Abstract
p204 is a member of the interferon-inducible p200 family proteins in mice. The p200 family has been reported to be multifunctional regulators of cell proliferation, differentiation, apoptosis and senescence. Interferon-inducible protein 16 (IFI16) is regarded as the human ortholog of p204 in several studies. This is possibly due to the similarity of their structures. However the consistency of their functions is still elusive. Currently, an emerging focus has been placed upon the role of the p200 proteins as sensors for microbial DNA in innate immune responses and provides new insights into infections as well as autoimmune diseases. This review specially focuses on IFI16 and p204, the member of p200 family in human and murine respectively, and their pathophysiological roles in innate immune responses, cell differentiation and proliferation.
Keywords: DNA sensor, IFI16, Innate immune response, Multifunctional regulator, p204
Introduction
p204 is a multifunctional interferon-inducible murine protein in the p200 family (also known as PYHIN or HIN-200 proteins). The first member of p200 family, p202, was identified in 1982, on the basis of the inducibility of its mRNA by interferon. p200 proteins have been linked to the cell cycle, proliferation, and differentiation and are all characterized by at least one carboxy-terminal HIN domain and an amino-terminal pyrin (PYD) domain. The p200 family proteins include p204, p202a, p202b, p203, MNDAL, and AIM2 in mice and additional four members in humans, IFI16, AIM2, MNDA, IFIX. There are common characteristics of the molecular structure of p200 family proteins. The N-terminus of p200 proteins contains a PYD domain except p202a and p202b. The PYD domain has approximately 90 amino acid motif that can form homotypic interactions with other PYD-containing proteins to form higher complexes, playing roles in inflammation, apoptosis, and the cell cycle.1
Previous studies have reported that the p200 family proteins can regulate: cell proliferation,2, 3, 4, 5, 6 differentiation,7, 8, 9, 10, 11, 12, 13, 14, 15 apoptosis16, 17, 18 and senescence.19, 20 Moreover, as an emerging focus, the role of the p200 proteins as receptors for microbial DNA in innate immune responses provides new insights into infectious as well as autoimmune diseases (Fig. 1). The innate immune response is the first line of defense against infection from exogenous microbial pathogens. These immune systems include: the Toll-like receptors (TLR), the retinoic acid inducible gene-like receptors (RLR), the nucleotide oligomerization domain-like receptors (NLR), C-type lectin receptors (CLR) and DNA-sensing molecules. The p200 proteins have recently emerged as sensors to bind microbial DNA and form downstream inflammation or type I IFN introduction. The Interferon-inducible protein 16 (IFI16) was identified in THP-1 cells directly bound to the IFN-β-stimulating viral DNA and transfected DNA but not RNA.21 Also IFI16 was identified as a host DNA sensor required for CD4 T cell death due to HIV infection.22 Another p200 protein, AIM2 (absent in melanoma 2), is a sensor for cytosolic DNA for the signaling pathway that activates caspase-1, leading to inflammasomes and release of interleukin 1β.23, 24, 25, 26 Thus, the p200 proteins were regarded as a new family of innate DNA sensors called ‘AIM2-like receptors’ (ALRs).21
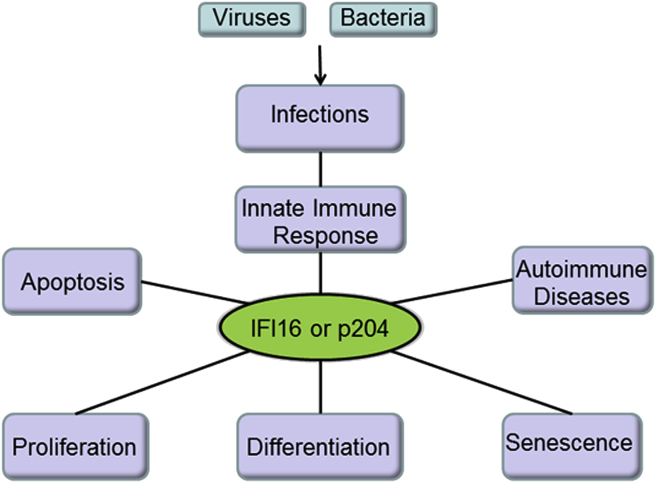
Figure 1.
A schematic representation of the multiple functions of p204 and IFI16.
Molecular structures
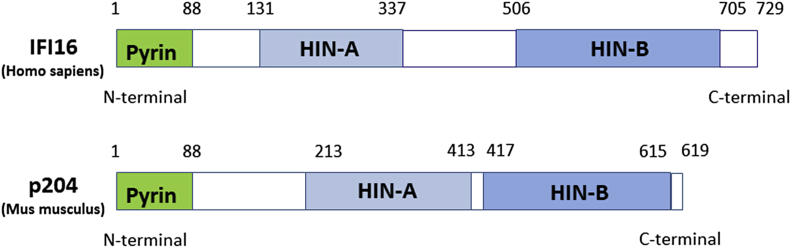
The C-terminus portion of the p200 proteins contains one or two copies of a conserved HIN-200 domain, which may be related to protein- and/or DNA binding. These have been characterized into three subtypes termed A, B, and C according to consensus motifs.27 Both p204 and IFI16 contain A-type and B-type HIN-200 domain (Fig. 2), MNDA, MNDAL, and IFIX include a single A-type HIN-200 domain, whereas AIM2 has a single C-type HIN-200 domain. The A-type and B-type domains each contain one copy of an MF/LHATVAT/S type sequence which is conserved in all 200 X-type domains.28 This sequence was reported to be involved in protein/protein interactions. In p204 the A-type domain contains two types of pRb-binding motifs: an IXCXE sequence and an LXCXE sequence, whereas the B-type domain contains only a single pRb-binding LXCXE sequence. The p204 protein amino acid sequence contains potential sites for phosphorylation by various protein kinases including: cAMP-dependent kinase, protein kinase C, calmodulin-dependent protein kinase, MAP kinase, ATM, Cdk2 and casein-kinase 2.29 p204 has been shown to be a phosphoprotein,30 and its phosphorylation is apparently involved in the translocation of p204 from the nucleus to the cytoplasm.10, 31 The structure of HIN-200 domain was relative to DNA binding because there were two oligonucleotide/oligosaccharide (OB) folds located within the domain.32 This was also reported in the solved structure of the IFI16 HIN-A domain.33 The two OB folds of the HIN-A domain of IFI16 had a greater affinity for single-stranded DNA (ssDNA) than double-stranded DNA (dsDNA), and could wrap, stretch and form oligomers with ssDNA.34 And the IFI16 HIN-B domain alone was able to bind to dsDNA, which was enhanced when both HIN-200 domains were present.21
Figure 2.
Molecular structure of p204 and IFI16. Both p204 and IFI16 contain N-terminal pyrin (PYD), HIN-A, and HIN-B domains.
