the original α-peptide in T-cell activation. These experiments focused on five α/β-peptide analogues of 7, including one with the ααβαααβ pattern (8) and all four with the αααβ pattern (9a–d) (Figure 1). Flow cytometry was used to probe for a population of CD8+ T-cells that produce IFN-γ upon exposure to 7 or an α/β analogue (Figure 5). Such a population was readily detected when the splenocytes were exposed to α-peptide 7, but not after exposure to any of the α/β-peptides. Thus, although studies by other groups have shown that some T-cell epitope peptide analogues containing a single α → β3 replacement can form MHC I complexes that are recognized by a cognate TCR,21−23 our results indicate that peptide analogues containing a higher α → β3 replacement density cannot mimic the role played by the α-peptide epitope GP33 in the GP33 + MHC I + TCR complex.
Figure 5.
α/β-Peptide homologues of the GP33 peptide do not induce IFNγ production in CD8+ cells from LCMV-infected mice. (a) Representative flow cytometry dot plots examining IFNγ production from CD8+ splenocytes of LCMV-infected mice stimulated directly ex-vivo with the GP33 α-peptide (7) or α/β homologues (8, 9a–d) . Plots are gated from total CD8+ lymphocyte populations. (b) Graphic representation of quantified flow cytometry data for IFNγ production from panel a. Data represent mean ± sd from three independent replicates. *p < 0.05 vs medium using Mann–Whitney t test.
Our observations are consistent with two alternative hypotheses: (1) α/β-peptides 8 and 9a–d do not bind to the MHC I that displays 7, or (2) the α/β-peptide+MHC I complexes form but are not recognized by the appropriate TCR. To distinguish between these two hypotheses we evaluated the binding of α-peptide 7 and α/β-peptides 8 and 9a–d to H2-Db MHC I using a competition-based assay.35 This MHC I subclass binds GP33 and is partially responsible for displaying the GP33 peptide epitope in vivo in C57BL/6 mice.31 Control experiments confirmed that α-peptide 7 potently inhibits binding of a tracer peptide to purified H2-Db (IC50 = 2.4 nM, Table 1). α/β-Peptides 8 and 9a–d bind less tightly to H2-Db than does α-peptide 7 (Table 1). The largest losses in affinity are seen in α/β-peptides 9b and 9d, which contain α → β replacements at “anchor” positions (positions 3, 5, and 9). Side chains from residues at these positions are known to form close contacts with H2-Db.36 α/β-Peptides that contain only α residues at anchor positions (8, 9a, and 9c) show stronger binding relative to α/β-peptides 9b and 9d, but α/β-peptide 8, 9a, and 9c nevertheless bind more weakly than does α-peptide 7. Circular dichroism studies of α-peptide 7 and α/β-peptides 8 and 9a–d show that none of these molecules adopt secondary structure in solution (Supporting Information Figure S1), suggesting that alterations in secondary structural propensities are likely not responsible for the weakened binding of α/β-GP33 analogues. Alternatively, the impaired binding of all α/β-peptides tested relative to prototype α-peptide 7 may reflect subtle disruptions to energetically important contacts between peptide backbone atoms and H2-Db caused by the backbone alteration.37 α/β-Peptides 8, 9a, and 9c would be considered “H2-Db binders” by some criteria,38,39 but if they form MHC I complexes under our assay conditions, then the complexes do not productively engage cognate T-cell receptors, as indicated by the lack of IFN-γ production. α/β-Peptides 8, 9a, and 9c contain α → β replacements at positions within the GP33 epitope that would interact with a cognate TCR,31 which may explain the lack of productive TCR engagement.
