Abstract
Smyd1/Bop is an evolutionary conserved histone methyltransferase previously shown by conventional knockout to be critical for embryonic heart development. To further explore the mechanism(s) in a cell autonomous context, we conditionally ablated Smyd1 in the first and second heart fields of mice using a knock-in (KI) Nkx2.5-cre driver. Robust deletion of floxed-Smyd1 in cardiomyocytes and the outflow tract (OFT) resulted in embryonic lethality at E9.5, truncation of the OFT and right ventricle, and additional defects consistent with impaired expansion and proliferation of the second heart field (SHF). Using a transgenic (Tg) Nkx2.5-cre driver previously shown to not delete in the SHF and OFT, early embryonic lethality was bypassed and both ventricular chambers were formed; however, reduced cardiomyocyte proliferation and other heart defects resulted in later embryonic death at E11.5-12.5. Proliferative impairment prior to both early and mid-gestational lethality was accompanied by dysregulation of transcripts critical for endoplasmic reticulum (ER) stress. Mid-gestational death was also associated with impairment of oxidative stress defense—a phenotype highly similar to the previously characterized knockout of the Smyd1-interacting transcription factor, skNAC. We describe a potential feedback mechanism in which the stress response factor Tribbles3/TRB3, when directly methylated by Smyd1, acts as a co-repressor of Smyd1-mediated transcription. Our findings suggest that Smyd1 is required for maintaining cardiomyocyte proliferation at minimally two different embryonic heart developmental stages, and its loss leads to linked stress responses that signal ensuing lethality.
Introduction
Cardiac organogenesis initiates in the cardiac crescent at embryonic day (E) 7.75 and gives rise to the linear heart tube by E8.0 [1]. Cardiac progenitor cells, termed the anterior/second heart field (SHF), originate in splanchnic mesoderm and contribute to the linear heart tube as it undergoes morphogenesis into a full four chambered heart [1]. A spectrum of congenital heart anomalies result in humans from perturbation of SHF formation, including atrial and conotruncal defects [2]. An expanding network of transcription factors, such as ISL LIM Homeobox 1 (Isl1), Hand 2 [3], and Myocyte Enhancer Factor 2C (Mef2c), as well as intercellular signaling pathways control SHF deployment by controlling cardiac progenitor cell proliferation in pharyngeal mesoderm, progenitor cell differentiation, and cardiac progenitor cell patterning in the dorsal pericardial wall [1]. Coordination of these three processes in the early embryo drives progressive heart tube elongation during cardiac morphogenesis.
The subsequent maturation of the heart and outflow tract (OFT) is a tightly regulated process in which oxygen tension plays a vital role [4]. It is now well established that the developing embryo is very sensitive to oxidative stress by the production of reactive oxygen species (ROS) which may induce embryonic death and malformations, such as fetal intrauterine growth retardation [5]. The accumulation of misfolded or unfolded proteins in response to ROS-mediated oxidative stress can trigger endoplasmic reticulum (ER) stress as an adaptive cellular response. Cation transport regulator homolog 1 (Chac1) and cEBP homologous protein (Chop/GADD153) are considered to be vital initiators of the ER stress response [6,7]. Oxidative stress as well as a variety of other signals that induce ER stress can stimulate either apoptosis or autophagy in order to clear damaged cells [6]. It is unclear what regulates the balance between these two clearance processes. At midgestation, anti-oxidant defense is initiated by expression of Myoglobin (Mb) and associated factors that contribute to oxygen storage and delivery [8]. Skeletal NAC (skNAC), a skeletal and heart pre-mRNA splice variant of nascent polypeptide-associated complex alpha subunit (αNAC), was shown to act as a muscle-specific transcriptional activator of Mb [9], and ∼50% of skNAC-deficient mice die between E9.5 and E12.5 due to cardiac malformations. At the molecular level, these mutant embryos have decreased expression of Mb and other anti-oxidative stress factors.
SET-MYND-domain 1 (Smyd1/Bop) encodes an evolutionary conserved histone methyltransferase containing a split SET (Su(var), Enhancer-of-zeste, Trithorax) domain interrupted by a MYND (Myeloid, Nervy and DEAF-1) domain [10] and physically interacts with skNAC [11,12]. This interaction occurs through the zinc finger MYND domain, a well characterized protein-protein interaction motif known best for its association with co-repressor complexes [13,14,15]. Expression of the Smyd1 gene is restricted to muscle tissues in human, fish, frog, chicken and mouse [10,16,17,18]. Global knockdown of Smyd1a and Smyd1b in zebrafish resulted in the disruption of myofibril formation and an absence of beating in the heart [16]. The zebrafish mutant flatline (fla), which harbors a nonsense mutation in the Smyd1b gene, exhibits a major defect in thick filament assembly causing both cardiac and skeletal muscle dysfunction [19]. Conventional null Smyd1 mice die in utero at E9.5 due to heart defects, including disrupted maturation of ventricular cardiomyocytes and malformation of the right ventricle (RV) [10]. Supporting a role for Smyd1 in muscle development, Smyd1 induced myocyte differentiation when expressed in C2C12 myoblasts [18].
Smyd1 has been shown to function through multiple mechanisms, any or all of which could be causal for the above described phenotypes. Firstly, Smyd1 functions as repressor through interaction of its MYND domain with co-repressors HDAC1–3, NCoR and SMRT [11]. Secondly, Smyd1 functions as a transcriptional activator by catalyzing trimethylation of H3K4 [16], a histone modification associated with transcriptionally active loci [20]. Lastly, Smyd1 interacts and colocalizes with skNAC in skeletal and heart muscle [11,12], but the relevance of the interaction is unknown. While the highly related paralogue Smyd2 methylates a number of non-histone targets [21,22,23], such has not been described for Smyd1.
During mouse embryonic development, expression of Smyd1 is detected as early as E7.75 in the cardiac crescent and the SHF, and later expression is found in the developing heart tube and OFT [10]. Expression continues throughout development and into adulthood. Thus, we reasoned that Smyd1 regulates gene program(s) central to both early and late heart development. To test this hypothesis, we generated mice harboring a conditional knockout (CKO) allele for Smyd1 (Smyd1 flox) and crossed them to two different cardiomyocyte-specific Nkx2.5-cre mouse lines: (1) a knock-in, in which the cre open reading frame was inserted into the first exon of the Nkx2.5 gene (Ki-Nkx2.5 cre/+) previously shown to delete in the SHF [24] and (2) a transgenic, in which cre is expressed under the control of the Nkx2.5 basal promoter and cardiac enhancer (Tg-Nkx2.5-cre) shown to delete subsequent to SHF and OFT development [25]. Early deletion using Ki-Nkx2.5 cre/+ resulted in E9.5 death, with defects consistent with impaired expansion of the SHF and OFT. Deletion with Tg-Nkx2.5-cre led to delayed lethality (E11.5–12.5) and to defects akin to those observed in skNAC knockout mice [12]. Proliferative defects underlying both phenotypes led to induction of ER and oxidative stress programs. Among the up-regulated genes, a sensor of both stress responses and a Smyd1-interacting protein, Tribbles3/TRB3 [26,27], was directly methylated by Smyd1. Methylated TRB3, in turn, functions as a Smyd1 co-repressor, suggestive of a potential feedback loop for stress response transcription.
Materials and Methods
Animals
Smyd1 flox mice were generated using endogenous sequence obtained from a Lambda FixII Vector 129SV Mouse Genomic Library (Stratagene). The targeting construct was designed in the Osdupdel vector, a gift from William A Kuziel, in order to introduce loxP sites flanking the second and the third exons of Smyd1 followed by a neomycin cassette that is flanked by a third loxP site. The targeting vector was electroporated into 129S6 ES cells. After selecting with G418 and gancyclovir, surviving clones were screened for homologous recombination by Southern blot after digestion either with Bgl II and Sal I or Bgl II and Kpn I, and hybridization with probe 2 and probe 3, respectively. ES clones showing correct targeting in both arms were injected into C57BL/6 blastocysts to create chimeric mice. The chimeric mice were mated to C57BL/6 females to create a germline knock-in referred to as Smyd1 FloxNeo. Smyd1 FloxNeo mice were crossed to EIIa-cre mice, in which cre is expressed at the 2-cell stage of development (The Jackson Laboratory, #003724) and progeny were screened for deletion of the neomycin cassette by Southern blot and back-crossed in order to remove cre resulting in germline Smyd1 Flox mice. From these crosses, some mice were also generated that had ubiquitous and germline deletions of the Smyd1 FloxNeo locus. These animals were termed Smyd1 Floxdel and used to confirm that the germline deletion of Smyd1 phenocopies the original conventional mutation (Smyd1 KO). Knock-in (Ki)-Nkx2.5 cre and transgenic (Tg)-Nkx2.5-cre mice were previously described [24,25]. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committees of the University of Houston or the University of Texas at Austin.
