Abstract
Currently available therapies for adult onset neurodegenerative diseases provide symptomatic relief, but are not disease modifying. We explore here a new neuroprotective approach based on drugs targeting chaperone-directed protein quality control. Critical target proteins that unfold and aggregate in these diseases, such as the polylglutamine androgen receptor (spinal and bulbar muscular atrophy), huntingtin (Huntington’s disease), α-synuclein (Parkinson’s disease) and tau (Alzheimer’s disease) are client proteins of Hsp90, and their turnover is regulated by the protein quality control function of the Hsp90/Hsp70-based chaperone machinery. In protein quality control Hsp90 and Hsp70 have opposing effects on client protein stability; Hsp90 stabilizes the clients and inhibits their ubiquitination, whereas Hsp70 promotes CHIP-dependent ubiquitination and proteasomal degradation. We discuss how drugs that modulate proteostasis by inhibiting Hsp90 function or by promoting Hsp70 function enhance the degradation of the critical aggregating proteins and ameliorate toxic symptoms in cell and animal disease models.
Keywords: Neurodegeneration, protein aggregation, ubiquitination, CHIP, proteasome
INTRODUCTION
The adult onset neurodegenerative disorders include a diverse collection of chronic progressive diseases that show selective vulnerability of distinct neuronal populations and accumulate abnormally processed or mutant proteins that misfold and aggregate. Among these disorders are Alzheimer’s disease (AD), Parkinson’s disease (PD), and the polyglutamine expansion disorders, such as Huntington’s disease (HD) and spinal and bulbar muscular atrophy (SBMA). Critical proteins that unfold and aggregate in these diseases, such as tau (AD), α-synuclein (PD), huntingtin (HD), and the expanded glutamine androgen receptor (polyQ AR) (SBMA), are “client” proteins of the abundant and ubiquitous protein chaperone Hsp90. The Hsp90/Hsp70-based chaperone machinery that dynamically assembles client protein-Hsp90 heterocomplexes (1) is also part of the cellular defense against unfolded proteins (2).
In this protein quality control function of the machinery, Hsp90 and Hsp70 have opposing effects on client protein stability. Hsp90 stabilizes client proteins, and when cycling with Hsp90 is blocked by specific Hsp90 inhibitors, like geldanamycin or radicicol, the client proteins undergo rapid degradation through the ubiquitin-proteasome pathway (3). In contrast, Hsp70 along with its co-chaperone Hsp40 is required for the degradation of many proteins (2). Opposing roles of Hsp90 and Hsp70 also regulate the turnover of critical proteins that aggregate in adult onset neurodegenerative disorders. This suggests that targeting the protein quality control function of the Hsp90/Hsp70-based chaperone machinery with small molecules that inhibit Hsp90 or promote Hsp70 function may be an effective way to alter disease progression.
In this review, we first discuss how the Hsp90/Hsp70-based chaperone machinery acts in protein quality control and how small molecules affect Hsp90 and Hsp70 function, and then we review the evidence supporting the notion that modifying the function of the machinery with small molecules provides a rational avenue of drug discovery for treatment of the adult onset neurodegenerative disorders.
THE CHAPERONE MACHINERY AND PROTEIN QUALITY CONTROL
The function and turnover of a wide variety of proteins are regulated by Hsp90 (1). In the chaperone machinery, Hsp90, Hsp70 and their co-chaperones function together as a multiprotein complex (1, 4). In contrast to the classic model of individual chaperones interacting with unfolded proteins to facilitate their refolding, the Hsp90/Hsp70-based chaperone machinery instead acts on pre-folded proteins in their native (or near native) conformations to assist in the opening and stabilization of ligand binding clefts, as illustrated in Figure 1 (5, 6). These Hsp90 client proteins constantly undergo cycles of Hsp90 heterocomplex assembly and disassembly in the cytoplasm and nucleoplasm (1).
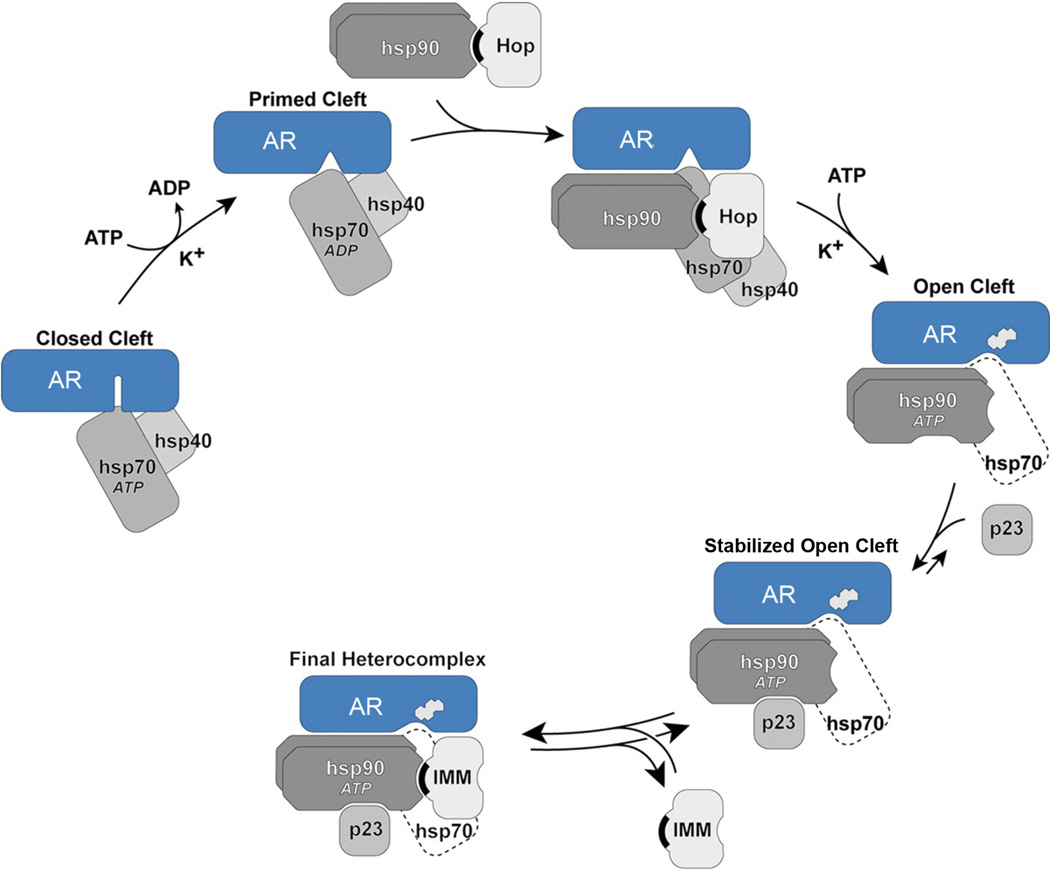
Figure 1. Mechanism of ligand binding cleft opening and AR-Hsp90-immunophilin heterocomplex assembly.
The ATP-dependent conformation of Hsp70 binds initially to the AR (polyQ AR), and in an ATP-, K+-, and Hsp40-dependent step, an AR-Hsp70 complex is formed that is primed to interact with Hsp90. After Hsp90 binding, there is a second ATP- and K+-dependent step that is rate limiting and leads to opening of the steroid binding cleft, enabling access of the steroid (indicated by the steroid structure). K+ is required for Hsp70 ATPase activity, implying active participation of Hsp70 in both steps of cleft opening. During receptor-Hsp90 heterocomplex assembly in cells and cell lysates, Hop (Hsp organizing protein) and some of the Hsp70 dissociate during or at the end of the cleft opening step. The dashed line for Hsp70 indicates it is present in amounts that are substoichiometric with respect to the receptor at this step. The receptor-bound Hsp90 is now in its ATP-dependent conformation and can be bound by p23, which stabilizes the chaperone in that conformation, preventing disassembly of the receptor-Hsp90 heterocomplex. When Hop dissociates, TPR domain immunophilins (IMM), such as FKBP52, Cyp40 or PP5 (protein phosphatase 5, an immunophilin homolog) can bind reversibly to the TPR acceptor site on receptor-bound Hsp90. TPR domains are indicated by black crescents. Details of Hsp90 heterocomplex assembly are reviewed in Pratt and Toft (1).