IFI16 acts as DNA sensor in innate immune response
IFI16 is identified as a sensor to exogenous DNA from pathogenic microbes in innate immune responses. Unterholzner et al21 reported that small interfering RNA (siRNA) targeting IFI16 or p204 inhibited the activation of IRF3 and NF-κB, and IFN-β induction, induced by DNA and herpes simplex virus type 1 (HSV-1). In THP-1 cells, cytosolic IFI16 was detected by immunoblot analysis in a complex with immobilized transferred ssDNA or dsDNA, which could be bound to the HIN domain of IFI16. IFI16 has been mainly localized to the nucleus when overexpressed in HEK293 cells.25 The co-localization of endogenous cytoplasmic IFI16 with IFN-β inducing viral DNA and its direct and cooperative binding to this DNA in vitro suggested that IFI16 might be a DNA sensor that mediates IFN-β induction. Within the human body IFI16 is the most similar protein to p204 and some researchers view it as a p204 homolog. They suggest that both IFI16 and p204 may function similarly due to the similarity of their domain structures,25, 35 however, more evidence is required.
A recent study suggests that IFI16 is a DNA sensor to HIV virus infection and required for CD4 T cell death due to AIDS.22 Monroe et al assessed the expression of various known innate immune sensors candidate by immune-blotting cytosolic lysates from tonsillar CD4 T cells, including the presence of IFI1636, 37; AIM223, 24, 25, 26; DAI38; STING 39, 40, 41; DNPK-142; NLRP343, 44, 45; and IFIX. Meanwhile, cyclic guanosine monophosphate adenosine monophosphate synthase (cGAS)46, 47, 48, 49 was detected neither at the protein level in tonsillar CD4 T cells nor in the affinity chromatography-mass spectrometry experiments, which may suggest that cGAS was not involved in the immune responses of human CD4 T cells during the abortive HIV infection. Originally they discovered that IFI16 was effectively bound to dsDNA in the tonsillar CD4 T cells, which was consistent with published data.21, 50 Then they used three independent shRNAs targeting IFI16 to knockdown IFI16 protein expression in mCherry-positive CD4 T cells and the results showed that all three IFI16 shRNAs rescued the depletion of mCherry-positive CD4 T cells infected by HIV-1 (Biotinylated HIV Nef DNA). These findings suggest that IFI16 is required to sense the infected HIV DNAs that accumulate in the cytoplasm of abortively infected T cells. Their accumulation leads to caspase-1 activation, which activates cellular signaling downstream causing pyroptosis, producing subsequent cell death.22
DNA has been proposed to be a potent inducer of immune responses and involved in the pathogenesis of psoriasis, a common immune-mediated inflammatory disease affecting the skin and joints. In psoriasis there is extensive DNA replication in keratinocytes, which is associated with the accumulation of dsDNA and the activation of the inflammasome. Chiliveru et al described the up-regulation of IFI16 in human psoriasis lesions and why IFI16 was essential for the DNA-driven innate immune responses in primary human epidermal keratinocytes in an inflammatory environment.51 They demonstrated the manner in which inflammatory cytokines enable keratinocytes to respond to cytoplasmic DNA with a potent innate immune response. The transfection of synthetic dsDNA into keratinocytes did not evoke significant expression of CCL20 nor CXCL10. Notably, a very potent induction of CCL20, CXCL10, IFN- β and IFIT1 was induced when keratinocytes were transfected by a synthetic dsDNA or human genomic DNA, together with TNF-α or IL-1β.51 It is important to note that the DNA-mediated induction of CCL20 and CXCL10 in TNF-α treated cells was significantly reduced in the IFI16 knockdown cells. The treatment of keratinocytes with inflammatory cytokines induces translocation of a small pool of IFI16 into the cytoplasm, which enables the cells to assemble several signaling complexes: IFI16, STING and TBK1. They then isolated skin biopsies from non-lesional and lesional psoriasis skin. The found that IFI16 upregulates both mRNA and protein levels in the lesional psoriasis samples when compared to the non-lesional samples.51
Roles of IFI16 in viral infection
During coevolution with their hosts, viruses have acquired effective mechanisms for blocking host immune signaling that involves IFI16. The human cytomegalovirus (HCMV) major tegument protein pUL83 is thought to play critical roles in immune evasion. It was shown to block antigen presentation and activation of natural killer cells and to suppress the induction of antiviral cytokines. Using a mass spectrometry-based approach during HCMV infection in fibroblast, Cristea et al reported that the nuclear pUL83 interacts with two interferon-inducible proteins from the p200 family, IFI16 and IFIX.52 They confirmed these interactions in all three IFI16 isoforms and pUL83 from the cell extracts that were infected with virus by co-immunoprecipitation and the co-localization within the nucleus and nucleolus by immunofluorescent analysis. They also found that the knockdown of IFI16 by shRNA in primary human fibroblasts resulted in decreased levels of immediate-early transcripts in the infection of wild type HCMV.52
However, the mechanisms involved in HCMV immune evasion remain to be further elucidated. A recent publication from the same group53 defined mechanisms underlying HCMV immune evasion, identifying a critical molecular hub that determines the outcome of host defense and viral pathogenesis. Similar to herpes viruses, DNA is protected by viral capsids in the cytoplasm prior to nuclear exposure, nuclear sensing is a critical aspect of defense against these nuclear replicating viruses. In endothelial cells, nuclear IFI16 can assemble inflammasomes during infection with Kaposi sarcoma-associated herpesvirus (KSHV), leading to secretion of pro-inflammatory interleukins.54 In U2OS cells and fibroblasts, nuclear IFI16 senses herpes simplex virus-1 (HSV-1) DNA, triggering type I interferon (IFN) response55, 56 and inflammasome formation.57 In macrophages, where capsids may be vulnerable to degradation, leaked HSV-1 DNA can be detected by cytosolic IFI16.58 Cytosolic IFI16 also detects transfected DNA, inducing type I IFN in an STING-dependent manner.21
The signaling pathways involving IFI16 and p204
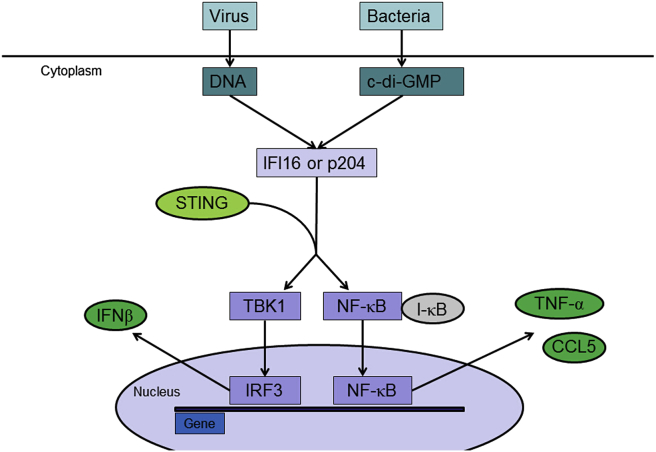
It is well-known that exogenous DNA derived from pathogens can provoke expression of a large number of antiviral and inflammatory genes, including IFNs. Medzhitov et al59 demonstrated the existence of a TLR-independent DNA response, which they called the ISD (immune stimulatory DNA) pathway, in mice lacking myeloid differentiation factor 88 (MyD88) or TRIF. However, this novel pathway converges on the inhibitor of nuclear factor κB (NF-κB) kinase (IKK)-related kinases TBK1 and IKKɛ, which phosphorylate and activate IRF3. IRF3 collaborates with NF-κB and ATF-2-c-Jun to then drive transcription of type I IFN genes. TBK1 also interacts with the endoplasmic reticulum (ER) adapter stimulator of interferon genes (STING), which plays a central role driving IFN not only in response to synthetic dsDNAs but also following HSV infection.40, 41 STING (TMEM173) is a signaling protein known to be required for TBK1-dependent IFN-β responses to viruses and DNA.40, 60 Infection of dsDNA caused a DNA-dependent association between endogenous STING and IFI16 in THP-1 cells. IFI16 from the DNA-stimulated THP-1 cells associated with immobilized Myc-tagged human and mouse STING. It was proven that STING was recruited to IFI16 after viral DNA stimulation and mediated the downstream IFN-β induction in cells infected by dsDNA or HSV, as in BMDMs lacking STING, IFN-β secretion from cells transfected with the viral DNAs was completely inhibited. Besides, other cytokines related to NF-κB, such as CCL5 and tumor necrosis factor α (TNFα), were also decreased due to impaired translocation of the p65 NF-κB subunit to the nucleus in p204 and IFI16 knockdown cells.21 These above results indicate a role for IFI16 and p204 as upstream cytosolic dsDNA receptors, which through their interaction with STING in response to dsDNA, activate TBK1-IRF3/NF-κB axis, drive induction of type I IFN and pro-inflammatory cytokines (Fig. 3).
