Highlights
-
•
Cryptosporidium diversity was investigated in a BTRW as part of a recovery programme.
-
•
Faecal samples from captive bred, supplemented and wild wallabies were screened.
-
•
Cryptosporidium isolates were identified at three gene loci using PCR.
-
•
Diverse species of Cryptosporidium were identified across populations.
-
•
Both specific, C. fayeri, and broad host species, C. meleagridis, were identified.
Keywords: Cryptosporidium, Molecular detection, Brush-tailed rock-wallaby, Conservation management
Graphical Abstract

Abstract
Host–parasite relationships are likely to be impacted by conservation management practices, potentially increasing the susceptibility of wildlife to emerging disease. Cryptosporidium, a parasitic protozoan genus comprising host-adapted and host-specific species, was used as an indicator of parasite movement between populations of a threatened marsupial, the brush-tailed rock-wallaby (Petrogale penicillata). PCR screening of faecal samples (n = 324) from seven wallaby populations across New South Wales, identified Cryptosporidium in 7.1% of samples. The sampled populations were characterised as captive, supplemented and wild populations. No significant difference was found in Cryptosporidium detection between each of the three population categories. The positive samples, detected using 18S rRNA screening, were amplified using the actin and gp60 loci. Multi-locus sequence analysis revealed the presence of Cryptosporidium fayeri, a marsupial-specific species, and C. meleagridis, which has a broad host range, in samples from the three population categories. Cryptosporidium meleagridis has not been previously reported in marsupials and hence the pathogenicity of this species to brush-tailed rock-wallabies is unknown. Based on these findings, we recommend further study into Cryptosporidium in animals undergoing conservation management, as well as surveying wild animals in release areas, to further understand the diversity and epidemiology of this parasite in threatened wildlife.
1. Introduction
Disease emergence presents a significant risk to the conservation of endangered wildlife. The risks of disease are leading to growing concern of the cost–benefit efficiency of the supplementation strategy (Kock et al., 2010). Species recovery actions such as the supplementation of dwindling populations with captive bred animals may introduce parasites atypical to the recovery species or exacerbate prevalence of existing pathogens due to stress and immune status of captive bred individuals, which may spread these pathogens into its new environment (Moberg, 1985; Cunningham, 1996). Control of disease risks requires a sound understanding of host–parasite interactions, both in threatened species and of hosts that may contribute to disease emergence. Further, parasites specific to the target species may not survive translocation or other conservation processes, thereby unbalancing the natural host–parasite relationship (Moir et al., 2012).
Cryptosporidium, a protozoan parasite with a broad vertebrate host range and variable host specificity, represents a potential indicator of disease risks associated with conservation management. This research strategy is particularly applicable to threatened Australian marsupials where the occurrence of human derived Cryptosporidium species has not been conclusively determined (Hill et al., 2008; Ng et al., 2011; Dowle et al., 2013).
Of the 26 described Cryptosporidium species (reviewed in Ryan et al., 2014), twelve have been reported in both humans and other hosts: C. parvum, C. hominis, C. ubiquitum, C. andersoni, C. bovis, C. cuniculus, C. muris, C. canis, C. felis, C. meleagridis, C. suis and C. fayeri (Xiao et al., 2001; Gatei et al., 2002; Xiao, 2002; Leoni et al., 2006; Robinson et al., 2010; Waldron et al., 2010). Each of the Cryptosporidium species reported in humans have been found in the Australian environment (Ryan and Power, 2012; Abeywardena et al., 2013; Nolan et al., 2013), though human infections in Australia are predominantly C. parvum and C. hominis (Waldron et al., 2011).
Despite Cryptosporidium being identified in 16 marsupial species from 7 families (reviewed in O'Donoghue, 1995 and Power, 2010), identifications of Cryptosporidium to species level is limited to recent studies employing molecular tools (Warren et al., 2003; Hill et al., 2008; Power and Ryan, 2008; Ryan et al., 2008; Yang et al., 2011). Following molecular identification, marsupials were found to be susceptible to two host-adapted Cryptosporidium species, C. fayeri and C. macropodum (Power and Ryan, 2008; Ryan et al., 2008). Several other host-specific genotypes have also been described in marsupials including brushtail possum genotype I from brushtail possums (Trichosurus vulpecula) (Hill et al., 2008) and kangaroo genotype I from western grey kangaroos (Macropus fuliginosus) (Yang et al., 2011).
