Abstract
Establishment, maintenance, and exit from pluripotency require precise coordination of a cell’s molecular machinery. Substantial headway has been made in deciphering many aspects of this elaborate system, particularly with respect to epigenetics, transcription, and noncoding RNAs. Less attention has been paid to posttranscriptional regulatory processes such as alternative splicing, RNA processing and modification, nuclear export, regulation of transcript stability, and translation. Here, we introduce the RNA binding proteins that enable the posttranscriptional regulation of gene expression, summarizing current and ongoing research on their roles at different regulatory points and discussing how they help script the fate of pluripotent stem cells.
Introduction
Embryonic stem cells (ESCs), which are derived from the inner cell mass of the mammalian blastocyst, are remarkable because they can propagate in vitro indefinitely while retaining both the molecular identity and the pluripotent properties of the peri-implantation epiblast. Consequently, ESCs provide a biologically relevant and experimentally tractable model system for studying regulators of cell fate and cell fate transitions in early development. Understanding the molecular mechanisms of ESC maintenance and differentiation is critically important not just scientifically but also clinically, because an improved knowledge of pluripotency and embryonic development will allow ESCs to be more effectively utilized as an in vitro platform for disease modeling, drug discovery, and tissue regeneration.
While the transcriptional, signaling, and epigenetic regulation of these cells have been the primary focus of research efforts in recent years (reviewed in Ng and Surani, 2011; Young, 2011; Watanabe et al., 2013), posttranscriptional and translational mechanisms of control remain relatively unexplored, despite evidence that they play a dominant role in driving ESC fate decisions. Indeed, posttranscriptional regulation has been reported to account for nearly 75% of the changes in protein levels after differentiation induced by knockdown of the transcription factor Nanog (Lu et al., 2009), and it was recently demonstrated that control over translational initiation by the eIF4e binding proteins dramatically influences the efficiency of reprogramming somatic cells to induced pluripotent stem cells (iPSCs) (Tahmasebi et al., 2014). The cell controls protein levels posttranscriptionally using a large collection of tools that includes noncoding RNAs and RNA binding proteins (RBPs). Recent work elucidating the functions of microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) in ESCs has been comprehensively reviewed elsewhere (Greve et al., 2013; Ghosal et al., 2013; Wright and Ciosk, 2013; Flynn and Chang, 2014). Similarly, posttranslational regulation of protein levels through the addition of covalent modifications also has been discussed recently (Wang et al., 2014b). The purpose of this Review is specifically to address the roles of RBPs in the maintenance and differentiation of ESCs.
RBPs are responsible for every event in the life of an RNA molecule, including its capping, splicing, cleavage, nontemplated nucleotide addition, nucleotide editing, nuclear export, cellular localization, stability, and translation (Keene, 2007). Overall, little is known about RBPs: most are classified based on computationally predicted similarities to proteins with known RNA binding domains, and until recently, few of these predictions have been verified in a cellular context in vivo. The recent introduction of a technique termed “mRNA interactome capture,” which enables the identification of proteins bound to polyadenylated RNAs in vivo, has been a significant development for the field (Baltz et al., 2012; Castello et al., 2012). Using this method, several groups were able to create a comprehensive catalog of RBPs in different mammalian cells, including 555 RBPs in mouse ESCs (Kwon et al., 2013). However, the functions of these RBPs in ESCs and their changes in ESC differentiation have yet to be addressed. Indeed, the mechanism of action of only a small number of RBPs has been examined in any great detail in the context of pluripotency. Here, we summarize current knowledge of RBP contribution to posttranscriptional and translational regulation in ESCs, following the approximate order that each regulatory event would be encountered as a transcript born in the nucleus migrates into the cytoplasm and is translated into a polypeptide (diagrammed in Figure 1). Throughout, we also discuss potential directions of future inquiry that will allow us to more fully appreciate the scope of RBP-mediated posttranscriptional and translational regulation in pluripotency.
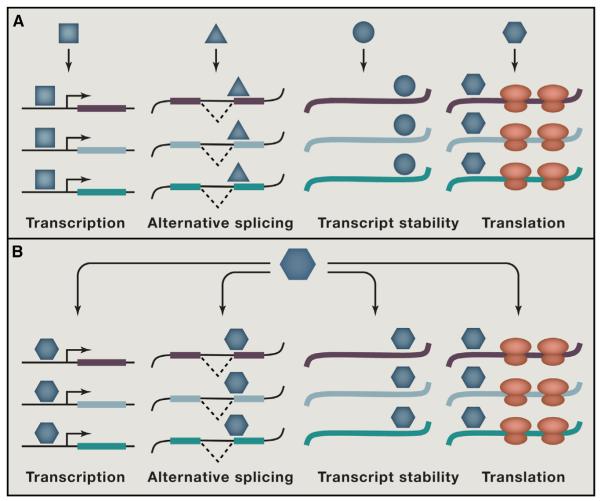
Figure 1. RBPs Involved in Pluripotency Act at Many Different Regulatory Steps.

Summary of the RBPs and the events they regulate in the maintenance and exit from pluripotency as discussed in this Review. Starting in the nucleus, RBPs regulate splicing (FOX2, SON, SFRS2, MBNL1, and MBNL2) and alternative polyadenylation (FIP1) simultaneously with transcription. RBPs then regulate export of transcripts (THOC2 and THOC5). RBPs also can induce modifications to RNAs including nucleotide changes (ADAR, METTL3, and METTL14 in nucleus) and nucleotidyl transfer (LIN28A in association with the TUTases ZCCHC6 and ZCCHC11 in the cytoplasm), which in turn influence mRNA stability and translation. In the cytoplasm, the binding of RBPs to the 3′UTRs of transcripts directly regulates mRNA stability and translation (TRIM71, PUM1, and BRF1). Translation is also influenced by RBPs that bind the 5′UTR of transcripts (NAT1, RBM35A, and PTBP1). Blue circles indicate RBPs. RBP genes in red are positive regulators of pluripotency. RBP genes in green are negative regulators of pluripotency. Black circles indicate the protein products of the genes whose expression levels are affected by RBPs.