Whole-mount X-gal staining
Whole-mount staining for β-galactosidase (lacZ) activity was performed according to a standard protocol [28].
Whole-mount RNA in situ hybridization
Whole-mount RNA in situ hybridization was carried out as previously described [29]. Isl1 and Mef2c probes were kindly provided by Dr. James Martin (Baylor College of Medicine, Houston, TX). Complementary RNA probes for Anf, Hand2, Myh7 (β-MHC) and Smyd1 were generated previously [30]. Detailed probe information is available upon request.
Real-time PCR and microarrays
Hearts and pharyngeal arches were collected from E9.5 control and Smyd1 Ki-CKO embryos. 20 hearts were pooled for each experiment. Individual hearts were collected and assayed from E10.5 control and Smyd1 Tg-CKO embryos. RNA was isolated with a Micro RNeasy kit (Qiagen, Venlo, Netherlands), and treated with RNase-free DNase I (Roche, Basel, Switzerland) to remove genomic DNA. RNA (1 μg or more) was sent to Phalanx Biotech (Palo Alto, CA) for Ambion dual amplification followed by genome-wide gene expression analysis. The raw data were normalized and analyzed using OneArray Studio software (Phalanx Biotech). Ratios were determined from three independent experiments and each sample was hybridized to duplicate microarrays. Complementary DNA was prepared from 1 ug total RNA using AMV Reverse Transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. Real-time PCR analysis for Isl1, Gapdh and Mef2c was performed using Taqman probes. Real-time PCR analysis for Hand2, Myh7 (β-Mhc), Gapdh and Anf was carried out using Brilliant qPCR reagent and the Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA). Primers were generated in our lab and sequences are available upon request. Ratios were determined from three independent experiments, each evaluated in triplicate by real-time PCR.
Histological analysis
Fixed tissues were cryopreserved and embedded in OCT before staining with H&E using standard techniques or anti-BrdU (DSHB clone G3G4; 1:250) and anti-mouse–DAB secondary sera. Paraffin embedded sections were stained with H&E using standard protocols or with anti-phospho-histone 3 S10 (H3S10ph) (Millipore, 1:100) and anti-rabbit-FITC secondary sera.
Expression plasmids
Mammalian expression plasmids were constructed in the backbone vector PCDNA3.1 containing either a MYC or FLAG tag. Construction of the following TRB3 protein truncations were constructed by PCR using EcoRI and XhoI restriction sites: TRB3 (1–80); TRB3 (1–149); TRB3 (1–214); TRB3 (1–295); TRB3 (81–355); TRB3 (150–355); TRB3 (215–355); TRB3(296–355). All PCR amplifications were performed with high fidelity Pfu Turbo DNA polymerase (Stratagene) and sequenced for accuracy. Construction of the catalytic mutant Smyd1-Y234 and TRB3 mutants K16/R, K17R, K33R and P45/G were constructed by QuikChange Site-Directed Mutagenesis (Stratagene). GAL4-Smyd1 and SV40 promoter-driven 5X-GAL4DBD-luciferase were previously reported [10]. For bacterial expression (6X-His)-tagged-full-length Smyd1, TRB3 and their respective catalytic, deletion and point mutations were cloned into Gateway (Invitrogen) pET-DEST42. High-level expression was induced by IPTG in E. coli strains MG232 (Scarab LTM) or Hsp90PlusTM (Expression Technologies Inc.). Proteins were purified to near homogeneity by Ni-bead chromatography as detailed in the Scarab LTM protocol.
Immunoprecipitation and western blotting
Cell lysates were incubated with 10 ml of 50% slurry of protein-A/G sepharose beads for 1 h to preclear. One microgram of polyclonal MYC A14 or monoclonal FLAG M2 (Sigma) antibody was added to the supernatant for a minimum of 4 hours at 4°C, followed by a 1 hour incubation with 20 ml protein-A/G sepharose beads (Sigma). After four washes with lysis buffer, the precipitated proteins were analyzed by SDS-PAGE and transferred to PVDF membrane. Membranes were probed with the monoclonal FLAG M2 or monoclonal MYC9E10. Western blots were developed with the enhanced chemiluminescence (ECL) analysis system (GE Life Sciences).
Cell culture and transient transfections
Cos7 cells (ATCC) were grown in DMEM (Life Technologies, Frederick, MD) supplemented with 10% fetal bovine serum at 37° C in an atmosphere of 5% CO2 in air. The cells were transfected using FuGENE6 reagent (Roche) or Lipofectamine Plus (Invitrogen). For luciferase assays, the total DNA concentration was held constant by adding the empty PCDNA3.1 expression plasmid. Cells were harvested 30–36 hours after transfection using the lysis buffer contained in the Promega Luciferase Kit. For immunoprecipitation experiments, Cos7 cells were transfected with FLAG-tagged Smyd1 or with FLAG-tagged Smyd1 and MYC-tagged TRB3 deletion constructs. Cells were lysed in buffer (10 mM Tris pH 7.4, 1mM EDTA, pH 8.0, 0.5% Triton X100 in PBS) containing protease inhibitors (Roche) and PMSF 30–36 hours after transfection.
Luciferase assays
Cell lysates were analyzed with a Rosys Anthos Lucy2 luminometer. Relative light units obtained from the pGL2-5XGAL4-SV40 reporter (Luciferase Assay Kit, Promega) were normalized by spectrophotometric analysis of cells co-transfected with the RSV-LacZ expression plasmid. Activation was determined in relation to the pGL2-5XGAL4-SV40 reporter alone. Three independent experiments were performed to calculate the mean and standard error.
Lysine methyltransferase assays
The in vitro reactions were performed as previously described [31]. Briefly, ∼μ1 g bacterially purified 6XHis-Smyd1, 6XHis-Smyd1or 6XHis-Set2 (the kind gift of Dr. Danny Reinberg) were incubated with 1 μg of TRB3 wild type and mutants. Approximately 2 μCi S-adenosyl-L–[methyl-3H] methionine (SAM; GE Biosciences) was included as a methyl donor. All reactions were carried out in 40μl HMT reaction buffer (10mM dithiothreitol, 100mM NaCl, 4mM MgCl2, and 50mM Tris-HCl at pH 8.8) at 30°C for 3 hours. A 12% SDS-PAGE gel was used to resolve the samples and fluorography was used to visualize positive methylation. Substrate loading was visualized by Coomassie blue staining.
In vivo methylation of TRB3 was performed with a procedure modified from the method of Liu et al. [32]. 48 hr post transfection of Smyd1 or Smyd1-Y234/F, three plates (100-mm-diameter) of C2C12 cells were incubated with cycloheximide (100 mg/ml) and chloramphenicol (40 mg/ml) in standard media. L-[methyl-3H]methionine was added to a concentration of 10 mCi per ml. Cells were then incubated for an additional 3 hr, and lysed in RIPA buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1% NP-40, 2 mM NaF,2 mM NaOV4, protease inhibitor cocktail tablets [Roche Diagnostics] with 2x concentration). Endogenous TRB3 was immunoprecipitated with anti-TRB3 polyclonal antibody (Sigma-Aldrich, T8076), and immunoprecipitated proteins were resolved on 8% SDS-PAGE. [3H]Methyl labeling was detected by fluorography in 22% PPO solution. Dried gels were exposed to Kodak MRX film for 1 week.
Computational analyses
A Student’s t test was used for statistical comparisons when appropriate. A p-value of < 0.05 was considered statistically significant. Global microarray analyses employed Nimblegen chips and were analyzed using Java Treeview and DAVID [32].
Results
Expression of Smyd1 during early heart development
To investigate the temporal and spatial expression of Smyd1 during embryogenesis, we performed whole mount in situ hybridization on embryos recovered at E8.5 and E9.5. Results of this analysis demonstrated that at E8.5 and E9.5, robust expression of Smyd1 was observed throughout the looped heart tube, including the ventricles, OFT and venous pole (Fig. 1A). To define which cell type(s) express Smyd1, we performed in situ hybridization on transverse sections at E10.0 and immunofluorescence on transverse sections at E13.5. Smyd1 mRNA was enriched in the myocardium and undetectable in the endocardium or epicardium (Fig. 1B). Smyd1 protein was very specifically localized to cardiomyocytes and undetectable in the endocardium, epicardium or coronary vasculature. Within cardiomyocytes, Smyd1 protein was localized to the sarcoplasm, showing a sarcomeric pattern similar to that reported for zebrafish (Fig. 1C).
