Abstract
The diversity of innate lymphoid cells (ILCs) is rapidly expanding. Three ILC classes have emerged, ILC1, ILC2, and ILC3, with ILC1 and ILC3 including several subsets. The classification of some subsets is unclear and it remains controversial whether NK cells and ILC1 are distinct cell types. To address these issues, we analyzed ILCs and NK cells gene expression within mouse small intestine, spleen, and liver, as part of the Immunological Genome Project. Results identify unique gene-expression patterns for some ILCs and overlapping patterns between ILC1 and NK cells, whereas few ILC subsets remain indistinguishable. A transcriptional program shared by small intestine ILCs and a core ILC signature is identified. Transcripts that suggest novel ILC functions and developmental paths are revealed and discussed.
Keywords: innate lymphoid cell, ILC, NK cell, ImmGen
The Immunological Genome (ImmGen) Project is a collaborative effort of immunologists and computational biologists that seek, through rigorously controlled protocols for data generation and analysis, to dissect gene expression patterns of cell types across the immune system to better understand the immune response and comprehensively define its regulatory networks1. In this context, we interrogated the global gene-expression profiles of innate lymphoid cells (ILC) from mice following ImmGen’s stringent standards1.
ILC are non-T, non-B lymphocytes, present throughout the body and enriched in frequency at mucosal surfaces2,3. Developing from an Id2+ common helper-like ILC progenitor (CHILP)4 three classes of ILCs have emerged that mirror helper T cells in both their cytokine production profiles and transcriptional circuitry, now known as ILC1, ILC2, and ILC32,3. Functionally, T-bet+ ILC1 respond to IL-12, IL-15, and IL-18 to produce interferon γ (IFN-γ); GATA-3+ ILC2 react to IL-33, IL-25, and thymic stromal lymphopoietin (TSLP) to produce type 2 cytokines including IL-5 and IL-13; and Rorγt+ ILC3 are activated by IL-1 and IL-23 to produce IL-22 and/or IL-17. As innate sources of distinct cytokines, ILCs have roles in the early defense against infections, modulation of the adaptive immune response, lymphoid tissue development, and are implicated in tissue repair and homeostasis2,3.
While shared transcription factors (TF), cell-surface markers, and functional properties of cytokine production define classes, deviations in one or more categories by subpopulations define subsets of ILCs within the larger class. In mouse, ILC2 appear to be the most homogenous class thus far, defined by expression of the IL-7 receptor (IL-7R, CD127), Sca-1, and the IL-33 receptor ST2. In comparison, at least five subsets of mouse ILC3 have been reported, four of which are found in highest numbers during steady-state in the adult small intestine and one in the adult large intestine. These include CD4+ and a CD4− subsets of Nkp46− Rorγt+ lymphoid tissue inducer-like (LTi-like) cells5; Rorγt+, T-bet+, Notch-dependent Nkp46+ ILC35–9; a potential ILC3→ILC1 transitional subset with downregulated Rorγt, which expresses T-bet and higher amounts of NK1.1 and produces IFN-γ (‘ex-RORγt+ ILC3’)4; and IL17-producing Rorγt+ Nkp46− ILC3 in the large intestine10.
ILC1 have the most complicated and controversial distinction of both the ILC class itself and between subsets within the class. ILC1 and NK cells have functional similarities, primarily IFN-γ production, and share expression of T-bet and many cell-surface markers, such as NKp46 and NK1.1. However, NK cells are currently thought to have more cytotoxic potential than ILC1. IL-7R and CD49a surface expression are used to distinguish ILC1 from NK cells in many but not all mouse tissues, as there is considerable diversity of ILC1 subsets between tissues2. Lineage-tracing experiments have shown that ILC1 and NK cells originate from distinct progenitors4,11, and mature NK cells are dependent on the transcription factor Eomesodermin (Eomes) while ILC1 are not. However, immature NK cells also share many ILC1 markers12 and lack expression of Eomes12,13.
The breadth of polarization and relationship among ILC subsets remain incompletely understood. To better understand the functional differences between ILC classes and reported subsets within a class, we discriminated 7 populations of ILCs in the small intestinal lamina propria (siLP) (NK cells, ILC1, ILC2, and 4 subsets of ILC3), 3 additional subsets of ILC1 from liver and spleen, and small intestinal intraepithelial lymphocytes (siIEL), and 2 NK cell subsets from liver and spleen. Our findings provide a molecular definition of ILC classes and subsets, as well as identify a core signature in ILCs compared to NK cells. We also identify novel targets for future investigation and generate a comprehensive high quality and publically available resource of ILC transcriptomes.
Results
Analysis of ILC frequency and diversity
We isolated all reported ILC subsets in the siLP from 6-week-old C57BL/6 male mice. ILC2 were isolated from wild-type C56BL/6 mice and all other subsets were isolated using RorγteGFP reporter mice. These subsets included CD127–NK cells, CD127+ ILC1, CD4− and CD4+ NKp46− LTi-like ILC3, NKp46+ Rorγthi ILC3, and NKp46+ Rorγtlo ILC3 (Fig. 1a). Notably, NKp46+ Rorγthi and Rorγtlo ILC3 cannot be discriminated by intracellular Rorγt staining in wild-type C56BL/6 mice, in which only one NKp46+ Rorγt subset is detectable; thus, the use of RorγteGFP reporter mice provided the unique opportunity to profile Rorγtlo and Rorγthi subsets, as previously described14. As the Rorγtlo ILC3 subset expresses higher amounts of NK1.1 compared to Rorγthi ILC3 (Supplemental Fig 1a–c), we reasoned this would be enriched for converted or actively converting ‘ex-RORγt+ ILC3’. We also profiled CD49b+TRAIL− NK cells and CD49b−TRAIL+ ILC1 in the liver, and CD127− NK cells from CD127+ ILC1 in the spleen, the last of which has been reported but has not previously been called ILC115. Small intestine intraepithelial (siIEL) ILC1 were isolated from the intestinal epithelium as NK1.1+NKp46+. They are phenotypically distinct from NK cells due to TGF-β imprinting16, although their developmental origin and transcriptional relationship to siLP ILC subsets remains unclear.
Figure 1. Analysis of ILC frequency and diversity.