Alternative Splicing
Alternative splicing of mRNA transcripts is probably the best-studied area of RBP-driven posttranscriptional control in ESCs, both in terms of the maintenance of the pluripotent state as well as in the artificial induction of pluripotency in somatic cells. A number of key pluripotency factors have been shown to exist as multiple isoforms that vary in stability, function, and intracellular localization due to differences in either coding exons or untranslated regulatory sequences. For example, Oct4 has two isoforms: Oct4A is the canonical pluripotency transcription factor expressed in ESCs and embryonal carcinoma cells, while Oct4B is a cytoplasmic protein with unknown functions expressed in nonpluripotent cells (Atlasi et al., 2008). Intriguingly, Oct4 was identified in the HeLa mRNA interactome (Castello et al., 2012), raising the possibility that certain isoforms of Oct4 could be acting as RBPs in particular cell types. Other important alternatively spliced genes affiliated with the ESC state include Sall4, which has two isoforms that play collaborative roles in maintaining pluripotency; Tcf3, whose two isoforms inhibit Nanog and Oct4 transcription to different degrees (reviewed in Cheong and Lufkin, 2011); Nanog, whose three isoforms maintain ESC pluripotency with varying efficacies (Das et al., 2011); and DNMT3B, whose ~40 isoforms are each differentially expressed in pluripotent cells and various distinct tissue types (Gopalakrishnan et al., 2009). An especially well-developed story is that of Foxp1, which possesses several isoforms, including one that is uniquely expressed in ESCs. This particular transcript contains an exon variant that encodes an altered forkhead domain, yielding an ESC-specific Foxp1 with a DNA binding motif different from the canonical Foxp1 consensus motif. Demonstrating the dramatic impact of this change in sequence specificity, only the ESC form of Foxp1 can stimulate the transcription of pluripotency genes and repress ESC differentiation genes (Gabut et al., 2011). Thus, this interesting example shows how alternative splicing can be an integral part of the switch that regulates the choice between self-renewal and differentiation.
Transcript splicing is mediated by a core set of proteins and small nuclear RNAs that comprise the spliceosome, but additional RBPs are critical for dictating the spatial and temporal specificity of splicing events needed to drive tissue-specific expression of mRNA isoforms. Despite the obvious importance of this posttranscriptional phenomenon, the RBPs that enable the establishment and maintenance of the ESC splicing signature have only begun to be deciphered. Thus far, both positive (FOX2, SON, and SFRS2) and negative (MBNL1 and MBNL2) regulators of the ESC-specific splicing signature have been identified; the list of RBPs involved is sure to increase as research in this area expands.
In human ESCs (hESCs), FOX2 (RBM9 and FXH) promotes exon exclusion when bound to the upstream flanking intron and exon inclusion when bound to the downstream flanking intron, as demonstrated by crosslinking immunoprecipitation with high-throughput sequencing (CLIP-seq). Interestingly, many FOX2 targets are splicing factors as well, which suggests that FOX2 is an upstream master regulator of splicing regulators. FOX2 activity is critical for pluripotency, because depletion of the gene leads to hESC differentiation and death, thus highlighting the functional importance of active maintenance of the ESC splicing program (Yeo et al., 2009).
Another RBP critical for maintaining the hESC-specific complement of transcript variants is the spliceosome-associated factor SON. A genome-wide RNAi screen found that knockdown of SON caused hESC differentiation. Sequencing of wild-type and SON-depleted hESCs demonstrated that SON specifically targets short introns with GC-rich weak splice sites, and in the absence of SON, the transcripts of a number of key pluripotency genes like OCT4 and PRDM14 have aberrant inclusion of introns, leading to their degradation by nonsense-mediated decay (Lu et al., 2013). Like FOX2, then, SON helps create a splicing signature important for specifying and maintaining the pluripotent state.
A third splicing factor that positively regulates the self-renewing state of hESCs was described in a recent report (Lu et al., 2014). Sifting through transcriptome and proteome data comparing hESCs and hiPSCs with fibroblasts, these authors identified SFRS2 as the most enriched splicing factor in pluripotent cells and MBD2 as the most differentially spliced transcript. This methyl-CpG binding protein exists as three isoforms (MBD2a–MBD2c), with MBD2c enriched in pluripotent stem cells and MBD2a in fibroblasts. Stable knockdown of SFRS2 in hESCs reduces expression of pluripotency genes and is associated with an isoform switch from MBD2c to MBD2a due to SFRS2 binding upstream of an exon unique to MBD2c. While overexpression of MBD2a promotes hESC differentiation, overexpression of MBD2c enhanced reprogramming of fibroblasts to hiPSCs. Interestingly, MBD2 had previously been identified as one of the miR-302 mRNA targets partially underlying the ability of this family of miRNAs to promote somatic cell reprogramming (Lee et al., 2013; Subramanyam et al., 2011). Here, the authors show that miR-302 specifically targets MBD2a, but not MBD2c, suggesting that the effect of MBD2 on pluripotency is isoform dependent. In sum, SFRS2 provides an exciting example of how multiple regulatory pathways can cooperate to promote the pluripotent state.