Table 1. H2-Db Binding.
| peptide | H2-Db (IC50, nM)a |
|---|---|
| 7 | 2.37 |
| 8 | 124 |
| 9a | 138 |
| 9b | 5670 |
| 9c | 249 |
| 9d | 18 500 |
IC50 represents the concentration of peptide needed to inhibit 50% of tracer peptide binding as determined using previously developed methods.35
Although analogues of a 9-mer GP33 epitope with two or three α → β replacements fail to stimulate IFN-γ production in splenocytes from mice that have recovered from LCMV infection, these observations do not indicate whether α/β-peptides can induce activation and proliferation of peptide-specific CD8+ T-cells under other conditions. To probe this question we injected C57BL/6 mice lacking prior exposure to LCMV with either α-peptide 7 or α/β-peptide 8 and analyzed splenocytes from these animals to look for populations of CD8+ T-cells that produce IFN-γ in response to ex vivo peptide pulsation. Injection of mice with α-peptide 7 yields splenocytes that show a significant population of CD8+ T-cells that produce IFN-γ upon ex vivo pulsation with α-peptide 7 but not with α/β-peptide 8 (Figure 6). Similar experiments using splenocytes from mice injected with α/β-peptide 8 reveal the absence of a significant population of CD8+ T-cells that produce IFN-γ in response to pulsation with α/β-peptide 8, which suggests that this α/β-peptide is unable to stimulate expansion of peptide-specific CD8+ T-cells in mice. α-Peptide 7 also fails to induce IFN-γ production in splenocytes from mice injected with α/β-peptide 8, confirming that injection of α-peptide 7 was responsible for the increased frequency of CD8+ T-cells that can be induced to produce IFN-γ in response to ex vivo pulsation with 7. Analogous studies in which Bim BH3-derived α-peptide 1 or α/β-peptide 2 were injected into C57/BL6 mice showed that neither of these molecules induces expansion of peptide-specific CD8+ T-cells (Supporting Information Figure S5). The negative result from α-peptide 1 is consistent with the predicted weak affinity40 of appropriately sized fragments from α-peptide 1 for H2-Kb and H2-Db.
Figure 6.
α-Peptide 7, but not α/β-peptide 8, stimulates the expansion of peptide-specific CD8+ T-cells when injected in mice. These peptide-specific CD8+ T-cells produce IFNγ in response to α-peptide 7, but not 8, ex vivo. (a, b) Representative flow cytometry plots depicting the frequencies of IFNγ responses of CD8+ cells to ex vivo stimulation with 7 or 8, respectively. Flow plots are gated from the total CD8+ lymphocyte population. (c) Graphic representation of IFNγ responses from splenocytes of mice injected with α-peptide 7 to exogenously administered GP33 analogues at indicated doses as assessed by flow cytometry. (d) Graphic representation of IFNγ response of splenocytes from mice injected with α/β-peptide 8. Peptides were injected into three to four mice each, and splenocytes from these were pooled for experiments. Data represent mean ± sd from three to four replicates. Medium represents the frequency of IFNγ-positive CD8+ T-cells observed with no peptide stimulation for that population of splenocytes. *p < 0.05 vs medium using Mann–Whitney t test.
Conclusions
Oligomers that contain combinations of α- and β-amino acid residues constitute an enormous pool of molecular diversity. Given the wide range of biological functions manifested by conventional peptides (l-α-amino acid residues only), one can anticipate that many α/β-peptides will display useful and specific biological properties, some that mimic functions of natural polypeptides and others without natural precedent. As examples of biologically active α/β-peptides multiply, it becomes increasingly important to understand how this type of unnatural oligomer is “perceived” by the immune system. The handful of previous relevant studies have focused almost exclusively on α/β-peptides that contain just one β residue; these precedents suggest that MHC-based and antibody-based recognition that developed to target a particular α-peptide is often retained for analogues containing a single α → β3 or α → aza-β3 replacement.20−23 The results reported here expand our understanding because we have considered α/β-peptides containing multiple α → β3 replacements, examples chosen to display β3 residue densities and distributions that enable functional mimicry of α-helical epitopes and strongly suppress proteolysis.13−19
The α/β-peptides we have examined retain the side chain sequence of the α-peptide that inspired them, but the presence of multiple α → β3 replacements is seen to block recognition by polyclonal antibodies that bind strongly to the parent α-peptide or to prevent participation in a TCR + MHC I + peptide complex that accommodates the parent α-peptide epitope. In addition, we show that antibodies can be raised against α/β-peptide as haptens (i.e., conjugated to proteins that contain helper T-cell epitopes) but that these antibodies do not strongly cross-react with homologous α-peptides. Parallel experiments show that a d-α-peptide can serve as a hapten as well, but the resulting antibodies do not cross-react with the enantiomer (l-α-peptide). Our observations suggest that the immune system will not necessarily initiate unintended immune responses following exposure to α/β-peptides.