Although there are reports of C. parvum and C. hominis in marsupials, these are based only upon a molecular signature from a faecal DNA sample, and an infection has never been confirmed using other methods such as parasite isolation (Hill et al., 2008; Ng et al., 2011; Dowle et al., 2013). The molecular detection of C. parvum and C. hominis in marsupial hosts has also been associated with an inability to confirm at greater than a single locus, namely the 18S rRNA. Passage of C. parvum of C. hominis oocysts through the marsupial gut is the likely reason for identifications of these Cryptosporidium species in marsupials (Dowle et al., 2013). The only confirmed case of Cryptosporidium infection in a marsupial that was not host specific was an infection of C. muris in captive greater bilbies (Macrotis lagotis) being bred for release into natural habitat (Warren et al., 2003).
Here we use molecular methods to detect and identify Cryptosporidium in the brush-tailed rock-wallaby (BTRW), Petrogale penicillata. This species is listed as ‘endangered’ in New South Wales, Australia (NSW Threatened Species Conservation Act 1995) and ‘near threatened’ on the IUCN Red List of Threatened Species across eastern Australia (IUCN, 2013). There is an approved NSW Recovery Plan for the species (DECC, 2008), as well as an approved National Recovery Plan (Menkhorst and Hynes, 2010). These plans identify supplementation of small colonies with captive bred individuals as an important recovery strategy and over the last few years several translocations of individuals between captive breeding facilities and wild populations have occurred (Menkhorst and Hynes, 2010). As rock-wallaby populations have experienced variable levels of human intervention, studying their parasites provides a platform to examine the effect of conservation management on the host–parasite relationship. Hence, our aim was to detect and identify Cryptosporidium species infecting wild, captive bred, and supplemented brush-tailed rock-wallaby populations.
2. Methods
2.1. Sample collection and sites
Brush-tailed rock-wallabies were once abundant in south-eastern Australia but are now reduced to fragmented populations in New South Wales and Victoria (Eldridge and Close, 2005). Dispersal between populations, which are located in steep, rocky habitats, is rare (Browning et al., 2001). For this study, seven BTRW sites were sampled between March 2010 and July 2013 (Table 1). Sample collection dates were spread evenly across three seasons (Autumn, Summer and Winter), with ~10 samples collected in Spring (2010 and 2012), spread evenly across the four years. The origin of each population varied and included three categories: one site with a BTRW population kept in a captive breeding facility (captive bred), sites where free-ranging populations had been supplemented with captive bred individuals (supplemented) and two pristine sites with only free-ranging animals (wild). Fresh faecal samples were collected in vials containing silicon beads from each site opportunistically from unknown individuals during routine colony management by the Office of Environment and Heritage staff and were then stored at 4 °C until further processing. The highest number of samples was obtained from Square Top in Warrumbungle National Park since this was a major release site.
Table 1.
The rate of Cryptosporidium detected at the different loci per screened site and site category. All sites are in New South Wales; the precise location is withheld for some sites for the safety of the animals. KV means Kangaroo Valley. Samples at the loci (18S rRNA, actin and gp60) were deemed as positive after DNA sequencing.
| Site | Population category | No. of samples | 18S rRNA (298 bp) | 18S rRNA (825 bp) | Actin | gp60 |
|---|---|---|---|---|---|---|
| KV Mountain | Wild | 55 | 7 | 7 | 2 | 3 |
| KV River | Supplemented | 43 | 2 | 1 | 0 | 1 |
| KV Creek | Supplemented | 10 | 4 | 3 | 0 | 0 |
| Nattai | Wild | 30 | 3 | 3 | 1 | 1 |
| Square Top | Supplemented | 123 | 5 | 4 | 0 | 0 |
| Waterfall Springs | Captive breda | 39 | 2 | 2 | 1 | 2 |
| Jenolan Caves | Supplemented | 24 | 0 | 0 | 0 | 0 |
Wallabies in a captive breeding facility.
2.2. DNA extraction and PCR screening
Genomic DNA was extracted from faecal material (~150 mg) using the ISOLATE Fecal DNA kit (Bioline, London, UK) following manufacturer's instructions. The extracted DNA was stored at −20 °C until further analysis. Directly prior to each PCR, the DNA samples were treated with GeneReleaser (BioVentures, Inc., TN, USA) by combining equal volumes of DNA and GeneReleaser, and subjecting the mixture to 7 min in a 500 W microwave.