(a) Sorting strategy for array analysis after gating on live CD45+ and CD3−CD19− cells. (b) Cytospins of cells using the indicated sorting strategy for each population. Each individual panel is 14.6μm in length. (c) Flow cytometry analysis of ILC percentage in each tissue among the lymphocyte population. To differentiate CD3+ T cells, CD19+ B cells, and ILCs within the same sample, the same gating strategy in (a) was used except: the siLP ILC2 marker KLRG1 and spleen ILC1 marker CD27 were excluded and liver ILCs were distinguished using NKp46, CD49b, and CD49a. Data from two independent experiments and n = 2–4 mice per tissue are shown. (d) PCA of gene expression by ILC and NK cells subsets. Numbers in parentheses indicate relative scaling of the principal variables. ImmGen sample nomenclature is used: spleen NK = NK.CD127−.Sp, liver NK = NK.CD49b+.Lv, siLP NK = NK.CD127−.SI, siIEL ILC1 = ILC1.IEL.SI, spleen ILC1 = ILC1.CD127+.Sp, liver ILC1 = ILC1.CD49b−.Lv, siLP ILC1 = ILC1.CD127+.SI, siLP NKp46+ Rorγtlo ILC3 = ILC3.NKp46+.Rorγtlo.SI, siLP NKp46+ Rorγthi ILC3 = ILC3.NKp46+.Rorγthi.SI, siLP NKp46− CD4− LTi-like ILC3 = ILC3.NKp46−.4−.SI, siLP NKp46− CD4+ LTi-like ILC3 = ILC3.NKp46−.4+.SI, and siLP ILC2 = ILC2.SI. (e) Hierarchical clustering of ILC and NK cells subsets based on the 10% of genes with the greatest variability. Data are combined from three to seven experiments per sample, with cells pooled from three to fifteen mice each.
Cytospins demonstrated that ILC subsets were morphologically pure lymphoid populations (Fig. 1b). We assessed ILC frequency with regard to these 12 subsets within the lymphocyte population of small intestine, liver, and spleen in naive mice (Fig. 1c). ILCs were most abundant in the siLP, followed by liver and spleen; siIEL contained the lowest frequency of ILCs. Next, we sorted these populations following ImmGen’s standardized protocol for data generation and analyzed gene expression using whole mouse genome array. A principal component analysis showed a greater degree of diversity generated by ILC2 and ILC3 subsets than ILC1 (Fig. 1d and Supplemental Fig. 1e). By hierarchical clustering, three pairs of ILC subsets were computationally indistinguishable (Fig. 1e): splenic and liver NK cells, Rorγthi and Rorγtlo ILC3, and CD4+ and CD4− NKp46–ILC3 were intermixed. All ILC1 subsets clustered separately from NK cells from the same tissue. However, siIEL ILC1, siLP NK cells, and siLP ILC1 clustered in a separate branch of the dendrogram from liver and spleen NK cells and ILC1 subsets. We conclude that beyond classical polarizations, there are environmental factors in the small intestine that differentiate subsets from liver and spleen.
Unique transcripts of individual ILC subsets
We assessed the unique gene-expression profile of each sorted subset (Fig. 2a–h). We found the greatest number of unique transcripts in the siLP ILC2, with 100 transcripts expressed more than 2-fold (Supplemental Table 1) and 34 transcripts expressed more than 4-fold higher compared to other subsets (Fig. 2a). These included expected transcripts such as Il13, Il5, IL4, Il9r, and the IL-25 receptor Il17rb,17–19 as well as several novel transcripts, including Rxrg, Pparg, Mc5r, Dgat2, and Alox5. The transcriptional repressor retinoid X receptor gamma (RXRG), which binds the Vitamin A metabolite 9-cis retinoic acid,20 has not been previously described in ILC2. Vitamin A is known to directly inhibit ILC2 differentiation by an unknown mechanism,21 suggesting RXRG may mediate this effect. We also find previously unrecognized expression of peroxisome proliferator-activated receptor gamma (PPARγ), which forms heterodimers with RXRs and transcriptionally mediates lipid homeostasis.20 ILC2 expressed several other genes implicated in lipid metabolism. These include DGAT2, which mediates the final reaction of triglyceride synthesis, MC5R, a receptor that causes lipid mobilization in adipocytes, and ALOX5, a lipoxygenase that catalyzes leukotriene A4 synthesis. While ILC2 are known to regulate immune cell function in visceral adipose tissues,22 our data suggest that ILC2 may also directly sense lipids as well as produce lipid mediators.
Figure 2. Unique transcripts of individual ILC subsets.
(a–i; and full gene lists in Supplemental Table 1) mRNA transcripts upregulated more than 4-fold (red font) or 2-fold (black font) in individual subsets (a,d,e,f,g), two similar subsets (b,c,h), or all subsets from siLP (i) were generated based on pairwise comparisons between the index subset(s) labeled in bold above the heat map compared to all other subsets. Between similar ILC3 subsets consisting of NKp46− CD4+ LTi-like and CD4− LTi-like (b) or NKp46+ Rorγthi and Rorγtlo(c) cells, some transcripts were expressed more than 4-fold (b) or 2-fold (c) higher in one subset, but not the other, in which case shared transcripts expressed by both subsets are indicated in blue. CD4+ LTi-like ILC3 expressed 4 transcripts 2-fold more than CD4− LTi-like ILC3 and 4-fold more than all other subsets, shown in gray (b).
NKp46− LTi-like ILC3 subsets expressed the second greatest number of unique transcripts. As suggested by clustering analysis (Fig. 1e), CD4+ and CD4− LTi-like had overlapping gene expression patterns. Only 4 transcripts were 2-fold uniquely upregulated in CD4+ LTi-like cells, one of which was CD4, and none were uniquely expressed in CD4− LTi-like (Fig. 2b). However, 65 transcripts were expressed by both subsets more than 2-fold higher (Supplemental Table 1) and 9 transcripts expressed more than 4-fold higher compared to other subsets (Fig. 2b). Eight additional transcripts were upregulated at least 4-fold by either CD4+ LTi-like or CD4− LTi-like ILC3, with differences in expression between subsets likely due to replicate variation (Fig. 2b). Unique transcripts shared by both LTi-like subsets included those encoding the chemokine receptor CCR6, which has previously been used as a marker for LTi-like ILC3,7 and CXCR5. We also find several novel genes, including Gucy1a3, Cntn1, Slc6a7, Cacna1g, and Nrp1. Gucy1a3 encodes the alpha subunit of the soluble guanylate cyclase receptor, which transduces signals from nitric oxide; however, we found no expression in LTi-like ILC3 of the other gunaylate cyclase receptor components (data not shown). Interestingly, immune expression of CNTN1, a GPI-linked member of the immunoglobulin family best known for its role in regulating axonal guidance and neural system development was also detected. The L-proline transporter SLC6A7 and voltage gated calcium channel CACNA1G are similarly atypical, and not expressed in other immune cells (www.immgen.org). We conclude LTi-like ILC3 specifically express several unique transcripts compared with the entire immune system, including putative factors involved in neural crosstalk.
The remaining ILC subsets had less candidate unique markers, likely due to multiple comparisons with other subsets in the same class. As indicated by PCA, siLP NKp46+ Rorγthi ILC3 and siLP NKp46+ Rorγtlo ILC3 had overlapping gene expression profiles. When compared to all other profiled ILCs except NKp46+ Rorγtlo ILC3, 16 transcripts were expressed 2-fold higher in NKp46+ Rorγthi ILC3, though a heat map reveals that most of these genes are also expressed by other siLP subsets at lower levels (Fig. 2c). Liver and splenic ILC1 each expressed some unique transcripts with greater than 2-fold change compared to all other subsets (Fig. 2d–e), but we found no unique transcripts expressed by siLP ILC1. This suggests there are few unique factors expressed by ILC1 subsets among all ILCs and NK cells. Surprisingly, the only unique transcript expressed by IEL ILC1 was Itgae, which encodes CD103 (Fig. 2f). CD103 protein is expressed in human IEL ILC1,16 but was not previously known to be expressed by mouse IEL ILC1, which lack surface protein CD103 expression, suggesting post-transcriptional regulation in the IEL. Focusing on NK cells, we identified 4 genes that were expressed 2-fold higher in siLP NK cells than in all ILCs (Fig. 2g). Splenic and liver NK cells expressed no unique transcripts. However, when both liver and spleen NK cells were compared as a group to all other subsets, 25 transcripts were 2-fold more highly expressed, although siLP NK, IEL ILC1, and splenic ILC1 show lower expression of these genes (Fig. 2h). We conclude that ILC2 and LTi-like ILC3 mRNA profiles are quite unique, while NKp46+ ILC3, ILC1, and NK cells profiles show considerable overlap.