Fig 1. Expression of Smyd1 during early heart development.
A. Whole mount in situ hybridization for Smyd1 in E8.5 (top panel) and E9.5 embryos (bottom panel) are shown from the left and right side. Red arrows denote the boundaries of Smyd1 expression at the atrial and venous poles. Scale bar = 500 μm. B. In situ hybridization for Smyd1 in transverse sections of E10.0 hearts. The box in the left panel is enlarged in the right panel. Smyd1 mRNA was enriched in the myocardium and not detectable in either the epicardium or endocardium. Black arrow = epicardium; Black arrowhead = endocardium. C. Immunolocalization of Smyd1 protein in the heart at E13.5. Smyd1 was specifically detected in cardiomyocytes. No Smyd1 protein was detectable in the endocardium (red arrowhead), epicardium (yellow arrowhead) or coronary vasculature (white arrowhead). Scale bar = 50 μm.
Generation of Smyd1 CKO mice
In order to analyze the role of Smyd1 specifically in the heart, we generated a Smyd1 CKO allele using the targeting strategy schematized in S1A Fig. The Smyd1 gene encodes three isoforms (Smyd1a, -b, and -c); Smyd1c utilizes an alternate first exon. Therefore, we designed this conditional allele with loxP sites flanking exons 2 and 3, the first 2 exons in common among all three isoforms and the same exons that were deleted in the conventional knockout [10]. Thus, in the presence of Cre recombinase, exons 2 and 3 are deleted resulting in a frameshift and a premature stop. Following validation of correct ES cell targeting and germline transmission (S1B-C Fig.), we confirmed that our deletion strategy was sufficient to replicate the conventional knockout by crossing Smyd1 flox/flox with EIIA-cre [33], which is expressed in pre-implantation zygotes and leads to germline deletion (data not shown).
Next, we performed a cardiac-specific Smyd1 CKO using the Ki-Nkx2.5 cre/+ strain previously shown to robustly delete in the first and SHF from E7.5 [24]. Examination of 21 litters from Ki-Nkx2.5 cre/+; Smyd1 +/flox x Smyd1 flox/flox revealed that 100% of Ki-Nkx2.5 cre/+; Smyd1 flox/flox (henceforth referred to as Smyd1 Ki-CKO) embryos died prior to E10.5. In contrast, control littermates of all other genotypes survived with no apparent abnormalities. No transcripts from the deleted exons were detectable from Smyd1 Ki-CKO hearts at E9.5 (S1D Fig.). We also performed western blots for Smyd1 using an antibody raised against the C-terminus of Smyd1 and observed no protein product in heart extracts from Smyd1 CKO embryos at E9.5 (S1E Fig.).
Smyd1 Ki-CKO embryos exhibit malformation of SHF derivatives and alterations in SHF gene expression
At a gross morphological level, Smyd1 Ki-CKO embryos appeared to be slightly growth retarded at E9.5 (data not shown). The length of the OFT and width of the RV appeared truncated (Fig. 2A). Therefore, we measured the length of the OFT (Fig. 2B) and the width of the RV (Fig. 2C). Both were significantly smaller in the Smyd1 Ki-CKO.
Fig 2. Loss of Smyd1 using Ki-Nkx2.5 cre/+ disrupts looping morphogenesis and chamber formation through perturbation of the SHF and activation of ER stress.
A. Gross morphological comparison of control (Cx: Nkx2.5 +/+; Smyd1 Flox/Flox) and Smyd1 Ki-CKO (CKO: Nkx2.5 cre/+; Smyd1 Flox/Flox) hearts at E9.5. Scale bar = 200 μm. B, C. The lengths of the outflow tract (OFT) (B) and right ventricle (RV) (C) were significantly reduced in Ki-CKO embryos at E9.5 (n = 6/group). D. Representative results of microarray gene expression comparison of transcripts critical to SHF and chamber formation at E9.5. Data are presented as expression values of 2 independent biological replicas of each genotype averaged from 2 technical replicas. E. Confirmation of microarray for deregulated transcripts critical to SHF and chamber formation by real-time PCR using RNA from E9.5 heart/pharyngeal mesoderm (n = 9/group). F. Whole mount in situ hybridization comparison of selected SHF and chamber formation transcripts deregulated and/or mislocalized in CKO hearts at E9.5 (n = 3/group). Arrows denote areas of differential expression. G. Comparison of cell proliferation in the outflow tract of Control (Cx) and Ki-CKO by BrdU immunohistochemistry. PE, Pharyngeal Endoderm. OFT, outflow tract. V, ventricle. Scale bar = 100 μm. H. Quantification of anti-BrdU staining in the outflow tract (n = 6/group). I. Loss of Smyd1 leads to deregulation of genes critical to anti-proliferative responses to ER stress. Data are presented as a heat map with expression values of 2 independent biological replicas of each genotype averaged from 2 technical replicas plotted as log2 expression values. For B, C, E and H, data were analyzed by Student’s t-test (*P < 0.05).
Truncation of the OFT and RV indicated that Smyd1 may be necessary for the expansion and maintenance of the SHF. To assess molecular mechanisms underlying these defects, we compared gene expression profiles of WT and Smyd1 Ki-CKO hearts at E9.5 by microarray. Consistent with the phenotypic results, loss of Smyd1 led to reduction in the expression of several factors required for proper development of SHF derivatives, including Isl1, Mef2c, Hand2/dHAND, Tbx5, Tbx1, Shox2 and Eya1 (Fig. 2D, top panel). Reduction of Hand2 expression is consistent with that of reported for the conventional Smyd1 knockout [10]. Fgf8 was modestly upregulated in Smyd1 Ki-CKO hearts.
We also observed significant upregulation of genes involved in chamber formation and/or cardiac stress response, including Myh7, Bmp7, Anf, Egr1, Acta1 and Fos. In contrast, Myh6 (α-MHC) and two genes crucial for establishing atrial-ventricle morphology and sidedness, Shh and Pitx2 [34,35] were down-regulated in the Smyd1 Ki-CKO (Fig. 2D, lower panel).
We confirmed these microarray results by real-time PCR using RNA extracted from E9.5 heart tissues. Expression of SHF transcription factor genes Isl1, Mef2c and Hand2 was decreased. Expression of chamber-specific genes Myh7 and Anf was increased (Fig. 2E).
Alterations in SHF and chamber gene expression were further assayed by whole mount in situ hybridization. Expression of Isl1, Mef2c and Hand2 was reduced in Smyd1 Ki-CKO hearts. Expression of Anf and Myh7 was expanded into the AV canal and atrium (Fig. 2F). These data support the idea that Smyd1 plays a critical role in regulating the gene expression program governing early heart development.
Loss of Smyd1 at E9.5 deregulates genes critical to anti-proliferative responses to ER stress
To determine whether the smaller OFT and RV in Smyd1 Ki-CKO embryos resulted from altered proliferation of SHF progenitors, we performed immunostaining for BrdU. At E9.5, Smyd1 Ki-CKO embryos appeared to have less BrdU+ cells throughout the heart (Fig. 2G). As an indicator of SHF cell expansion, we quantified the percentage of BrdU+ cells in the OFT and found a significant decrease in cell proliferation for the Ki-CKO (Fig. 2H).
We further analyzed our microarray data for proliferation-related genes. These analyses indicated that cardiac ablation of Smyd1 resulted in an increase in anti-proliferative genes, particularly those known to respond to ER stress by activating autophagic responses (Fig. 2I). These included BCL2/adenovirus E1B 19kDa interacting protein 3 (Bnip3), Activation Transcription Factor 5 (Atf5), and Chop1/GADD153. Simultaneously, genes canonically down-regulated during autophagy induced by ER-stress [36,37], such as Isocitrate Dehydrogenase 1 (Idh1) and Cell Cycle and Apoptosis Regulatory Protein 1 (Ccar1), were down-regulated. Furthermore, genes known to be unregulated in cardiomyocytes by both ER stress and hypoxia, such as Prolyl hydroxylase domain 1 and 3 (PHD/Egln1/3), Jun dimerization protein 2 (Jundm2/JDP2), adrenomedullin (Adm), and Tribbles 3 (TRB3) [26,27,38,39,40,41], are overexpressed in the Smyd1 Ki-CKO (Fig. 2I and S1 Table).
