Abstract
Analysis of protein phosphorylation remains a significant challenge due to the low abundance of phosphoproteins and the low stoichiometry of phosphorylation, which requires effective enrichment of phosphoproteins. Here we have developed superparamagnetic nanoparticles (NPs) whose surface is functionalized by multivalent ligand molecules that specifically bind to the phosphate groups on any phosphoproteins. These NPs enrich phosphoproteins from complex cell and tissue lysates with high specificity as confirmed by SDS-PAGE analysis with a phosphoprotein-specific stain and mass spectrometry analysis of the enriched phosphoproteins. This method enables universal and effective capture, enrichment, and detection of intact phosphoproteins towards a comprehensive analysis of the phosphoproteome.
Protein phosphorylation, one of the most common and important post-translational modifications, plays a pivotal role in the control of many biological processes, including cell growth, division, and signaling;1 and aberrant phosphorylation has been implicated in the pathogenesis of human diseases.2 Therefore, a comprehensive analysis of phosphoproteins is essential for understanding cellular biology and disease mechanisms. However, analysis of protein phosphorylation is challenging due to the low abundance of phosphoproteins and the low stoichiometry of phosphorylation as well as the extremely high dynamic range and the complexity of the proteome.3 Mass spectrometry (MS) has quickly become the method of choice for the analysis of protein phosphorylation.3–4 But it is nearly impossible to analyze phosphoproteins directly from the complex proteome by MS without a specific enrichment procedure.
In the past decade, a plethora of methods have been developed for the enrichment and MS analysis of phosphopeptides from protein digests.3, 4b, 5 This digestion step significantly increases the overall complexity since each protein is digested into hundreds of peptides. Enriched intact phosphoproteins can then be analyzed by top-down MS for comprehensive characterization of protein phosphorylation.2b, 6 However, very few methods7 are available for the enrichment of phosphoproteins and each has different major drawbacks. Although phospho-specific anti-phosphotyrosine antibodies have high affinity7a, the enrichment of phosphoserine/threonine containing proteins (which represent >99% of all phosphoproteins)3–4 remains a challenge due to the low affinity and specificity of phospho-Ser/Thr antibodies.4a, 7a The enrichment strategies employing immobilized metal ion affinity chromatography (IMAC)7a, 7c can enrich proteins with phosphorylated Ser, Thr, and