It is impossible for a single study to provide a comprehensive answer to the question, “how are α/β-peptides ‘perceived’ by the immune system?” The results reported here suggest that immune receptors that recognize specific α-peptides are not necessarily cross-reactive with homologous α/β-peptides. These observations encourage further exploration of oligomers with αααβ or ααβαααβ backbones for biomedical applications; perhaps it will be possible to design α/β-peptide immunogens that mimic specific α-helical epitopes. Further studies will be required to determine whether the conclusions drawn here, regarding a lack of cross-reactivity between homologous α- and α/β-peptides, are general.
Methods
Peptide Synthesis
Peptides were synthesized as C-terminal carboxylic acids with uncapped N-termini. Previously reported microwave-assisted solid-phase conditions were utilized.18 Detailed methods for peptide synthesis are found in the Supporting Information. See Supporting Information Figure S6 for sequences and Supporting Information Table 1 for analytical HPLC retention times, purity data and a list of observed masses from MALDI-TOF-MS. See Supporting Information Figure S7 for HPLC chromatograms and MALDI-TOF mass spectra.
Enzyme Linked Immunosorbent Assays (ELISAs): Commercial Polyclonal Antibody
Assays were conducted using 96-well flat bottom Nunc Maxisorp plates. Peptides were prepared as DMSO stock solutions at a concentration of 10 mg mL–1 and diluted in phosphate buffered saline containing 0.05% Tween-20 (PBST). Assay plates were coated with peptide via application of peptide solutions to assay wells for 2 h at room temperature (RT). Following peptide immobilization, and between each of the subsequent steps, assay plates were washed (4×) with PBST. Unoccupied sites on the surface of the assay plate were blocked via application of a solution of 1% bovine serum albumin (weight by volume) in PBST overnight at 4 °C. A solution containing a 1:200 dilution of anti-Bim BH3 polyclonal rabbit IgG 1° antibody (Abgent, AP1308a) was applied to assay plates for 2 h at RT. A 1:2000 dilution of alkaline phosphatase-conjugated antirabbit-IgG 2° antibody (Promega, S3731) was then applied for 2 h at RT. Following the final washing, Tris buffered solution containing p-nitrophenyl phosphate and magnesium chloride (Sigma-Aldrich, N1891) was added to enable evaluation of the extent of peptide-1° antibody formation via quantification of p-nitrophenolate formation resulting from alkaline phosphatase-catalyzed hydrolysis. Plates were read by recording absorbance at 405 nm on a Synergy 2 plate reader (BioTek) 3 h after addition of p-nitrophenyl phosphate-containing buffer.
Indirect ELISA: Commercial Polyclonal Antibody
Nunc Maxisorp assay plates were coated with aqueous solutions containing indicated peptides at varied concentrations for 2 h at RT. Subsequent steps of these ELISAs were run according to the protocol described above. Dose–response curves relating the magnitude of the absorption at 405 nm to peptide concentrations used in the assay plate coating step are found in Figure 2.