In contrast to the positive regulators of ESC self-renewal described above, MBNL1 and MBNL2 repress ESC splicing patterns. Knockdown of either gene causes both human and mouse somatic cell lines to shift toward an ESC-like splicing pattern, including the acquisition of the ESC isoform of Foxp1. Moreover, depletion of Mbnl1 and Mbnl2 improved the efficiency of mouse somatic cell reprogramming of MEFs. Splicing code analysis in human and mouse ESCs suggests that the presence of Mbnl binding sites upstream of an exon is associated with its inclusion, while downstream binding is associated with exclusion—a pattern of binding opposite to that of FOX2 (Han et al., 2013).
Together, the examples of FOX2, SON, SFRS2, and MBNL1/2 reveal a very active role for RBPs that regulate splicing in both self-renewal and differentiation conditions. Determining how RBPs regulate splicing programs in cell fate choices encountered during the development of the three embryonic germ layers as well as the germline will be an important area of future pursuit.
Alternative 3′UTR Cleavage and Polyadenylation
In addition to varying the combination of coding exons that are included in an mRNA molecule, posttranscriptional regulatory machineries influence the processing of the 3′ ends of transcripts. Many mammalian genes contain more than one polyadenylation site that can be recognized for cleavage and addition of the poly(A) tail, and current estimates suggest that upward of 50%–70% of mammalian RNAs are subject to alternative polyadenylation (APA), leading to transcripts with varying 3′UTR lengths (Shepard et al., 2011; Derti et al., 2012). APA appears to be closely linked with cell state: for example, ESC differentiation and embryonic development is associated with 3′UTR lengthening (Ji et al., 2009; Shepard et al., 2011), while proliferation and somatic cell reprogramming are accompanied by 3′UTR shortening (Sandberg et al., 2008; Ji and Tian, 2009). This is thought to occur because the length of the 3′UTR can affect the availability of RBP and miRNA binding sites that are critical for determining transcript stability, localization, and translation. Indeed, there have been a few notable examples of how mRNA isoforms with shorter 3′UTRs can be more stable (Mayr and Bartel, 2009, Boutet et al., 2012) and more efficiently translated (Sandberg et al., 2008) than their longer counterparts due to differential inclusion of repressive miRNA binding sites. However, the biological roles of APA remain unclear at the global level, because sequencing studies, including one recently conducted in NIH 3T3 fibroblasts, have shown that long and short 3′UTR mRNA isoforms of the same genes have comparable stability and translation levels (Spies et al., 2013). These observations suggest, among other possibilities, that the effects of APA on transcript stability and translation may be specific to certain subsets of biologically important genes, while for the majority of transcripts, APA primarily serves some other regulatory purpose, such as intracellular localization (Spies et al., 2013).
Transcript cleavage and polyadenylation occur through the cooperation of several protein complexes: cleavage and polyadenylation specificity factor (CPSF) recognizes the polyadenylation signal (PAS) upstream of the cleavage site, while cleavage stimulating factor (CSTF) recognize downstream sequence elements (DSEs), and this leads to the recruitment of cleavage factor I and II, poly(A) binding proteins, and poly(A) polymerase (reviewed in Elkon et al., 2013). Interestingly, one of the protein subunits of CPSF, Fip1, was recently described to play a role in creating the APA profile of ESCs (Lackford et al., 2014). Consistent with its being important in pluripotency, Fip1 knockdown in ESCs promoted differentiation. Deep sequencing revealed that Fip1 depletion alters the APA profile of 374 genes, with most displaying a lengthening of their 3′UTR by shifting to the more distal PAS (proximal-to-distal shift [PtoD]). The genes showing PtoD were generally more highly expressed in self-renewing conditions than in differentiation conditions, and knockdown of a representative set partially recapitulated the Fip1 knockdown phenotype. Individual nucleotide crosslinking and immunoprecipitation followed by high-throughput sequencing (iCLIP-seq) showed that Fip1 binds U-rich sequences upstream of PAS sequences. Interestingly, when the PASs were located far apart, Fip1 bound upstream of the proximal PAS and promoted cleavage and polyadenylation at that site. However, when the PASs were close together, Fip1 promoted usage of the distal site. Given these data, the authors propose a model in which Fip1 promotes cleavage and polyadenylation at all PASs with upstream U-rich regions. However, when Fip1 is highly expressed, as it is in ESCs, it will bind and promote cleavage of the weaker proximal PAS, if there is a sufficient delay in transcription between the two PAS sites. This interesting model demonstrates how a general CPSF factor can regulate pluripotency by linking 3′UTR length with production of critical self-renewal factors. It will be important to investigate whether these results are unique to Fip1 or whether there are additional factors in ESCs that are critical for determining PAS usage in other sets of genes.
RNA Modification
Posttranscriptional RNA modification offers another exciting means for improved control over transcript levels and even increased proteomic diversity without the need to make changes to the genome. In regulatory regions, sequence alterations can affect transcript stability and translation efficiency, while in transcript coding regions, nucleotide changes can increase protein product diversity. RNA modifications can be separated into two types: the nontemplated addition of nucleotides and the chemical modification of templated bases. A well-established example of the former is the addition of nucleotides to the 3′ ends of RNA molecules. For instance, addition of uridines (uridylation) to pre-let-7 miRNA leads to its degradation in ESCs. In particular, the 3′ terminal uridylyl transferases (TUTases), Zcchc11 and Zcchc6, are directed by the RBP Lin28a to add a string of approximately 11 uridines to the pre-miRNA (Heo et al., 2008; Hagan et al., 2009). Oligouridylated pre-let-7 is then recognized by the DIS3L2 exoribonuclease, which catalyzes transcript degradation (Ustianenko et al., 2013). Interestingly, the addition of a single uridine has the opposite effect, promoting the processing of let-7 to its mature form (Heo et al., 2012). The remarkable number of pathways regulating let-7 activity befits its important role in promoting ESC differentiation and blocking reprogramming of somatic cells to iPSCs (Melton et al., 2010).