Tyr residues without bias, however, the IMAC-based agarose phosphoprotein enrichment methods have low specificity, efficiency, and poor reproducibility.8 Thus, the effective capture and universal enrichment of phosphoproteins from complex protein mixtures remains a significant challenge. Herein, we have designed and synthesized functionalized multivalent superparamagnetic nanoparticles (NPs) to universally and effectively enrich phosphoproteins from highly complex mixtures. The effectiveness of NP-enabled phosphoprotein enrichment from complex cell and tissue lysates was demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis with a phosphoprotein-specific stain, and furthermore by top-down MS analyses of the enriched phosphoproteins (Figure S1).
In our approach, we utilized superparamagnetic iron oxide (magnetite, Fe3O4) NPs with a small diameter ranging from 4 to 6 nm (Figure 1). The small size of the NPs gives them a high surface-to-volume ratio, which allows for easy modification of the NPs’ surface with multivalent ligand molecules properly designed to have a specific interaction with target biomolecules (i.e. proteins).9 Furthermore, the size of NPs is comparable to the size of proteins, therefore enable the NPs to: i) penetrate better in protein mixtures, leading to a higher binding rate; ii) reduce the probability that proteins are denatured, and iii) have good solubility.9b, 9c, 10 Furthermore, Fe3O4 NPs of this small size (below 20 nm) are superparamagnetic,11 therefore, unlike bulky micron-sized magnetic beads, the NPs are not spontaneously magnetic without external magnetic field. This property prevents the NPs from aggregating by themselves due to mutual magnetic attraction, but enables them to aggregate and precipitate readily under applied magnetic fields. Crucial to the specific capture, enrichment, and release of the phosphoproteins is the use of a dinuclear Zn (II)-dipicolylamine (Zn-DPA) complex as an affinity ligand to specifically bind to the phosphate groups (Figure 1A). The Zn-DPA coordination complex has a vacancy on two Zn metal ions that the phosphate dianion (ROPO32−) can access to form the complex ROPO32−–(2Zn-DPA)3+, and therefore is known to exhibit a high affinity toward anionic phosphorylated chemical species (against SO42−, CH3COO−, and Cl−) in aqueous solution at a neutral pH.12 Based on this attribute, acrylamides containing co-polymerized M2+-DPA complexes (so called “phos-tag™”) have been used for phosphate-affinity gel electrophoresis and visualization of proteins with phosphorylated Ser, Thr, and Tyr residues without bias.12b, 12c
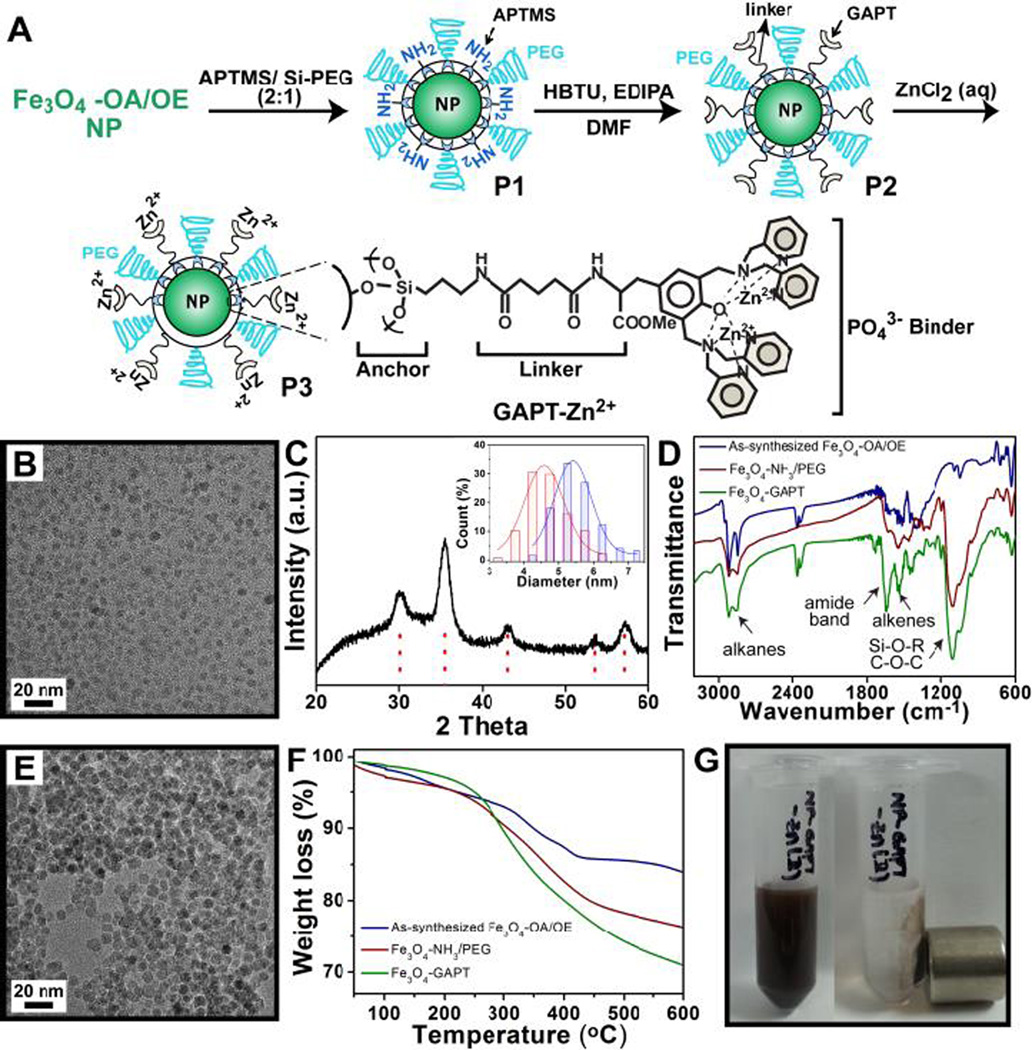
Figure 1.