A transcriptional signature shared by all siLP subsets
We next asked whether there are any uniquely expressed transcripts between profiled tissues of siLP, liver, and spleen. Given that siLP NK cells and ILC1 cluster further from liver and splenic subsets by hierarchical clustering and PCA, we focused on the siLP (Fig. 1d–e) Pairwise comparisons of all subsets from the siLP to remaining subsets from liver and spleen revealed that all siLP subsets expressed a core 35-transcript signature (Fig. 2i), which included several transcription factors such as Rora, Atf3, Nr4a1, Maff, Epas1, Bhlhe40, and Per1. Furthermore, all siLP-resident ILCs expressed high transcript levels of the activation marker Cd69 and varied levels of Csf2, which encodes GM-CSF. Although production of GM-CSF by ILC2,18 ILC3,23,24 and NK cells is known,25 production by ILC1 has not been reported. Thus, ILC subsets in the siLP appear to be more activated than ILCs in other tissues, likely reflecting their constant exposure to varied environmental signals from the microbiome and incoming nutrients, including well-documented transcriptional activators such as Vitamin A21,26 and AhR ligands6,27.
TF, cytokines, and chemokines of ILC subsets
To address broad patterns of gene expression between ILC subsets and classes, we interrogated expression of previously reported and novel transcription factors (TFs), chemokines, cytokines, and other secreted factors. The TFs expressed at the highest relative levels included well-documented ILC-defining Id2, ILC3 class-defining Rorc, ILC1 and NKp46+ ILC3-defining Tbx21 and NK cells-defining Eomes (Fig 3a). ILC2-defining TFs Gata3 and Rora were enriched in ILC2 but also expressed in all ILCs, consistent with an early role in ILC development, at least for GATA-3.19 Nfil3, which is required for NK cells28,29, ILC3, and ILC230,31 development, was expressed most highly in siLP subsets; NKp46+ ILC3, ILC1, and NK cells showed highest transcript levels, followed by LTi-like ILC3 and ILC2, and much lower transcript levels were present in the liver and spleen ILC1 and NK cells. Furthermore, TFs identified in the siLP signature Atf3 and Nr4a1 (Fig. 2i), were expressed at levels similar to lineage-defining TFs. Collectively, these data suggest a marked role of the intestinal microenvironment in the expression of certain TF, which subsequently can have diverging roles between ILC classes. Analysis of chemokines and chemokine receptors (Fig. 3 b), as well as cytokines and cytokine receptors (Fig. 3c), revealed shared and distinct expression patterns. Beyond known signature cytokine and chemokine circuitries, we identified a novel candidate feed-forward loop for ILC2, which expressed both CCR8 and its ligand CCL1. We also identified the novel ILC2 expression of Bmp7 and Bmp2, the latter of which is known to modulate intestinal peristalsis by binding the BMP receptor on enteric neurons32. Interestingly, we found Il2 expression in several ILC populations, which suggests that ILCs may be able to activate T cells or other ILCs through IL2R signaling.
Figure 3. Spectrum of unique and shared transcriptional profiles between siILC subsets.
(a) Heat map of selected transcription factors, normalized by expression within the entire heat map. (b–c) Heat map of selected chemokine receptors and ligands (b) and cytokine receptors and ligands (c), normalized by expression within each row. (d; and full gene lists in Supplemental Table 2) Among selected siLP ILCs, transcripts upregulated by a single subset or 2 subsets are indicated, as described in the color-coded key above the plot. Numbers within the plot indicate transcripts upregulated at least by a 2-fold change in the indicated color-coded subset(s). (e) Volcano plot of Rorγthi NKp46+ ILC3 (n = 3 replicates) compared to ILC1 (n = 2 replicates), with transcripts significantly upregulated at least 2-fold quantified in corners. P ≤ 0.05 (t-test).
Shared transcriptional profiles between siILC subsets
We next focused our analysis of transcriptional profiles to the four major CD127+ ILC subsets within the siLP: ILC1, ILC2, NKp46+ Rorγthi ILC3, and CD4− LTi-like ILC3 (Fig 3d). Comparison of siLP ILC subsets revealed that ILC subsets had overlapping patterns of gene expression that were not identified in unique signatures (Supplemental Table 2). For example, ILC2 and LTi-like ILC3 shared 17 transcripts including Arg1 and Ret. Arginase-1, encoded by Arg1, which marks fetal and adult ILCs and facilitates the identification of developing ILCs in the siLP.33 The receptor tyrosine kinase Ret is also expressed by fetal CD11c+ lymphoid tissue initiator (LTin) cells and is required for Peyer’s patch development34, but has not been previously reported to be expressed by fetal or adult ILCs, including LTi or LTi-like ILC3. Together, these results suggest that, at least in fetal mice, ILC2 and LTi-like ILC3 share a common progenitor, 33 though their functional relevance in adult siLP ILCs remains to be investigated. ILC1 and ILC2 shared 19 transcripts, including Ets2. Ets-1 and Ets-2 interact with Id proteins, and while Ets-1 has been implicated in early NK cell development35, Ets-2 has not. Thus, Ets-2 may be relevant in ILC1 and ILC2 development or maintenance.
NKp46+ and LTi-like ILC3 have long been recognized to share many characteristics due to mutual production of IL-22 and expression of Rorγt. Yet NKp46+ ILC3 shared an even greater number of transcripts with NKp46+ ILC1 than with LTi-like ILC3 (Fig. 3d, Supplemental Table 2). These comprised Tbx21, Ifng, and Il12rb transcripts. While T-bet is required for NKp46+ ILC3 development,8–10 these cells produce little IFN-γ protein in response to IL-12 and IL-23 (data not shown). Because IFN-γ is well documented to be post-transcriptionally regulated38, finding the Ifng transcript in Nkp46+ ILC3 suggests that T-bet may be sufficient to induce transcription, but other factors are needed for protein production, such as bacterial infections in vivo4. In addition to shared transcripts, NKp46+ ILC3 and NKp46+ ILC1 showed 213 genes differentially expressed, with the NKp46+ ILC3 upregulated genes including many genes shared with LTi-like ILC3 (Fig. 3e). Thus, NKp46+ ILC3 have a transcriptional profile with intermediate characteristics between those of NKp46− LTi-like ILC3 and NKp46+ ILC1, which may enable functional plasticity. Their functional polarization towards ILC3 or ILC1 most likely depends on the tissue microenvironment.