Ligand binding clefts are hydrophobic clefts that must be open to allow access of ligands, such as steroids or ATP, to their binding sites within the interior of proteins. In the absence of the chaperone machinery, ligand binding clefts are dynamic, shifting to varying extents between closed and open states. When clefts open, hydrophobic residues of the interior of proteins are exposed to solvent, and continued opening may progress to protein unfolding. Therefore, the extent to which ligand binding clefts are open determines ligand access and thus protein function, but clefts are inherent sites of conformational instability. By this model, the stability of the open state of the cleft is modulated by the Hsp90/Hsp70-based chaperone machinery, and when cleft opening is extensive enough such that Hsp90 can no longer interact with it, protein unfolding proceeds and the client protein is degraded (4–6).
Although it is clear that the ubiquitin-proteasome pathway is the major route of client protein degradation, it has not been clear how damaged or aberrant proteins are selected for ubiquitination. The prevailing view has been that E3 ubiquitin ligases perform the role of protein substrate recognition and bring the ubiquitin-charged E2 enzyme to the substrate (7). However, in the case of damaged or aberrant proteins that are unfolding, chaperones appear to be responsible for substrate recognition, and chaperone-dependent E3 ligases, like CHIP (carboxyl terminus of Hsc70-interacting protein) target the ubiquitin-charged E2 enzyme to the substrate (8).
CHIP
Overexpression of CHIP increases ubiquitination and proteasomal degradation of many classic Hsp90 client proteins, like the glucocortiooid receptor (GR), p53, and ErbB-2 (4, 9, 10). As we will detail later, CHIP also promotes degradation of the Hsp90 client proteins that aggregate in neurodegenerative diseases. Parkin is another E3 ligase that is targeted to substrate by Hsp70, and there is functional redundancy between CHIP and parkin against some substrates, such as polyQ ataxin-3 (11, 12). It is likely that other E3 ligases are functionally redundant with CHIP, as Hsp90 client proteins like the GR, the estrogen receptor and the polyQ AR, are degraded at the same rate in CHIP−/− cells as in CHIP+/+ cells (12, 13). Nevertheless, CHIP is generally regarded as the most important E3 ligase involved in chaperone-dependent ubiquitination and degradation of damaged and aberrant proteins (8, 14).
CHIP is a 35-kDa E3 ligase that binds via an amino-terminal tetratricopeptide repeat (TPR) domain to both Hsc/Hsp70 and Hsp90 (8). CHIP possesses a carboxy-terminal U-box that interacts with the UBCH5 family of E2 ubiquitin conjugating enzymes (8). Because CHIP binds with roughly the same affinity to TPR acceptor sites on Hsp70 and Hsp90, it was originally thought that both chaperones could target CHIP to the substrate (8, 9). However, it is clear that the two chaperones have opposing effects on CHIP-dependent ubiquitination. Hsp90 inhibits substrate ubiquitination and degradation, whereas Hsp70 promotes ubiquitination and degradation (6, 15).
Protein Triage
The opposing effects of the two chaperones can account for the triage of damaged and aberrant proteins. Both Hsp90 and Hsp70 bind selectively to domains of client proteins that contain ligand binding clefts. For example, both chaperones interact with the ligand binding domains of steroid receptors (16) and with the oxygenase domain of neuronal nitric-oxide synthase (nNOS), which is the domain containing the heme/substrate binding cleft (6). Hsp90 also interacts with the catalytic domains containing the ATP binding clefts of protein kinase clients, such as v-Raf (17) and ErbB-2 (18). Modulation of ligand binding clefts triggers CHIP-dependent ubiquitination and proteasomal degradation of proteins that cycle with Hsp90. For example, site-specific inactivators trigger ubiquitination of nNOS (15), ErbB-2 (19), and the estrogen receptor (20), and CHIP serves as an E3 ligase for ubiquitination of each (10, 21, 22). In some cases Hsp90 client proteins, such as huntingtin, α-synuclein and tau, do not bind any known ligands, and the domain of Hsp90 binding is unknown. Ligand binding is not a requirement for this model, as a major protein folding cleft that is unstable would provide the same requirements for interaction with and stabilization by Hsp90.
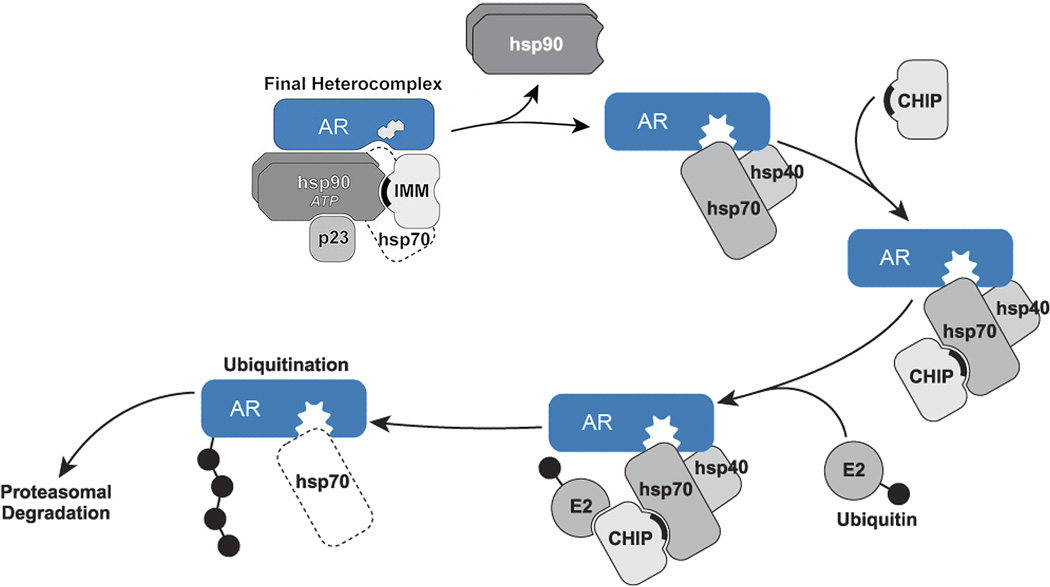
A model of triage of damaged or aberrant proteins that cycle with Hsp90 is presented in Figure 2. Site-specific inactivation serves as an example of toxic damage that is targeted to the ligand binding cleft and triggers protein ubiquitination. As the chaperoned protein undergoes such damage, the ligand binding cleft opens as the initial step in unfolding of the protein (4–6). As long as Hsp90 can cycle even transiently with the opening cleft, ubiquitination by Hsp70-depedent ubiquitin ligases, like CHIP, is inhibited. However, a point is reached where unfolding of the cleft progresses to a state that cannot cycle with Hsp90, and ubiquitination by the Hsp70-dependent E3 ligase is unopposed. Because it is substrate-bound Hsp70 that is mediating CHIP-dependent ubiquitination, one can get the impression that Hsp70 makes the triage decision. But we propose it is the Hsp90 interaction with the unfolding substrate that determines whether ubiquitination will proceed at any moment or not, and the opposing effects of the two chaperones on ubiqitination determine protein quality control by the chaperone machinery.
Figure 2. Regulation of polyQ AR degradation.
Loss of cycling with Hsp90, such as following the addition of small molecule Hsp90 inhibitors or steroid-dependent conformational change of the polyQ AR, permits unfolding of the mutant protein (indicated by jagged steroid binding cleft). Substrate-bound Hsp70 then recruits chaperone dependent ubiquitin ligases such as CHIP to promote ubiquitination with subsequent degradation by the proteasome. (Modified from ref. 45.)