(A) Schematic illustration of the synthesis and functionalization of Fe3O4 NPs. GAPT-Zn chelate groups have strong binding to phosphate groups. Molecules and NPs are not drawn to scale. (B) TEM image of as-synthesized Fe3O4-OA/OE NPs, (C) PXRD pattern of Fe3O4-OA/OE NPs. Inset shows the size distributions of the Fe3O4-OA/OE (red) and Fe3O4-GAPT-Zn (blue) (D) FTIR of Fe3O4-OA/OE (blue), Fe3O4-NH3+/PEG (red), and Fe3O4-GAPT (green) NPs. (E) TEM image of the functionalized Fe3O4-GAPT-Zn NPs. (F) TGA analysis of Fe3O4-OA/OE(blue), APTMS and Si-PEG (red), and GAPT coupled to APTMS (green). (G) Functionalized Fe3O4-GAPT-Zn NPs well-dispersed (left) and collected by a magnet (right) in aqueous solutions.
We designed and synthesized Fe3O4 NPs functionalized with Zn-DPA ligands that are linked via glutaric acid (hereafter referred to as GAPT), as illustrated in Figure 1A. First, oleic acid (OA) and oleylamine (OE) ligand capped Fe3O4 NPs were synthesized as reported previously (see SI for detailed procedures).13 Transmission electron microscopy (TEM) imaging revealed that the size of as-synthesized Fe3O4-OA/OE NPs was 4.6 ± 0.66 nm (Figure 1B and the red size distribution histogram in the inset of Figure 1C). Powder X-ray diffraction (PXRD) confirmed the Fe3O4 NPs to be magnetite (Figure 1C). Next, the hydrophobic OA and OE ligands on these intermediate Fe3O4 NPs were exchanged with 3-aminopropyl trimethoxy silane (APTMS) and 2-methoxy (polyethyleneoxy) propyl trimethoxysilane (hereafter referred to as Si-PEG), to produce Fe3O4-NH3+/PEG (Figure 1A, P1).14 These PEG groups, which contain a polyethyleneoxy moiety, were added to the surface of the NPs to decrease non-specific interactions between proteins and the NPs as well as to increase the solubility of the NPs in aqueous buffers.15 Subsequently, the Zn-chelating ligand molecules, GAPT, which were synthesized following a previous report,16 were coupled to the free amino groups of the APTMS molecules on the NPs. Finally, the Fe3O4-GAPT NPs (Figure 1A, P2) were activated with 10 mM ZnCl2 (aq) to generate Fe3O4-GAPT-Zn NPs (Figure 1A, P3). TEM image analysis showed that the average size of Fe3O4-GAPT-Zn NPs increased to 5.4 ± 0.62 nm (the blue size distribution histogram in Figure 1C inset) except that its aggregation was slightly increased (Figure 1E). Fourier transform infrared spectroscopy (FT-IR) was used to confirm the proper functionalization of NPs with the ligand molecules. The broad and strong band around 1022 cm−1 and peaks around 1200 cm−1 of Fe3O4-NH3/PEG NPs indicated the existence of Si-O-R and C-O-C, EO CH2,14a and the sharp peak around 1630 cm−1of Fe3O4-GAPT NPs proved the existence of the amide bond (Figure 1D). In addition, thermogravimetric analysis (TGA) of the as-synthesized Fe3O4, Fe3O4-NH3+/PEG, and Fe3O4-GAPT NPs showed a different weight loss (%) for each of the surface-modified NPs that was commensurate with the corresponding ligand molecules (Figure 1F). After activating Fe3O4-GAPT NPs with Zn ions, we used X-ray photon spectroscopy (XPS) to observe the characteristic Zn 2p peak at 1020 eV (Figure S2) and confirm the presence of Zn on the NP surface. The resulting NPs can be dispersed in aqueous solution and easily gathered using a magnet (Figure 1G).
After the NP synthesis and characterization, we performed phosphoprotein enrichment experiments using the functionalized Fe3O4-GAPT-Zn NPs (Figure 2A) as follows: (1) mixing and agitation of the protein mixture with the NPs, (2) separation of the unbound non-phosphoproteins by removing the supernatant solution as flow-through, and (3) elution of the phosphoproteins. First, we tested the phosphoprotein enrichment from standard protein mixtures containing either β-casein or pepsin (both of which are phosphoproteins) and bovine serum albumin (BSA, a non-phosphoprotein) in the same mass-to-volume ratio. The best enrichment results were obtained using a buffer solution containing 50 mM HEPES and 150 mM NaCl (pH 7.7) for binding and the subsequent washing, and a buffer containing 100 mM sodium phosphate (Na2HPO4) and 200 mM NaCl (pH adjusted to 7.3 with HCl) for elution. The large excess of phosphate ions in the elution buffer out-competes the phosphate groups on the captured phosphoproteins for binding to the NPs, thereby releasing the phosphoproteins.