Competition ELISA: Commercial Polyclonal Antibody
Nunc Maxisorp assay plates were coated with aqueous solutions of peptide 1 at a concentration of 44.3 μM for 2 h at RT. Assay plate washing and blocking steps were performed as described above. Prior to addition of the anti-Bim BH3 1° antibody to the assay plate, the antibody-containing solution was mixed and incubated with varied concentrations of free (nonimmobilized) peptide in a separate 96 well plate for >10 min. Following this incubation step, the 1° antibody/peptide solution was added to the assay plate for 2 h. Subsequent steps in this assay were performed as described above.
Preparation of Carrier Protein Conjugated Peptides
The sequence for mouse sPLA2-IB (sPLA2) was sourced from pubmed.gov protein: NP_035237.1 and was based on sequence analysis from ref (41). One milligram of each peptide (Bim BH3 or sPLA2) was conjugated to one milligram bovine gamma globulin (BGG) (Sigma-Aldrich, G5009) or ovalbumin (OVA) (Sigma-Aldrich, A5503) using gluataraldehyde (8 mM) in sodium acetate buffer (100 mM, pH 7.0). Excess glutaraldehyde was quenched through addition of glycine and peptide-carrier protein conjugates were then dialyzed against phosphate buffered saline overnight in 12 000–14 000 molecular weight dialysis membrane tubing (Fisher Scientific, 21-152-8). Conjugates were then dispensed in aliquots and frozen at −80 °C until ready for injection into the laying hens or ELISA.
Chicken Studies Using sPLA2 or Bim BH3 Peptides
All procedures involving chickens were approved by the University of Wisconsin, College of Agricultural and Life Sciences Animal Care and Use Committee. Single comb white leghorn laying hens were subcutaneously injected (500 μg peptide/chicken) with sPLA2-BGG or Bim BH3-BGG peptide conjugates emulsified with Freund’s Complete Adjuvant (Fisher Scientific, PI-77140). Control antibodies were taken from chickens injected with adjuvant only. Chickens were given a single booster injection with each peptide (500 μg peptide/chicken) 1 week after the initial injection with the peptide conjugate emulsified in Freund’s Incomplete Adjuvant (Fisher Scientific, PI-77145). Eggs were collected beginning 2 weeks after the initial injection. Egg yolks from each peptide were physically separated from albumin, combined, lyophilized, and stored at RT until needed. Enzyme linked immunosorbant assays (ELISAs) were then used to determine the titer of the antibody.
Enzyme Linked Immunosorbent Assays (ELISAs): Chicken Polyclonal Antibody
Assays were conducted using 96-well flat bottom Nunc Maxisorp plates. Assay plates were coated with 1:100 dilutions of previously prepared peptide-OVA conjugates in PBS via application of peptide solutions to assay wells overnight at 4 °C. Following peptide immobilization, and between each of the subsequent steps, assay plates were washed (4×) with PBST. Unoccupied sites on the surface of the assay plate were blocked via application of a solution of 1% bovine serum albumin (weight by volume) for 1 h. Egg yolk or lyophilized egg powder was first diluted 1:10 in acidified PBS (pH = 5) and centrifuged before further dilutions in PBS and addition to the assay plate. A 1:1000 dilution of alkaline phosphatase-conjugated antichicken-IgG 2° antibody (Sigma-Aldrich, A9171) was then applied for 1 h at RT. Following the final washing, Tris buffered solution containing p-nitrophenyl phosphate and magnesium chloride was added to enable evaluation of the extent of peptide-1° antibody formation via quantification of p-nitrophenolate formation resulting from alkaline phosphatase-catalyzed hydrolysis. Development of color in experiment wells was halted by addition of 1 N NaOH after color development progressed sufficiently to be easily observed by eye (5–60 min). Plates were read by recording absorbance at 405 nm on a Synergy 2 plate reader (BioTek).
Indirect ELISA: Chicken Polyclonal Antibody
ELISAs were run according to the protocol described above. During the primary antibody incubation step, serial dilutions of the egg primary antibody were then prepared in PBS in a separate polypropylene dilution plate (Costar EW-01728-80) before application to the assay plate. Subsequent steps in this assay were performed as described above.