2.3. PCR screening at the 18S rRNA locus
DNA samples were initially screened for Cryptosporidium using nested PCR to amplify a partial fragment of the 18S rRNA. The primary reaction followed the methodology of Xiao et al. (1999) but with a lower MgCl2 concentration (2 mM). The secondary reaction comprised the primers 18S IF and 18S IR and followed the method of Morgan et al. (1997). PCRs were performed using Red Hot Taq DNA polymerase (ThermoFisher Scientific, Waltham, MA, USA) as previously described (Hill et al., 2008). Both reactions were modified to increase specificity for Cryptosporidium by lowering the concentration of dNTPs to 50 µM.
Longer 18S rRNA fragments were generated for samples testing positive for Cryptosporidium using the 18S IF and 18s IR primer set. The longer fragments were amplified using the primers of Xiao et al. (1999) for both primary and secondary reactions, following conditions as previously described by Waldron et al. (2011), inclusive of dNTPs and MgCl2 concentrations as described above.
2.4. PCR amplification at confirmatory loci
To confirm 18S rRNA positives, DNA samples were screened at two additional loci, actin and glycoprotein 60 (gp60). For the actin locus, a nested protocol (Sulaiman et al., 2002) was performed with minor modifications. To improve specificity for Cryptosporidium, the concentration of MgCl2 was lowered to 2 mM, dNTPS to 50 µM, and the annealing temperature raised to 54 °C in the secondary reaction. All amplifications were performed using Red Hot Taq DNA Polymerase.
Amplification of the gp60 locus was achieved using a nested protocol with primary amplification achieved using the primers outF and outR (Power et al., 2009) and secondary reactions using ATGF and StopR (Waldron et al., 2009). Red Hot Taq was used for both amplifications. All PCR reactions performed included a negative control (H2O) and a positive control of DNA extracted from purified oocysts of C. parvum.
2.5. Sequencing of positive samples
All amplicons generated for 18S rRNA, actin and gp60 were sequenced to enable Cryptosporidium species identification. Amplicons from each of the four PCRs which contained a band of the expected size when resolved by electrophoresis (2% agarose in TBE with SYBR Green staining) were purified using the QIAQuick PCR purification kit (Qiagen, Venlo, the Netherlands). Purified amplicons were sequenced in both directions (Macrogen, Seoul, Korea) using appropriate primers for amplifications, with the exception of the short fragment of the 18S rRNA, which were only sequenced with the primer 18S IF (Morgan et al., 1997).
2.6. Sequencing and phylogenetic analyses
The sense and antisense sequence fragments for each locus were aligned with Geneious (version 6.1.7, Biomatters LtD, New Zealand) and manually examined for quality and read errors. Consensus sequences for each positive sample were extracted and searched against GenBank using BlastN function in Geneious. To enable species identification within a phylogenetic framework, samples positive for 18S rRNA (~825 bp) were trimmed to the same length and aligned with Cryptosporidium reference sequences from GenBank using Clustal W (Larkin et al., 2007). A phylogenetic tree was constructed based on this alignment using neighbour-joining. Sequences generated in this study have been submitted to GenBank under accession numbers KP730299-KP730329.
2.7. Statistical analysis
To test differences of Cryptosporidium detection rate between sites and site categories, samples were tested at the 18S rRNA (~298 bp) locus for presence or absence and checked for significant differences with a chi-square test in Minitab (version 17.1.0, Minitab Inc.).
3. Results
3.1. Cryptosporidium screening
DNA was extracted from 324 samples and screened for Cryptosporidium using 18S rRNA PCR. Of the 324 screened faecal samples, 43 contained the expected amplicon. DNA sequencing and Blast searches identified 23 samples as being Cryptosporidium, giving a total positive identification rate of 7.1% in BTRW. Cryptosporidium positive samples were obtained from three site types (captive bred, supplemented and wild). Positives were found to be present across most study sites except for Jenolan Caves. There was no significant difference in Cryptosporidium detection between captive-bred, wild and supplemented as categories (χ2 = 3.811, DF = 2, p = 0.149). However, there was a significant difference between the sites (χ2 = 23.6, DF = 6, p < 0.001). Kangaroo Valley Creek had the highest rate of positive samples (40%), but this site had the lowest amount of samples tested (n = 10; Table 1).
3.2. Species identification at the 18S rRNA locus
From the initial positive samples (n = 23), 20 samples yielded sequence data for the larger 18S rRNA fragment (825 bp), which was used to generate a phylogeny (Fig. 1). Four samples from supplemented sites and three samples from wild sites clustered with the C. parvum and C. hominis. Three samples from supplemented sites, one from a captive bred site and one from a wild site grouped with the marsupial-specific species C. fayeri and C. macropodum. A further four samples from a wild site and one from a captive-bred site grouped with C. meleagridis. Two samples from a wild site were grouped with C. ubiquitum.