Figure 3.
Signaling pathways regulated by IFI16 or p204 in DNA sensing. IFI16 or p204 acts as a DNA sensor during the innate immune responses.
Notably, a negative feedback loop has been identified between the levels of IFI16 and STING during the cytosolic sensing of bacterial second messenger cyclic-di-GMP (c-di-GMP).61 The activation of c-di-GMP-induced signaling in human and murine cells increase the steady-state levels of IFI16 and p202 proteins, which is c-di-GMP concentration- and time-dependent. Whereas the constitutive knockdown of IFI16 or p202 expression in cells increases steady-state levels of STING and results in activation of the TBK1/IRF3 axis. Accordingly, increased levels of the IFI16 or p202 protein in cells decrease STING levels. Together, these observations identify a novel negative feedback loop between c-di-GMP-induced levels of IFI16 and p202 cytosolic DNA sensors and the adaptor protein STING.
Additionally, knockdown of IFI16 decreased caspase-1 activation in the mCherry-positive CD4 T cells infected by HIV-1,22 indicating that IFI16 is required to sense incomplete DNA reverse transcripts that accumulate in abortively infected cells, which leads to caspase-1 activation and the subsequent death of these cells via pyroptosis.62 In another study,54 IFI16 was showed to interact with the adaptor molecule ASC and procaspase-1 to form a functional inflammasome in endothelial cells during Kaposi Sarcoma-Associated Herpesvirus (KSHV) infection. This complex was initially detected in the nucleus and subsequently in the perinuclear area. Caspase-1 activation by KSHV was reduced by IFI16 and ASC silencing. These findings demonstrate that IFI16 as a nuclear pathogen sensor to form the inflammasome by recruiting ASC, resulting in the activation of caspase-1 and induction of proinflammatory cytokines.
IFI16 distinguishes foreign and host DNA
The molecular mechanism of IFI16 distinguishing self from non-self DNA in the innate immunity is an outstanding question, which attracts much attention from the researcher community. A recent study63 showed that cooperatively assembling into filaments on dsDNA may serve as an integral mechanism by which IFI16 engages foreign DNA. IFI16 cooperatively binds dsDNA in a length-dependent manner and clusters into distinct protein filaments even in the presence of excess dsDNA. Consequently, the assembled IFI16-dsDNA oligomers are clearly different from the conventional non-interacting entities resembling beads on a string. The isolated DNA-binding domains of IFI16 engage dsDNA without forming filaments and with weak affinity, and it is the non-DNA-binding pyrin domain of IFI16 that drives the cooperative filament assembly. The surface residues on the pyrin domain that mediate the cooperative DNA binding are conserved, suggesting that related receptors use a common mechanism. These results suggest that IFI16 clusters into signaling foci (e.g., inflammasome) in a switch-like manner and that it is capable of using the size of naked dsDNA as a molecular ruler to recognize self DNA from non-self.63
They also proposed two features of foreign DNA that are much more conducive to filament formation by IFI16. First, the DNA genome from infective pathogens is exposed naked immediately after the invasion. Second, although the viral genome packages into chromatin with the host histones, it is less dense and much more loosely packed than the nuclear self-DNA.63 The electron micrographs demonstrate the filaments formation by IFI16 corresponding to the length of dsDNA. IFI16 prefers to bind to a longer-bp non-self dsDNA rather than a short dsDNA from the host. What is important to note is the relative binding affinity of IFI16 to various dsDNA fragments changes cooperatively not only with increasing IFI16 concentrations but also with increasing number of binding sites, suggesting that IFI16 is capable of amplifying its clustering behavior in a switch-like manner.63 Thus, these results suggest that IFI16 can clearly define an “on” or “off” state with respect to its concentration and the length of dsDNA.63 Additionally, we also suppose that the host nuclear DNA may avoid the p204-mediated recognition in mice because of the protection by histones or other protective host proteins, or the tight fold of the genome, according to the unpublished findings about the binding of p204 to Sin3a (a multifunctional agent promoting histone deacetylation by HDAC), which remains to be explored.
Acetylation regulates IFI16 subcellular localization
Cellular localizations, targets specificities, and downstream signaling pathways define pathogen-associated molecular patterns functions. In the case of IFI16, its targets and downstream pathways are starting to be defined, but there are important unanswered questions regarding its cell type-dependent and dual subcellular localization. IFI16 is a predominantly nuclear protein in lymphoid, epithelial, endothelial, and fibroblast tissues and also has been reported a cytoplasmic localization in macrophages. Its dual localization confers IFI16 the ability of colocalize with transfected vaccinia virus (VACV) dsDNA in the cytoplasm of differentiated THP-1 monocytes11 and colocalize with KSHV DNA during early infection in the nucleus, consequently activating inflammasome formation.41
Cristea et al55 has provided evidence for a two-signal model for the function of IFI16 as a pathogenic DNA sensor. Firstly, they defined an evolutionary conserved multipartite nuclear localization signal (NLS), consisting of two essential motifs and two accessory motifs, required for IFI16 sensing of HSV-1 viral DNA in the nucleus. Herpesviridae are characterized for replicating their dsDNA genomes in host nuclei as well as their ability to evade host immunity. These new findings55 demonstrated that upon HSV-1 infection, the diffused nuclear localization of IFI16 was drastically altered, becoming more concentrated within viral DNA-containing nuclear bodies. They confirmed that IFI16 recognized HSV-1 DNA primarily in the nucleus and the detection of nuclear viral DNA required a functional NLS. Their findings also explained that a cytoplasmic localization of a DNA sensor, such as IFI16, as described in macrophages and lymphocytes, might maximize immune system sensitivity to DNA viruses.