Smyd1 Tg-CKO embryos exhibit mid-gestational proliferative defects, upregulation of ER/oxidative stress genes and lethality
In order to identify potential defects in later cardiomyocyte development, we generated a CKO of Smyd1 using a transgenic (Tg)-Nkx2.5-cre line that initiates deletion primarily in differentiated ventricular cardiomyocytes [25]. Embryos were recovered at various gestational time points to record gross abnormalities and viability. By E12.5, all Tg-CKO embryos were either dead or abnormal. No live Tg-CKOs were recovered after E15.5 (Fig. 3A). RT-PCR using mRNA extracted from hearts at E10.5 showed residual Smyd1 expression, likely the result of Smyd1 expression in cells outside of the cre-expressing zone, such as OFT myocardium (Fig. 3B). Smyd1 mRNA expression in the heart at E10.5 was also assayed by real-time PCR. Smyd1 mRNA levels were significantly reduced in the Tg-CKO (Fig. 3C).
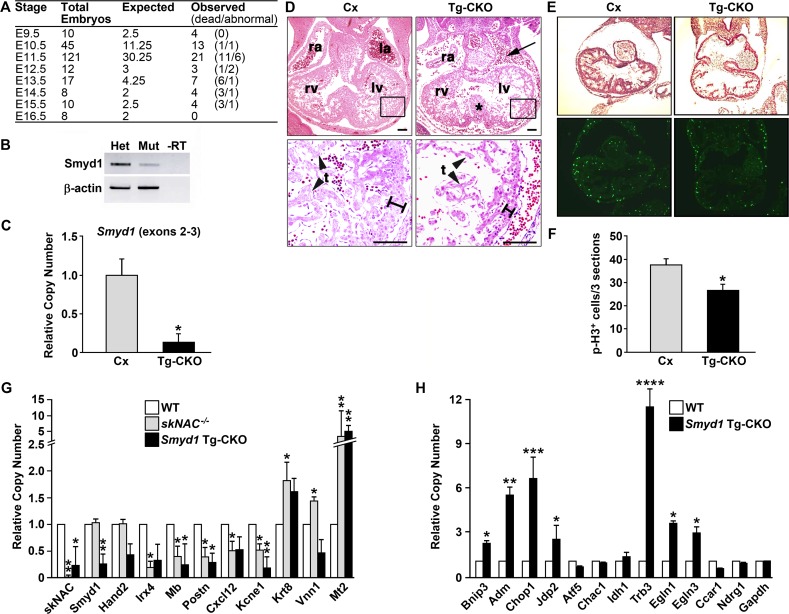
Fig 3. Deletion of Smyd1 by Tg-Nkx2.5-cre leads to a delayed embryonic lethal cardiac phenotype.
A. Smyd1 flox/flox; Tg-Nkx2.5-cre (Tg-CKO) embryos die at midgestation. Table numbers are total recovered embryos of each genotype. The number of dead or abnormal embryos is given in parentheses. B, C. Smyd1 mRNA expression at E10.5 assayed by RT-PCR (B) and real-time PCR (C). D. H&E-stained transverse sections of E11.5 control (Cx) and Tg-CKO embryos showing pericardial edema, thinned pericardium and decreased trabeculation. E. Decreased proliferation was observed in the hearts of E10.5 Smyd1 Tg-CKO embryos. Representative images of Cx and Tg-CKO hearts stained with H&E (upper panels) and the mitosis marker phospho-histone H3 serine 10 (p-H3) (lower panels). F. Quantification of p-H3 positive cells within the heart from three sections for three independent embryos (n = 3). G. Comparison of skNAC knockout and Smyd1 Tg-CKO heart gene expression by real-time PCR at E11.5. Data, focused primarily on oxidative response deregulation, are presented as mean for each genotype (skNAC -/-, n = 5; Smyd1 Tg-CKO, n = 4). Error bars indicate SEM. H. Genes encoding mediators of ER stress are deregulated by loss of Smyd1. Real-time PCR data represents average of 3 biological replicates each with 3 technical replicates; error bars indicate SEM. Data were analyzed by Student’s t-test (*P < 0.05, **P <0.01, ***P < 0.001, ****P < 0.0001).
Histological analyses were conducted using embryos with detectable heartbeats upon dissection at E11.5. Serial transverse sections were used for H&E staining. Compared to control embryos, all Tg-CKO embryos (n = 3) exhibited pericardial edema, thinned pericardium and decreased trabeculation in similar heart sections (Fig. 3D). As observed for the Ki-CKO at E9.5, E11.5 Smyd1 Tg-CKO embryos exhibited a significant decrease in proliferation within the heart as determined by immunofluorescence staining for the mitosis marker phospho-histone H3 S10 (Fig. 3E, F).
The observed mid-gestation Smyd1 Tg-CKO phenotype (thinned myocardium and poor trabeculation) is similar to that resulting from the knockout of skNAC, a Smyd1-interacting transcription factor exclusively expressed in striated muscle and initially characterized as a positive regulator of Mb [9]. Smyd1 directly associates with skNAC via binding of the skNAC PXLXP motif to the Smyd1 MYND domain [11,12]. Thus, we hypothesized that the Smyd1 Tg-CKO and skNAC KO exhibit similar dysregulation of gene expression. To this end, we analyzed expression of genes previously shown to be deregulated in skNAC knockouts at E11.5 [12] (Fig. 3G). Three of the 5 genes down-regulated in the skNAC knockout were also down-regulated in the Smyd1 Tg-CKO. Of the 3 genes upregulated in the skNAC knockout, only Metallothionein (Mt2) was significantly upregulated. The most significantly deregulated category was oxidative stress response [4,5,8,9,12]. Strong upregulation of Mt2 in the Smyd1 Tg-CKO may be in response to reduced expression of Mb and Vnn1. High upregulation of Mt2 in both animal models suggests that impaired oxidative stress defense systems may have contributed to the similar phenotype and timing of death. Expression of skNAC was decreased in the Smyd1 Tg-CKO, but loss of skNAC did not affect expression of Smyd1. Thus, Smyd1 is necessary for proper expression of skNAC, but not vice versa. Hand2 was down-regulated in the Smyd1 Tg-CKO, but unaffected in the skNAC -/-. This result supports the idea that Smyd1 and skNAC work in both complementary and independent pathways [12].
Based on previously observed similarities in genes modulated by stress response to both ER and oxidative stress [26,27,39,40,41,42], we analyzed potential deregulation in the Smyd1 Tg-CKO of the ER stress program activated in Ki-CKO hearts (Fig. 2I). As shown in Fig. 3H, real-time RT-PCR confirmed upregulation of Bnip3, Adm, Chop1, Jdp2, TRB3, Egln1 and Egln3. TRB3, a sensor for both ER and oxidative stress, exhibited the highest fold change. As with Ki-CKO hearts, we observed no apoptosis (data not shown) associated with Tg-CKO proliferative loss, and accordingly, Ccar1 was down-regulated (p<.061) [36,37]. In contrast to the Ki-CKO ER stress response, expression of Atf5, Chac1, Idh1 and Ndrg1 was unaffected. These data support the idea that both the ER and the oxidative stress response is initiated in the absence of Smyd1 at both gestational stages.
Smyd1 methylation of TRB3 provides a potential co-regulatory circuit for cardiac ER/oxidative stress transcriptional control
TRB3 is a kinase domain-containing multifunctional protein expressed broadly within the cytoplasm and nucleus of numerous cell types. While it was first characterized as a suppressor of AKT signaling [26,27], TRB3 expression is induced by several ER and oxidative stressors [26,27], and its transcription is activated by Chop1 [43], which as with TRB3, is strongly up-regulated by both Smyd1 Ki-CKO and Tg-Smyd1 CKO (Fig. 2I and 3H). TRB3 was of further interest because we had previously identified it as a putative Smyd1-binding partner in yeast two-hybrid assays (data not shown). Co-immunoprecipitation experiments confirmed that Smyd1 associates with TRB3 (Fig. 4A) and that the TRB3 far N-terminal region (residues 1–80) is both necessary and sufficient for the interaction (Suppl. S2 Fig.). This previously uncharacterized N-terminal region contains a conserved MYND domain binding motif (45P-X/A-L-X/L-P49) [13,14,15]. A similar motif within skNAC is essential for its interaction with SMYD1 [11].