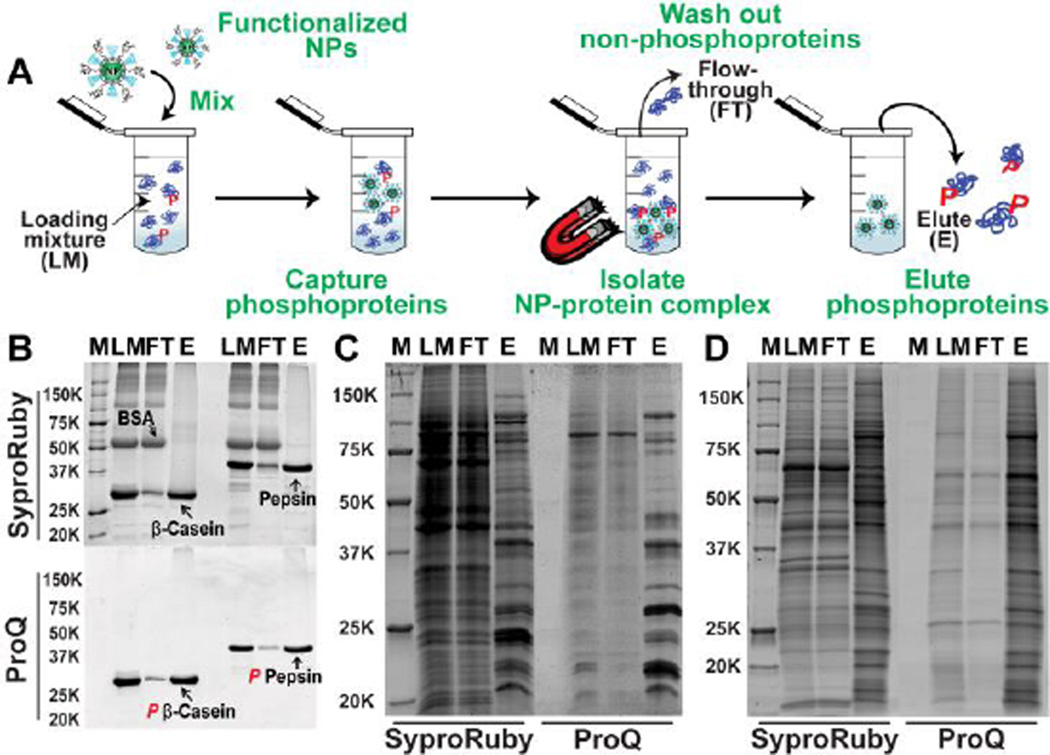
Figure 2.
(A) Workflow of the capture and enrichment strategy using the functionalized NPs. (B) SDS-PAGE analysis with Sypro Ruby (top) and Pro-Q Diamond (bottom) staining confirmed the highly specific enrichment of phosphoproteins (β-casein and pepsin) from standard protein mixtures containing non-phosphoprotein (BSA). M, molecular markers; LM, loading mixture before enrichment; FT, flow-through; and E, elution after enrichment. (C–D) Global enrichment of phosphoproteins from (C) HEK 293 cell lysate (equal amount loading, 15 µg) and (D) swine heart tissue extract (equal amount loading, 10 µg) as demonstrated by SDS-PAGE analysis with Sypro Ruby (left side) and Pro-Q Diamond staining (right side).