Although a pairwise comparison failed to identify unique siLP ILC1 transcripts, when compared to other ILC subsets within the siLP, ILC1 demonstrated higher cytotoxic capacity, as evidenced by expression of Gzma and Prf1, encoding granzyme A and perforin. It is possible this is due to imperfect distinction between siLP ILC1 and NK cells (see below). ILC1 also strongly expressed Il21r, a member of the common gamma chain cytokine family (Fig. 3d). When an additional comparison between ILC1 and NK cells in the siLP was included, ILC1 expressed only 4 unique transcripts, including Gpr55, Trat1, Mmp9, and Cpne7 (Supplemental Table 2). Thus, while siILC1 are more NK-like than other siILC subsets, they show no obvious unique markers when NK cells are included in the comparisons using our sorting strategy.
Defining novel transcripts within ILC3
Pairwise comparison of all ILC3 subsets versus other profiled cells revealed a 42-transcript ILC3 signature (Fig. 4a). This signature included well-studied transcripts such as Il23r, Rorc, and Il22, as well as several novel molecules such as the highly and quite specifically expressed retinoic acid target and JNK activator, Pram1. This result supports a role for retinoic acid in ILC3 development21,26. Remarkably, Il17 was not among the transcripts with higher than 2-fold expression in ILC3 than in other ILCs, nor was it expressed uniquely by any individual ILC3 subset (Fig. 2b–c). This result suggests that IL-17 is not a major ILC3 product in the small intestine, at least in adult mice at steady-state.
Figure 4. ILC3-specific transcriptional programs and cell surface markers.
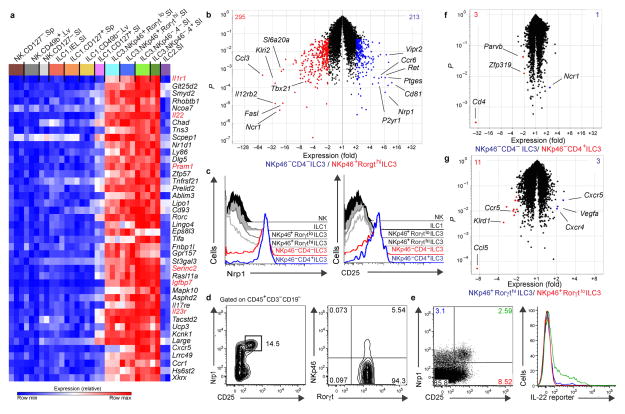
(a) Heat map of mRNA transcripts upregulated more than 4-fold (red font) or 2-fold (black font) in Rorγt+ ILC3 subsets compared to all other subsets. (b; and full gene lists in Supplemental Table 3) Volcano plots comparing NKp46+ Rorγthi ILC3 to CD4− LTi-like ILC3 with transcripts significantly upregulated at least 2-fold quantified in corners and color-coded according to key below plot. P ≤ .05 (t-test) (n = 3 replicates each) (c) Expression of Nrp1 and CD25 compared to other siLP ILC subsets from the RorγteGFP reporter mice. (d) Unbiased selection of Nrp1+ CD25+ cells gating only on CD45+ and CD3−CD19− cells, with percent enrichment of LTi-like in by intracellular Rorγt staining from C56BL/6 wt mice. (e) Enrichment of IL-22 production from naive IL-22 reporter mice in Nrp1+CD25+ cells compared to other quadrants, gating on CD45+, CD3−CD19−, and NKp46− cells. (f–g; Supplemental Table 3) Volcano plots comparing CD4+ LTi-like (n = 2 replicates) to CD4− LTi-like ILC3 (n = 3 replicates) (f) and NKp46+ Rorγthi ILC3 to NKp46+ Rorγtlo ILC3 (n = 3 each) (g and Supplemental Table 3). P ≤ .05 (t-test). Data are representative of 3 experiments.
Comparison of the major adult LTi-like population, CD4− LTi-like ILC3, and NKp46+ Rorγthi ILC3 revealed 508 total significant differences greater than 2-fold (Fig. 4b, Supplemental Table 3). Among the transcripts most highly and most significantly upregulated by CD4− LTi-like ILC3, we found Nrp1. Also identified as part of our LTi-like ILC3 signature (Fig. 2b), Nrp1 is a co-receptor for several ligands, including VEGF, TGFβ1, and semaphorins, and may have functional roles in negatively regulating the immune response in part through enhanced Treg cell survival37. It is also one of a few markers that differentiate natural Treg cells from peripherally generated mucosal derived iTreg cells38,39. However, Nrp1 has never previously been reported to be expressed by LTi-like ILC3. We confirmed by flow cytometry that Nrp1 is expressed at higher amounts on LTi-like ILC3 than other siLP subsets (Fig. 4c). We also found CD25 protein expression is higher on LTi-like ILC3 than other siLP subsets (Fig. 4c). Gating on Nrp1+ CD25+ cells within CD3− CD19− siLP lymphocytes results in a cell population highly enriched for LTi-like ILC3 (Fig. 4d) and IL-22 production from naïve IL-22 reporter mice40 (Fig. 4e). Between LTi-like ILC3, distinctions between CD4+ versus CD4− subsets were not robust and unlikely to have functional significance (Fig 4f), although they may reflect different developmental lineages5,9. Similarly, comparison of siLP NKp46+ Rorγtlo ILC3 to siLP NKp46+ Rorγthi ILC3 revealed substantial overlap with few differences (Fig. 4g, Supplemental Table 3). Because siLP Rorγtlo ILC3 expressed more NK cells-like transcripts such as Ccl5 and Klrd1 than siLP Rorγthi ILC3, the siLP Rorγtlo ILC3 may include a minor ILC3 population ‘converting’ into ‘ex-RORγt+ ILC3’. Robust identification of ‘converting’ ILC3 can be established only using fate mapping experiments, as recently described.4
Transcriptional differences between ILC1 and NK cells
One area that has been particularly controversial is the difference between ILC1 and NK cells, in part due to a lack of unique and consistently expressed markers between NK cells and ILC1 in different organs. As such, we used different sorting strategies in each tissue to discriminate between ILC1 and NK cells, consistent with what has been reported previously2,41. We achieved the best separation of subsets in the liver and the worst in the siLP (Fig. 5a). Comparisons of NK cells to ILC1 in liver, spleen, and siLP reflected the degree of separation between populations during sorting, with the most number of significant differences in the liver and the least in the siLP. ILC1 replicates clustered together and were distinct from NK cells in liver, spleen, and siLP (Fig. 5b, Supplemental table 4). As expected, Eomes expression was significantly different with greater than 2-fold expression between NK cells and ILC1 in all analyzed tissues. Levels of transcripts encoding proteins of cytotoxic machinery were generally higher in NK cells than in ILC1. However, in the siLP, perforin transcripts were only 1.7-fold increased compared to ILC1, while in the liver only granzyme K was differentially expressed between NK cells and ILC1. In fact, granzyme A and C were expressed more highly in liver ILC1 than in liver NK, consistent with the reported cytolytic activity of liver ILC142. Liver and spleen ILC1 selectively expressed transcripts for known cell surface markers. In liver ILC1, these transcripts included Itga1 and Tnfrsf10 encoding Cd49a and TRAIL, respectively, which have been used for separating liver ILC1 from NK cells39. Splenic ILC1 highly expressed IL2ra and IL7r, which encode cytokine receptors that collectively enable this population to escape Treg cell regulation15. The most highly and significantly expressed transcripts in siLP ILC1 compared to siLP NK included only previously uncharacterized transcripts such as Tmem64, Npas2, and Lmo4. However, these transcripts were expressed in other siILC, and therefore are unlikely to provide useful markers. A comparison between the splenic and liver ILC1 subsets revealed that splenic ILC1 expressed markers of immature and mature NK cells, including CXCR4, c-Kit, and Eomes13,41 (Fig. 5c). These differences in transcription could be explained by the finding that our sorting strategy based on CD127 and CD27 included an Eomes+ subset within the splenic ILC1 (Fig. 5d). We also noted that the siLP NK cells subset identified using cell surface markers contained a large population of Eomes− cells. Thus, NK cells and ILC1 cannot be discriminated by CD127 alone and/or CD27 in the spleen and siLP. Use of EomeseGFP reporter mice may be useful in the future to better discriminate NK cells from ILC1, as recently described.4,41
Figure 5. Transcripts differentially expressed between NK cells and ILC1.