Although the existence of base modifications has been known for some time, their potential regulatory roles in mammalian cell fate decisions are just beginning to be uncovered. For example, the RNA editing enzyme Adar (Adar1) catalyzes the conversion of adenosine to inosine, which is subsequently read as guanosine by the translation machinery. Adar can target both mRNAs and dsRNA precursors of endo-siRNAs and miRNAs. In hESCs, ADAR was shown to drive A-to-I editing across the transcriptome, particularly in Alu repetitive elements. Furthermore, transient ADAR knockdown with an siRNA decreased A-to-I editing and led to an increase in the mRNA expression of genes associated with differentiation and development, although the ADAR-depleted ESCs did not exhibit significant morphological changes compared to wild-type (Osenberg et al., 2010). These results were based on the effects of a single transiently transfected siRNA, so it will be important to further examine the role of Adar by multiple different siRNAs, stable shRNA knockdown, and/or genetic deletion to rule out the possibility of off-target effects and to separate the effects of acute versus chronic loss of Adar. Moreover, the direct downstream consequences of A-to-I editing on targeted transcripts in ESCs remain to be elucidated. It is important to note that Adar has been found to have a number of different roles in other cellular systems. In a complex with Elavl1 (HuR), for example, Adar modulates mRNA stability (Wang et al., 2013a). Adar can also affect miRNA processing both negatively and positively, editing pri-miRNA transcripts to prevent Microprocessor cleavage (Wu et al., 2011) or dimerizing with Dicer in an RNA-dependent way to promote pre-miRNA cleavage (Ota et al., 2013). But because these studies were conducted in non-ESC cell lines, it would be meaningful to investigate whether these other diverse functions of Adar are also relevant in ESCs.
While ADAR-driven A-to-I base conversion is clearly important, methylation of adenosine to form N6-methyladenosine (m6A) is the most prevalent chemical modification of eukaryotic mRNA. Enriched at stop codons and 3′UTRs, the presence of m6A marks appears to influence many aspects of RNA metabolism. Indeed, recent evidence suggests that methylation at internal positions within transcripts is associated with transcript destabilization, while methylation at the transcriptional start site enhances translation (Schwartz et al., 2014). Biochemical studies recently revealed that these m6A marks are primarily deposited by the methyltransferases Mettl14 and Mettl3, which function together as a heterodimer (Liu et al., 2014). In mESCs, stable shRNA knockdown of either Mettl3 or Mettl14 greatly reduces m6A levels and impairs ESC self-renewal (Wang et al., 2014a). Interestingly, m6A-immunoprecipitation and sequencing (meRIP-seq) revealed that mRNAs of epigenetically poised developmental regulators (marked by H3K4me3 and H3K27me3 bivalent chromatin domains) are normally highly methylated, and these genes are upregulated in Mettl3- or Mettl14-depleted cells due to increased transcript half-life. These data suggest that m6A destabilizes transcripts. Further examining the mechanistic connection between methylation and RNA stability, the authors found that Mettl3/Mettl14 knockdown is associated with increased binding of the RBP HuR near m6A sites and leads to increased transcript stability, suggesting that HuR competes with Mettl3/Mettl14 and that their differential expression dynamically regulates RNA half-life. While it will be important to confirm these results in a genetic knockout system and further investigate the mechanism of methylated RNA destabilization, this study is intriguing for raising the idea that, like DNA methylation and chromatin modifications, “epigenetic” modification of RNA can be critical for determining cell identity.
Finally, the connections to HuR seen with both Adar- and Mettl3/Mettl14-mediated RNA editing are tantalizing, because HuR is also known to regulate transcript stability by either cooperating with or antagonizing miRNAs through a variety of different, context-dependent mechanisms. Additionally, HuR expression is itself modulated by miRNAs (reviewed in Simone and Keene, 2013). Together these studies exemplify how RNA modifications, RBPs, and miRNAs can function together to control steady-state mRNA expression levels.
Nuclear Export
In addition to directly altering the transcript sequences through alternative splicing, alternative polyadenylation, and nucleotide modifications, RNA fate can be influenced by the regulation of transport between the nucleus and cytoplasm, which controls protein levels by modulating access to the translation machinery. An exciting recent study has shown that such regulation can play a significant role in ESCs (Wang et al., 2013b). In an RNAi screen for factors that are required for ESC self-renewal, Hu and colleagues identified Thoc2 and Thoc5, two members of the THO complex, which provide an interface between nuclear transcription and RNA export. The authors show that knockdown of Thoc2 and Thoc5 does not alter overall transcript levels but rather leads to a nuclear accumulation of a subset of pluripotency mRNAs, including Nanog, Sox2, Klf4, and Esrrb. Immunoprecipitation followed by deep sequencing further showed that Thoc2 binds a subset of mRNAs in a Thoc5-dependent manner. Interestingly, Thoc5 is believed to be an adaptor protein, and its expression is downregulated with ESC differentiation. Overexpression of Thoc5 delays ESC differentiation, while knockdown promotes ESC differentiation and blocks somatic cell reprogramming. Therefore, the downregulation of Thoc5 that occurs during normal development appears to be critical for silencing parts of the pluripotency program and thus promoting differentiation. These findings uncover a new aspect of posttranscriptional control in pluripotency and may be just the first of many more examples to come of transcript-specific nuclear export influencing early embryonic cell fate decisions.