DRUGGING THE CHAPERONE MACHINERY
By this model of protein quality control, there are two principal ways to promote the degradation of Hsp90 client proteins that aggregate in the neurodegenerative disorders. The first, and the one that has been examined most extensively, is to inhibit their stabilization by inhibiting client protein-Hsp90 heterocomplex assembly with specific Hsp90 inhibitors. The second is to develop drugs that promote Hsp70-dependent ubiquitination and degradation. There are fundamental differences in these two approaches that must be considered in formulating a rationale for long-term treatment of a neurodegenerative disorder as opposed to relatively short course treatment of cancer with drug combinations. The rationale in cancer treatment is to eliminate or decrease the level of many Hsp90 client oncoproteins that are critical in the genesis and maintenance of malignancy by treating with Hsp90 inhibitors (23). This approach inhibits the cycling of the not-yet-unfolded client protein with Hsp90 to promote its degradation. In the case of the neurodegenerative disorders, the goal is to eliminate the already-unfolded client protein present in oligomers that are thought to be responsible for the toxic gain-of-function that generates the pathology. It is reasonable that these disorders are better approached with drugs that promote Hsp70-dependent degradation.
Hsp90 Inhibition
Hsp90 was connected to ubiquitin-dependent degradation in studies of the ansamycin class of natural products that are quite specific Hsp90 inhibitors. The first of these compounds, herbimycin A, was found to reverse v-Src transformation, and it was then used as a protein-tyrosine kinase inhibitor (4). v-Src was the first protein identified as an Hsp90 client, and in 1994, it was shown that the target of herbimycin A and geldanamycin is Hsp90, not v-Src (24). Treatment with geldanamycin inhibited v-Src-Hsp90 heterocomplex formation and reduced the level of v-Src protein (24). Shortly thereafter, it was shown that ansamycin-induced degradation of several receptor tyrosine kinases occurred via the ubiquitin-proteasome pathway (25, 26). Before 1994, there were only about a dozen known Hsp90 client proteins, but the number increased exponentially thereafter (1).
Geldanamycin binds in the nucleotide binding pocket near the N-terminus of Hsp90. This ATP binding site is structurally unique to the GHKL (Gyrase, Hsp90, Histidine Kinase, MutL) family whose ATP binding domains contain four common motifs that define a “Bergerat fold” (27). As most of these proteins are only found in prokaryotes, geldanamycin’s effects are quite specific for inhibition of Hsp90 family proteins in eukaryotes. Essentially, geldanamycin acts as a nucleotide mimic in occupying the ATP/ADP-binding site of Hsp90, inhibiting the intrinsic ATPase activity of the chaperone, which is essential for client protein-Hsp90 heterocomplex assembly and Hsp90 function in vivo (28). Although geldanamycin was important for understanding the mechanism of Hsp90 inhibition, it was too toxic for use in the clinic. Subsequently, a variety of compounds were developed as less toxic nucleotide mimics, and, to date, 17 compounds have entered clinical trial for cancer treatment (29).
A different line of reasoning has evolved to explain how geldanamycin prevents formation of protein aggregates in models of neurodegenerative disorders. Some time ago, it was reported that overexpression of Hsp70 or its co-chaperone Hsp40 decreases the level of abnormal proteins and improves viability in cellular models of several neurodegenerative disorders, such as PD, HD and SBMA (30–32). Treatment of cells with geldanamycin also increases levels of Hsp70 and prevents formation of protein aggregates. Hsp70 increases because cycling of heat shock factor 1 (HSF1) with Hsp90 maintains it in an inactive state, and treatment of cells with geldanamycin activates HSF1, which induces the stress proteins (33). Thus, it is generally assumed that geldanamycin alleviates the phenotype and accumulation of misfolded proteins in neurodegenerative disease models by inducing a stress response (34–37). When applied to Hsp90 client proteins, however, this explanation is incomplete. Geldanamycin inhibits formation of aggregates and promotes proteasomal degradation of polyglutamine AR in Hsf1−/− cells that cannot mount a stress response (38). The increase in Hsp70 that occurs with the stress response would facilitate client protein ubiquitination that occurs when cycling with Hsp90 is prevented by geldanamycin, but geldanamycin-induced degradation of the client protein is not primarily due to the generation of a stress response. Rather, geldanamycin inhibits cycling of the client protein with Hsp90, allowing Hsp70-dependent ubiquitination to proceed unopposed. In contrast, proteasomal degradation of client protein fragments (e.g. mutant huntingtin exon 1) that do not cycle with Hsp90 is entirely due to HSF1-mediated stress response, as we will discuss later.
In considering the possible use of Hsp90 inhibitors for treatment of neurodegenerative disorders, it is important to emphasize that Hsp90 interacts with the not-yet-unfolded client protein and not with the unfolded-and-already-aggregated client protein. Most of the protocols examining geldanamycin action have shown that it prevents the formation of oligomers and aggregates, which reflects degradation of the soluble client protein via the ubiquitin-proteasome pathway before aggregation can occur. Thus, it has been shown in a cellular model of PD, for example, that geldanamycin treatment reduces formation of α-synuclein aggregates and α-synuclein-induced toxicity, but geldanamycin treatment of cells with pre-existing inclusions did not result in reduction in the number of cells with inclusions (39).
If one could prevent the formation of protein aggregates with Hsp90 inhibitors, then the primary effect of these drugs in the clinical setting might be to slow disease progression. However, in those cells where aggregation-induced toxicity has not proceeded too far, some reversal might occur. In the protein aggregation neurodegenerative disorders, the inclusions that form contain Hsp70, ubiquitin, CHIP, E1 and E2 ubiquitinating enzymes, and even proteasomes. If the ubiquitin-proteasome system is functional in these inclusions, even at a reduced rate, then it is possible that, over time, as the generation of oligomers and aggregates is blocked, some reversal of pre-existing toxicity may occur. This process may also require the disaggregase function of an Hsp110-Hsp70-Hsp40 machinery to extract unfolded proteins from aggregates prior to Hsp70/CHIP-dependent ubquitination (40).
Patients with neurodegenerative disorders would require treatment over a span of years, and this is not likely to be possible with drugs that cause the degradation of hundreds of Hsp90 client proteins. However, the experience gained from clinical trials in cancer treatment may support patient tolerance of Hsp90 inhibitors over limited timespans. As the neurodegenerative diseases are slowly progressive, one might be able to treat with an Hsp90 inhibitor for a period of time, followed by a drug holiday. Such periodic treatment might be tolerated in lieu of long term, continuous treatment.
Modulating Hsp70
A variety of Hsps are expressed at higher levels in cancers, probably as a stress response to conditions of hypoxia and low pH generated in tumors (23). Hsp70 interacts with key proteins in the apoptotic pathway to inhibit apoptosis, and small molecule inhibitors of Hsp70 are being developed to target its anti-apoptotic function for cancer therapy (41). Inhibition of Hsp70 interaction with target proteins seems counterintuitive as an approach to treatment of late-onset neurodegenerative diseases. Inhibition of Hsp70 would inhibit Hsp70/CHIP-dependent ubiquitination and proteasomal degradation, as was shown in a cellular model of SBMA (42). A more rational approach would be to promote Hsp70/CHIP-dependent ubiquitination with small molecule activators of Hsp70.
Hip (Hsc70 interacting protein) is a co-chaperone of Hsp70 that prevents the accumulation of polyglutamine inclusions in a cellular model (43). Inasmuch as Hip stabilizes Hsp70 in its ADP-bound conformation, the conformation that recognizes unfolded or damaged substrates with high affinity (44), it was hypothesized that Hip overexpression might facilitate Hsp70-dependent ubiquitination and client protein degradation. Overexpression of Hip increases ubiquitination of nNOS (45), a well characterized Hsp90 client protein (5, 6) whose ubiquitination is dependent upon Hsp70 (6, 15, 42) and mediated by CHIP (21). Hip overexpression also promoted polyQ AR clearance in a cellular model and alleviated toxicity in a Drosophila model of SBMA (45). Hip overexpression also decreased the formation of fibrils by α-synuclein, and its knock-down in C. elegans increased α-synuclein aggregation (46). These results support a model in which increased affinity of Hsp70 for substrate proteins promotes their ubiquitination and degradation.