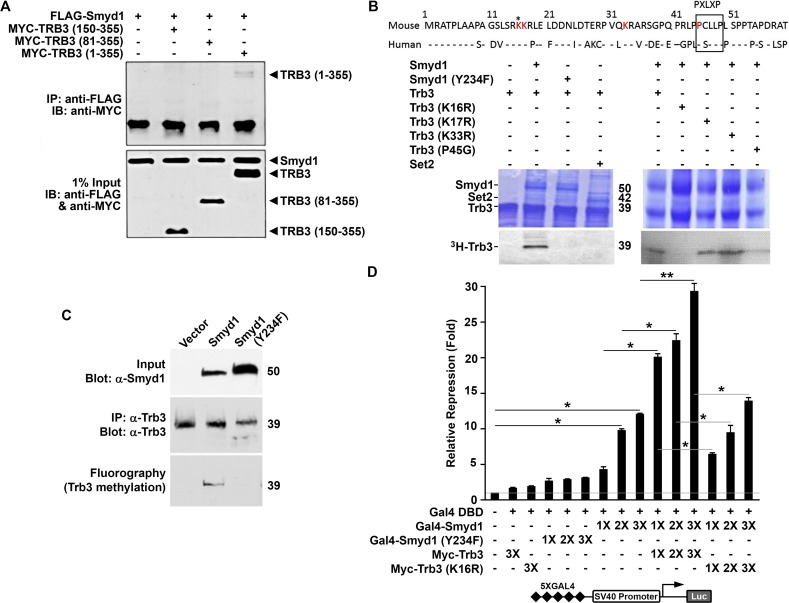
Fig 4. Smyd1 interacts with and methylates the ER and oxidative stress sensor Trb3 to regulate to regulate transcription.
A. Smyd1 interacts with the Trb3 N-terminus. MYC-Trb3 and FLAG-Smyd1 were co-transfected into Cos7 cells, and cell lysates were immunoprecipitated using a monoclonal anti-FLAG M2 antibody. Following SDS-PAGE, proteins were analyzed by western blot using a polyclonal anti-MYC A14 antibody. Input is shown in the lower panel. B. Smyd1 in vitro methylation of TRB3 at K16 requires an intact MYND interaction domain. Upper panel: Comparison of N-terminal amino acid sequences of human and mouse Trb3 (dashes indicate identity) to illustrate conserved lysines (in red) and the PXLXP motif (boxed). Asterisk (*) denotes the site of methylation. Lower panels: In vitro methylation assays were performed using, as enzymes, recombinant 6XHis-Smyd1, 6XHis-Smyd1 catalytic mutant (Y234F) or 6XHis-Set2. Substrates were wild-type 6XHis-TRB3 or the indicated K/R substitution mutant. S-[methyl-3H]adenosyl-L-methionine was employed as the methyl donor. Above: Coomassie blue-staining after fractionation by 12% SDS-PAGE. Molecular weights (kD) are indicated to the side of the image. Below: Fluorography-visualized methylation of Trb3 and mutants other than Trb3-K16/R or Trb3-P45/G by Smyd1 but not Smyd1Y234F or Set2. C. Smyd1 methylates Trb3 in vivo. COS7 cells were transiently transfected with MYC-Smyd1WT or MYC-Smyd1Y234F. Two days after transfection, cells were labeled with L-[methyl-3H]-methionine in the presence of protein synthesis inhibitors. Trb3 was immunoprecipitated by anti-Trb3 antibody resolved by SDS-PAGE, pull-down confirmed by anti-Trb3 western blot (middle panel) and its methylation was detected by fluorography (lower panel). Top panel: Western blot for Smyd1 using 10% of the input used for immunoprecipitation. D. Methylation-competent Trb3, but not a methylation-deficient mutant (Trb3-K16/R) enhances repression by Smyd1. Cos7 cells were co-transfected with an SV40-driven luciferase reporter containing 5X-GAL4 DNA-binding domain elements, GAL4-Smyd1, RSV-lacZ and increasing amounts of the indicated MYC-Trb3 or MYC-Trb3-K16/R expression plasmid. Results were normalized to β-galactosidase activity and fold-repression (y-axis) was determined relative to SV40-driven luciferase activity alone. Data were analyzed by Student’s t-test (*P < 0.05, **P <0.01).
Smyd1 was initially characterized as a histone H3K4-specific HMTase [16]; however, non-histone substrates are also likely because Smyd1 localizes to both the nucleus and sarcoplasm [11] (Fig. 1C). Non-histone, lysine-methylated targets for its highly related paralogue, Smyd2, include the proliferation suppressors, Rb and p53 [21,22,23]. These observations prompted us to carry out in vitro methylation assays, employing bacterially-expressed and purified 6X-His-Smyd1 and 6XHis-TRB3 along with several TRB truncations using 3H-S-adenosyl-methionine as a methyl donor. Smyd1, but not an HMTase SET domain catalytic mutant (Smyd1-Y234F), methylated full-length TRB3(1–355) as well as the TRB3(1–80) N-terminus but not further C-terminal truncations (S3 Fig.). TRB3(1–80) contains three lysines (K16,17 and 33), and the former two are conserved among human and other mammals (Fig. 4B, top panel and data not shown). We mutated each of these residues as well as the P45 of the PXLXP motif within the full length TRB3(1–355) backbone and tested the purified 6X-His-proteins for methylation. Catalytically active Smyd1, but not Smyd1-Y234/F nor an unrelated HMTase SET2, robustly methylated each of the TRB3 substrates with the exception of TRB3-K16 and P45 (Fig. 4B). Thus, Smyd1 methylation of TRB3 at K16 requires PXLXP-mediated interaction of the enzyme and substrate.
To confirm that Smyd1 methylates endogenous TRB3 within mammalian cells, we utilized an assay based on the fact that the methyl group of the methyl donor S-adenosyl-methionine is derived from free methionine in the cell [32]. Two days after transfection of Smyd1-negative Cos7 cells with Smyd1 or Smyd1-Y/234/F, cells were labeled with L-[methyl-3H]-methionine in the presence of protein synthesis inhibitors. TRB3 was immunoprecipitated, and the immunoprecipitates were resolved by SDS-PAGE, blotted with the corresponding antibodies, and methylated proteins were detected by fluorography. As shown in Fig. 4C, wild type, but not catalytically inactivate Smyd1-Y/234/F, methylates TRB3 in vivo.
To test if there is a link between Smyd1 modulation of gene expression and its methylation of TRB3, we utilized a GAL4 two-hybrid assay previously established for Smyd1-mediated repression by Sims et al [11]. Transfection of GAL4-Smyd1 led to robust, dose-dependent repression of SV40-driven 5x-GAL4-DBD reporter activity when analyzed 48 hr following transfection into Cos7 cells. Co-transfection of Myc-tagged-TRB3 full-length, but not the Myc-TRB3-K16/R methylation-deficient substitution mutant, led to significantly enhanced repression in a dose dependent manner (Fig. 4D). Thus, methylated TRB3, at least in cultured cells, functions as a specific co-repressor for Smyd1, suggesting a possible feedback loop to control TRB3 transcription and putatively other genes underlying the stress response to Smyd1 loss.
Discussion
To gain insight into the physiological functions of Smyd1 in the developing heart, we conditionally inactivated the Smyd1 gene using two different Nkx2.5-cre drivers, which become active in early cardiomyocyte progenitors (Ki-Nkx2.5-cre) or primarily in differentiated ventricular cardiomyocytes (Tg-Nkx2.5-cre). Our findings indicate that Smyd1 is a critical regulator of cardiac development at both the stage at which the SHF gene program is initiated and at later stages of cardiac morphogenesis. Both CKOs exhibit impaired cell proliferation and activation of stress pathways, which may be consequence of proliferative failure. A component of the underlying molecular mechanism of Smyd1 is upregulation and subsequent methylation of TRB3, a sensor for both ER and oxidative stress and suppressor of Akt signaling [26,27]. Smyd1 and TRB3 act synergistically to repress transcription in a methylation-dependent manner.