Secondly, they identified NSL acetylation as a mechanism underlying the dynamic localization of IFI16 that extended its range of DNA surveillance. They identified phosphorylation clusters within two regions of IFI16: the linker region (S95, S106, S153, S168 and S174) and C-terminus (S724) and lysine acetylation sites within Pyrin (K45) or HIN (K214, K542 and K558) domains or between HIN domains (K444, K451, and K505). They also found that the two major NLS motifs, Δmotif-1 and Δmotif-2, contained acetylations at K99 and K128. They found the phosphorylations within the linker region (S95, S106 and S153) had only minor roles in IFI16 localization, as shown by transient transfections of IFI16-EGFP mutants in U2Os cells. Since both lysine sites are highly conserved among IFI16 homologs and HIN-200 family members, they suggest that acetylation as a common regulator of subcellular localization.55
IFI16 cellular distribution and sensing functions are modulated by a combination of genetically encoded NLS and post-transcriptional acetylation mechanisms. In addition, p204 was found to interact with Sin3A, a multifunctional agent promoting histone deacetylation by HDAC. Thus, it would be interesting to analyze whether acetylation/deacetylation modification also play an important role in regulating the subcellular localization of p204, and whether this modification is also critical for its functions in pathogenic immune responses.
IFI16 is an antiviral restriction factor in host defense against HCMV
IFI16 has been shown to bind to and function as a pattern recognition receptor (PRR) of virus-derived intracellular DNA, and trigger the expression of antiviral cytokines via the STING-TBK1-IRF3 signaling pathway.21, 22, 37, 52, 53, 54, 56, 57, 58, 64, 65 Besides, its role as an antiviral restriction factor (RF) in host defense against HCMV was recently reported by Dell'oste V et al66 Restriction factors are some constitutively expressed proteins mediating a frontline antiviral defense response in the intrinsic immune system.67, 68 One evasion strategy that viruses may use is to exploit the effects of an RF for its own purposes, or to generate an interfering protein that neutralizes the effect of an RF. Another strategy involves the virus hijacking an RF during its phase of maturation to guarantee protection.69, 70 They proposed the evidence supporting such a role of IFI16 as follows: (a) small interfering RNA (siRNA)-mediated depletion of IFI16 in primary human embryonic lung fibroblasts (HELF) significantly increases HCMV replication efficiency as a result of augmented viral DNA synthesis; (b) similarly, viral plaque formation is enhanced in the presence of an exogenous dominant-negative IFI16 mutant that competes with the endogenous IFI16; (c) overexpression of functional IFI16 in HCMV-infected HELFs decreases both virus yield and viral DNA copy number; and (d) early and late, but not immediate-early viral mRNAs and proteins are strongly down-regulated under these same conditions, suggesting that IFI16 exerts its main antiviral effect at the level of viral genome synthesis.66
In this study, Dell'oste V et al examined the mechanisms of HCMV overcoming the antiviral activity of the nuclear restriction factor IFI16.66 Consistent with its property as a pathogenic DNA sensor, detailed kinetics studies exploiting immunofluorescence show that in the early phases of infection, IFI16 binds to viral DNA, also confirmed by FISH combined with Western blot analysis. These results are in line with previous studies showing that following HCMV infection IFI16 binds viral DNA and triggers the expression of antiviral cytokines via the STING-TBK1-IRF3 signaling pathway.53 During a late phase post infection, however, IFI16 levels decreases inside the nucleus and this is accompanied by a parallel increase in its presence in the cytoplasmic virus assembly complex (AC). This nucleo-cytoplasmic egress of IFI16 in HCMV-infected cells is driven, at least in part, by the viral protein kinase pUL97, which binds and phosphorylates nuclear IFI16. This can be seen in the acetylation of the nuclear localization sequence promotes the cytoplasmic accumulation of IFII6 by inhibiting its nuclear import.55 In HCMV-infected cells, IFI16 interacts with viral pUL97 and undergoes in vitro phosphorylation. Moreover, the nuclear accumulation of IFI16 can be observed upon treatment with Gö6976, an inhibitor of pUL97 phosphorylation.71, 72 Subsequently, the IFI16-AC complex mediates its incorporation into newly assembled virions. IFI16 mislocalization and assembly into mature virions appears to be regulated by the ESCRT machinery through its sorting and trafficking into multivesicular bodies. Together, these results suggest that HCMV has evolved mechanisms to mislocalize and hijack IFI16, trapping it within mature virions.66
AIM2 acts as DNA sensor and interacts with IFI16
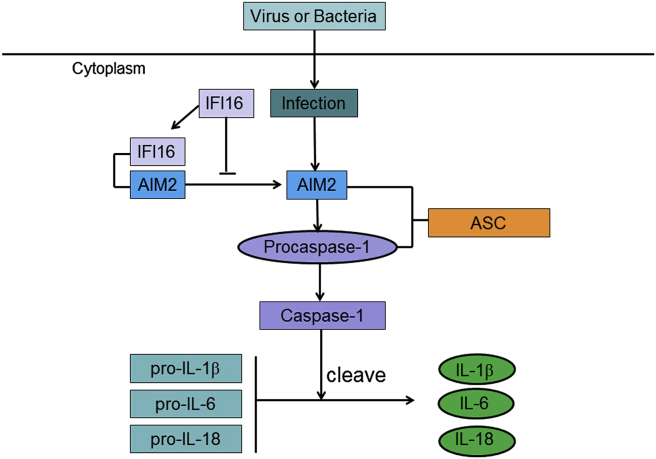
Both AIM2 and IFI16 contain C-terminal DNA-binding HIN domain(s) and an N-terminal Pyrin (PYD) domain that belongs to the death domain superfamily of signaling modules, and thus were also named as the PYHIN family of receptors or the AIM2-like receptors.21, 25, 73 AIM2 is predominantly a cytosolic protein that responds to dsDNA from both host and pathogens to interact with ASC and activate caspase-1 via CARD-CARD (caspase recruitment and activation domain) interaction and form inflammasomes.74, 75, 76 The activated caspase-1 cleaves pro-IL-1β and pro-IL-18 into the mature proinflammatory cytokines.50 IL-1β and IL-18 are very essential in antiviral defense both in innate and adaptive responses. AIM2 drives the maturation of IL-18, which is particularly important in early antiviral defenses in murine cytomegalovirus. The production of IL-18 and natural killer cell–dependent production of interferon-γ, events critical in the early control of virus replication, were dependent on AIM2 during mouse cytomegalovirus infection in vivo76 (Fig. 4).