(a) Gating strategy of liver, spleen, and siLP ILC1 and NK cells after the first round of sorting from pooled samples. (b–c; and full gene lists in Supplemental Table 4) Volcano plots comparing liver ILC1 to liver NK cells (n = 3 replicates each), spleen ILC1 to spleen NK cells, siLP ILC1 to siLP NK cells (b), and liver ILC1 to spleen ILC1 (c), with transcripts significantly upregulated at least 2-fold quantified in corners and color-coded according to the key below plot. P ≤ .05 (t-test). (d) Expression of CD127 and Eomes in ILC1 and NK cells compared to isotype-matched control Ig staining from liver, spleen, and siLP, color-coded according to the key on the left. Histograms are representative of 2 experiments.
A core, NK-distinct, ILC signature
If ILC1 are different from NK cells as a class, we reasoned there should be shared transcripts between ILC1 from all tissues compared to NK cells and visa versa (Fig. 6a). Spleen and liver ILC1 shared expression of reported ILC1 genes, such as Tnfsf10, Tnf, and Il2, although Il2 was filtered out following our methods due to variability between replicates (Supplemental Table 5). Meanwhile, NK cells transcripts were enriched for Eomes, Itgam (encoding CD11b), and members of the Klra family of genes (encoding Ly49 receptors) (Supplemental Table 5). This NK specific signature is consistent with that previously identified in a more limited dataset by comparing NK cells to ILC1 and ‘ex-RORγt+ ILC3’ within the siLP4. Visualization of genes differentially expressed in liver, spleen, and siLP in the entire dataset revealed that with few exceptions, genes upregulated in ILC1 over NK cells were even more highly expressed in many other ILC subsets (Fig 6b). Genes upregulated in all NK cells were consistently not expressed in other ILCs (Fig 6b), except for low transcript levels in ILC1. Unexpectedly, the transcripts most highly expressed by ILCs compared to NK cells (Fig. 6a) were Tcrg-V3, Tmem176a, Tmem176b, Il7r, and CXCR6 (Fig. 6b, Supplemental Table 6). We validated by flow cytometry that the TCRδ chains were not expressed in any of our cell types, either extracellularly or intracellularly (Fig. 6c; Supplemental Fig. 2a), suggesting that they lacked a functional TCRγδ receptor. Furthermore, from a published RNAseq dataset that included liver ILC1, liver NK cells, and spleen NK cells from Rag1−/− mice42, we found high levels of the 3′ end of the TCRγ locus, annotated as Tcrg C4 transcript, present in liver ILC1 but not NK cells from liver or spleen (Fig. 6d). We finally confirmed that TCRγ–V3 transcript was a germline transcript by PCR (data not shown). As STAT5-activating cytokines IL-7 and IL-15 are well known to mediate germline TCRγ expression,43–44 we conclude that ILCs have open chromatin at the Tcrγ locus and germline transcription, likely due to signaling through IL-7. We also assessed the ability to use CXCR6 as an ILC marker using a CXCR6eGFP reporter mouse. Although we found statistically significant differences between ILC1 and NK cells in the percentage of CXCR6+ cells, it failed to label all ILCs (Fig. 6e–f; Supplemental Fig. 2b). Additionally, in steady state, CXCR6 was not required for ILC development or tissue homing, as we found no difference in ILC frequency between CXCR6eGFP/+ and CXCR6eGFP/eGFP mice (Fig. 6g).
Figure 6. Generation of a core ILC signature distinct from NK cells.
(a) Visualization of transcripts differentially expressed by ILC1 compared NK cells from different tissues. Transcripts upregulated by 2 or 3 subsets are indicated, as described in the color-coded key above the plot. Numbers within the plot indicate transcripts significantly expressed with at least a 2-fold change between liver and spleen ILC1 and NK cells (n = 3 replicates each) or with an additional filter for at least a 2-fold change between siLP ILC1 and NK cells (n = 2 replicates each). P ≤ .05 (t-test). (b; and full gene lists in Supplemental Table 5–6) Heat map showing gene signatures generated and color-coded in (a); transcripts upregulated ≥ 4-fold in all 3 comparisons between ILC1 and NK cells are shown in red. (c–h): ILCs were discriminated as in Fig. 1c, except spleen was positively selected using anti-CD49b-coated beads and Rorγt was stained intracellularly in siLP samples (c; and Supplemental Figure 2). Representative plots from ILCs and positive control IEL γδ T cells stained extracellularly or intracellularly for TCRδ by flow cytometry; for intracellular stain, PE-conjugated anti-TCRδ mAb was used to stain positive control IEL extracellularly before intracellular staining with FITC-conjugated anti-TCRδ (n = 2 independent experiments). Color-coded gates demonstrate percentage of indicated subsets positive for anti-TCRδ, except for control IEL γδ cells, which are shown as a percentage of CD45+ cells. (d) RNAseq data from a previous dataset (GEO accession GSE52043), selecting the 3′ end of the TcRγ locus between 19426 kb and 19446 kb on Chromosome 13. (e) Quantification of percentage of CXCR6+ cells within each ILC subset from CXCR6eGFP/+ reporter mice. (1-way ANOVA with multiple comparisons; error bars mean +/− SEM; * P ≤ .05). (f) Quantification of ILC frequency in CXCR6eGFP/+ and CXCR6eGFP/eGFP mice (2-way ANOVA with multiple comparisons; error bars mean +/− SEM; all comparisons are not significant). (e–f) Data are from 3–4 mice from each genotype per tissue. (g) Visualization of transcripts upregulated between siLP ILC1, siLP NK, and siIEL ILC1 by 1 or 2 subsets are indicated, as described in the color-coded key above the plot. (n = 2–3 replicates each).