Different phenotypes with different Nkx2.5-cre drivers
It is of particular interest that the Smyd1 CKO phenotypes using two different Nkx2.5-cre drivers were dissimilar. Ki-CKO embryos died by E10.5 with impaired expansion of the SHF. Tg-CKO embryos survived until E11.5 or later, showed normal development of SHF-derived structures and exhibited myocardial abnormalities in both ventricles. The knock-in Nkx2.5-cre disrupts the Nkx2.5 open reading frame [24]. As a consequence, the Smyd1 Ki-CKO is also heterozygous for Nkx2.5. It is possible that loss of one Nkx2.5 allele amplifies the loss of Smyd1 phenotype; however, the difference in phenotype between the two CKOs is more likely due to differences in spatial and temporal expression of the two Nkx2.5-cre drivers. The transgenic Nkx2.5-cre [25] is fused to an enhancer that directs expression primarily in ventricular cardiomyocytes, whereas the knock-in Nkx2.5-cre employs the full range of endogenous enhancer/promoter elements and is active as early as E7.5 in the cardiac crescent [24]. Since the enhancer driving Tg-Nkx2.5-cre has a more restricted expression pattern than the endogenous Nkx2.5 gene [44], it is likely that Tg-CKO embryos survive longer due to delayed deletion of Smyd1 or lack of deletion in SHF progenitors and OFT myocardium.
Smyd1 Ki-CKO hearts exhibited similarities with those of the conventional Smyd1 knockout [10], whereas the Smyd1 Tg-CKO phenotype was more similar to that of the skNAC knockout [12]. The conventional Smyd1 knockout failed to form a RV, showed reduced expression of SHF genes and died before E10.5. Likewise, the Smyd1 Ki-CKO showed reduced size of SHF-derived structures (OFT and RV), a reduction is SHF gene expression and lethality by E10.5. The striated muscle-restricted transcription factor skNAC is one of the few known Smyd1-interacting proteins [9,11]. The skNAC knockout and Smyd1 Tg-CKO both exhibited myocardial defects in both ventricles, including thinning of the compact myocardium and decreased trabeculation. Furthermore, many of the same genes dysregulated in the skNAC knockout were similarly dysregulated in the Smyd1 Tg-CKO. These observations support the idea that the Smyd1-skNAC interaction regulates cardiomyocyte gene expression and is important for proper heart development.
Alterations in SHF-related gene expression
Among the 803 annotated transcripts significantly deregulated by Smyd1 elimination at E9.5, 324 were down-regulated, while 499 were upregulated. Smyd1 loss altered gene expression of a number of pathways (S1 Table), but particularly relevant to the early phenotype were genes regulating SHF development/maintenance. Prominent among these is the SHF transcription factor Isl1, whose expression defines the SHF and is critical for its proper differentiation [1,2]. Mice lacking Isl1 expression die in utero due to severe heart malformation, lacking all derivatives of the SHF [45,46,47]. Thus, reduction of Isl1 gene expression in Smyd1 Ki-CKO hearts could explain why the OFT and RV were reduced in size. Isl1 drives expression of core SHF cardiac transcription factors, including Mef2c [48]. The Smyd1 gene is a direct target of Mef2c [49]. Reduction in Isl1 expression suggests a positive feedback loop from Isl1 to Mef2c to Smyd1 and back to Isl1. In support, expression of Mef2c was also reduced in Smyd1 Ki-CKO hearts. Since residual Isl1 remained restricted to the pharyngeal mesoderm, additional factors not regulated by Smyd1 must control its spatial expression pattern.
Although expression of Hand2, Mef2c, Shox2, Tbx1 and Isl1 were reduced in Smyd1 Ki-CKO embryos, none were completely silenced. Thus, the severe SHF phenotype resulting from loss of Smyd1 may result from the combined effects of reducing expression of each of these genes as well as other genes comprising pathways not restricted to SHF. For example, we observed deregulation of chamber-specific factors (e.g. Myh7, Bmp7, Anf, Acta1, Egr1 and Fos) that are also upregulated in adult cardiac hypertrophy and/or heart failure [50,51,52,53]. While hypertrophy is unlikely to be detectable in early embryonic cardiomyocytes, induced deletion of Smyd1 in adult hearts leads to marked hypertrophy with upregulation of the same markers (Franklin et al, in review).
Alterations in expression of ER and oxidative stress genes
In the absence of Smyd1, cardiomyocyte proliferation is reduced concurrent with upregulation of genes involved in ER and oxidative stress responses. Gene expression profiling at E9.5 uncovered an apparent attempt to resolve ER stress via autophagy. Cells exposed to ER stress undergo the unfolded protein response in an attempt to avoid apoptosis, but may also activate autophagy [6,7]. While dysregulation of autophagy has been implicated in multiple pathological conditions, including cardiovascular diseases [54,55], little is known as to regulation of this pathway in the developing heart. In the Smyd1 Ki-CKO, three key trans-activators of the autophagic pathway were upregulated, including BCL2/adenovirus E1B 19-kDa interacting protein 3 (Bnip3), Activation Transcription Factor 5 (Atf5), and Chop1/GADD153 [6,7,56]. Atf5 directly activates another gene up-regulated in the Smyd1 Ki-CKO, the hallmark autophagy marker, cation transport regulator homolog 1 (Chac1) [6,7,56]. Chac1 and Idh1 are known to be differentially expressed in response to autophagic vs. apoptotic signals [36,37]. Isocitrate Dehydrogenase 1 (Idh1) and Cell Cycle and Apoptosis Regulatory Protein 1 (Ccar1) were down-regulated in the Smyd1 Ki-CKO. These genes are canonically down-regulated during autophagy induced by ER-stress, but are upregulated in ER stress-induced apoptosis [36,37]. This observation is consistent with the non-apoptotic phenotype observed in Smyd1 Ki-CKO. Prolyl hydroxylase domain 1 and -3 (PHD/Egln1/3) are known to be upregulated in cardiomyocytes by both ER stress and hypoxia [38], as are several additional genes which are overexpressed in the Smyd1 Ki-CKO including, Jun dimerization protein 2 (Jundm2/JDP2), adrenomedullin (Adm), and Tribbles 3 (TRB3) [26,27,39,40,41].
Although the above gene expression deregulation suggests initiation of autophagic resolution to ER stress, Smyd1 Ki-CKO cardiomyocytes did not show morphological signs of cellular degradation characteristic of the execution phase of this mechanism. This second stage is mediated by the autophagic PERK/eIF2α axis which, at least by transcript profiling, was unaffected in Smyd1 Ki-CKO (data not shown) [57]. This apparent paradox may be rationalized by the parallel strong upregulation of N-myc down-regulated gene 1 (Ndrg1) [58] in the Smyd1 Ki-CKO. Ndrg1 over-expression inhibits the ER stress-mediated autophagic pathway by suppressing PERK/eIF2α leading to a pro-survival phenotype [59]. As a further link between the observed early and late phenotypes of ER and oxidative stress, Idh1 reduction as well as Egln1 and Egln3 upregulation leads to decreased intracellular NADPH and, thus, to an increase in intracellular ROS [38]. Such triggers may underlie the array of oxidative stress response mediators shared by Smyd1 and skNAC-deficiencies.
In response to hypoxia and numerous stresses, tissues implement mechanisms to enhance oxygen delivery, including the activation of angiogenesis [52]. Ndrg1 and Adm, upregulated targets discussed above in the context of ER stress, are also strong inducers of angiogenesis. Several additional genes upregulated by Smyd1 Ki-CKO belong to the angiogenesis pathway. These include VegFα, Plek, and CxCl4 (S1 Table).
A potential Smyd1-TRB3 feedback loop for stress response regulation
We identified TRB3 as a non-histone methylation target of Smyd1. TRB3 and its family members function at the intersection of multiple stress-activated pathways including the mammalian target of rapamycin (mTOR), ER stress, oxidative stress and autophagy [60]. Consistent with a functional role in Smyd1 deficiency-mediated anti-proliferative responses, TRB3 transcript levels were significantly up-regulated in both Ki-CKO and Tg-CKO hearts. We confirmed TRB3 to be a Smyd1-interacting protein and found that Smyd1 methylates TRB3 in vitro and in vivo. Methylation of TRB3 at a single conserved lysine in its previously uncharacterized N-terminal 80 amino acids requires an intact PXLXP motif, which is known to mediate interactions with MYND domains. Thus, Smyd1 interacts with TRB3 through its MYND domain and methylates TRB3 through its SET domain. Methylated, but not unmethylated, TRB3 functions as a co-repressor for Smyd1.
Collectively these data indicate that Smyd1 regulates oxidative and ER stress responses at two levels. First, Smyd1 activates TRB3 transcription, perhaps indirectly via activation of Chop1, which encodes an established trans-activator of TRB3 [43]. Second, Smyd1 converts TRB3 into a co-repressor via direct methylation. The feasibility of this feedback loop is strengthened by the previously observed nucleo-cytoplasmic shuttling of Smyd1 during myocyte differentiation [11]. It remains an important extension of this work to determine to what extent target gene dysregulation in the absence of Smyd1 is due to direct epigenetic targeting of gene regulatory regions or due to methylation of non-histone proteins.