Regulation of mRNA Stability
Whether a molecule of RNA is stable or is quickly degraded in the cytoplasm governs, to a large extent, its ultimate protein expression levels. Transcript stability, which varies among cell types (Neff et al., 2012), is strongly influenced by RBP and miRNA binding, particularly at the 3′UTR. Three RBPs that have been implicated in this mode of regulation are Trim71 (mLin41), Pum1, and Brf1 (zfp36l1).
The role of Trim71 is noteworthy because it demonstrates an interconnection among multiple protein complexes. miRNAs, which typically regulate transcript stability and translation by binding to 3′UTRs, are critical for enabling the rapid proliferation of ESCs by shortening the G1 phase of the cell cycle (Wang et al., 2008, 2013c), and Gregory and colleagues found that Trim71 is important for facilitating this activity (Chang et al., 2012). Using immunoprecipitation followed by mass spectrometry, they found that Trim71 binds to miRNA-containing Ago2 complexes. Through this molecular interaction, Trim71 cooperates with ESC-specific miR-290 and miR-302 to bind to the 3′UTR of Cdkn1a and inhibit its translation. Because Cdkn1a is a negative regulator of the G1-S transition, the miRNA-dependent 3′UTR activity of Trim71 promotes the cell cycle structure necessary for ESC self-renewal. Importantly, Trim71 function is tightly incorporated into a larger intracellular network because it is a target of the prodifferentiation miRNA let-7, whose biogenesis is inhibited by the RBP Lin28a (see the subsection “Multifunctional RBPs,” below). Adding to the complexity of this regulatory circuit, another study found that Trim71 is also an E3 ubiquitin ligase for Ago2 and showed through luciferase assays that Trim71 cooperates with Lin28a to inhibit let-7 activity (Rybak et al., 2009). Although Chang et al. (2012) did not detect a change in Ago2 levels upon perturbation of Trim71 expression, the work from these two groups clearly establishes Trim71 as a key player in several dimensions of miRNA-mediated ESC regulation. Intriguingly, more recent studies suggest that Trim71 also associates directly with some mRNAs at their 3′UTRs, independent of miRNA involvement, and that this interaction drives transcript degradation and inhibits translation (Loedige et al., 2013, Wor-ringer et al., 2014). Filipowicz and colleagues used luciferase assays to demonstrate Trim71 binding to the 3′UTRs of a subset of prodifferentiation genes, which led to a decrease in mRNA and protein levels in an Ago2-independent fashion (Loedige et al., 2013). Yamanaka and colleagues showed that upregulation of Trim71 enhanced human somatic cell reprogramming in part by directly binding to the mRNA of the fibroblast-enriched transcription factor EGR1 and decreasing its protein levels (Wor-ringer et al., 2014). Using deletion mutants, both of these groups showed that these direct effects on transcript repression do not involve the E3 ubiquitin ligase domain of Trim71. From all of these studies, it is clear that the story of Trim71 is highly complex, and further investigations will likely expand the scope of this RBP’s involvement in the regulation of pluripotency.
In contrast to Trim71, Pum1 appears to be a negative regulator of the ESC state. Pum1 is a member of the evolutionarily conserved PUF family of RBPs first characterized in Drosophila and C. elegans as inhibitors of translation and mRNA stability at the 3′UTR. A transposon-mediated mutagenesis screen in haploid ESCs found that Pum1 disruption promoted self-renewal in an ESC-like state when the cells were cultured in conditions permissive to differentiation (Leeb et al., 2014). This phenotype appeared to be mediated by Pum1 binding to its canonical eight-nucleotide motif in the 3′UTRs of multiple pluripotency genes, such as Tfcp21l, Esrrb, Klf2, and Sox2, which was associated with a decrease in their transcript levels, presumably through increased degradation of the mRNA. In addition to establishing a new role for Pum1 in ESCs, this study exemplifies the importance of RBP-3′UTR interactions in the maintenance of a unique cell state. Lastly, Pum1 has a closely related PUF family member, Pum2, which is a translational repressor in germ cell development (Moore et al., 2003). Because Pum2 is highly expressed in ESCs (Moore et al., 2003), it will be important to determine how these two PUF proteins genetically interact in the context of ESC self-renewal and differentiation.
Another RBP that destabilizes pluripotency transcripts is the AU-rich element mRNA-binding protein (AUBP) Brf1. AUBPs are known to be developmentally important regulators of splicing, stability, translation, and localization, and Brf1 is critical in embryogenesis: knockout mice die by E10.5 (Stumpo et al., 2004). A recent study found that activation of the FGF-Erk signaling pathway, which stimulates mouse ESC differentiation, leads to upregulation of Brf1 (Tan and Elowitz, 2014). Brf1 overexpression under pluripotency conditions impaired ESC proliferation, and under differentiation conditions it promoted the acquisition of mesendoderm fate. RNA immunoprecipitation and sequencing (RIP-seq) showed that Brf1 binds to AU-rich elements (AREs) in a number of pluripotency mRNAs, including Nanog, Klf2, Kdm4c, and Zfp143. Moreover, FGF stimulation led to degradation of Nanog transcripts in an ARE-dependent fashion, implicating Brf1 in mRNA destabilization and ESC differentiation. Future work will likely reveal more roles for AREs and other AUBPs in pluripotency and early embryonic development.
With all of these RBPs involved in determining the stability of different mRNA transcripts, it will be interesting to explore to what extent their targets overlap and whether they cooperate to coregulate a particular subset of transcripts. For those mRNAs with binding sites for several RBPs, it would furthermore be worthwhile to examine how the relative RBP concentrations and motif binding affinities affect transcript levels.