MKT-077 and YM-1 are rhodocyanines that cause selective death of cancer cells and bind with low micromolar affinity to the nucleotide binding domain of ADP- but not ATP-bound Hsp70, stabilizing the ADP-bound state (47). Both MKT-077 and YM-1 substitute for Hip in promoting Hsp70-dependent nNOS maturation (48). YM-1 increases the binding of purified Hsp70 to unfolded luciferase by stabilizing the ADP conformation of the chaperone, and the binding of YM-1 to Hsp70 is blocked by purified Hip (45). Treatment of cells with YM-1 increased CHIP-dependent ubiquitination of nNOS. YM-1 promoted polyQ AR clearance by the ubiquitin-proteasome system in a cellular model of SBMA and alleviated toxicity in a Drosophila model of SBMA (45). Importantly, YM-1 markedly decreased polyQ AR oligomers and detergent-insoluble aggregates while having little effect on soluble polyQ AR and no effect on other Hsp90 client proteins (45).
Thus, YM-1 is an appropriate platform for developing drugs that promote the degradation of Hsp90 client proteins after they have unfolded and are no longer cycling with Hsp90. This approach would be focused on accelerating the degradation of the oligomers and aggregates of the client protein that are deemed responsible for neurodegeneration. Inasmuch as neither Hip (1) nor YM-1 (45) affects client protein-Hsp90 heterocomplex assembly, cycling with Hsp90 proceeds normally to stabilize the soluble, properly folded client protein. This avoids the broad depletion of client proteins that would occur with Hsp90 inhibitors and would seem to be more amenable to the continuous or intermittent long-term treatment of neurodegenerative disorders. Also, the use of drugs like YM-1 that promote Hsp70/CHIP-dependent ubiquitination and degradation in combination with an Hsp90 inhibitor might shorten the treatment periods required to deplete aggregated client protein, thus reducing toxicity from Hsp90 inhibition. This model is an important shift in the way we think of using the protein quality control system to treat neurodegenerative diseases. In the next sections, we discuss how this model might be applied in SBMA, HD, PD and AD.
POLYGLUTAMINE DISEASES
Nine hereditary neurodegenerative disorders result from expansion of CAG repeats coding for polyglutamine tracts in the respective proteins: they include SBMA, HD, dentatorubropallidoluysian atrophy and six forms of spinocerebellar ataxia (49, 50).
Spinal and Bulbar Muscular Atrophy
SBMA is a progressive neuromuscular disorder that affects only men and is characterized by proximal limb and bulbar muscle weakness, atrophy and fasciculations. The causative mutation in SBMA is an expansion of a CAG repeat in the first exon of the AR gene (51). The glutamine tract encoded by this repeat is polymorphic in length in the normal population, containing between 9 and 37 residues. Pathologic expansions of this tract to 38 or more glutamines cause SBMA (51).
The expanded glutamine tract promotes hormone-dependent AR unfolding and oligomerization. Expanded polyQ AR unfolds and aggregates in vitro in a repeat length-dependent manner reflecting the disease threshold seen in vivo (52). The aggregates appear first as soluble oligomers, and, as with other polyQ disorders, there is evidence that these oligomers underlie the pathology (53). These oligomers then form large inclusions that may be localized in the cytoplasm or nucleus in a variety of tissues, with large intranuclear inclusions accumulating preferentially in motor neurons (54). The inclusions are thought to be cytoprotective in that they promote polyQ AR degradation and sequester the protein into an insoluable compartment in which it does not exert toxic effects (55). A variety of cellular processes are affected to produce toxicity, including transcription, RNA splicing, axonal transport, and mitochondrial function. It is reasonable to propose that oligomeric polyQ AR is ubiquitinated and becomes enmeshed with ubiquitinating enzymes, chaperones and proteasomes as oligomers pass through protein quality control sites during retrograde transport to form inclusions, much as aggresomes are formed during dynein-dependent movement of mutant CFTR and other missfolded proteins along microtubules (55, 56).
The AR is a classic Hsp90 client protein (16). Immunoadsorption of polyQ AR from cytosols is accompanied by co-immunoadsorption of Hsp90 and the co-chaperone p23, showing that biochemically stable heterocomplexes form in cells (12, 38, 57). Hsp90-binding immunophilins such as FKBP52 (52 kDa FK506 binding protein) and protein phosphatase 5 (PP5), an immunophilin homolog, link Hsp90-bound client proteins to the dynein/dynactin motor complex (58). Dynein, PP5 and FKBP52 co-localized with AR112Q aggregates in cell culture, consistent with the entry of AR112Q into aggregates as trafficking complexes (38). Hsp90 inhibitors promote proteasomal degradation of polyQ AR in cellular models of SBMA (38, 57), and ameliorate polyglutamine-mediated motor neuron impairment in mouse models of SBMA (57, 59).
In both cellular and animal models of SBMA, polyQ AR aggregates stain for Hsp70 and Hsp40 as well as ubiquitin and proteasome components (60, 61). Overexpression of Hsp70 or Hsp40 enhances the proteasomal degradation of the polyQ AR in cellular models of SBMA (32, 62), and overexpression of Hsp70 in a transgenic mouse model of SMBA reduces nuclear polyQ AR aggregates and ameliorates the phenotype (63). Both CHIP and Parkin co-localize with polyQ AR aggregates in SBMA cell and mouse models but only co-expression of CHIP reduced the level of AR112Q in cells (12). Overexpression of CHIP in a mouse model of SBMA inhibited neuronal nuclear accumulation of the mutant AR and ameliorated motor symptoms (64).
These observations show that the polyQ AR is an Hsp90 client protein that is regulated by the Hsp90/Hsp70-based chaperone machinery and undergoes Hsp70/CHIP-dependent ubiquitination and proteasomal degradation. The reports showing that Hsp90 inhibitors ameliorate the phenotype in a mouse model of SBMA (57, 59) and that a small molecule activator of Hsp70 reduces toxicity in a fly model (45) provide strong support for the continued development of small molecule modulators of the chaperone machinery as a potential approach to therapy of SBMA.
Huntington’s Disease
Huntington’s disease is an autosomal dominant, neurodegenerative disorder characterized by chorea and dystonia, cognitive decline and psychiatric disturbances, leading to progressive dementia and death 15–20 years after disease onset. The neuropathological changes consist of prominent neuron loss and atrophy in the caudate nucleus and putamen, although loss of neurons in other brain regions occurs. The mutation is an expanded CAG repeat in the gene encoding the protein huntingtin (Htt), encoding a polyglutamine tract at the amino-terminus of the protein. Normal individuals possess 35 or fewer repeats, and most adult onset cases have 40–50 CAG repeats. Although Htt is required for normal brain development, it does not play an essential non-redundant role after development. Details of the genetics and pathogenesis of HD have been the subject of review (65).
The function of wild-type Htt is unknown. Htt is a very large (348-kDa) multidomain protein with 28–36 predicted HEAT motifs, which comprise degenerate ∼50-amino-acid sequences that are usually involved in protein-protein interactions (65). Wild-type Htt is mostly cytoplasmic, although it clearly undergoes nucleocytoplasmic trafficking (66). One proposal is that Htt functions as a scaffold protein involved in intracellular trafficking and signaling, and it has been shown to facilitate transport along microtubules (67–69). As proposed for the polyQ AR, inhibition of axonal trafficking in HD could be a key event leading to disruption in downstream pathways (70). For example, it has been shown in HD model flies that mutant Htt (mHtt) fragments inhibit fast axonal transport of vesicles (67, 71). As with other polyQ diseases, there is evidence that mHtt inclusions are not responsible for neuronal toxicity, which is likely to result from soluble oligomeric species (72–75). Although HD reflects a polyQ-length-dependent toxic gain-of-function, it is not clear how much of the toxicity is due to full-length Htt or to amino-terminal fragments generated by proteases. Expression of amino-terminal, exon 1 fragments in animal models yields nuclear inclusions and an HD-like phenotype, as is seen in R6/2 mice (76, 77).