Next, we demonstrated the selective and effective enrichment of phosphoproteins by the functionalized NPs using SDS-PAGE (Figure 2B). The gel was first stained by Pro-Q Diamond fluorescent dye to visualize phosphoproteins and then by Sypro Ruby dye to observe the profile of total proteins (including both non-phosphoproteins and phosphoproteins) in the gel. After enrichment, in the elution (E) solutions the bands corresponding to β-casein and pepsin were far more prominent whereas the non-phosphoprotein, BSA, was nearly absent (Figure 2B, top). In contrast, the flow-through (FT) solution contained significantly higher amount of BSA. Moreover, Pro-Q Diamond dye stained only β-casein or pepsin, not BSA (Figure 2B, bottom), confirming that the eluted β-casein and pepsin are indeed phosphoproteins. These results show that the Fe3O4-GAPT-Zn NPs can selectively bind to phosphoproteins in mixtures containing non-phosphoproteins. To confirm the importance of the metal-ion chelating group GAPT-Zn for the specific binding to phosphate groups, we compared protein binding using three different control NPs: 1) control NP-1, Fe3O4-GAPT NPs that were not activated by Zn2+; 2) control NP-2, Fe3O4-NH3+/PEG NPs that were functionalized with positively charged ligands, but without metal-ion chelating ligands, and 3) control NP-3, Fe3O4-PEG NPs that were functionalized only with PEG groups. All of the control NPs showed poor affinity toward phosphoproteins in the same enrichment experiments carried out with protein mixtures containing BSA and β-casein in equal mass ratio (Figure S3). These control experiments unequivocally confirmed that the specific binding of phosphoproteins occurs via interactions between the phosphate group and the GAPT-Zn ligand complex on the surface of the Fe3O4-GAPT-Zn NPs.
To further evaluate the specificity of the phosphoprotein enrichment, we systematically increased the mass ratio of BSA: β-casein from 9: 1 to 99: 1 while holding the amount of β-casein constant at 200 µg. The β-casein was clearly enriched even in the mixture containing an overwhelming amount of BSA (BSA: β-casein = 99: 1) (see Figure S4A, B). We determined the percentage of protein recovery and the enrichment factor (defined as the gain in the relative ratio of the phosphoprotein to non-phosphoprotein) to assess the performance of phosphoprotein enrichment (Figure S4C, D). For a mixture of 99:1 BSA to β-casein, the enrichment factor was over 140 folds (Figure S4D), which could almost be considered as "purification" of phosphoproteins. The enrichment performance of Fe3O4-GAPT-Zn NPs was also compared with an IMAC-based phosphoprotein enrichment kit (Thermo). For the same enrichment experiment of a mixture of 99:1 BSA to β-casein, our NPs showed significantly reduced non-specific binding and greatly outperformed this IMAC-based material (Figure S5).
We further assessed the enrichment of phosphoproteins from human embryonic kidney (HEK) 293 cell lysate (Figure 2C) and swine heart tissue extract (Figure 2D), two highly complex mixtures, using the Fe3O4-GAPT-Zn NPs. We loaded equal amount of proteins before and after enrichment on the SDS-PAGE gel, stained with Pro-Q diamond (for phosphoprotein detection only) first, de-stain and re-stain the same gel with Sypro Ruby (for total protein detection). Despite the equal amount loading as confirmed by the similar total intensities of the loading mixture before enrichment (LM), flow-through (FT), and the elution after enrichment (E) lanes stained by Sypro Ruby, the Pro-Q diamond stained LM and FT lanes showed significantly lower intensity than that of E lane, suggesting that most of the proteins in the LM and FT solutions are non-phosphoproteins. Furthermore, the banding patterns of the E lane stained with both Pro-Q diamond and Sypro Ruby are highly similar; in contrast to the LM and FT lanes which are very different between the Pro-Q diamond and Sypro Ruby stains; indicating the enriched proteins are predominantly phosphoproteins. These SDS-PAGE analyses clearly indicate that the Fe3O4-GAPT-Zn NPs can specifically and effectively enrich phosphoproteins from complex biological samples with high affinity and efficiency.
To demonstrate our NP enrichment strategy is compatible with top-down MS, we have examined the intact proteins present in the complex swine heart tissue extracts before and after enrichment by LC-MS. The LM (before enrichment) and the elution solution (after enrichment) from swine heart tissue extract without digestion was desalted, concentrated, and separated by reverse phase chromatography. Subsequent MS analysis of the LM before enrichment revealed that most of the detected proteins are highly abundant blood proteins, such as hemoglobin subunit α (see Figure S6) or β, and myoglobin. However, these blood proteins were not detected or dramatically decreased by MS in the elution after enrichment (Figure S7A). This suggests that these highly abundant non-phosphoproteins were not captured by the NPs and, consequently, were removed during the washing step.