Finally, we asked whether intestinal intraepithelial ILC1 should be classified as an ILC1 or NK cell population, as this population has unique transcription factors and cell surface markers, preventing it from fitting clearly into an ILC1 or NK cell designation16. Comparisons between IEL ILC1, siLP ILC1, and siLP NK cells revealed that IEL ILC1 expressed transcripts characteristic of both NK cell and ILC1 populations. While NK-signature transcripts Eomes and Klra3 (Fig. 6b) were more highly expressed by IEL ILC1 than in siLP ILC1, ILC1-signature transcripts Tcrg-V2 and Tcrg-V3 (Fig. 6b) were more abundant in IEL ILC1 than in siLP NK (Fig. 6g). It remains unclear whether IEL ILC1 are a single unique subset with distinct developmental and functional characteristics, or whether siLP NK cells and ILC1 both traffic to the epithelium, where they become phenotypically indistinguishable, possibly in response to tissue factors in the epithelium.
Discussion
Here we have provided the first comprehensive transcriptional analysis of the spectrum of ILC subsets reported in siLP, liver, and spleen. The transcriptional programs we found will allow a better definition of the individual ILC classes, as well as ILC subsets, within a class. ILC2 was the most homogeneous and distinguishable ILC class, which expressed the most unique genes, with well-documented factors as well as novel genes including the nuclear receptors Rxrg and Pparg, and several others involved in lipid metabolism. Within ILC3, LTi-like ILC3 expressed several unique genes, previously unknown to be expressed in immune cells, including Cntn1, Slc6a7, Cacna1g. They were distinct from NKp46+ ILC3 and were effectively marked by the previously unreported LTi-like factor Nrp1, as well as CD25. The definition of ILC1 was the most problematic, because compared to all ILCs and NK cells, we found no unique markers for siLP ILC1 and few for liver and spleen ILC1. CD103 was a surprising unique transcript for siIEL ILC1, given that it was not found as a protein in this subset in mouse but only in human16. Among siLP subsets, we also found a previously unrecognized “intestinal” gene signature composed of activation markers like Cd69 and many transcription factors including Rora, Atf3, Nr4a1, and Maff, which likely reflects continuous environmental exposures.
Besides unique factors, ILC classes also shared many transcripts. For example, siLP NKp46+Rorγt+ ILC3 shared many transcripts with siLP ILC1, including Ifng and Il12rb2. These data, which are consistent with previous reports,4,14 provide a basis for the functional plasticity of siLP NKp46+Rorγt+ ILC3, which become similar to ILC1 in certain conditions yet to be defined. Furthermore, ILC1 and ILC2 shared expression of the TF Ets-2, which suggest new transcriptional pathways shared by ILC1 and ILC2. Interestingly, ILC2 and LTi-like ILC3 expressed genes that suggest a function in neural and glial crosstalk. ILC2 expressed Bmp2, which was recently found to modulate enteric motility in response to the microbiota32; given the importance of motility in clearing helminth infections, this observation might reveal a novel mechanism of innate defense. We discovered that both ILC2 and LTi-like ILC3 also expressed Ret, encoding a receptor for the glial derived neurotrophic family (GDNF) of molecules, known to drive Peyer’s patch development36, but unstudied in ILCs. Thus, in the siLP, ILCs may engage in crosstalk with neurons and glia during steady-state and an immune response.
We generated a core ILC signature that includes 17 genes, with highest expression of TCRγ germline transcripts, Cxcr6, Tmem176a, Tmem176b, and Il7r. Surprisingly, the most highly expressed gene by all ILCs compared to NK cells was TCRγ-V3 germline transcript, which has not previously been reported in ILCs but was recently reported in a putative ILC3 cell line.45 We postulate that this reflects signaling by IL7R, which is expressed by all ILCs but not by mature NK cells43. In our study, we validated that all ILCs express CXCR6 at higher amounts than NK cells. Interestingly, two recent studies interrogated CXCR6 expression in ILCs. The first study demonstrated that a CXCR6+ early progenitor gave rise to both NK cells and ILC, but not T cells46. The second study found that CXCR6 deficiency preferentially affects NKp46+ ILC3 frequency and function by preventing appropriate interactions with CD11b+ intestinal dendritic cells47. However, we found that CXCR6 neither marks the entire population nor were any ILC frequencies affected by its loss, though it is possible that CXCR6 loss affects ILC function, which should be tested across ILC classes. Notably, Tmem176b was previously reported to be a marker of innate lymphocytes as one of only 3 transcripts shared between NK cells, NKT cells, and gamma delta T cells48, though in our dataset, we find that ILC1, ILC2, and ILC3 express significantly higher levels of Tmem176b than do NK cells. These data suggest that the core ILC signature identified here may also extend to other innate and/or tissue resident lymphoid cells.
IFN-γ-producing ILC1 and NK cells can develop from different progenitors, and are independent or dependent of Eomes, respectively 4,11. However, in tissues they show overlapping phenotypes and functional programs. While NK cells are well known to have cytolytic capacity, liver ILC1 express granzyme A and C and were also shown to be cytolytic49. While liver ILC1 were identified by TRAIL and CD49a, other cell surface markers such as CD127 and CD27 were insufficient to discriminate Eomes− ILC1 from Eomes+ NK cells in siLP and spleen. Thus, transcriptional data generated using Eomes reporter mice will be a useful comparison to our dataset.4,41 Moreover, the induction of cytokines like IL-15 and/or IL-2 in certain pathologic conditions may further increase the phenotypic and functional similarity of ILC1 with NK cells. Thus, it remains unclear whether ILC1 and NK cells are truly distinct lineages or a spectrum of cells within the same lineage, which includes ILC1, immature NK cells and mature NK cells.
Our data offers the most complete transcriptional profile of ILCs and NK cells to date and provides a comprehensive view of the relationship between ILC subsets in the steady state. Our findings will help define new avenues of research and should aid in the production of new tools to study ILCs, especially with the identification of several molecular targets expressed at high levels among all ILCs. Finally, they will also be a valuable resource to the scientific community, with accessibility to our data set and comparisons to other published datasets generated under the same rigorous conditions provided by the ImmGen Project.
Methods
Mice
Consistent with ImmGen standards, six-to-eight week old male C57BL/6J mice and male B6.129P2(Cg)-Rorctm2Litt/J (with the sequence encoding enhanced green fluorescent protein (EGFP) inserted into the locus encoding Rorγt) obtained from Jackson Laboratories and maintained at Washington University School of Medicine (WUSM) under specific pathogen-free conditions were used for cell sorting and validation experiments. B6.129P2-Cxcr6tm1Litt/J mice with the sequence encoding EGFP inserted into the locus encoding CXCR6 obtained from Jackson Laboratories were used for validation experiments in which mice were littermates or age-matched, gender-matched, and co-housed when possible. Il-22 tdTomato reporter BAC transgenic mice were used for validation experiments.40 The WUSM animal studies committee approved all experiments.