Most of the evidence for Hsp90 regulation of Htt has been derived from effects of Hsp90 inhibitors on truncated (exon 1) Htt. However, a very careful study of full-length Htt has shown that both wild-type (Q25) and mutant (Q72) Htt co-immunoprecipitate with Hsp90 (78). They also co-immunoprecipitate with p23, and in both cases co-immunoprecipitation is abrogated by treatment with an Hsp90 inhibitor. Thus, both full-length mHtt and wild-type Htt behave as classic Hsp90 client proteins, and mHtt degrades in the presence of an Hsp90 inhibitor in a dose-dependent manner (78). mHtt is ubiquitinated upon Hsp90 inhibition and is degraded by proteasomes in a manner that does not require induction of Hsp70 through activation of HSF1 (78). Thus, all of the criteria for full-length Htt being regulated by the quality control function of the Hsp90/Hsp70-based chaperone machinery have been met.
One important unanswered question is where Hsp90 binds Htt. As emphasized previously, in those cases where Hsp90 binding has been examined, the chaperone does not bind in a promiscuous manner; one dimer of Hsp90 is bound per client protein (1). Hsp90 inhibitors reduce mHtt exon 1 aggregate formation and ameliorate toxicity in cellular, fly and mouse models of HD (36, 37, 79–81). The changes noted were uniformly interpreted as due to HSF1-mediated increases in chaperone expression, and in one case knockdown of HSF1 abolished the induction of molecular chaperones and the therapeutic effect of the Hsp90 inhibitor (80). Thus, it seems that beneficial effects of Hsp90 inhibitors in exon 1 models of HD are entirely due to HSF1-mediated increases in chaperone expression. Direct binding of Hsp90 to the exon 1 protein fragment has not been demonstrated, and it is likely that the fragment is not a client protein of Hsp90. In human HD, where a full-length mHtt is an Hsp90 client protein, the primary effect of the inhibitors would be to prevent cycling with Hsp90, thus promoting its Hsp70-dependent ubiquitination and proteasomal degradation (78).
mHtt inclusions contain Hsp70, Hsp40, proteasomes and ubiquitin (31, 36). Overexpression of Hsp70 or Hsp40 suppresses mHtt fragment aggregation in yeast and cellular models of HD (31, 37, 82–84). Transgenic expression of Hsp40 in a fly model of HD suppresses toxicity (85), but overexpression of Hsp70 in R6/2 mice produces only very modest delay in aggregate formation and disease progression (36, 86). However, deletion of Hsp70.1 and Hsp70.3 in R6/2 mice increases the size of mHtt exon 1 inclusion bodies and exacerbates physical, behavioral and neuropathological measures (87), showing that endogenous Hsp70s are critical components of the cellular defense against the toxic effects of mHtt exon1.
Both CHIP and parkin have been implicated as E3 ligases in mHtt degradation. CHIP co-immunoprecipitates with mHtt exon 1 fragments and CHIP overexpression increases mHtt ubiquitination and degradation (11). Parkin colocalizes with mHtt in brain sections from HD transgenic mice and full-length polyQ-expanded Htt co-immunoprecipitates with parkin (88). Hsp70 enhances parkin binding and ubiquitination of a polyQ-expanded fragment in vitro, consistent with the chaperone recruiting parkin to the unfolded substrate (88).
Taken together, there is good evidence that polyQ-expanded, full-length huntingtin is an Hsp90 client protein that is regulated by the Hsp90/Hsp70-based chaperone machinery and undergoes Hsp70/CHIP/parkin-dependent ubiquitination and proteasomal degradation. Because much of the HD work has been performed with cellular and animal models expressing mHtt exon 1, a fragment that is very unlikely to be an Hsp90 client, a great deal of emphasis has been placed on explaining effects of Hsp90 inhibitors in terms of an HSF1-mediated stress response. In contrast, there has been limited focus on the protein quality control function of the chaperone machinery directly on the aberrant mHtt client protein (78).
PARKINSON’S DISEASE
Parkinson’s disease is a progressive neurodegenerative disorder that is due to selective degeneration of dopaminergic cells in the substantia nigra. The characteristic features of PD are tremor at rest, bradykinesia, rigidity and postural instability, which respond to therapy with dopaminergic drugs (89, 90). The diagnosis is a clinical one, and no neuroprotective therapy currently exists. No single cause of PD has been defined. A variety of mutations linked to the disease have been reported, including mutations in SNCA, which encodes α-synuclein, and mutations in PARK2, which encodes parkin (91). Although PD is a disorder with a complex, multifactorial aetiology, deposition of aggregated α-synuclein in Lewy bodies in cells of the brainstem and cortex is a pathological hallmark. In addition to α-synuclein, Lewy bodies contain ubiquitin, ubiquitin activating enzyme, Hsp70 and proteasome subunits. Like aggresomes, α-synuclein aggregates are formed from soluble oligomers in a microtubule-dependent manner (92), and it is thought that oligomers are the toxic species (93).
α-Synuclein is a 140 amino acid protein of unknown function that is often described as a natively unfolded 14 kDa monomer in biochemical studies of bacterially expressed protein. However, a recent careful study of endogenous α-synuclein reveals that α-synuclein occurs physiologically as an α-helically folded, ∼58 kDa homotetramer (94). As the tetramers undergo little or no aggregation, it is proposed that destabilization must occur to yield monomers that misfold and aggregate in PD and other human synucleinopathies (94). α-Synuclein is predominantly localized to presynaptic terminals in the central nervous system, where it is loosely associated with synaptic vesicles (95). Native, cell-derived, tetrameric α-synuclein binds lipid (95), and membrane association is viewed as a principal functional property of α-synuclein. There is very good evidence that unfolded α-synuclein plays an integral role in the onset of PD and related disorders (91, 95, 96), and α-synclein should be a major focus for neuroprotective therapy.
α-Synuclein co-immunoprecipitates with Hsp90 and Hsp70 (97), and Hsp90 suppresses α-synuclein toxicity in yeast (98). Treatment with Hsp90 inhibitors decreases α-synuclein oligomerization and alleviates toxicity in yeast, cellular and Drosophila models of PD (35, 39, 99–101). Taken together, these observations support the notion that α-synuclein is a client protein of Hsp90. Overall, the evidence for α-synuclein being an Hsp90 client protein is not as substantial as that for the polyQ AR or full-length huntingtin, and more direct biochemical studies of α-synuclein interaction with Hsp90 are required. For example, α-synuclein monomers can assume multiple partially folded conformations that undergo multiple forms of processing (reviewed by Dev et al. (102)), and it is not known what form or forms interact with Hsp90.
Overexpression of Hsp70 reduces high molecular weight forms of α-synuclein and ameliorates toxicity in cellular, fly and mouse models of PD (30, 103–107). Overexpression of Hsp40 produces similar effects in cellular PD models (104, 108). CHIP colocalizes with α-synuclein and Hsp70 in α-synuclein inclusions, and overexpression of CHIP inhibits α-synuclein inclusion formation and reduces α-synuclein protein levels (109). CHIP is a component of Lewy bodies in the human brain where it colocalizes with α-synuclein and Hsp70 (109). α-Synuclein is ubiquitinated by CHIP both in vitro and in cells (110), and α-synuclein and Hsp70 both co-immunoprecipitate with CHIP, suggesting the existence of a terniary complex (109).