Figure 4.
The interaction between AIM2 and IFI16. IFI16 binds to AIM2 and inhibits the role of AIM2 in mediating the inflammasome-caspase-1 pathway.
Besides sensing virus DNA, AIM2 is essential for inflammasome activation in response to bacterial infection in the immune system. Previous studies have reported that AIM2 is a critical inflammasome sensor during the infection of Francisella tularensis and Listeria monocytogenes.77, 78, 79, 80 Also AIM2 showed the role as a sensor to mediate imflammasomes and regulate IL-1β release and survival during acute CNS Staphylococcus aureus infection. ASC and caspase-1/11 KO animals were exquisitely sensitive, with approximately 50% of mice succumbing to infection within 24 h. Unexpectedly, the survival of NLRP3 KO mice was similar to WT animals, suggesting the involvement of an alternative upstream sensor, which was later identified as AIM2 based on the similar disease patterns between AIM2 and ASC KO mice. These studies demonstrate the role of AIM2 as an inflammasome sensor during acute CNS S. aureus infection.81
In addition, AIM2 physically interacts with IFI16. It was observed that IFI16 protein was bound to FLAG-tagged AIM2 protein in both cytoplasmic and nuclear fractions in HEK-293T cells (Fig. 4). More AIM2 protein bound to the IFI16 protein in the cytoplasmic fraction than the nuclear fraction. Importantly, increased expression of the IFI16 protein in HEK-293T cells reduced the activation of caspase-1 as determined by a reduction in the levels of the activated caspase-1. These observations indicated that the IFI16 protein binds to AIM2 protein in the cytoplasmic fraction and increased expression of IFI16 protein in transfected cells can inhibit the AIM2-ASC-mediated activation of caspase-1.82 Moreover, the knockdown of the IFI16 expression moderately increased the basal levels of AIM2 and P-CASP1 proteins in THP-1 cells.82 Importantly, the knockdown increased the basal levels of the activated caspase-1 as determined by increases in the p20 protein band.83 These findings suggest that the expression of IFI16 protein in THP-1 cells decreases the basal levels of the AIM2 and PCASP-1, and the activation of CASP-1.
IFI16 as a possible target in autoimmune diseases
Systemic autoimmune diseases, including Sjogren's Syndrome (SjS), Systemic Lupus Erythematosus (SLE), Systemic Sclerosis (SSc) and Rheumatoid arthritis (RA), are characterized by self antigen-driven immune responses that target host tissues and organs for damage.84 Although these autoimmune diseases differ from primary tissues that are targeted by autoantibodies, these diseases share certain common features and mechanisms. For example, exhibiting increased serum levels of proinflammatory cytokines such as: tumor necrosis factor-α (TNF-α), interleukin-1 (IL-1), and interferons (IFNs). It was suggested that increased levels of proinflammatory cytokines are the result of an abnormal activation of the innate immune response that is initiated by innate immune sensors.84 These sensors include DNA-dependent activator of IFN-regulatory factors (DAI; also referred to as ZBP1), DExD/H box helicases (DHX9 and DHX36), murine absent in melanoma 2 (Aim2), human AIM2, RNA polymerase III (Pol III), leucine-rich repeat (in Flightless I) interacting protein-1 (Lrrfip1), murine p204, and human IFI16.
Genomics studies also have revealed that type I IFN inducible genes are markedly overexpressed in the peripheral blood of patients with systemic autoimmune diseases, including SLE, SSc, and SjS. According to this, anti-IFI16 autoantibodies have been present in the serum of patients affected by SLE, SSc and SjS. Interestingly, Caneparo et al85 suggest that the development of anti-IFI16 antibodies may result in beneficial functional properties rather than being pathogenic, as IFI16-positive patients tend to exhibit a reduced risk of: (a) C3 hypocomplemetemia, and (b) proteinuria, indicating a decreased renal involvement that actually is one of the most frequent and serious complications in SLE. Although more studies are warranted to understand the mechanisms of a possible beneficial function of anti-IFI16 antibodies and validate its clinical significance, these antibodies hold the potential to serve as a new biomarker in the diagnosis and assessment of disease activity in systemic autoimmune diseases.
Regulation of cell differentiation by p204
Besides the roles of p204 and IFI16 in immune responses, the differentiation and proliferation of cells in various systems are also regulated by p204. The differentiation of skeletal muscle myoblast was regulated by p204. The p204 level was strongly increased during the differentiation of cultured C2C12 myoblasts to skeletal muscle-type myotubes.10 In addition, p204 was required for the differentiation of the myoblasts: its overexpression accelerated the process, whereas decreasing its level inhibited it. The increase of p204 levels during myoblast differentiation was due to transcription by the muscle-specific transcription factors MyoD and myogenin.10 p204 enabled the differentiation, partly by overcoming the known inhibition of MyoD and myogenin activity by the Id (inhibitor of differentiation as well as of DNA-binding) proteins.86 This inhibition is known to block the binding of MyoD and myogenin to DNA and thus interferes with the synthesis of numerous muscle proteins, thereby blocking the differentiation.87, 88, 89 p204 was found to overcome this inhibition by (1) binding to the Id proteins, (2) thereby promoting their nuclear export signal-dependent translocation from the nucleus to the cytoplasm, consequently separating the Id proteins from the transcription factors which, in the nucleus, they inhibited, and (3) accelerating their ubiquitination, and thereby their degradation in the cytoplasm by proteasomes.9, 86
p204 was involved in the differentiation of beating cardiac myocyte. The expression p204 increased strongly in the course of the differentiation of cultured P19 embryonal carcinoma stem cells to beating cardiac myocytes in response to dimethyl sulfoxide (DMSO) treatment.8 Furthermore, ectopic p204 could substitute for DMSO in inducing the differentiation, whereas p204 antisense RNA blocked the process. In this study, p204 expression was synergistically transactivated by the cardiac Gata4, Nkx2.5, and Tbx5 transcription factors.8 The Id proteins (Id1, Id2, or Id3) inhibited the differentiation of P19 cells as a result of their binding to Gata4 and Nkx2.5 and by inhibiting the binding of these transcription factors to each other and to DNA. This, in turn, blocked the activation of expression of numerous cardiac proteins.9 Similarly to its action in skeletal muscle differentiation, in heart differentiation, p204 overcomes the inhibition by Id proteins by binding to them, promoting their translocation from the nucleus to the cytoplasm, and enhancing their degradation by ubiquitination followed by proteasomal digestion.9, 86