Antibodies and flow cytometry
Anti-CD3e (145-2C11; 145-2C11), anti-CD19 (eBio1D3), anti-CD27 (LG.7F9), anti-CD49b (HMa2; DX5), anti-gamma delta (UC7-13D5; eBioGL3), anti-CD127 (A7R34; eBioSB/199), anti-CD49a (HMa1), anti-Nkp46 (29A1.4), anti-CD304 (3DS304M), anti-CD45 (30-F11), anti-NK1.1 (PK136), anti-Sca-1 (D7), anti-ST2 (RMST2-2), anti-TRAIL (eBioN2B2), anti-CD4 (GK1.5), anti-KLRG1 (2F1), anti-Rorγt (AFKJS-9), anti-Eomes (Dan11mag), SAv-PE/Cy7, and isotype-matched control monoclonal antibodies were obtained from eBioscience. Antibody to CD25 (PCB1) and 7-AAD were obtained from BD Biosciences. Antibody to CD45 (30-F11) was obtained from Miltenyi Biotec. LIVE/DEAD Fixable Aqua and SAv-APC were obtained from Life Technologies. Monoclonal anti-NKp46 (Cs96, rat IgG) was produced and biotinylated in our lab. Fc receptors were blocked before surface staining using supernatant from anti-CD32 mAb-producing hybridoma cells (HB-197, ATCC). For intracellular staining, the FOXP3 staining kit (eBioscience) was used. Data were acquired on a BD FACSCanto II and analyzed using FlowJo software (Treestar).
Cell identification, isolation, and microscopy
All cells were stained and sorted by following the published standard operations protocol on the ImmGen website (http://www.immgen.org/Protocols/ImmGenCellprepandsortingSOP.pdf). Cells were isolated from between 3 to 15 mice per sample. For siLP and siIEL isolation, small intestine beginning half a centimeter after the pylorus and ending half a centimeter before the cecum with Peyer’s patches removed was dissociated using the Miltenyi Lamina Propria Dissociation kit, which includes 2 DTT/EDTA washes and an enzymatic digestion step of 30 minutes. For liver and spleen isolation, mice were perfused with 20 mL cold PBS and tissue was dissociated through a 70 μm cell strainer. Lymphocytes were enriched at the interface between a gradient of 40% and 70% Percoll in HBSS. siIEL were further pre-enriched by negative selection with anti-CD8 MicroBeads (Miltenyi). Liver and spleen were pre-enriched by negative selection with anti-CD19 and anti-CD4 MicroBeads (Miltenyi). Cells were double-sorted directly into TRIzol with a Becton-Dickinson FACSAria II. The data browser of the ImmGen Project website shows flow cytometry plots from gating strategies and purity from the first round of sorting. Cytospins were prepared following the same ImmGen standards, except from only 1 to 3 mice; splenic samples were positively pre-enriched by using DX5-coated beads (Miltenyi). Sorted cells were spun onto slides, left to dry overnight, and then fixed and stained with Diff-Quik. Pictures were taken at 60× with oil-immersion using a Nikon Eclipse E800 and Leica Application Suite software.
Microarray and data analysis
Two to three replicates of RNA were obtained from each sample that passed quality control. RNA amplified and hybridization to the Affymetrix Mouse Gene 1.0 ST array was carried out by ImmGen following a standardized TRIzol extraction protocol available at www.immgen.org. Data generation and quality control documentation also followed the ImmGen protocol, methods for which can be found at www.immgen.org along with QC data, replicate information, and batch information from each sample. Data were analyzed using GenePattern50 software (Broad Institute). Raw data were normalized using RMA. Differences in gene expression were identified using the Multiplot Studio function of GenePattern, from a filtered subset of genes with coefficient of variation less than <0.1 in all samples and expression of at least 120 relative units in one subset by the class mean functions, a value that corresponds to a 95% confidence of true expression across the ImmGen dataset. For gene expression signatures of individual subsets, pairwise comparisons were made between subsets, filtering for fold-changes of 2 or 4-fold, as indicated. For comparisons between 2–4 samples, probe sets were considered to have differences of expression for transcripts expressed >120 relative units, with > 2-fold change and P value of <0.05 (Student’s t-test), except between siLP ILC1 and siLP NK cells where P value was not considered. Volcano plots and fold-change-by-fold-change plots were produced in Multiplot, and degree of overlapping genes between subsets was calculated in MATLAB. Heat maps were generated using Gene-E (http://www.broadinstitute.org/cancer/software/GENE-E/). Data were log2-transformed, visualized by ‘Relative’ expression per row or ‘Global’ expression in the heat map, as indicated, and rows were clustered using the Hierarchical Clustering function with the Pearson correlation as a metric. Where more than one probe set with the same gene annotation was found, the probe set with highest average expression was used. For PCA, the top 10% of most variable probe sets was calculated using PopulationDistances PCA program (Scott Davis, Harvard Medical School, Boston MA). This program identifies differentially expressed genes by ANOVA using the geometric standard deviation of populations to weight genes that vary in multiple populations. The top 10% of most variable genes dataset was log2-transformed in MATLAB and used to generate a PCA with the functions pca and scatter3. Hierarchical clustering of sample replicates was carried out in Gene-E from the same dataset using the Pearson correlation as a metric. For RNAseq analysis, data from GEO accession GSE52043 were visualized in IGV and aligned to the mm9 genome, as described previously.42
Statistics
Prism (GraphPad Software) was used for statistical analysis of flow cytometry data. Data were tested using either one-way or two-way ANOVA, as appropriate. Error bars are shown as mean +/− SEM.
Supplementary Material
Acknowledgments
We thank our colleagues in the ImmGen consortium, especially C. Benoist and L. Lanier, for input and discussion; the core ImmGen team, K. Rothamel and A. Rhodes, for contributions and technical assistance; M. Artyomov, G. Krishnan, and J. Siegel for computational assistance; D. Sojka for discussion; E. Lantelme and D. Brinja for sorting assistance; P. Wang for microscopy assistance; and eBioscience and Affymetrix for support of the ImmGen Project. The Colonna laboratory is supported by grants from the National Institutes of Health (1U01AI095542, R01DE021255, and R21CA16719). The MSTP T32 GM07200 supported M.L.R. The Infectious Disease Training Grant T32 AI 7172-34 supported V.S.C.
The members of the Immunological Genome Consortium are: Adam J. Best,1 Jamie Knell,1 Ananda Goldrath1, Vladimir Jojic2, Daphne Koller2, Tal Shay3, Aviv Regev3, Nadia Cohen4, Patrick Brennan4, Michael Brenner4, Francis Kim5, Tata Nageswara Rao5, Amy Wagers5, Tracy Heng6, Jeffrey Ericson6, Katherine Rothamel6, Adriana Ortiz-Lopez6, Diane Mathis6, Christophe Benoist6, Natalie A Bezman7, Joseph C Sun7, Gundula Min-Oo7, Charlie C Kim7, Lewis L Lanier7, Jennifer Miller8, Brian Brown8, Miriam Merad8, Emmanuel L Gautier8,9, Claudia Jakubzick8, Gwendalyn J Randolph8,9, Michelle L Robinette9, Marco Colonna9, Paul Monach10, David A Blair11, Michael L Dustin11, Susan A Shinton12, Richard R Hardy12, David Laidlaw13, Jim Collins14, Roi Gazit15, Derrick J Rossi15, Nidhi Malhotra16, Katelyn Sylvia16, Joonsoo Kang16, Taras Kreslavsky17, Anne Fletcher17, Kutlu Elpek17, Angelique Bellemare-Pelletier17, Deepali Malhotra17, Shannon Turley17.