Competition ELISA: Chicken Polyclonal Antibody
ELISAs were run according to the protocol described above. Prior to addition of the 1° egg antibody to the assay plate, antibody-containing solution was mixed and incubated with varied concentrations of free (nonimmobilized) peptide in a separate 96 well plate for >10 min. Free peptides were prepared as DMSO stock solutions at a concentration of 10 mg mL–1 and diluted in PBS. Subsequent steps in this assay were performed as described above.
Mouse Studies with the GP33 Viral Epitope: LCMV Infection
C57/BL6 mice between the ages of 6 and 8 weeks were infected with 2 × 105 plaque forming units of lymphocytic choriomeningitis virus (LCMV) Armstrong strain by intraperitoneal (IP) injection. After 30 days, when a competent memory CD8+ T cell population is established, mice were sacrificed, and single cell suspensions of splenocytes were prepared, as described previously.32All animal experiments were performed in accordance with protocols approved by the University of Wisconsin School of Veterinary Medicine Institutional Animal Care and Use Committee (IACUC).
Mouse Studies with the GP33 Viral Epitope or Bim BH3: Peptide Injections
C57/BL6 mice between the ages of 6 and 8 weeks were injected subcutaneously with 50 μg of peptide [GP33 (7), α/β-GP33 (8), Bim BH3 (1), or α/β-Bim BH3 (2)] emulsified in Freund’s incomplete adjuvant (3 to 4 mice per peptide). After 30 days, mice were sacrificed and single cell suspensions of splenocytes were prepared, as described previously.32 Splenocytes from animals in each injection group were pooled for flow cytometry analysis. All animal experiments were performed in accordance with protocols approved by the University of Wisconsin School of Veterinary Medicine Institutional Animal Care and Use Committee (IACUC).
Flow Cytometry
To quantify intracellular cytokine production, 1 × 106 splenocytes were stimulated for 5 h at 37 °C in a 96 well microtiter plate with α-GP33, α-Bim BH3, α/β-GP33 or α/β-Bim BH3 peptide analogues in the presence of Brefeldin A.32 Following stimulation, cells were incubated with antibody for the surface marker CD8 (BD Biosciences). Next, cells were permeabilized and fixed using the Cytofix/Cytoperm kit (BD Biosciences) and subsequently stained for the intracellular cytokine IFNγ using the Cytofix/Cytoperm kit (BD Biosciences, Franklin Lakes, NJ). Cells were fixed with 2% paraformaldehyde in PBS, and data were acquired on a FACSCalibur flow cytometer. Data were analyzed using FlowJo analysis software (Treestar), and the frequency of IFNγ -producing CD8+ T cells was determined.
MHC Purification and Peptide-Binding Assays
Purification of H2-Db class I MHC molecules by affinity chromatography, and performance of assays to quantitatively measure peptide affinity based on the inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules, has been detailed elsewhere.35 Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values.42,43 Each competitor peptide was tested at six different concentrations covering a 100 000-fold dose range and in three or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
Circular Dichroism (CD) Spectroscopy
An Aviv 420 Circular Dichroism Spectrophotometer was used to acquire all CD data. Wavelength scans were acquired at 20 °C at a step value of 1 nm and an averaging time of 3.0 s from 260 to 200 nm. A 0.1 mm cell was used for all spectra. All samples were prepared at a peptide concentration of 50 μM in 10 mM sodium phosphate buffer, pH 7.0. Peptide concentrations were calculated by measuring absorption at 280 nm after dissolving lyophilized peptide samples in sodium phosphate buffer (assuming ε(Trp) = 5690 M–1 cm–1 and ε(Tyr) = 1280 M–1 cm–1 at 280 nm).