Fig. 1.
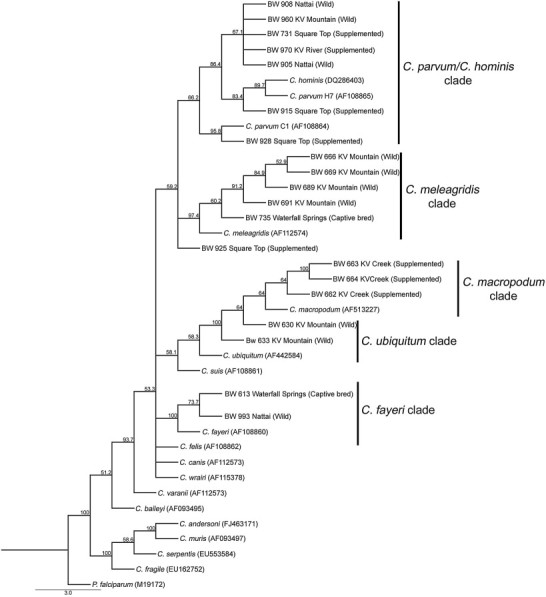
Samples were identified within a phylogenetic framework with the tree constructed using neighbour-joining with bootstrap test (1,000 replicates, displayed at nodes) using the 18S rRNA locus (878 bp). KV denotes Kangaroo Valley.
3.3. Species confirmation using actin and gp60
Sequence analysis at the actin and gp60 loci resulted in amplicons and sequence data from only eight samples across all loci (Table 2). At the actin locus, although 15 samples generated a band of the expected size (~1066 bp), only four were identified using BlastN searches as Cryptosporidium, with two samples being C. fayeri and two being C. meleagridis. For gp60, seven samples generated an amplicon with three samples assigned to C. fayeri and four to C. meleagridis (Table 2). However, the four samples from Kangaroo Valley Mountain identifying as C. meleagridis may represent the same individual sampled twice over two time points. For those samples identified at the 18S rRNA as C. hominis and C. parvum, neither actin nor gp60 could be amplified. An exception was sample 973 identified as C. hominis at the 18S rRNA and C. fayeri by gp60 sequencing.
Table 2.
Species identification across three loci (18S rRNA, actin and gp60) for the samples positive at the 18S rRNA locus for C. fayeri and C. meleagridis. Samples identified at the 18S rRNA locus as other Cryptosporidium species (Fig. 1) were omitted from this table since they could not be amplified at other loci. NP indicates no product was amplified. KV means Kangaroo Valley.
| Label BW # | Location | Site type | 18S rRNA (298 bp) | % Similarity | 18S rRNA (825 bp) | % Similarity | Actin | % Similarity | gp60 | % Similarity |
|---|---|---|---|---|---|---|---|---|---|---|
| 613 | Waterfall Springs | Captive bred | C. fayeri | 99.6% | C. fayeri | 99.5% | C. fayeri | 99.9% | C. fayeri (subtype A10) | 99.7% |
| 666 | KV Mountain | Wild | C. meleagridis | 99.2% | C. meleagridis | 99.9% | NP | NP | NP | NP |
| 669 | KV Mountain | Wild | C. meleagridis | 99.8% | C. meleagridis | 99.6% | C. meleagridis | 99.9% | C. meleagridis (subtype IIIgA) | 91.4% |
| 689 | KV Mountain | Wild | C. meleagridis | 99.6% | C. meleagridis | 99.9% | C. meleagridis | 99.7% | C. meleagridis (subtype IIIgA) | 91.2% |
| 691 | KV Mountain | Wild | C. meleagridis | 99.2% | C. meleagridis | 99.7% | NP | NP | C. meleagridis (subtype IIIgA) | 91.6% |
| 735 | Waterfall Springs | Captive bred | C. meleagridis | 99.1% | C. meleagridis | 99.8% | NP | NP | C. meleagridis (subtype IIIbA) | 88.8% |
| 973 | KV River | Supplemented | C. hominis | 100.0% | NP | NP | NP | NP | C. fayeri (subtype A10) | 99.9% |
| 993 | Nattai | Wild | C. fayeri | 99.6% | C. fayeri | 99.4% | C. fayeri | 99.4% | C. fayeri (subtype A7) | 99.8% |
4. Discussion
The level of detection of Cryptosporidium in BTRW (7.1%) is consistent with observations of Cryptosporidium in other marsupials which range between 6.7% and 12.2% (Power et al., 2004; Hill et al., 2008; Yang et al., 2011; Dowle et al., 2013). Here, Cryptosporidium detection in BTRW is based on sequence identifications using a 298 bp fragment of the 18S rRNA. PCR is commonly employed for detection of Cryptosporidium in faecal samples as this approach has greater sensitivity than microscopy, both in detection and identification of species (Fall et al., 2003; Power et al., 2003; Ryan et al., 2008; Dowle et al., 2013). In our study, a larger fragment (~825 bp) failed to amplify three samples confirmed as Cryptosporidium using the smaller fragment, indicating that selection of optimal amplification methods should be considered when undertaking molecular detection of this parasite.