Concluding remarks
In summary, we have shown that Smyd1 is essential for proper expression of the cardiac gene program. Smyd1 null mutants exhibit malformation of SHF derivatives owing, at least in part, to reduced expansion of the progenitor pool. Smyd1 also regulates response to ER and oxidative stress. In the absence of Smyd1, a gene expression program consistent with a non-apoptotic autophagic response is initiated. These gene expression changes are consistent with observed reduction in cell proliferation and the absence of apoptosis. We also have shown that later in development, Smyd1 continues to play an essential role in response to oxidative stress, through both interaction with skNAC and dysregulation of non-skNAC target genes. As Smyd1 is continuously expressed into adulthood, it is likely involved with these pathways in both normal and pathologic conditions. For example, during ischemia as well as development and progression of maladaptive myocardial remodeling and failure, there is significant nutrient and oxygen starvation which leads to the induction of a number of ER and oxidative stress response genes—several of whom we observed here to be deregulated by Smyd1 deficiency [61,62,63]. Further analysis of Smyd1 function in adult tissues could lead to a better understanding of these mechanisms and allow development of novel and effective therapeutic strategies against heart failure.
Supporting Information
A. Illustration of the gene targeting strategy to generate the Smyd1 conditional knockout allele. The neo cassette was subsequently removed via breeding to EIIA-cre and selecting for loss of neo and retention of exons 2–3 (Smyd1 flox allele). B. Confirmation of correct Smyd1 gene targeting by Southern blot. Southern blots were performed using probes flanking the left and right homology arms (probes 2 and 3) to distinguish the 13.7 kb and 4.5 kb wild type (WT) allele fragments from the targeted allele (7.5 and a 7.0 kb), respectively. C. Smyd1 flox and WT alleles were distinguished by PCR. D, E. No Smyd1 RNA (D) or protein (E) was detectable in Smyd1 flox/flox;Ki-Nkx2.5-cre (Ki-CKO) hearts at E9.5.
(TIF)
C-terminal MYC-Trb3 deletion constructs were tested by co-immunoprecipitation with FLAG-Smyd1 to map the region of Trb3 that interacts with Smyd1. Cos7 cells were co-transfected FLAG-Smyd1 and the MYC-Trb3 deletion constructs, incubated for 30 h and lysed. Cell lysates were immunoprecipitated using a monoclonal anti-FLAG M2 antibody. The immunoprecipitates were separated by SDS-PAGE and analyzed by western blot using a polyclonal anti-MYC A14 antibody. Input is shown in the lower panels. The far N-terminal deletion (1–80) was sufficient for interaction with Smyd1.
(PDF)
6X-His-tagged wild-type Trb3(1–355) and the indicated truncation mutants were tested as substrates for methylation by full-length Smyd1. Methods are detailed in Materials and Methods section and legend to Fig. 4B.
(TIF)
Annotated genes were organized based on gene ontology using DAVID software [64]. These data were derived from two independent biologic replicas and at least 2 technical replicas of each for wild type (wt: Nkx2.5 +/+; Smyd1 Flox/Flox) and mutant (mut: Smyd1 Ki-CKO : Nkx2.5 cre/+; Smyd1 Flox/Flox) hearts at E9.5. Down-regulated transcripts indicated in yellow.
(PDF)
Acknowledgments
We thank Deborah Surman, Chhaya Das, June Harriss and Maya Ghosh for technical assistance. We thank the CPRIT Core Facility Support group for assistance with microarray analysis. We acknowledge the late Paul Gottlieb and Shanna Mika who initiated the project prior to their untimely loss to cancer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grants from the National Institutes of Health (NIH)(CA-31534), The Norman Hackerman Advanced Research Program (NHARP-003658-0149), Cancer Prevention Research Institute of Texas (CPRIT RP100612), and the Maria Betzner Morrow Endowment to HOT. Additional funding for this work came from grants from the American Heart Association (10BGIA4250008 and 12BGIA11860006) to MDS; by grants from the Natural Science Foundation of China (31140021 and 81260032) to YM; and by NIH grant HL-089902 to RJS. DS was supported by grants from the National Heart Lung and Blood Institute/NIH and the California Institute for Regenerative Medicine. Microarray analysis at the University of Texas is supported by CPRIT (CPRIT-RP120348). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Francou A, Saint-Michel E, Mesbah K, Theveniau-Ruissy M, Rana MS, Christoffels VM, et al. (2013) Second heart field cardiac progenitor cells in the early mouse embryo. Biochim Biophys Acta 1833: 795–798. 10.1016/j.bbamcr.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 2. Gittenberger-de Groot AC, Bartelings MM, Poelmann RE, Haak MC, Jongbloed MR (2013) Embryology of the heart and its impact on understanding fetal and neonatal heart disease. Semin Fetal Neonatal Med 18: 237–244. 10.1016/j.siny.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 3. Tsuchihashi T, Maeda J, Shin CH, Ivey KN, Black BL, Olson EN, et al. (2011) Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev Biol 351: 62–69. 10.1016/j.ydbio.2010.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J (2007) Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 39: 44–84. [DOI] [PubMed] [Google Scholar]
- 5. Dennery PA (2007) Effects of oxidative stress on embryonic development. Birth Defects Res C Embryo Today 81: 155–162. [DOI] [PubMed] [Google Scholar]
- 6. Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7: 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quick QA, Faison MO (2012) CHOP and caspase 3 induction underlie glioblastoma cell death in response to endoplasmic reticulum stress. Exp Ther Med 3: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hendgen-Cotta UB, Kelm M, Rassaf T (2014) Myoglobin's novel role in nitrite-induced hypoxic vasodilation. Trends Cardiovasc Med 24: 69–74. 10.1016/j.tcm.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 9. Yotov WV, St-Arnaud R (1996) Differential splicing-in of a proline-rich exon converts alphaNAC into a muscle-specific transcription factor. Genes Dev 10: 1763–1772. [DOI] [PubMed] [Google Scholar]
- 10. Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, et al. (2002) Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31: 25–32. [DOI] [PubMed] [Google Scholar]
- 11. Sims RJ 3rd, Weihe EK, Zhu L, O'Malley S, Harriss JV, Gottlieb PD (2002) m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem 277: 26524–26529. [DOI] [PubMed] [Google Scholar]
- 12. Park CY, Pierce SA, von Drehle M, Ivey KN, Morgan JA, Blau HM, et al. (2010) skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc Natl Acad Sci U S A 107: 20750–20755. 10.1073/pnas.1013493107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masselink H, Bernards R (2000) The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene 19: 1538–1546. [DOI] [PubMed] [Google Scholar]
- 14. Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T, et al. (2007) Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell 11: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lutterbach B, Sun D, Schuetz J, Hiebert SW (1998) The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol Cell Biol 18: 3604–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan X, Rotllant J, Li H, De Deyne P, Du SJ (2006) SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci U S A 103: 2713–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawamura S, Yoshigai E, Kuhara S, Tashiro K (2008) smyd1 and smyd2 are expressed in muscle tissue in Xenopus laevis. Cytotechnology 57: 161–168. 10.1007/s10616-008-9128-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li D, Niu Z, Yu W, Qian Y, Wang Q, Li Q, et al. (2009) SMYD1, the myogenic activator, is a direct target of serum response factor and myogenin. Nucleic Acids Res 37: 7059–7071. 10.1093/nar/gkp773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Just S, Meder B, Berger IM, Etard C, Trano N, Patzel E, et al. (2011) The myosin-interacting protein SMYD1 is essential for sarcomere organization. J Cell Sci 124: 3127–3136. 10.1242/jcs.084772 [DOI] [PubMed] [Google Scholar]
- 20. Strahl BD, Ohba R, Cook RG, Allis CD (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A 96: 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632. [DOI] [PubMed] [Google Scholar]
- 22. Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, et al. (2011) Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol 3: 301–308. 10.1093/jmcb/mjr025 [DOI] [PubMed] [Google Scholar]
- 23. Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, et al. (2010) Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 285: 37733–37740. 10.1074/jbc.M110.137612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ (2001) Embryonic expression of an Nkx2–5/Cre gene using ROSA26 reporter mice. Genesis 31: 176–180. [DOI] [PubMed] [Google Scholar]
- 25. McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN (2005) The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132: 189–201. [DOI] [PubMed] [Google Scholar]