Data Calculations
Data were processed by using the Microsoft Excel and GraphPad Prism 4.0 software packages. Graphical representations of ELISA data depict experimental means ± standard deviations, with each condition run in duplicate. Graphical representations of flow cytometry data depict experimental means ± standard deviations, with each condition run in triplicate or quadruplicate. Mann–Whitney tests were used to generate p-values in flow cytometry experiments.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grant R01 GM-056414 (S.H.G.) and R21 AI101976 (M.S.). This research was also supported by NIH-NIAID contract numbers HHSN272201400045C, HHSN272200900042C, and HHSN272201200010C (A.S.). R.W.C. was supported in part by a Biotechnology Training Grant from NIGMS (T32 GM008349), J.A.S. was supported by a postdoctoral fellowship from the American Heart Association, J.M.S. was supported by the Wisconsin Alumni Research Foundation, and T.W. was supported in part by a Hilldale Undergraduate Research Fellowship from UW—Madison.
Supporting Information Available
Contains detailed methods as well as Supporting Information Figures 1-7 and Supporting Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): S.H.G. is a co-founder of Longevity Biotech, Inc., which is pursuing biomedical applications of α/β-peptides.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Alberts B. (1998) The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 92, 291–294. [DOI] [PubMed] [Google Scholar]
- Hunter T. (2000) Signaling—2000 and beyond. Cell 100, 113–127. [DOI] [PubMed] [Google Scholar]
- Overall C. M.; Blobel C. P. (2007) In search of partners: Linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 8, 245–257. [DOI] [PubMed] [Google Scholar]
- Jung T.; Catalgol B.; Grune T. (2009) The proteasomal system. Mol. Aspects Med. 30, 191–296. [DOI] [PubMed] [Google Scholar]
- Janeway C. (2005) Immunobiology: The Immune System In Health And Disease; Garland Science Taylor & Francis Group, New York. [Google Scholar]
- Wells J. A.; McClendon C. L. (2007) Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Leader B.; Baca Q. J.; Golan D. E. (2008) Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discovery 7, 21–39. [DOI] [PubMed] [Google Scholar]
- Diao L.; Meibohm B. (2013) Pharmacokinetics and pharmacokinetic–pharmacodynamic correlations of therapeutic peptides. Clin. Pharmacokinet. 52, 855–868. [DOI] [PubMed] [Google Scholar]
- Wade D.; Boman A.; Wahlin B.; Drain C. M.; Andreu D.; Boman H. G.; Merrifield R. B. (1990) All-D amino acid-containing channel-forming antibiotic peptides. Proc. Natl. Acad. Sci. U.S.A. 87, 4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongsiriwatana N. P.; Patch J. A.; Czyzewski A. M.; Dohm M. T.; Ivankin A.; Gidalevitz D.; Zuckermann R. N.; Barron A. E. (2008) Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 105, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. M.; Hamilton A. D. (2007) Benzoylurea oligomers: Synthetic foldamers that mimic extended α helices. Angew. Chem., Int. Ed. 46, 8614–8617. [DOI] [PubMed] [Google Scholar]
- Porter E. A.; Wang X. F.; Lee H. S.; Weisblum B.; Gellman S. H. (2000) Antibiotics—Non-haemolytic β-amino-acid oligomers. Nature 404, 565–565. [DOI] [PubMed] [Google Scholar]