During the differentiation of osteoblasts from mesenchymal C2C12 cells, the level of p204 was differentially induced, and this induction is mediated by the Smad transcription factor complex. p204, similar to pRb, acts as a co-factor of Cbfa1 transcription factor and enhances Cbfa1 activated osteogenesis.11, 12 Id proteins are involved in osteogenesis, and are upregulated by BMP2.90, 91, 92 However, Wnt, which triggers osteoblast differentiation, also promotes a rapid decrease in Id level.93 This rapid decrease is required because Id proteins bind to Cbfa1, thereby inhibiting its DNA binding and transcription of the alkaline phosphatase and osteocalcin genes that are involved in the differentiation.91 p204 overcomes the inhibition of Id proteins in osteogenesis in the same manner as it does in skeletal muscle myoblast and cardiac myocyte differentiation.13 Thus, in osteogenesis, p204 boosts the Cbfa1-dependent transcription pathway in two ways: (a) by boosting the transcription, forming a ternary complex p204-pRb-Cbfa1; (b) by overcoming the inhibition of Cbfa1 by Id proteins.11, 13
Chondrocyte differentiation is crucial for the development of long bones. It involves the cessation of the proliferation of chondrocytes and their conversion into hypertrophic chondrocytes.94 The overexpression of p204 accelerates the hypertrophy of chondrocyte, whereas the knockdown of p204 abolishes this process. In the process, p204 associates with Cbfa1.15 The resulting p204-Cbfa1 complex promotes the differentiation of chondrocytes by (a) decreasing the expression of parathyroid hormone/parathyroid hormone-related peptide receptor (PTHrP), an agent promoting the proliferation and delaying the differentiation of chondrocytes, and (b) stimulating the expression of Indian hedgehog protein (Ihh), an agent promoting the differentiation.15
Mature monocyte and macrophage cells express p204. Moreover, macrophage colony stimulating factor (M-CSF) or leukemia inhibitory factor (LIF) has been found to induce p204 expression as well as the differentiation to macrophages of the myeloid progenitor line FD-Fms. The constitutive expression of p204 strongly decreases the M-CSF (and interleukin 3) dependent proliferation of the FD-Fms cell line, whereas it promoted its M-CSF-induced differentiation to macrophages.7 The p204 mRNA level is lower in less mature (CD4+ CD8+ double positive) thymocytes than in more mature (CD4+ or CD8+ single positive) thymocytes, which may indicate a role for p204 in lymphocytic differentiation.95
Regulation of cell proliferation by p204
p204 can play an antiproliferative role in cell cycle via its HIN-200b domain.96 The inhibition of cell proliferation by p204 may occur through multiple mechanisms. p204 can inhibit the proliferation of a human osteosarcoma cell line (U2OS) with active p53 and pRb, and also of a second osteosarcoma line (Saos2), which is lacking active p53 and pRb.97 This indicates that the antiproliferative activity of p204 does not have to depend on active p53 or pRb. A difference between the modes of inhibition by p204 of the proliferation of the above two osteosarcoma cell lines was also revealed by the finding that p204 induction resulted in a partial G2/M arrest in Saos2 cultures, but had little effect on cell cycle distribution in U2OS cultures. Furthermore in U2OS cultures, but not in Saos2 cultures, the expression of p204 significantly increased the levels of pRb and its active hypo-phosphorylated form. The same report which found that Saos2 proliferation is not inhibited by p204, also states that p204 inhibits the proliferation of wild-type MEF cells, but not of MEF cells in which the pRb gene was inactivated.98 However, p204 from which the C-terminal 72 amino acid segment was deleted in order to remove the pRb-binding sequence (LXCXE) from the b segment, without or together with the mutation of the LXCXE sequence in the a segment was shown not to decrease, but actually to increase cell proliferation as detected by focus forming assays.98 It remains to be established whether this increase in the rate of cell proliferation is a consequence of the loss of pRb-binding activity, or of other, as yet uncharacterized consequences of the deletion of the C-terminal segment.
p204 directly associates with the ribosomal RNA-specific UBF-1 transcription factor in vivo and in vitro.4, 99 The inhibition of ribosomal RNA synthesis by p204 may be due to the inhibition by p204 of the specific DNA binding of UBF-1. A direct interaction between p204 and UBF1 was revealed in vitro in pull-down assays, and in vivo by co-immunoprecipitation from cell extracts previously. UBF1 bound strongly to at least two regions of p204: the N-terminal segment linked to the conserved 200 amino acid a segment, and the conserved 200 amino acid b segment. Cleavage of the a or b segments into two segments (encoded by single exons) resulted in a strong decrease or loss of binding. Thus, p204 serves as a mediator of the inhibition of rRNA transcription by interferon (4). This inhibition of ribosomal RNA synthesis is likely to contribute to the antiproliferative and differentiation promoting activities of p204.
Summary and future directions
Growing evidences demonstrate the essential role of the p200 family proteins as sensors of the infection of virus or bacteria. IFI16 or p204 may act as upstream cytosolic dsDNA receptors to interact with STING, then activate TBK1-IRF3/NF-kB axis, drive induction of type I IFN and pro-inflammatory cytokines. However, there is a negative feedback loop between IFI16 and STING during the cytosolic sensing of bacterial second messenger cyclic-di-GMP (c-di-GMP). IFI16 has also been identified as a nuclear pathogen sensor that interacts with the adaptor molecule ASC and procaspase-1 to form a functional inflammasome in endothelial cells during Kaposi Sarcoma-Associated Herpesvirus (KSHV) infection. This interaction results in the caspase-1 activation and proinflammatory cytokines production. IFI16 protein also interacts with AIM2 in the cytoplasmic fraction and increased expression of IFI16 protein in transfected cells can inhibit the AIM2-ASC-mediated activation of caspase-1.11 The knockdown of the IFI16 expression moderately increases the basal levels of AIM2 and activated caspase-1. AIM2 is the first member of p200 protein family defined as a sensor of DNA in the innate immune defense. AIM2 binds to DNA and interacts with ASC, which could activate caspase-1. AIM2, ASC and caspase-1 form the inflammasomes which mediate the downstream proinflammatory pathways and cleave both IL-1β and IL-18, which are important in early antiviral defenses. It is important to note that AIM2 is essential for inflammasome activation in response to bacterial infection such as Francisella tularensis and Listeria monocytogenes and S. aureus infection.
The exact mechanisms by which IFI16 regulates the immune defenses during infection of virus or bacteria are not fully understood. It has been established that IFI16 can detect foreign DNA in the nucleus but prevent interacting with the cellular DNA. A recent study63 shows that cooperative assembly into filaments on the dsDNA may serve as an integral mechanism by which IFI16 engages foreign DNA, and it is capable of using the size of naked dsDNA as a molecular ruler to distinguish host from foreign DNA. More studies are required to determine the interaction between IFI16/p204 and other sensors of DNA as well as the molecular events involved.
The further exploration of the functions of p204 would be greatly facilitated by the generation of mice in which p204 formation could be inhibited by inducible tissue-specific and/or embryo developmental stage-specific knockout.55 The availability of such mice would open the door to new research possibilities with p204, including the testing of conclusions drawn from experiments with cultured cells. Future research will advance our understanding of the roles of both IFI16 and p204 in the immune system and other pathophysiological processes.