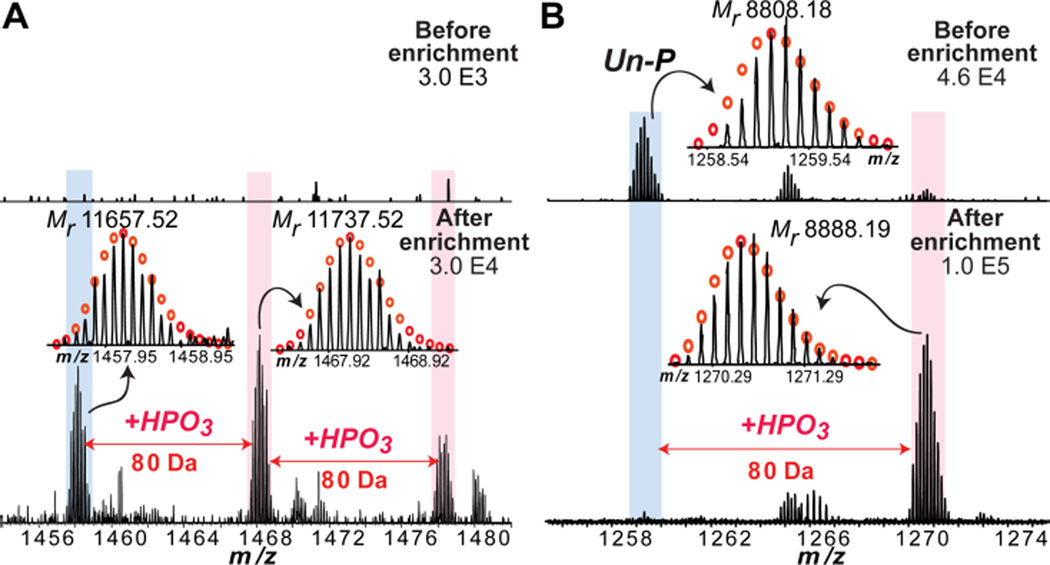
Importantly, the top-down MS data clearly showed that phosphoproteins in swine heart tissue extracts were enriched by the Fe3O4-GAPT-Zn NPs, even in the presence of highly abundant blood proteins (vide supra). Many of the detected phosphoproteins have very low abundance in comparison to non-phosphoproteins and/or low stoichiometry (low phosphorylation occupancy) in the pre-enrichment samples; however, these phosphoproteins and their respective phosphorylated forms were significantly enriched in the post-enrichment samples, some with more than one phosphorylation detected for the same protein (mass increases of multiples of 80 Da) (Figure 3 and Figure S7B). As a representative example, after enrichment, a protein with Mr 11, 657.52 (Mr, most abundant molecular weight) was detected by top-down MS together with two additional peaks with 80 Da mass increases (labeled as + HPO3) (Figure 3A, bottom panel), which correspond to multiple phosphorylated forms of the protein, respectively. However, none of these peaks were detected in the MS of the swine heart tissue extract before enrichment (Figure 3A, top) implying they are all phosphorylated protein forms that have low abundance. Another representative MS of the original protein mixture before enrichment displayed a protein with Mr 8, 808.18 (Figure 3B, top panel; labeled as Un-P). After enrichment, a protein with Mr 8, 888.19 with a mass increase of 80 Da was detected in the MS instead (Figure 3B, bottom panel; labeled as + P). This clearly shows that the relative abundance of the phosphorylated species (Mr 8, 888.19) is significantly increased compared to the non-phosphorylated species (Mr 8, 808.18) after enrichment. It should also be noted that a few Zn2+-binding proteins, such as a parathymosin-like protein17 (Figure S8), were also detected in the elution fractions due to its high affinity to GAPT-Zn ligands on the NPs. Thus, top-down MS analysis confirmed that the number and amount of phosphoproteins in the elution fraction was significantly increased compared to the loading mixture, which was dominated by overwhelmingly abundant blood proteins. The fact that no high mass phosphoproteins were identified in this top-down MS study is mostly likely due to the exponential decay in the signal-to-noise ratio that occurs with increasing mass in high-resolution MS18 and the use of Q-Exactive mass spectrometer, which is not optimized for high-mass intact protein detection.19 However, high-mass proteins (from 20 kDa to 150 kDa) were clearly detected by SDS-gel stained with SyproRuby and Pro-Q Diamond of the elution fractions after phosphoenrichment from HEK293 cell lysates (Figure 2C) and swine heart tissue extracts (Figure 2D). This confirms that high mass phosphoproteins were indeed enriched by the Fe3O4-GAPT-Zn NPs.