In summary, the response to Hsp90 inhibitors supports the notion that α-synuclein is an Hsp90 client that is degraded in an Hsp70-dependent manner. It should be noted that the co-chaperone Hip stabilizes Hsp70-α-synuclein complexes in vitro, and knockdown of Hip in a C. elegans model of α-synuclein inclusion formation increases the number of inclusions over 2-fold (46). This suggests that drugs, like YM-1, that act like Hip to increase Hsp70-dependent ubiquitination should be tested in PD model systems.
ALZHEIMER’S DISEASE
Alzheimer’s disease is the most common form of late onset dementia, characterized by a progression from memory problems, language disturbances and executive dysfunction to a slow general decline in cognitive function, with death occurring roughly a decade after diagnosis (111, 112). Pathological hallmarks are senile plaques comprising extracellular deposits of amyloid-β (Aβ) and intracellular neurofibrillary tangles (NFTs) comprised of filaments of hyperphosphorylated tau. Both lesions are located in brain regions involved in cognition and memory.
Presently, there is no neuroprotective therapy for AD, and programs to develop disease modifying drugs focus on both Aβ and tau (113). According to the amyloid cascade model, accumulation of Aβ in the brain is considered to be the primary influence driving AD pathogenesis, and the rest of the disease process, including tau tangle formation, results from an imbalance between Aβ production and Aβ clearance (114). Although the genetics of AD favor the involvement of Aβ in pathogenesis, the clinicopathological correlations favor a disease based on tau NFTs (115). The tau proteins are comprised of six isoforms generated by alternative splicing from a single gene, and they are present predominantly in neurons (116). A hyperphosphorylated filamentous tau is the main component of NFTs. Under cell-free conditions, tau promotes microtubule assembly and stability, an effect that does not appear to be a critical function of tau in vivo (116). Tau NFTs are also part of the pathology of other neurodegenerative disorders, such as Pick’s disease, which do not manifest extracellular deposits of Aβ and are collectively considered as tauopathies (117).
The physiological function of tau is unknown, and complete ablation of tau in knockout mice does not cause premature mortality or major neurological deficit (116). This may reflect a partial functional redundancy of tau with microtubule-associated protein 1B (MAP1B) and perhaps other MAPs (118). A reasonable speculation is that, physiologically, tau plays some role in trafficking along microtubules. Overexpression of tau, for example, inhibits trafficking of vesicles and organelles (119, 120), and tau differentially modulates dynein and kinesin mobility (121). Soluble cytosolic forms of hyperphosphorylated tau appear to be the toxic species (116, 122). Efforts to actively reduce tau in AD would seem to be beneficial. For example, reducing endogenous tau levels prevented behavioral deficits in transgenic mice expressing human amyloid precursor protein without altering their high Aβ levels (123).
Hsp90 co-immunoprecipitates with tau, indicating that tau forms biochemically stable complexes that are typical for Hsp90 clients (124–126). Treatment with an Hsp90 inhibitor reduces levels of phospho-tau in cells in culture and in AD model mice (125, 127, 128) in a manner independent of HSF1 activation (128). Taken together, these observations establish tau as an Hsp90 client protein, but the exact forms of tau that are bound by Hsp90 are not clear.
Overexpression of Hsp70 decreases the levels of detergent-insoluble tau aggregates in cells (129). Tau lesions in postmortem human tissue are immunopositive for CHIP (129), and CHIP ubiquitinates phosphorylated tau (129–131). CHIP overexpression increases tau degradation and rescues tau-induced cell death (130, 131), and deletion of CHIP in mice leads to the accumulation of hyperphosphorylated tau (132, 133). Taken together, there is good evidence that some state (or states) of tau is regulated by the Hsp90/Hsp70-based chaperone machinery and undergoes Hsp70/CHIP-dependent ubiquitination and proteasomal degradation. It is important to note that YM-1 reduces tau levels in cellular and primary neuronal models of tauopathy (134), supporting further development of drugs that promote Hsp70/CHIP-dependent ubiquitination for treatment of AD.
CONCLUSIONS
Although the clinical features of the adult onset neurodegenerative disorders that we have reviewed here are quite different, there are commonalities both in their general mechanisms of pathogenesis and in the pathways of elimination of the critical proteins that unfold and aggregate. The target proteins in SBMA, HD, PD and AD all unfold and form soluble oligomers before they move into aggregates to form inclusions. There is good evidence in all cases that it is the soluble oligomer that is neurotoxic. Although a variety of processes are affected that undoubtedly contribute to neurotoxicity, there is the common theme that the target protein oligomers inhibit axonal trafficking, and this may be a critical lesion in the toxic pathway.
Despite the different structures and functions (known or potential) of the target proteins, there is good evidence that all of them are Hsp90 client proteins (Table 1). In all cases treatment of cell or animal models of the disease with Hsp90 inhibitors results in proteasomal degradation of the target protein and amelioration of neurotoxicity. In all cases Hsp70 and CHIP play a key role in target protein ubiquitination and degradation. The common roles played by Hsp90 in target protein stabilization and Hsp70/CHIP in target protein degradation via the ubiquitin-proteasome pathway suggest that drugging the Hsp90/Hsp70-based chaperone machinery could provide a common neuroprotective treatment for all four of these neurodegenerative disorders.
Table 1.
References presenting evidence that key target proteins in the adult onset neurodegenerative disorders are regulated by the protein quality control function of the Hsp90/Hsp70-based chaperone machinery.
| Disorder Target protein |
SBMA Poly Q AR |
HD Huntingtin |
PD α-Synuclein |
AD Tau |
|---|---|---|---|---|
| Hsp90 binding1 | 12, 38, 57 | 78 | 97 | 124–126 |
| Hsp90 inhibition2 | 38, 57, 59 | 37, 78–81 | 35, 39, 99–101 | 125, 127–129 |
| Hsp70 overexpression3 | 32, 62, 63 | 31, 37, 82–84 | 30, 103–107 | 129 |
| Hsp40 overexpression3 | 32, 60, 62 | 31, 37, 83, 85 | 104, 108 | |
| CHIP overexpression3 | 12, 64 | 11 | 109 | 130, 131 |
Direct demonstration of target protein-Hsp90 complexes by co-immunoprecipitation.
Treatment of cellular or animal disease models with Hsp90 inhibitor decreases the level of target protein.
Overexpression of Hsp70 or its co-chaperone Hsp40 or the Hsp70-binding ubiquitin E3 ligase CHIP decreases the level of the target protein.
One can envision both monotherapy and combination drug therapy administered in intermittent protocols that allow for recovery from side effects and alternation of treatments with different side effects. The combination of an Hsp90 inhibitor with an Hsp70 promoter may be synergistic in yielding target protein degradation. Either class of drug would likely be synergistic with a deubiquitinase inhibitor. Early results at the cellular level suggest that combination treatment regimens may allow lowering the dosage of one drug, permitting the development of clinical protocols that reduce drug-specific side effects. The authors’ intent in this review is to enable a mechanistic thinking about neuroprotection that is based on understanding the protein quality control function of the Hsp90/Hsp70-based chaperone machinery rather than the assumption that the therapeutic effects reflect an HSF1-mediated stress response.
ACKNOWLEDGEMENTS
The authors’ work was funded by the National Institutes of Health (NS055746, NS32214 to APL; NS059690 to JEG; GM077430 to YO), the Muscular Dystrophy Association (MDA238924 to APL), and the McKnight Foundation (to APL). The authors are all participants in The University of Michigan Medical School’s FastForward Protein Folding Diseases Initiative.