Despite no significant difference in the detection of Cryptosporidium between captive bred and free ranging animals, the identity of Cryptosporidium species in BTRW determined by sequencing raises concern for the health status of captive and wild BTRW. Cryptosporidium fayeri has previously been identified in six marsupial hosts including the related yellow-footed rock-wallaby P. xanthopus (Morgan et al., 1997; Power et al., 2003, 2009; Ryan et al., 2008; Power, 2010; Yang et al., 2011; Nolan et al., 2013). Cryptosporidium fayeri does not appear to cause disease in marsupials (Ryan et al., 2008). Cryptosporidium meleagridis has been identified in a range of vertebrates, including avian and mammalian hosts, as well as humans (Akiyoshi et al., 2003; Xiao et al., 2004). While C. meleagridis is the most common infection of Cryptosporidium in humans after C. parvum and C. hominis (Elwin et al., 2012), human infections are rare in Australia (Waldron et al., 2011). Consequently, it is unlikely that the captive bred animals were infected from human sources, but by other host species, such as free ranging birds, inhabiting the captive breeding site. The wild site, where most of the C. meleagridis isolates were found, is secluded from humans and thus transmission between humans and BTRW is unlikely. This is supported by the gp60 analysis.
The C. meleagridis gp60 sequences from BTRW isolates displayed greater genetic similarity to gp60 sequences from avian hosts (Stensvold et al., 2014), yet they were distinct from described sequences, indicating a new gp60 C. meleagridis subtype. This finding is the first report where a zoonotic species of Cryptosporidium was confirmed across multiple loci in a wild marsupial host. As such, much is unknown about the diversity and pathogenicity of C. meleagridis in wild marsupials and thus further study is required to understand the extent to which this species has penetrated marsupial hosts and likely transmission routes.
Cryptosporidium parvum and C. hominis were also identified in BTRW samples; however, these identifications were only possible at a single locus, the 18S rRNA. Only one of these samples could be amplified at one of the two confirmatory loci where it was typed as C. fayeri. Both C. parvum and C. hominis have been reported in a range of marsupials but similar to this study, other studies also failed to confirm identifications at loci other than the 18S rRNA (Hill et al., 2008; Ng et al., 2011; Dowle et al., 2013). Some isolates were inferred to be C. ubiquitum and C. macropodum through a GenBank match at the 18S rRNA locus but failed to amplify at subsequent loci (Fig. 1). While C. macropodum is specific to marsupials, particularly macropods (Power and Ryan, 2008), C. ubiquitum is typical to cattle but is commonly identified in humans as well (Fayer et al., 2010). So far, no report has been made of C. ubiquitum in marsupials (Ryan and Power, 2012).
Failure to amplify C. parvum and C. hominis isolates from marsupials at other loci has been attributed to low numbers of oocysts and the multi copy nature of the 18S rRNA locus compared to single copy confirmatory loci (Hill et al., 2008; Power et al., 2009; Ng et al., 2011). Indeed, oocyst counts in possums and bandicoots confirm low oocyst numbers (Hill et al., 2008; Dowle et al., 2013). The question remains if the presence of these human infective species is merely passage of oocysts through the marsupial gut or a true infection. Whole genome amplification could be employed to boost amplification of low oocyst numbers. This method has previously proved successful on clinical samples of C. parvum and C. hominis across three loci (Bouzid et al., 2010). Identification at the 18S rRNA locus alone has been found to underestimate mixed infections as this technique preferentially amplifies a predominant genotype (Reed et al., 2002). Mixed infections of Cryptosporidium were considered rare but they have only been studied so far in humans, mainly AIDS patients (Cama et al., 2006) and children (Xiao et al., 2001), and in calves (Tanriverdi et al., 2003). The role of mixed infections in Cryptosporidium pathology is still unclear. No study has so far described mixed infections in marsupials (Ryan and Power, 2012). The difficulty to amplify at discriminatory loci for genotypes such as C. parvum in marsupials highlights the need to identify Cryptosporidium using a multi-locus approach.