Figure 3.
Representative intact protein MS spectra, before and after enrichment, confirming the highly specific enrichment of phosphoprotein from a swine heart tissue extract. (A) shows a low abundance phosphoprotein that is not detectable before enrichment (top) is detected after enrichment (bottom); and (B) shows a phosphoprotein with very low phosphorylation occupancy that the un-phosphorylated form (un-P) is predominant before the enrichment but the phosphorylated form (+P) becomes dominant after the enrichment. Mr represents most abundant molecular weight. +HPO3, the covalent addition of a phosphate group (+80 Da).
To recapitulate, we have developed superparamagnetic NPs that are functionalized with multivalent ligand molecules to capture and enrich intact phosphoproteins with high specificity and efficacy. Such functionalized NPs can have many advantages for phosphoprotein enrichment, which include: a) High surface area per volume,9a–c, 10 which results from the small size the NPs. The NP diameter of ca. 5 nm (Figure 1C) corresponds to a surface area of more than 200 m2/g (see calculation in Table S1), which translates into high ligand density. b) NPs have comparable nanometer size to proteins and can be well-dispersed in aqueous solution (as shown in Figure 1G). This gives NPs similar kinetics to proteins and allow them to intermingle well with proteins as demonstrated previously,9b, 9c, 10 therefore allowing for effective capture of phosphoproteins. c) The NPs developed here have multiple flexible binding sites (see an estimate in Table S2) packed in a small dimension comparable to the size of proteins; this multivalency effect20 can increase the overall binding affinity and ensure the efficient capture of low concentration phosphoproteins. d) NPs modified with GAPT-Zn metal complex enrich all phosphoproteins regardless whether it is Ser, Thr, or Tyr phosphorylation12c to allow a global and comprehensive analysis of phosphoproteins. e) Unlike conventional bottom-up approach which requires prior digestion of proteins into peptides, NPs capture intact phosphoproteins in p conditions directly and reversibly. Circular Dichroism measurements revealed that the enriched protein maintained the same conformation as the original one (Figure S9). The enriched intact phosphoproteins can be analyzed by conventional biochemical methods such as Western blotting and more recently developed top-down MS technologies for comprehensive phosphoprotein characterization.2b, 6 f) In comparison to the development and production of new antibodies for capturing phosphoproteins, which are labor intensive, expensive, and difficult to scale up, the chemical synthesis of nanomaterials is simpler, faster and more scalable.
These advantages of the NP approach have enabled us to demonstrate the specific enrichment of phosphoproteins from standard protein mixtures and complex cell and tissue lysates, as confirmed by SDS-PAGE analysis with phosphospecific stain as well as top-down MS analysis of the enriched phosphoproteins. This simple, effective, and universal nanoparticle-based phosphoenrichment method enables a comprehensive characterization of the phosphoproteome.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH R21 EB013847 (to both S.J. and Y. G.). Y. G. also thanks the support by NIH R01 HL096971 and R01 HL109810. We would like to thank Ying-Hua Chang for critical reading of this manuscript and Yi Zhang for assistance in NPs synthesis.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Method details and additional supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Hunter T. Cell. 2000;100:113. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.(a) Tan CSH, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jørgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Sci. Signal. 2009:2. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]; (b) Zhang J, Guy MJ, Norman HS, Chen Y-C, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, Moss RL, Ge Y. J. Proteome Res. 2011;10:4054. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12867. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nita-Lazar A, Saito-Benz H, White FM. Proteomics. 2008;8:4433. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Mann M, Ong S-E, Grønborg M, Steen H, Jensen ON, Pandey A. Trends Biotechnol. 2002;20:261. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]; (b) Zhou H, Watts JD, Aebersold R. Nat. Biotechnol. 2001;19:375. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 5.(a) Tao WA, Wollscheid B, O'Brien R, Eng JK, Li X-j, Bodenmiller B, Watts JD, Hood L, Aebersold R. Nat. Methods. 2005;2:591. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]; (b) Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Nat. Methods. 2007;4:231. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]; (c) Nelson CA, Szczech JR, Dooley CJ, Xu Q, Lawrence MJ, Zhu H, Jin S, Ge Y. Anal. Chem. 2010;82:7193. doi: 10.1021/ac100877a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nelson CA, Szczech JR, Xu Q, Lawrence MJ, Jin S, Ge Y. Chem. Commun. 2009:6607. doi: 10.1039/b908788e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Siuti N, Kelleher NL. Nat Methods. 2007;4:817. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ge Y, Rybakova IN, Xu Q, Moss RL. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12658. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Schmidt SR, Schweikart F, Andersson ME. J. Chromatogr. B. 2007;849:154. doi: 10.1016/j.jchromb.2006.09.016. [DOI] [PubMed] [Google Scholar]; (b) Oda Y, Nagasu T, Chait BT. Nat. Biotechnol. 2001;19:379. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]; (c) Porath J, Carlsson JAN, Olsson I, Belfrage G. Nature. 1975;258:598. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 8.Kaur-Atwal G, Weston DJ, Bonner PLR, Crosland S, Green PS, Creaser CS. Curr. Anal. Chem. 2008;4:127. [Google Scholar]
- 9.(a) Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; (b) Gao J, Gu H, Xu B. Acc. Chem. Res. 2009;42:1097. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]; (c) Pan Y, Du X, Zhao F, Xu B. Chem. Soc. Rev. 2012;41:2912. doi: 10.1039/c2cs15315g. [DOI] [PubMed] [Google Scholar]; (d) Pan Y, Long MJC, Lin H-C, Hedstrom L, Xu B. Chem. Sci. 2012;3:3495. [Google Scholar]; (e) Erathodiyil N, Ying JY. Acc. Chem. Res. 2011;44:925. doi: 10.1021/ar2000327. [DOI] [PubMed] [Google Scholar]
- 10.Aubin-Tam M-E, Hamad-Schifferli K. Biomed. Mater. 2008;3:034001. doi: 10.1088/1748-6041/3/3/034001. [DOI] [PubMed] [Google Scholar]
- 11.Sun S. Adv. Mater. 2006;18:393. [Google Scholar]
- 12.(a) Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Mol. Cell. Proteomics. 2006;5:749. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]; (b) Kinoshita E, Kinoshita-Kikuta E, Koike T. Nat. Protoc. 2009;4:1513. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]; (c) Kinoshita E, Kinoshita-Kikuta E. Proteomics. 2011;11:319. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- 13.Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, Li G. J. Am. Chem. Soc. 2003;126:273. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 14.(a) De Palma R, Peeters S, Van Bael MJ, Van den Rul H, Bonroy K, Laureyn W, Mullens J, Borghs G, Maes G. Chem. Mater. 2007;19:1821. [Google Scholar]; (b) Jana NR, Earhart C, Ying JY. Chem. Mater. 2007;19:5074. [Google Scholar]
- 15.Karakoti AS, Das S, Thevuthasan S, Seal S. Angew. Chem., Int. Ed. 2011;50:1980. doi: 10.1002/anie.201002969. [DOI] [PubMed] [Google Scholar]
- 16.Ojida A, Honda K, Shinmi D, Kiyonaka S, Mori Y, Hamachi I. J. Am. Chem. Soc. 2006;128:10452. doi: 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- 17.Trompeter H-I, Blankenburg G, Brügger B, Menne J, Schiermeyer A, Scholz M, Söling H-D. J. Biol. Chem. 1996;271:1187. doi: 10.1074/jbc.271.2.1187. [DOI] [PubMed] [Google Scholar]
- 18.Compton PD, Zamdborg L, Thomas PM, Kelleher NL. Anal. Chem. 2011;83:6868. doi: 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiu L, Valeja SG, Alpert AJ, Jin S, Ge Y. Anal. Chem. 2014;86:7899. doi: 10.1021/ac501836k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mammen M, Choi S-K, Whitesides GM. Angew. Chem., Int. Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.