LITERATURE CITED
- 1.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 2.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 4.Pratt WB, Morishima Y, Peng HM, Osawa Y. Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp. Biol. Med. (Maywood) 2010;235:278–289. doi: 10.1258/ebm.2009.009250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng HM, Morishima Y, Pratt WB, Osawa Y. Modulation of heme/substrate binding cleft of neuronal nitric-oxide synthase (nNOS) regulates binding of Hsp90 and Hsp70 proteins and nNOS ubiquitination. J. Biol. Chem. 2012;287:1556–1565. doi: 10.1074/jbc.M111.323295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 9.Connell P, Ballinger CA, Jiang J, Wu Y, Thomson LJ, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P, Fernandes N, Dodge IL, Redd AL, Rao N, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 11.Jana NR, Dikshit P, Goswami A, Kotliarova S, Murata S, et al. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 12.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum. Mol. Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tateishi Y, Kawabe Y, Chiba T, Murata S, Ishikawa K, et al. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23:4813–4823. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng HM, Morishima Y, Clapp KM, Lau M, Pratt WB, Osawa Y. Dynamic cycling with Hsp90 stabilizes neuronal nitric oxide synthase through calmodulin-dependent inhibition of ubiquitination. Biochemistry. 2009;48:8483–8490. doi: 10.1021/bi901058g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 17.Stancato LF, Chow YH, Hutchinson KA, Perdew GH, Jove R, Pratt WB. Raf exists in a native heterocomplex with hsp90 and p50 that can be reconstituted in a cell-free system. J. Biol. Chem. 1993;268:21711–21716. [PubMed] [Google Scholar]
- 18.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, et al. Sensitivity of mature ErbB2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J. Biol. Chem. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 19.Citri A, Alroy I, Lavi S, Rubin C, Xu W, et al. Drug-induced ubiquitylation and degradation of ErbB2 receptor tyrosine kinases: implications for cancer therapy. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 21.Peng HM, Morishima Y, Jenkins GJ, Dunbar AY, Lau M, et al. Ubiquitylation of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J. Biol. Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- 22.Fan M, Park A, Nephew KP. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol. Endocrinol. 2005;19:2901–2914. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- 23.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 24.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v–src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepp-Lorenzino L, Ma Z, Lebwohl DE, Vinitsky A, Rosen N. Herbimycin A induces the 20 S proteasome- and ubiquitin-dependent degradation of receptor tyrosine kinases. J. Biol. Chem. 1995;270:16580–16587. doi: 10.1074/jbc.270.28.16580. [DOI] [PubMed] [Google Scholar]
- 26.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c–erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J. Biol. Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 27.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 28.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 29.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces alpha-synuclein aggregation and toxicity. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 31.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum. Mol. Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 32.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 33.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 34.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 35.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat. Med. 2002;8:1185–1186. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 36.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 37.Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum. Mol. Genet. 2001;10:1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 38.Thomas M, Harrell JM, Morishima Y, Peng HM, Pratt WB, Lieberman AP. Pharmacologic and genetic inhibition of hsp90-dependent trafficking reduces aggregation and promotes degradation of the expanded glutamine androgen receptor without stress protein induction. Hum. Mol. Genet. 2006;15:1876–1883. doi: 10.1093/hmg/ddl110. [DOI] [PubMed] [Google Scholar]
- 39.McLean PJ, Klucken J, Shin Y, Hyman BT. Geldanamycin induces Hsp70 and prevents alpha-synuclein aggregation and toxicity in vitro. Biochem. Biophys. Res. Commun. 2004;321:665–669. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J. Biol. Chem. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- 41.Powers MV, Jones K, Barillari C, Westwood I, van Monfort RLM, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9:1542–1550. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 42.Wang AM, Morishima Y, Clapp KM, Peng HM, Pratt WB, et al. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J. Biol. Chem. 2010;285:15714–15723. doi: 10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howarth JL, Glover CP, Uney JB. HSP70 interacting protein prevents the accumulation of inclusions in polyglutamine disease. J. Neurochem. 2009;108:945–951. doi: 10.1111/j.1471-4159.2008.05847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv. Protein Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4. [DOI] [PubMed] [Google Scholar]
- 45.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 2013;9:112–118. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roodveldt C, Bertoncini CW, Andersson A, van der Goot AT, Hsu ST, et al. Chaperone proteostasis in Parkinson's disease: stabilization of the Hsp70/alpha-synuclein complex by Hip. EMBO J. 2009;28:3758–3770. doi: 10.1038/emboj.2009.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rousaki A, Miyata Y, Jinwal UK, Dicky CA, Gestwicki JE, Zuiderweg ER. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 2011;411:614–632. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morishima Y, Lau M, Peng HM, Miyata Y, Gestwicki JE, et al. Heme-dependent activation of neuronal nitric oxide synthase by cytosol is due to an Hsp70-dependent, thioredoxin-mediated thiol-disulfide interchange in the heme/substrate binding cleft. Biochemistry. 2011;50:7146–7156. doi: 10.1021/bi200751t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 50.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 51.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 52.Merry DE, Kobayashi Y, Bailey CK, Taye AA, Fischbeck KH. Cleavage, aggregation and toxicity of the expanded androgen receptor in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 1998;7:693–701. doi: 10.1093/hmg/7.4.693. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Chevalier-Larsen ES, Merry DE, Diamond MI. Soluble androgen receptor oligomers underlie pathology in a mouse model of spinobulbar muscular atrophy. J. Biol. Chem. 2007;282:3157–3164. doi: 10.1074/jbc.M609972200. [DOI] [PubMed] [Google Scholar]
- 54.Adachi H, Katsuno M, Minamiyama M, Waza M, Sang C, et al. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain. 2005;128:659–670. doi: 10.1093/brain/awh381. [DOI] [PubMed] [Google Scholar]
- 55.Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 56.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 57.Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 58.Pratt WB, Galigniana MD, Harrell JM, DeFranco DB. Role of hsp90 and the hsp90-binding immunophilins in signaling protein movement. Cell Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Tokui K, Adachi H, Waza JM, Katsuno M, Minamiyama M, et al. 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum. Mol. Genet. 2009;18:898–910. doi: 10.1093/hmg/ddn419. [DOI] [PubMed] [Google Scholar]
- 60.Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum. Mol. Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 61.Abel A, Walcott J, Woods J, Duda J, Merry DE. Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum. Mol. Genet. 2001;10:107–116. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi Y, Kume A, Li M, Doyu M, Hata M, et al. Chaperones Hsp70 and Hsp40 suppress aggregate formation and apoptosis in cultured neuronal cells expressing truncated androgen receptor protein with expanded polyglutamine tract. J. Biol. Chem. 2000;275:8772–8778. doi: 10.1074/jbc.275.12.8772. [DOI] [PubMed] [Google Scholar]
- 63.Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, et al. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adachi H, Waza M, Takui K, Katsuno M, Minamiyama M, et al. CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J. Neurosci. 2007;27:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, et al. Huntington’s disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 66.Truant R, Atwal RS, Burtnik A. Nucleocycloplasmic trafficking and transcription effects of huntingtin in Huntington’s disease. Prog. Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 67.Gunawardena S, Her LS, Brusch RG, Layman RA, Niesman IR, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 68.Gauthier LR, Charrin BC, Borrell-Pages M, Dumpierre JP, Rangone H, et al. Huntington controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 69.Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caviston JP, Holzbaur ELF. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee WC, Yoshihara M, Littleton JJ. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 73.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 74.Mukai H, Iregawa T, Goyama E, Tanaka S, Bence NF, et al. Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 2005;102:10887–10892. doi: 10.1073/pnas.0409283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 76.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 77.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 78.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J. Biol. Chem. 2012;287:1406–1414. doi: 10.1074/jbc.M111.294801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herbst M, Wanker EE. Small molecule inducers of heat shock response reduce poly Q-mediated huntington aggregation. Neurodegen. Dis. 2007;4:254–260. doi: 10.1159/000101849. [DOI] [PubMed] [Google Scholar]