Conflicts of interest
We herein declare that we have no conflict of interest.
Acknowledgments
This work is supported partly by NIH Research Grants R01AR062207, R01AR061484, and R56AI100901, Disease Targeted Research Grants from the American College of Rheumatology Research and Education Foundation.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Park H.H., Lo Y.C., Lin S.C., Wang L., Yang J.K., Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choubey D., Li S.J., Datta B., Gutterman J.U., Lengyel P. Inhibition of E2F-mediated transcription by p202. EMBO J. 1996;15:5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 3.Lembo M., Sacchi C., Zappador C. Inhibition of cell proliferation by the interferon-inducible 204 gene, a member of the Ifi 200 cluster. Oncogene. 1998;16:1543–1551. doi: 10.1038/sj.onc.1201677. [DOI] [PubMed] [Google Scholar]
- 4.Liu C.J., Wang H., Lengyel P. The interferon-inducible nucleolar p204 protein binds the ribosomal RNA-specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 1999;18:2845–2854. doi: 10.1093/emboj/18.10.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min W., Ghosh S., Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xin H., D'Souza S., Fang L., Lengyel P., Choubey D. p202, an interferon-inducible negative regulator of cell growth, is a target of the adenovirus E1A protein. Oncogene. 2001;20:6828–6839. doi: 10.1038/sj.onc.1204844. [DOI] [PubMed] [Google Scholar]
- 7.Dauffy J., Mouchiroud G., Bourette R.P. The interferon-inducible gene, Ifi204, is transcriptionally activated in response to M-CSF, and its expression favors macrophage differentiation in myeloid progenitor cells. J Leukoc Biol. 2006;79:173–183. doi: 10.1189/jlb.0205083. [DOI] [PubMed] [Google Scholar]
- 8.Ding B., Liu C.J., Huang Y. p204 is required for the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes: its expression is activated by the cardiac Gata4, Nkx2.5, and Tbx5 proteins. J Biol Chem. 2006;281:14882–14892. doi: 10.1074/jbc.M511747200. [DOI] [PubMed] [Google Scholar]
- 9.Ding B., Liu C.J., Huang Y., Yu J., Kong W., Lengyel P. p204 protein overcomes the inhibition of the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes by Id proteins. J Biol Chem. 2006;281:14893–14906. doi: 10.1074/jbc.M511748200. [DOI] [PubMed] [Google Scholar]
- 10.Liu C., Wang H., Zhao Z. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Mol Cell Biol. 2000;20:7024–7036. doi: 10.1128/mcb.20.18.7024-7036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C.J., Chang E., Yu J. The interferon-inducible p204 protein acts as a transcriptional coactivator of Cbfa1 and enhances osteoblast differentiation. J Biol Chem. 2005;280:2788–2796. doi: 10.1074/jbc.M412604200. [DOI] [PubMed] [Google Scholar]
- 12.Luan Y., Yu X.P., Xu K. The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the Cbfa1 transcription factor thereby increasing its activity. J Biol Chem. 2007;282:16860–16870. doi: 10.1074/jbc.M610943200. [DOI] [PubMed] [Google Scholar]
- 13.Luan Y., Yu X.P., Yang N., Frenkel S., Chen L., Liu C.J. p204 protein overcomes the inhibition of core binding factor alpha-1-mediated osteogenic differentiation by Id helix-loop-helix proteins. Mol Biol Cell. 2008;19:2113–2126. doi: 10.1091/mbc.E07-10-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludlow L.E., Purton L.E., Klarmann K. The role of p202 in regulating hematopoietic cell proliferation and differentiation. J Interferon Cytokine Res. 2008;28:5–11. doi: 10.1089/jir.2007.0070. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Kong L., Carlson C.S., Liu C.J. Cbfa1-dependent expression of an interferon-inducible p204 protein is required for chondrocyte differentiation. Cell Death Differ. 2008;15:1760–1771. doi: 10.1038/cdd.2008.112. [DOI] [PubMed] [Google Scholar]
- 16.Aglipay J.A., Lee S.W., Okada S. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8938. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Liu C., Lu Y. The interferon- and differentiation-inducible p202a protein inhibits the transcriptional activity of c-Myc by blocking its association with Max. J Biol Chem. 2000;275:27377–27385. doi: 10.1074/jbc.M003409200. [DOI] [PubMed] [Google Scholar]
- 18.Yan D.H., Abramian A., Li Z. P202, an interferon-inducible protein, inhibits E2F1-mediated apoptosis in prostate cancer cells. Biochem Biophys Res Commun. 2003;303:219–222. doi: 10.1016/s0006-291x(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 19.Xin H., Curry J., Johnstone R.W., Nickoloff B.J., Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 20.Xin H., Pereira-Smith O.M., Choubey D. Role of IFI 16 in cellular senescence of human fibroblasts. Oncogene. 2004;23:6209–6217. doi: 10.1038/sj.onc.1207836. [DOI] [PubMed] [Google Scholar]
- 21.Unterholzner L., Keating S.E., Baran M. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monroe K.M., Yang Z., Johnson J.R. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burckstummer T., Baumann C., Bluml S. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 24.Roberts T.L., Idris A., Dunn J.A. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 25.Hornung V., Ablasser A., Charrel-Dennis M. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludlow L.E., Johnstone R.W., Clarke C.J. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 28.Deschamps S., Meyer J., Chatterjee G., Wang H., Lengyel P., Roe B.A. The mouse Ifi200 gene cluster: genomic sequence, analysis, and comparison with the human HIN-200 gene cluster. Genomics. 2003;82:34–46. doi: 10.1016/s0888-7543(03)00092-2. [DOI] [PubMed] [Google Scholar]
- 29.Pinna L.A., Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 30.Choubey D., Lengyel P. Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72-kD protein that is encoded by the Ifi 204 gene from the gene 200 cluster. J Cell Biol. 1992;116:1333–1341. doi: 10.1083/jcb.116.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding B., Lengyel P. p204 protein is a novel modulator of ras activity. J Biol Chem. 2008;283:5831–5848. doi: 10.1074/jbc.M709680200. [DOI] [PubMed] [Google Scholar]
- 32.Albrecht M., Choubey D., Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 33.Liao J.C., Lam R., Brazda V. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011;19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan H., Dalal K., Hon B.K., Youkharibache P., Lau D., Pio F. RPA nucleic acid-binding properties of IFI16-HIN200. Biochim Biophys Acta. 2008;1784:1087–1097. doi: 10.1016/j.bbapap.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Cridland J.A., Curley E.Z., Wykes M.N. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baeck S., Han E., Chung H., Pyo M. Effects of repeated hair washing and a single hair dyeing on concentrations of methamphetamine and amphetamine in human hairs. Forensic Sci Int. 2011;206:77–80. doi: 10.1016/j.forsciint.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 37.Singh V.V., Kerur N., Bottero V. Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J Virol. 2013;87:4417–4431. doi: 10.1128/JVI.03282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka A., Wang Z., Choi M.K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 39.Burdette D.L., Monroe K.M., Sotelo-Troha K. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper A., Garcia M., Petrovas C., Yamamoto T., Koup R.A., Nabel G.J. HIV integration and T cell death: additional commentary. Retrovirology. 2013;10:150. doi: 10.1186/1742-4690-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariathasan S., Weiss D.S., Newton K. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 44.Sutterwala F.S., Ogura Y., Szczepanik M. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Kanneganti T.D., Ozoren N., Body-Malapel M. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 46.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao D., Wu J., Wu Y.T. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., Wu J., Du F. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin T., Perry A., Jiang J. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiliveru S., Rahbek S.H., Jensen S.K. Inflammatory cytokines break down intrinsic immunological tolerance of human primary keratinocytes to cytosolic DNA. J Immunol. 2014;192:2395–2404. doi: 10.4049/jimmunol.1302120. [DOI] [PubMed] [Google Scholar]