Regulation of Translation
RBPs can also directly modulate protein translation, often through binding at the 5′UTR of RNA transcripts. In so doing, RBPs recruit translation initiation factors, adjust the accessibility of the RNA to ribosomes, create ribonucleoprotein structures conducive for cap-independent and internal ribosome entry site (IRES)-mediated translation, and regulate the movement of the ribosomes along the transcript. Given these many roles, it is no surprise that RBPs involved in these 5′UTR-related processes have also been linked to the control of ESC pluripotency. In particular, RBP-5′UTR interactions involving the proteins Nat1, Rbm35a (Esrp1), and Ptbp1 have been shown to regulate ESC differentiation and proliferation.
Nat1 was first identified in liver carcinomas as a general repressor of translation. In mouse ESCs, depletion of Nat1 does not affect proliferation and self-renewal in ESC growth conditions, but Nat1−/− ESCs exhibit a defect in retinoic acid (RA)-induced differentiation (Yamanaka et al., 2000). Previous studies in other cell lines have shown that Nat1 is homologous to the translation initiation factor eIF4G, is located in the cytoplasm and can autoregulate its own translation from an IRES in its mRNA; thus, the authors speculated that Nat1 binds to highly structured 5′UTRs and associates with translational initiation factors. However, their study did not detect a difference in cap-dependent or cap-independent translation in wild-type and Nat1−/− ESCs as measured by [35S]methionine incorporation and bicistronic luciferase assays, respectively. Despite the negative results, it is important to note that these experiments were conducted under steady-state pluripotency conditions, not under the differentiation conditions in which they observed the phenotype. Therefore, it remains plausible that Nat1 influences ESC differentiation through translational mechanisms, and further studies should address the role of Nat1 in transitions into and out of pluripotency.
Like Nat1, Rbm35a is a negative regulator of pluripotency. Knockdown of Rbm35a inhibits ESC differentiation and promotes somatic cell reprogramming by increasing expression of Oct4, Nanog, and Sox2. Indeed, Rbm35a immunoprecipitation and polysome profiling show that Rbm35a normally binds to the 5′UTR of Oct4 and Sox2 mRNAs, thus preventing them from being loaded into polysomes (Fagoonee et al., 2013). In addition, Rbm35a activity is not restricted to ESCs, because the RBP is a regulator of alternative splicing in epithelial cell lines (Warzecha et al., 2009) and has also been shown be expressed in tumor cells where it binds to highly structured GC-rich 5′UTRs of oncogenes and prevents their translation (Leontieva and Ionov, 2009). ESCs themselves form teratomas when injected into mice and share many properties with somatic tumors, including limitless replicative potential. Therefore, Rbm35a suppression may be an important component of maintaining ESC and tumor cell immortality through both shared and unique molecular targets present in these different cellular contexts.
Ptbp1, another RBP that binds the 5′UTR of gene transcripts, also appears to control ESC growth through its regulation of the cell cycle. Ptbp1 knockout ESCs have a proliferation defect with a prolonged G2/M phase. This phenotype appears to be at least in part secondary to problems with chromosomal segregation. Bicistronic luciferase assays show that Ptbp1 binds to the IRES of CDK11p58 and represses translation of this gene, high levels of which are associated with a prolonged telophase caused by chromosomal lagging. Overexpression of CDK11p58 in wild-type ESCs led to chromosome missegregation (Ohno et al., 2011). Nevertheless, it is unclear whether or not overexpression of CDK11p58 is the sole contributor to the Ptbp1 knockout phenotype—a question that could be addressed through global comparisons of mRNA and protein abundance in wild-type and knockout cells. Notably, Ptbp1 has also been extensively studied in other cell lines, where it has been implicated in alternative splicing, alternative polyadenylation, mRNA stability at the 3′UTR, and IRES-driven translation (Boutz et al., 2007; Castelo-Branco et al., 2004; Kosinski et al., 2003; Bushell et al., 2006). It would be useful and informative for future studies to examine whether these other well-known functions of Ptbp1 are also involved in pluripotency.
Multifunctional RBPs
In addition to Adar, Pum1, Rbm35a, and Ptbp1, other RBPs have been proposed to play pleiotropic roles. For example, while Lin28a clearly regulates let-7 biogenesis through directing uridylation of pre-let-7, it appears to have additional functions independent of its relationship with let-7 (reviewed in Shyh-Chang and Daley, 2013). Through binding to an AG-rich motif similar to its pre-let-7 binding motif (Wilbert et al., 2012; Cho et al., 2012), Lin28a both positively and negatively modulates the translation of hundreds of mRNAs in human and mouse ESCs (Wilbert et al., 2012; Cho et al., 2012; Peng et al., 2011; Xu et al., 2009). How Lin28a achieves this incredibly complex orchestration of its many targets remains to be determined.
Lin28a is an unusual example of an RBP attributed with multiple functions in a single cellular context. To date, most studies done on multifunctional RBPs, like Ptbp1 and Adar, only investigate a single function of that RBP in ESCs, which does not exclude the possibility that the RBP’s other proposed functions are also important in supporting the pluripotency network. In general, a number of RBPs, such Ilf2, Unr, and others, have been genetically deleted in ESCs and shown to have an effect on the ESC phenotype, but currently, the functions of these RBPs can only be inferred from work done in other cell types. As every study ascribes a different role to each of these RBPs, it is unclear which, if any, are actually involved specifically in the context of pluripotency.