- 80.Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J. Biol. Chem. 2008;283:26188–26197. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J. Clin. Invest. 2011;121:3306–3319. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl U. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan HYE, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 85.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 86.Hansson O, Nylandsted J, Castilho RF, Leist M, Jaattela M, Brundin P. Overexpression of heat shock protein 70 in R6/2 Huntington’s disease mice has only modest effects on disease progression. Brain Res. 2003;970:47–57. doi: 10.1016/s0006-8993(02)04275-0. [DOI] [PubMed] [Google Scholar]
- 87.Wacker JL, Huang SY, Steela AD, Aron R, Lotz GP, et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington’s disease. J. Neurosci. 2009;29:9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 89.Jankovic J. Parkinson’s disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry. 2007;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 90.Obeso JA, Rodriguez-Oroz MC, Guetz CG, Marin C, Kordower JH, et al. Missing pieces in the Parkinson’s disease puzzle. Nature Med. 2010;16:653–661. doi: 10.1038/nm.2165. [DOI] [PubMed] [Google Scholar]
- 91.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nature Rev. Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 92.Lee H-J, Lee S-J. Characterization of cytoplasmic α-synuclein aggregates. Fibril formation is tightly linked to the inclusion-forming process in cells. J. Biol. Chem. 2002;277:48976–48983. doi: 10.1074/jbc.M208192200. [DOI] [PubMed] [Google Scholar]
- 93.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 94.Bartels T, Chui JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee VM, Trojanowski JQ. Mechanisms of Parkinson’s disease linked to pathological α-synuclein: new targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 96.Gupta A, Dawson VL, Dawson TM. What causes cell death in Parkinson’s disease? Ann. Neurol. 2008;64:S3–S15. doi: 10.1002/ana.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uryu K, Richter-Landsberg C, Welch W, Sun E, Goldbaum O, et al. Convergence of heat shock protein 90 with ubiquitin in filamentous α-synuclein inclusions of α-synucleinopathies. Am. J. Pathol. 2006;168:947–961. doi: 10.2353/ajpath.2006.050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang J, Clark-Dixon C, Wang S, Flower TR, Williams-Hart T, et al. Novel suppressors of α-synuclein toxicity identified using yeast. Hum. Mol. Genet. 2008;17:3784–3795. doi: 10.1093/hmg/ddn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flower TR, Chesnokova LS, Froelich CA, Dixon C, Witt SN. Heat shock prevents alpa-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J. Mol. Biol. 2005;351:1081–1100. doi: 10.1016/j.jmb.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 100.Auluck PK, Meulener MC, Bonini NM. Mechanisms of suppression of α-synuclein neurotixicity by geldanamycin in Drosophila. J. Biol. Chem. 2005;280:2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 101.Putcha P, Danzer KM, Kranich LR, Scott A, Silinski M, et al. Brain-permeable, small-molecule inhibitors of Hsp90 prevent α-synuclein-induced toxicity. J. Pharmacol. Exptl. Ther. 2010;332:849–857. doi: 10.1124/jpet.109.158436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dev KK, Hofele K, Barbieri S, Buchman VL, van der Putten H. Part II: α-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacol. 2003;45:14–44. doi: 10.1016/s0028-3908(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 103.Auluck PK, Chan E, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 104.McLean PJ, Kawamata H, Shariff S, Hewett J, Sharma N, et al. Torsin A and heat shock proteins act as molecular chaperones: suppression of α-synuclein aggregation. J. Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 105.Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, et al. Analysis of α-synuclein-associated proteins by quantitative proteomics. J. Biol. Chem. 2004;279:39155–39164. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- 106.Yu F, Xu HT, Zhuo M, Sun LY, Dong AW, Liu XY. Impairment of redox state and dopamine level induced by α-synuclein aggregation and the preventive effect of hsp70. Biochem. Biophys. Res. Commun. 2005;331:278–284. doi: 10.1016/j.bbrc.2005.03.148. [DOI] [PubMed] [Google Scholar]
- 107.Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, et al. Formation of toxic oligomeric α-synuclein species in living cells. PLoS ONE. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fan GH, Zhou HY, Yang H, Chen SD. Heat shock proteins reduce α-synuclein aggregation induced by MPP+ in SK-N-SH cells. FEBS Lett. 2006;580:3091–3098. doi: 10.1016/j.febslet.2006.04.057. [DOI] [PubMed] [Google Scholar]
- 109.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates α-synuclein degradative decisions between proteasomal and lysosomal pathways. J. Biol. Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 110.Kalia LV, Kalia SK, Chau H, Lozano AM, Hymen BJ, McLeaen PJ. Ubiquitinylation of α-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5) PLoS ONE. 2011;6:e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cummings JL. Alzheimer’s disease. New Eng. J. Med. 2004;351:56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- 112.Burns A, Iliffe S. Alzheimer’s disease. Brit. Med. J. 2009;338:467–471. doi: 10.1136/bmj.b158. [DOI] [PubMed] [Google Scholar]
- 113.Citron M. Alzheimer’s disease: strategies for disease modification. Nat. Rev. Drug Disc. 2010;9:387–398. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- 114.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 115.Mesulam MM. Neuroplasticity failure in Alzheimer’s disease: bridging the gap between plaques and tangles. Neuron. 1999;24:521–529. doi: 10.1016/s0896-6273(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 116.Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 118.Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J. Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, et al. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer’s disease. J. Cell Biol. 1998;143:777–794. doi: 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stamer K, Vogel R, Thies E, Mandelkow E, Mandelkow EM. Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol. 2002;156:1051–1063. doi: 10.1083/jcb.200108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feuillette S, Miguel L, Frebourg T, Campion D, Lecourtois M. Drosophila models of human tauopathies indicate that tau protein toxicity in vivo is mediated by soluble cytosolic phosphorylated forms of the protein. J. Neurochem. 2010;113:895–903. doi: 10.1111/j.1471-4159.2010.06663.x. [DOI] [PubMed] [Google Scholar]
- 123.Roberson ED, Scearce-Lavie K, Palop JJ, Yan F, Cheng LH, et al. Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 124.Dou F, Natzer WJ, Tanemura K, Li F, Hartl FU, et al. Chaperones increase association of tau protein with microtubules. Proc. Natl. Acad. Sci. USA. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo W, Dou F, Rodina A, Chip S, Kim J, et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl. Acad. Sci. USA. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tortosa E, Santa-Maria I, Moreno F, Lim F, Perez M, Avila J. Binding of Hsp90 to tau promotes a conformational change and aggregation of tau potein. J. Alzheimers Dis. 2009;17:319–325. doi: 10.3233/JAD-2009-1049. [DOI] [PubMed] [Google Scholar]
- 127.Dicky CA, Dunmore J, Lu B, Wang JW, Lee WC, et al. HSP induction mediates selective clearance of tau phosphorylated at proline-directed Ser/Thr sites but not KXGS (MARK) sites. FASEB J. 2006;20:753–755. doi: 10.1096/fj.05-5343fje. [DOI] [PubMed] [Google Scholar]
- 128.Dicky CA, Kamal A, Lundgren K, Klosak N, Bailey RM, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 130.Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsp70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 131.Hatakeyama S, Matsumoto M, Kamura T, Murayama M, Chui DH, et al. U-box protein carboxyl terminus of Hsc70-interacting protein (CHIP) mediates poly-ubiquitination preferentially on four-repeat tau and is involved in neurodegeneration of tauopathy. J. Neurochem. 2004;91:299–307. doi: 10.1111/j.1471-4159.2004.02713.x. [DOI] [PubMed] [Google Scholar]
- 132.Dickey CA, Yue M, Lin WL, Dickson DW, Dunmore JH, et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J. Neurosci. 2006;26:6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sahara N, Murayama M, Mizoroki T, Urushitani M, Imai Y, et al. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J. Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 134.Abisambra J, Jinwal UK, Miyata Y, Rogers J, Blair L, et al. Alloteric heat shock protein 70 inhibitors rapidly rescue synaptic plasticity deficits by reducing aberrant tau. Biol. Psychiatry. 74:364–374. doi: 10.1016/j.biopsych.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]