- 52.Cristea I.M., Moorman N.J., Terhune S.S. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T., Chen J., Cristea I.M. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kerur N., Veettil M.V., Sharma-Walia N. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li T., Diner B.A., Chen J., Cristea I.M. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orzalli M.H., DeLuca N.A., Knipe D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson K.E., Chikoti L., Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horan K.A., Hansen K., Jakobsen M.R. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 60.Saitoh T., Fujita N., Hayashi T. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panchanathan R., Liu H., Xin D., Choubey D. Identification of a negative feedback loop between cyclic di-GMP-induced levels of IFI16 and p202 cytosolic DNA sensors and STING. Innate Immun. 2013;20:751–759. doi: 10.1177/1753425913507097. [DOI] [PubMed] [Google Scholar]
- 62.Doitsh G., Galloway N.L., Geng X. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proceedings of the National Academy of Sciences of the United States of America.111(1):E62–E71. [DOI] [PMC free article] [PubMed]
- 64.Ansari M.A., Singh V.V., Dutta S. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J Virol. 2013;87:8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berg R.K., Rahbek S.H., Kofod-Olsen E. T cells detect intracellular DNA but fail to induce type I IFN responses: implications for restriction of HIV replication. PLoS One. 2014;9:e84513. doi: 10.1371/journal.pone.0084513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dell'oste V., Gatti D., Gugliesi F. Innate nuclear sensor IFI16 translocates into the cytoplasm during early stage of in vitro HCMV infection and is entrapped in the egressing virions during late stage. J Virol. 2014;88:6970–6982. doi: 10.1128/JVI.00384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bieniasz P.D. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 68.Roy C.R., Mocarski E.S. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 69.Douglas J.L., Gustin J.K., Viswanathan K., Mansouri M., Moses A.V., Fruh K. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takeuchi H., Matano T. Host factors involved in resistance to retroviral infection. Microbiol Immunol. 2008;52:318–325. doi: 10.1111/j.1348-0421.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- 71.Milbradt J., Webel R., Auerochs S., Sticht H., Marschall M. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J Biol Chem. 2010;285:13979–13989. doi: 10.1074/jbc.M109.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marschall M., Stein-Gerlach M., Freitag M., Kupfer R., van Den Bogaard M., Stamminger T. Inhibitors of human cytomegalovirus replication drastically reduce the activity of the viral protein kinase pUL97. J Gen Virol. 2001;82:1439–1450. doi: 10.1099/0022-1317-82-6-1439. [DOI] [PubMed] [Google Scholar]
- 73.Schattgen S.A., Fitzgerald K.A. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 74.Davis B.K., Wen H., Ting J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 76.Rathinam V.A., Jiang Z., Waggoner S.N. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fernandes-Alnemri T., Yu J.W., Juliana C. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mariathasan S., Weiss D.S., Dixit V.M., Monack D.M. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim S., Bauernfeind F., Ablasser A. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warren S.E., Armstrong A., Hamilton M.K. Cutting edge: cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanamsagar R., Aldrich A., Kielian T. Critical role for the AIM2 inflammasome during acute central nervous system bacterial infection. J Neurochem. 2014;129:704–711. doi: 10.1111/jnc.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veeranki S., Duan X., Panchanathan R., Liu H., Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 84.Choubey D. DNA-responsive inflammasomes and their regulators in autoimmunity. Clin Immunol. 2012;142:223–231. doi: 10.1016/j.clim.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caneparo V., Cena T., De Andrea M. Anti-IFI16 antibodies and their relation to disease characteristics in systemic lupus erythematosus. Lupus. 2013;22:607–613. doi: 10.1177/0961203313484978. [DOI] [PubMed] [Google Scholar]
- 86.Liu C.J., Ding B., Wang H., Lengyel P. The MyoD-inducible p204 protein overcomes the inhibition of myoblast differentiation by Id proteins. Mol Cell Biol. 2002;22:2893–2905. doi: 10.1128/MCB.22.9.2893-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 88.Iavarone A., Garg P., Lasorella A., Hsu J., Israel M.A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 89.Norton J.D., Deed R.W., Craggs G., Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 90.Vinals F., Reiriz J., Ambrosio S., Bartrons R., Rosa J.L., Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 2004;23:3527–3537. doi: 10.1038/sj.emboj.7600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katagiri T., Yamaguchi A., Komaki M. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127(6 Pt 1):1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogata T., Wozney J.M., Benezra R., Noda M. Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc Natl Acad Sci U S A. 1993;90:9219–9222. doi: 10.1073/pnas.90.19.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng Y., Kang Q., Luo Q. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 94.Karsenty G., Wagner E.F. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 95.Deftos M.L., Huang E., Ojala E.W., Forbush K.A., Bevan M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gribaudo G., Riera L., De Andrea M., Landolfo S. The antiproliferative activity of the murine interferon-inducible Ifi 200 proteins depends on the presence of two 200 amino acid domains. FEBS Lett. 1999;456:31–36. doi: 10.1016/s0014-5793(99)00916-3. [DOI] [PubMed] [Google Scholar]
- 97.Asefa B., Dermott J.M., Kaldis P., Stefanisko K., Garfinkel D.J., Keller J.R. p205, a potential tumor suppressor, inhibits cell proliferation via multiple pathways of cell cycle regulation. FEBS Lett. 2006;580:1205–1214. doi: 10.1016/j.febslet.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 98.Hertel L., Rolle S., De Andrea M. The retinoblastoma protein is an essential mediator that links the interferon-inducible 204 gene to cell-cycle regulation. Oncogene. 2000;19:3598–3608. doi: 10.1038/sj.onc.1203697. [DOI] [PubMed] [Google Scholar]
- 99.Luan Y., Lengyel P., Liu C.J. p204, a p200 family protein, as a multifunctional regulator of cell proliferation and differentiation. Cytokine Growth Factor Rev. 2008;19:357–369. doi: 10.1016/j.cytogfr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]