- 26. Avery J, Etzion S, DeBosch BJ, Jin X, Lupu TS, Beitinjaneh B, et al. (2010) TRB3 function in cardiac endoplasmic reticulum stress. Circ Res 106: 1516–1523. 10.1161/CIRCRESAHA.109.211920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koh HJ, Toyoda T, Didesch MM, Lee MY, Sleeman MW, Kulkarni RN, et al. (2013) Tribbles 3 mediates endoplasmic reticulum stress-induced insulin resistance in skeletal muscle. Nat Commun 4: 1871 10.1038/ncomms2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagy A (2003) Manipulating the mouse embryo: a laboratory manual Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. x, 764 p. p. [Google Scholar]
- 29. Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, et al. (2007) Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci U S A 104: 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, et al. (2008) Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci U S A 105: 17824–17829. 10.1073/pnas.0805491105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Foreman KW, Brown M, Park F, Emtage S, Harriss J, Das C, et al. (2011) Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS One 6: e22290 10.1371/journal.pone.0022290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Q, Dreyfuss G (1995) In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol 15: 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, et al. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A 93: 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hildreth V, Webb S, Chaudhry B, Peat JD, Phillips HM, Brown N, et al. (2009) Left cardiac isomerism in the Sonic hedgehog null mouse. J Anat 214: 894–904. 10.1111/j.1469-7580.2009.01087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME (2008) Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 135: 1157–1167. 10.1242/dev.014563 [DOI] [PubMed] [Google Scholar]
- 36. Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, et al. (2012) D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 26: 2038–2049. 10.1101/gad.198200.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang Y, Puliyappadamba VT, Zhang L, Wu W, Wali A, Yaffe MB, et al. (2010) A novel mechanism of cell growth regulation by Cell Cycle and Apoptosis Regulatory Protein (CARP)-1. J Mol Signal 5: 7 10.1186/1750-2187-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sen Banerjee S, Thirunavukkarasu M, Tipu Rishi M, Sanchez JA, Maulik N, Maulik G (2012) HIF-prolyl hydroxylases and cardiovascular diseases. Toxicol Mech Methods 22: 347–358. 10.3109/15376516.2012.673088 [DOI] [PubMed] [Google Scholar]
- 39. Hill C, Wurfel A, Heger J, Meyering B, Schluter KD, Weber M, et al. (2013) Inhibition of AP-1 signaling by JDP2 overexpression protects cardiomyocytes against hypertrophy and apoptosis induction. Cardiovasc Res 99: 121–128. 10.1093/cvr/cvt094 [DOI] [PubMed] [Google Scholar]
- 40. Tanigawa S, Lee CH, Lin CS, Ku CC, Hasegawa H, Qin S, et al. (2013) Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis 4: e921 10.1038/cddis.2013.448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wetzel-Strong SE, Li M, Klein KR, Nishikimi T, Caron KM (2013) Epicardial-derived adrenomedullin drives cardiac hyperplasia during embryogenesis. Dev Dyn. [DOI] [PMC free article] [PubMed]
- 42. Fang BA, Kovacevic Z, Park KC, Kalinowski DS, Jansson PJ, Lane DJ, et al. (2014) Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim Biophys Acta 1845: 1–19. 10.1016/j.bbcan.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 43. Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN (1999) Control of early cardiac-specific transcription of Nkx2–5 by a GATA-dependent enhancer. Development 126: 75–84. [DOI] [PubMed] [Google Scholar]
- 45. Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. (2003) Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. (2006) Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127: 1151–1165. [DOI] [PubMed] [Google Scholar]
- 47. Kattman SJ, Huber TL, Keller GM (2006) Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11: 723–732. [DOI] [PubMed] [Google Scholar]
- 48. Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL (2004) Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131: 3931–3942. [DOI] [PubMed] [Google Scholar]
- 49. Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, et al. (2005) BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development 132: 2669–2678. [DOI] [PubMed] [Google Scholar]
- 50. Oka T, Akazawa H, Naito AT, Komuro I (2014) Angiogenesis and cardiac hypertrophy: maintenance of cardiac function and causative roles in heart failure. Circ Res 114: 565–571. 10.1161/CIRCRESAHA.114.300507 [DOI] [PubMed] [Google Scholar]
- 51. Iwaki K, Sukhatme VP, Shubeita HE, Chien KR (1990) Alpha- and beta-adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an alpha 1-mediated response. J Biol Chem 265: 13809–13817. [PubMed] [Google Scholar]
- 52. Komuro I, Kaida T, Shibazaki Y, Kurabayashi M, Katoh Y, Hoh E, et al. (1990) Stretching cardiac myocytes stimulates protooncogene expression. J Biol Chem 265: 3595–3598. [PubMed] [Google Scholar]
- 53. Komuro I, Kurabayashi M, Shibazaki Y, Katoh Y, Hoh E, Kaida T, et al. (1990) Molecular mechanism of cardiac hypertrophy. Jpn Circ J 54: 526–534. [DOI] [PubMed] [Google Scholar]
- 54. Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sridhar S, Botbol Y, Macian F, Cuervo AM (2012) Autophagy and disease: always two sides to a problem. J Pathol 226: 255–273. 10.1002/path.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lane DJ, Saletta F, Suryo Rahmanto Y, Kovacevic Z, Richardson DR (2013) N-myc downstream regulated 1 (NDRG1) is regulated by eukaryotic initiation factor 3a (eIF3a) during cellular stress caused by iron depletion. PLoS One 8: e57273 10.1371/journal.pone.0057273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sahni S, Bae DH, Lane DJ, Kovacevic Z, Kalinowski DS, Jansson PJ, et al. (2014) The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J Biol Chem 289: 9692–9709. 10.1074/jbc.M113.529511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cunard R (2013) Mammalian Tribbles Homologs at the Crossroads of Endoplasmic Reticulum Stress and Mammalian Target of Rapamycin Pathways. Scientifica (Cairo) 2013: 750871 10.1155/2013/750871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Groenendyk J, Sreenivasaiah PK, Kim do H, Agellon LB, Michalak M (2010) Biology of endoplasmic reticulum stress in the heart. Circ Res 107: 1185–1197. 10.1161/CIRCRESAHA.110.227033 [DOI] [PubMed] [Google Scholar]
- 62. Takahashi M, Konishi T, Maeda Y, Matsugu Y, Akazawa F, Eto T, et al. (1993) Use of the conjugate of disulphated ursodeoxycholic acid with p-aminobenzoic acid for the detection of intestinal bacteria. Gut 34: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsutsui H, Kinugawa S, Matsushima S (2011) Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol 301: H2181–2190. 10.1152/ajpheart.00554.2011 [DOI] [PubMed] [Google Scholar]
- 64. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Illustration of the gene targeting strategy to generate the Smyd1 conditional knockout allele. The neo cassette was subsequently removed via breeding to EIIA-cre and selecting for loss of neo and retention of exons 2–3 (Smyd1 flox allele). B. Confirmation of correct Smyd1 gene targeting by Southern blot. Southern blots were performed using probes flanking the left and right homology arms (probes 2 and 3) to distinguish the 13.7 kb and 4.5 kb wild type (WT) allele fragments from the targeted allele (7.5 and a 7.0 kb), respectively. C. Smyd1 flox and WT alleles were distinguished by PCR. D, E. No Smyd1 RNA (D) or protein (E) was detectable in Smyd1 flox/flox;Ki-Nkx2.5-cre (Ki-CKO) hearts at E9.5.
(TIF)
C-terminal MYC-Trb3 deletion constructs were tested by co-immunoprecipitation with FLAG-Smyd1 to map the region of Trb3 that interacts with Smyd1. Cos7 cells were co-transfected FLAG-Smyd1 and the MYC-Trb3 deletion constructs, incubated for 30 h and lysed. Cell lysates were immunoprecipitated using a monoclonal anti-FLAG M2 antibody. The immunoprecipitates were separated by SDS-PAGE and analyzed by western blot using a polyclonal anti-MYC A14 antibody. Input is shown in the lower panels. The far N-terminal deletion (1–80) was sufficient for interaction with Smyd1.
(PDF)
6X-His-tagged wild-type Trb3(1–355) and the indicated truncation mutants were tested as substrates for methylation by full-length Smyd1. Methods are detailed in Materials and Methods section and legend to Fig. 4B.
(TIF)
Annotated genes were organized based on gene ontology using DAVID software [64]. These data were derived from two independent biologic replicas and at least 2 technical replicas of each for wild type (wt: Nkx2.5 +/+; Smyd1 Flox/Flox) and mutant (mut: Smyd1 Ki-CKO : Nkx2.5 cre/+; Smyd1 Flox/Flox) hearts at E9.5. Down-regulated transcripts indicated in yellow.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.