Footnotes
Division of Biological Sciences, University of California San Diego, La Jolla, California, USA.
Computer Science Department, Stanford University, Stanford, California, USA.
Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Division of Rheumatology, Immunology and Allergy, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Joslin Diabetes Center, Boston, Massachusetts, USA.
Division of Immunology, Department of Microbiology & Immunobiology, Harvard Medical School, Boston, Massachusetts, USA.
Department of Microbiology & Immunology, University of California San Francisco, San Francisco, California, USA.
Icahn Medical Institute, Mount Sinai Hospital, New York, New York, USA.
Department of Pathology & Immunology, Washington University, St. Louis, Missouri, USA.
Department of Medicine, Boston University, Boston, Massachusetts, USA.
Skirball Institute of Biomolecular Medicine, New York University School of Medicine, New York, New York, USA.
Fox Chase Cancer Center, Philadelphia, Pennsylvania, USA.
Computer Science Department, Brown University, Providence, Rhode Island, USA.
Department of Biomedical Engineering, Howard Hughes Medical Institute, Boston University, Boston, Massachusetts, USA.
Program in Molecular Medicine, Children’s Hospital, Boston, Massachusetts, USA.
Department of Pathology, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
Dana-Farber Cancer Institute and Harvard Medical School, Boston, Massachusetts, USA.
Gene Expression Omnibus accession number: GSE37448
Author contributions: M.L.R. analyzed data; A.F., M.L.R., J.S.L., and Y.W. sorted cell subsets; M.L.R., A.F, and V.S.C. performed follow-up experiments and analyzed data; S.G. maintained mice; S.K.D. provided critical reagents; M.L.R., A.F., S.G. and M.C. designed studies. M.L.R. and M.C. wrote the paper. The ImmGen Project Consortium contributed to experimental design and data collection.
References
- 1.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 2.Diefenbach A, Colonna M, Koyasu S. Development, Differentiation, and Diversity of Innate Lymphoid Cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Klose CS, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 5.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;29:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 6.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d”Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-Rorγt+ innate lymphoid cells. Nature. 2013;494:262–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 8.Sciumé G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, Spencer SP, Wilhelm C, Poholek AC, Vahedi G, et al. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, McKenzie AN, Carotta S, Nutt SL, Belz GT. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34:573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;40:25–39. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynders A, Yessaad N, Vu Mahn TP, Dalod M, Fenis A, Aubry C, Nikitas G, Escalière B, Renauld JC, Dussurget O, et al. Identity, regulation and in vivo function of gut NKp46+RORγt+ and NKp46+RORγt–lymphoid cells. EMBO J. 2011;30:2834–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasteiger G, Hemmers S, Bos BD, Sun JC, Rudensky AY. IL-2-dependent adaptive control of NK cell homeostasis. J Exp Med. 2013;6:1179–1187. doi: 10.1084/jem.20122571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial Type 1 Innate Lymphoid Cells Are a Unique Subset of IL-12-and IL-15-Responsive IFN-g-Producing Cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner JE, Morrison PJ, Wlhelm C, Wilson M, Ahlfors H, Renauld JC, Panzer U, Helmby H, Stockinger B. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J Exp Med. 2013;210:2951–2965. doi: 10.1084/jem.20130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mjosber J, Bernink J, Korneliusz G, Karrich JJ, Peters CP, Blom B, te Vlede AA, Fokkens WJ, van Drunen CM, Spits H. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Yagi R, Zhong C, Northrup DL, Yu Ff, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, Belkaid Y, Zhao K, Zhu J. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF, Jr, Wang J, Ramalingam TR, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, Locksley RM. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med. 2013;210:535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–225. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal tolerance. Science. 2014;343:6178. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitt LJ, Nagler A, Lee F, Abrams J, Shatsky M, Thompson D. Production of granulocyte/macrophage-colony stimulating factor by human natural killer cells. Modulation by the p75 subunit of the interleukin 2 and by the CD2 receptor. J Clin Invest. 1991;88:67–75. doi: 10.1172/JCI115306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, Almeida FF, Ibiza S, Barbosa I, Goverse G, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 28.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 29.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres ML, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bando JK, Liang HE, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nature Immunol. doi: 10.1038/ni.3057. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veiga-Fernandes H, Coles MC, Foster KE, Patel A, Williams A, Natarajan D, Barlow A, Pachnis V, Kloussis D. Tyrosine receptor RET is a key regulator of Peyer’s patch organogenesis. Nature. 2007;446:547–441. doi: 10.1038/nature05597. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 37.Kumanogoh A, Kikutani H. Immunological functions of the neuropillins and plexins as receptors for semaphorins. Nat Rev Immunol. 2013;13:802–814. doi: 10.1038/nri3545. [DOI] [PubMed] [Google Scholar]
- 38.Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–174. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen W, Li W, Hixon JA, Bouladoux N, Belkaid Y, Dzutzev A, Durum SK. Adaptive immunity to murine skin commensals. PNAS. 2014;111:E2977–E2986. doi: 10.1073/pnas.1401820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daussy C, Faure F, Mayol K, Veil S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UZ, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye SY, Maki K, Kitamura T, Sunaga S, Akashi K, Domen J, Weissman IL, Honjo T, Ikuta K. Induction of germline transcription in the Tcrγ locus by Stat5: Implications for accessibility control by the IL-7 receptor. Immunity. 1999;11:213–223. doi: 10.1016/s1074-7613(00)80096-5. [DOI] [PubMed] [Google Scholar]
- 44.Zhao H, Nguyen H, Kang J. Interleukin 15 controls the generation of restricted T cell receptor repertoire of intraepithelial lymphocytes. Nat Immunol. 2005;6:1263–1271. doi: 10.1038/ni1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allan DS, Kirkham CL, Aguilar OA, Qu LC, Chen P, Fine JH, Serra P, Awong G, Gommerman JL, Zúñiga-Pflücker JC, Carlyle JR. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. doi: 10.1038/mi.2014.71. advance online publication. [DOI] [PubMed] [Google Scholar]
- 46.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell progenitor. eLife. 2014;10:e04406. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satoh-Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G, Di Santo JP. The Chemokine Receptor CXCR6 Controls the Functional Topograophy of Interluekin-22 Producing Intestinal Innate Lymphoid Cells. Immunity. 2014;41:776–788. doi: 10.1016/j.immuni.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL The Immunological Genome Project Consortium. ImmGen Report: Molecular definition of natural killer cell identity abd activation. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nature Genetics. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.