A notable example providing evidence that RBPs can be multifunctional within a single context comes from a recent study of the RNA helicases DDX5 and DDX17 not in pluripotent cells but in the differentiation of myoblast and epithelial cell lines (Dardenne et al., 2014). The authors showed that DDX5 and DDX17 cooperate with the hnRNP H/F splicing factors to promote a differentiation-specific splice profile. At the same time, DDX5 and DDX17 serve as transcriptional coregulators of key differentiation transcription factors as well as miRNAs that, in turn, negatively feed back on the expression of the RBPs themselves. It is unclear whether these two roles for DDX5 and DDX17 are physically linked; nevertheless, this story provides evidence that an RBP can simultaneously function at the level of DNA (transcription) and RNA (splicing).
Conclusions and Perspectives
It is without a doubt that posttranscriptional regulation by RBPs contributes extensively to the establishment and maintenance of, as well as exit from, the ESC state. The field remains relatively young and largely uncharted, however, and there are many opportunities for further inquiry and discovery.
As described above and summarized in Table 1, a number of studies have documented the significance of individual RBPs in pluripotency based on knockdown and knockout models demonstrating that perturbation of the RBP disrupts the wild-type ESC phenotype. However, few probe the actual mechanism by which the RBPs produce their effect in ESCs. Instead, RBP function has been examined mostly in cancer cell lines and cell-free biochemical assays, the in vivo relevance of which needs to be more clearly characterized. The studies that do delve into the molecular mechanisms in ESCs generally examine only a limited number of targets. While the RBPs discussed here are not unique to ESCs, cell context likely influences their downstream effects on cell fate and cell fate transitions significantly by providing a specific combination of intracellular pathways with which these RBPs interface. Thus, to reach a deep and unified understanding of the role of RBPs in pluripotency, we will need to make use of global approaches like those that have already been successfully applied to the field of epigenetics. In particular, we must dissect all RBP-RNA interactions systematically in ESC self-renewal and differentiation conditions so as to examine not only how RNA transcript levels change but also how the components of ribonucleoprotein (RNP) complexes are rearranged during transitions from one cell fate to another.
Table 1.
Summary of RBPs Associated with the Pluripotency Network
| Category of Posttranscriptional Regulation |
RBP | Positive or Negative Regulator of Pluripotency? |
Mechanism of Action in ESCs | Other Functions (Not Validated in ESCs) |
|---|---|---|---|---|
| Alternative splicing | FOX2 (RBM9, FXH) | positive (Yeo et al., 2009) | binds upstream of splice site for exon exclusion; binds downstream of splice site for exon inclusion |
|
| SON | positive (Lu et al., 2013) | binds short introns with weak splice sites |
||
| MBNL1, MBNL2 | negative (Han et al., 2013) | binds upstream of splice site for exon inclusion; binds downstream of splice site for exon exclusion |
||
| SFRS2 (SC35) | positive (Lu et al., 2014) | regulates splicing of MBD2 | ||
| Alternative Polyadenylation |
Fip1 | positive (Lackford et al., 2014) | promotes ESC-specific patterns of alternative polyadenylation |
|
| RNA modification | Zcchc11, Zcchc6 (TUTases) |
positive (Hagan et al., 2009) | inhibits let-7 biogenesis by uridylating the pre-miRNA |
|
| ADAR (ADAR1) | positive (Osenberg et al., 2010) | drives A-to-I editing | mRNA stability with Elavl1 (HuR); A-to-I editing of pri-miRNAs to prevent their processing; promote pre-miRNA cleavage in complex with Dicer |
|
| Mettl3, Mettl14 | positive (Wang et al., 2014a) | methylates adenosines in mRNA, reducing transcript stability |
||
| Nuclear export | Thoc2, Thoc5 (members of THO complex) |
positive (Wang et al., 2013b) | exports pluripotency gene transcripts to the cytoplasm for translation |
|
| RNA stability | Trim71 (mLin41) | positive (Chang et al., 2012; Rybak et al., 2009; Loedige et al., 2013; Worringer et al.,2014) |
binds Ago2 complexes, enhancing ESC miRNAs’ inhibition of Cdkn1a expression, which thus promotes ESC cell cycle structure; antagonizes Ago2 by ubiquitination; degrades prodifferentiation gene transcripts |
|
| Pum1 | negative (Leeb et al., 2014) | degrades pluripotency gene transcripts |
translation repressor | |
| Brf1 | negative (Tan and Elowitz, 2014) | degrades pluripotency gene transcripts |
||
| Translation | Nat1 | negative (Yamanaka et al., 2000) | unclear | translation repressor |
| Rbm35a | negative (Fagoonee et al., 2013) | prevents polysome loading of pluripotency gene transcripts |
alternative splicing | |
| Ptbp1 | positive (Ohno et al., 2011) | binds to IRES and represses translation of CDK11p58 |
alternative splicing; alternative polyadenylation; mRNA stability at 3′UTR |
|
| Multifunctional | Lin28a | positive (reviewed in Shyh-Chang and Daley, 2013) |
inhibits let-7 biogenesis by recruiting TUTases to uridylate pre-let-7; translational activation; mRNA degradation with Drosha |
Intriguingly, as mentioned above under the subsection “Multifunctional RBPs,” a number of RBPs appear to be involved in multiple aspects of RNA metabolism in both the nucleus and the cytoplasm. This observation may result from an RBP having different roles in different cell types, multiple roles in a single cell type, or some mix of the two. Regardless, it is almost certain that the particular combination of targets and cofactors that an RBP encounters influences its functions in context-specific ways—a notion that expands upon the “RNA regulon” model originally proposed by Jack Keene, in which an RBP binds multiple targets to effect changes in various cellular processes (Keene, 2007). In other words, it is possible that instead of regulating multiple targets through just a single mechanism, one RBP could simultaneously participate in several layers of RNA metabolism (Figure 2). In so doing, the RBP becomes part of increasingly complex regulatory modules, with many opportunities for feedback and crosstalk, where one aspect of metabolism such as translation can be linked to another such as splicing. Consequently, perturbation of the RBP could have a cascading effect on the molecular landscape of a cell and precipitate drastic switches in cellular identity. Also, it follows that disrupting subsets of the mRNA targets or cofactors of any one RBP could affect parallel pathways by shifting the RBP’s dominant activities to different genes or even different regulatory modules altogether. For example, a decrease in the levels of a cofactor that enables an RBP to regulate splicing could drive that RBP to shift its main activity to transcription.