- Haase H. S.; Peterson-Kaufman K. J.; Levengood S. K. L.; Checco J. W.; Murphy W. L.; Gellman S. H. (2012) Extending foldamer design beyond α-helix mimicry: α/β-Peptide inhibitors of vascular endothelial growth factor signaling. J. Am. Chem. Soc. 134, 7652–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloha R. C.; Maeda A.; Dean T.; Gardella T. J.; Gellman S. H. (2014) Backbone modification of a polypeptide drug produces analogs with altered duration of action in vivo. Nat. Biotechnol. 32, 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne W. S.; Johnson L. M.; Ketas T. J.; Klasse P. J.; Lu M.; Moore J. P.; Gellman S. H. (2009) Structural and biological mimicry of protein surface recognition by α/β-peptide foldamers. P. Natl. Acad. Sci. U.S.A. 106, 14751–14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M.; Gellman S. H. (2013) α-Helix mimicry with α/β-peptides. Method Enzymol. 523, 407–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne W. S.; Boersma M. D.; Windsor M. A.; Gellman S. H. (2008) Sequence-based design of α/β-peptide foldamers that mimic BH3 domains. Angew. Chem., Int. Ed. 47, 2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne W. S.; Price J. L.; Gellman S. H. (2008) Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 9151–9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma M. D.; Haase H. S.; Peterson-Kaufman K. J.; Lee E. F.; Clarke O. B.; Colman P. M.; Smith B. J.; Horne W. S.; Fairlie W. D.; Gellman S. H. (2012) Evaluation of diverse α/β-backbone patterns for functional α-helix mimicry: Analogues of the Bim BH3 domain. J. Am. Chem. Soc. 134, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard G.; Zerbib A.; Le Gal F. A.; Hoebeke J.; Connan F.; Choppin J.; Briand J. P.; Guillet J. G. (2000) Melanoma peptide MART-1(27–35) analogues with enhanced binding capacity to the human class I histocompatibility molecule HLA-A2 by introduction of a β-amino acid residue: Implications for recognition by tumor-infiltrating lymphocytes. J. Med. Chem. 43, 3803–3808. [DOI] [PubMed] [Google Scholar]
- Reinelt S.; Marti M.; Dedier S.; Reitinger T.; Folkers G.; de Castro J. A. L.; Rognan D. (2001) β-Amino acid scan of a Class I major histocompatibility complex-restricted alloreactive T-cell epitope. J. Biol. Chem. 276, 24525–24530. [DOI] [PubMed] [Google Scholar]
- Webb A. I.; Dunstone M. A.; Williamson N. A.; Price J. D.; de Kauwe A.; Chen W. S.; Oakley A.; Perlmutter P.; McCluskey J.; Aguilar M. I.; Rossjohn J.; Purcell A. W. (2005) T cell determinants incorporating β-amino acid residues are protease resistant and remain immunogenic in vivo. J. Immunol. 175, 3810–3818. [DOI] [PubMed] [Google Scholar]
- Dali H.; Busnel O.; Hoebeke J.; Bi L. R.; Decker P.; Briand J. P.; Baudy-Floc’h M.; Muller S. (2007) Heteroclitic properties of mixed α- and aza-β(3)-peptides mimicking a supradominant CD4 T cell epitope presented by nucleosome. Mol. Immunol. 44, 3024–3036. [DOI] [PubMed] [Google Scholar]
- Croft N. P.; Purcell A. W. (2011) Peptidomimetics: Modifying peptides in the pursuit of better vaccines. Expert Rev. Vaccines 10, 211–226. [DOI] [PubMed] [Google Scholar]
- Andersson I. E.; Batsalova T.; Haag S.; Dzhambazov B.; Holmdahl R.; Kihberg J.; Linusson A. (2011) (E)-Alkene and ethylene isosteres substantially alter the hydrogen-bonding network in Class II MHC A(q)/glycopeptide complexes and affect T-cell recognition. J. Am. Chem. Soc. 133, 14368–14378. [DOI] [PubMed] [Google Scholar]
- Unudurthi S. D.; Hotta K.; Kim C. Y. (2013) Engineering the polyproline II propensity of a Class II major histocompatibility complex ligand peptide. ACS Chem. Biol. 8, 2383–2387. [DOI] [PubMed] [Google Scholar]
- Letai A.; Bassik M. C.; Walensky L. D.; Sorcinelli M. D.; Weiler S.; Korsmeyer S. J. (2002) Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2, 183–192. [DOI] [PubMed] [Google Scholar]