Figure 2. The Multifunctional RBP: The RNA Regulon Revisited.
(A and B) Alternative models for RBP regulation of RNA metabolism. (A) In the classical view of the RNA regulon, an RBP (blue object) binds multiple transcripts to execute a single action on many RNAs (purple, blue, and green). This in turn can affect an array of cellular processes depending on the nature of the mRNAs targeted. (B) In an expanded version of the RNA regulon model, an RBP not only has multiple targets but also acts on those targets at multiple levels of intracellular RNA metabolism. An example RBP shown here regulates transcription, splicing, RNA stability, and mRNA translation of a common set of transcripts. While it is unlikely that such an RBP exists, we propose that many RBPs will have a subset of these functions on overlapping sets of targets.
Moving forward, it will be critical to investigate RBPs at the genomics level in biologically relevant cell types, focusing on those states in which the RBPs are demonstrated to play significant roles. Studying RBPs in ESCs provides an excellent starting context to achieve such a goal, because not only can ESCs provide unlimited untransformed material for large-scale genomic studies, they can also be differentiated down any cellular lineage to determine how context changes protein function. Thus, experimental tools and platforms developed in the ESC system can be used to study a multitude of cell types that comprise the mammalian body plan.
ACKNOWLEDGMENTS
We apologize to the authors whose work could not be in this Review due to limitations in space. This work was supported by funds to R.B. from the National Institutes of Health (R01: NS057221 and GM101180), the Leona M. and Harry B. Helmsley Charitable Trust (09PG-T1D002), and the California Institute of Regenerative Medicine (New Faculty Award: RN2-00906-1).
REFERENCES
- Atlasi Y, Mowla SJ, Ziaee SAM, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr., Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CY, Lufkin T. Alternative splicing in self-renewal of embryonic stem cells. Stem Cells Int. 2011:560261. doi: 10.4061/2011/560261. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, et al. RNA heli-cases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7:1900–1913. doi: 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Das S, Jena S, Levasseur DN. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J. Biol. Chem. 2011;286:42690–42703. doi: 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, Tolosano E, Provero P, Pandolfi PP, Silengo L, Altruda F. The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS ONE. 2013;8:e72300. doi: 10.1371/journal.pone.0072300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Van Emburgh BO, Shan J, Su Z, Fields CR, Vieweg J, Hamazaki T, Schwartz PH, Terada N, Robertson KD. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol. Cancer Res. 2009;7:1622–1634. doi: 10.1158/1541-7786.MCR-09-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve TS, Judson RL, Blelloch R. microRNA control of mouse and human pluripotent stem cell behavior. Annu. Rev. Cell Dev. Biol. 2013;29:213–239. doi: 10.1146/annurev-cellbio-101512-122343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kosinski PA, Laughlin J, Singh K, Covey LR. A complex containing polypyrimidine tract-binding protein is involved in regulating the stability of CD40 ligand (CD154) mRNA. J. Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM, et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Prasain N, Chae HD, Kim YJ, Mantel C, Yoder MC, Broxmeyer HE. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells. 2013;31:666–681. doi: 10.1002/stem.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontieva OV, Ionov Y. RNA-binding motif protein 35A is a novel tumor suppressor for colorectal cancer. Cell Cycle. 2009;8:490–497. doi: 10.4161/cc.8.3.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 2013;41:518–532. doi: 10.1093/nar/gks1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Göke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013;15:1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. http://dx.doi.org/10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, Dorfman DM, Pera RA. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff AT, Lee JY, Wilusz J, Tian B, Wilusz CJ. Global analysis reveals multiple pathways for unique regulation of mRNA decay in induced pluripotent stem cells. Genome Res. 2012;22:1457–1467. doi: 10.1101/gr.134312.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Ohno S, Shibayama M, Sato M, Tokunaga A, Yoshida N. Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11(p58) in mouse embryonic stem cells. Cell Cycle. 2011;10:3706–3713. doi: 10.4161/cc.10.21.17903. [DOI] [PubMed] [Google Scholar]
- Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob-Hirsch J, Keshet G, Amariglio N, Itskovitz-Eldor J, Rechavi G. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS ONE. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr. Opin. Genet. Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23:2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell. Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, et al. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell. 2014;14:606–616. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Tan FE, Elowitz MB. Brf1 posttranscriptionally regulates pluripotency and differentiation responses downstream of Erk MAP kinase. Proc. Natl. Acad. Sci. USA. 2014;111:E1740–E1748. doi: 10.1073/pnas.1320873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013a;5:849–860. doi: 10.1016/j.celrep.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013b;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Melton C, Li YP, Shenoy A, Zhang XX, Subramanyam D, Blelloch R. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep. 2013c;4:99–109. doi: 10.1016/j.celrep.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014a;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Peterson SE, Loring JF. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014b;24:143–160. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Yamada Y, Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120292. doi: 10.1098/rstb.2012.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JE, Ciosk R. RNA-based regulation of pluripotency. Trends Genet. 2013;29:99–107. doi: 10.1016/j.tig.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV, Jr., Iwao H, Innerarity TL. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000;19:5533–5541. doi: 10.1093/emboj/19.20.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]