- Czabotar P. E.; Lee E. F.; van Delft M. F.; Day C. L.; Smith B. J.; Huang D. C. S.; Fairlie W. D.; Hinds M. G.; Colman P. M. (2007) Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott D. L.; Yang M.; Gonzalez J.; Larson A. E.; Tepp W. H.; Johnson E. A.; Cook M. E. (2009) Egg yolk antibodies for detection and neutralization of Clostridium botulinum Type A neurotoxin. J. Food. Protect. 72, 1005–1011. [DOI] [PubMed] [Google Scholar]
- Dintzis H. M.; Symer D. E.; Dintzis R. Z.; Zawadzke L. E.; Berg J. M. (1993) A comparison of the immunogenicity of a pair of enantiomeric proteins. Proteins 16, 306–308. [DOI] [PubMed] [Google Scholar]
- Tissot A. C.; Ciatto C.; Mittl P. R. E.; Grutter M. G.; Pluckthun A. (2000) Viral escape at the molecular level explained by quantitative T-cell receptor/peptide/MHC interactions and the crystal structure of a peptide/MHC complex. J. Mol. Biol. 302, 873–885. [DOI] [PubMed] [Google Scholar]
- Sullivan J. A.; Kim E. H.; Plisch E. H.; Peng S. L.; Suresh M. (2012) FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoS Pathog. 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K. L.; Rothstein L.; Benacerraf B. (1992) Analysis of the association of peptides of optimal length to Class-I molecules on the surface of cells. Proc. Natl. Acad. Sci. U.S.A. 89, 8918–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A.; Bromley S. K.; Sumen C.; Davis M. M.; Shaw A. S.; Allen P. M.; Dustin M. L. (1999) The immunological synapse: A molecular machine controlling T cell activation. Science 285, 221–227. [DOI] [PubMed] [Google Scholar]
- Sidney J.; Southwood S.; Moore C.; Oseroff C.; Pinilla C.; Grey H. M.; Sette A. (2013) Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. 10.1002/0471142735.im1803s100Chapter 18; Unit 18.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achour A.; Michaelsson J.; Harris R. A.; Odeberg J.; Grufman P.; Sandberg J. K.; Levitsky V.; Karre K.; Sandalova T.; Schneider G. (2002) A structural basis for LCMV immune evasion: Subversion of H2-D(b) and H2-K(b) presentation of gp33 revealed by comparative crystal structure analyses. Immunity 17, 757–768. [DOI] [PubMed] [Google Scholar]
- Matsumura M.; Fremont D. H.; Peterson P. A.; Wilson I. A. (1992) Emerging principles for the recognition of peptide antigens by MHC Class-I molecules. Science 257, 927–934. [DOI] [PubMed] [Google Scholar]
- Sette A.; Vitiello A.; Reherman B.; Fowler P.; Nayersina R.; Kast W. M.; Melief C. J. M.; Oseroff C.; Yuan L.; Ruppert J.; Sidney J.; Delguercio M. F.; Southwood S.; Kubo R. T.; Chesnut R. W.; Grey H. M.; Chisari F. V. (1994) The Relationship between Class-I binding affinity and immunogenicity of potential cytotoxic T-cell. J. Immunol. 153, 5586–5592. [PubMed] [Google Scholar]
- Kotturi M. F.; Peters B.; Buendia-Laysa F.; Sidney J.; Oseroff C.; Botten J.; Grey H.; Buchmeier M. J.; Sette A. (2007) The CD8(+) T-cell response to lymphocytic choriomeningitis virus involves the L antigen: Uncovering new tricks for an old virus. J. Virol. 81, 4928–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Ponomarenko J.; Zhu Z.; Tamang D.; Wang P.; Greenbaum J.; Lundegaard C.; Sette A.; Lund O.; Bourne P. E.; Nielsen M.; Peters B. (2012) Immune epitope database analysis resource. Nucleic Acids Res. W525–W530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollie N. I.; Hui D. Y. (2011) Group 1B phospholipase A(2) deficiency protects against diet-induced hyperlipidemia in mice. J. Lipid. Res. 52, 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- Gulukota K.; Sidney J.; Sette A.; DeLisi C. (1997) Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 267, 1258–1267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.