Another difficulty encountered in this study was the potential for pseudo-replication. When working with an endangered species one encounters issues with sample collection and numbers available for stringent analyses. For instance, the Kangaroo Valley Mountain population is estimated to comprise less than 10 individuals. As we identified C. meleagridis in four samples from Kangaroo Valley Mountain collected over two sampling periods, it is possible that the same individual has been sampled multiple times. A possible solution to reduce bias relative to sampling would be to apply microsatellite (MSAT) analysis to identify individuals. This method has been widely applied to many species ranging from large carnivores to small marsupials using faecal DNA, to monitor threatened populations, analyse their genetic diversity and wide-scale demographics of large populations (Spencer et al., 1995; Dool et al., 2013; Wultsch et al., 2014).
The findings in this study suggested that there was no direct effect of captive breeding and translocation on Cryptosporidium in brush-tailed rock-wallabies. In Australia, translocation policies are developed by the representative State bodies, and veterinary screening is not mandated but is increasingly employed to monitor the health of captive bred animals before release (Short, 2009). Health screening and its relation to the success of a recovery program is further complicated by a diverse number of potential pathogens and a lack of baseline data on risks that selected pathogens may pose to wildlife species. If unusual parasite species atypical to the host group are found, such as C. meleagridis in BTRW, consideration as to whether the animal should be used for translocation or isolated from the population would form part of the management response. The pathology of Cryptosporidium in wild marsupials is also currently unknown (reviewed in Ryan and Power, 2012), making such a risk assessment difficult for BTRW. The identification of Cryptosporidium species with varying host specificity found in both captive bred and wild brush-tailed rock-wallabies indicates that further research is required into the diversity and pathology of this parasite in Australian wildlife.
Conflict of interest
The authors declared that there is no conflict of interest.
Acknowledgements
Funding for this research was provided by the Australian Research Council, Office of Environment and Heritage and the Australian Museum through the ARC Linkage program, project ID LP110200569. The animal ethics approval numbers for the collections are AEC 050207/02 and AEC 080728/01. The authors would like to thank Todd Soderquist, Celia Thomson, Melinda Norton and Juliet Dingle for collecting the BTRW faecal samples.
References
- Abeywardena H., Jex A.R., von Samson-Himmelstjerna G., Haydon S.R., Stevens M.A., Gasser R.B. First molecular characterisation of Cryptosporidium and Giardia from Bubalus bubalis (water buffalo) in Victoria, Australia. Infect. Genet. Evol. 2013;20:96–102. doi: 10.1016/j.meegid.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Akiyoshi D.E., Dilo J., Pearson C., Chapman S., Tumwine J., Tzipori S. Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infect. Immun. 2003;71:1828–1832. doi: 10.1128/IAI.71.4.1828-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid M., Heavens D., Elwin K., Chalmers R.M., Hadfield S.J., Hunter P.R. Whole genome amplification (WGA) for archiving and genotyping of clinical isolates of Cryptosporidium species. Parasitology. 2010;137:27–36. doi: 10.1017/S0031182009991132. [DOI] [PubMed] [Google Scholar]
- Browning T.L., Taggart D.A., Rummery C., Close R.L., Eldridge M.D.B. Multifaceted genetic analysis of the “Critically Endangered” brush-tailed rock-wallaby Petrogale penicillata in Victoria, Australia: implications for management. Conserv. Genet. 2001;2:145–156. [Google Scholar]
- Cama V., Gilman R.H., Vivar A., Ticona E., Ortega Y., Bern C. Mixed Cryptosporidium infections and HIV. Emerg. Infect. Dis. 2006;12(6):1025–1028. doi: 10.3201/eid1206.060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham A.A. Disease risks of wildlife translocations. Conserv. Biol. 1996;10:349–353. [Google Scholar]
- DECC . In: Recovery Plan for the Brush-Tailed Rock-Wallaby (Petrogale penicillata) NSW, D.o.E.a.C.C., editor. 2008. [Google Scholar]
- Dool S.E., Puechmaille S.J., Dietz C., Juste J., Ibáñez C., Hulva P. Phylogeography and postglacial recolonization of Europe by Rhinolophus hipposideros: evidence from multiple genetic markers. Mol. Ecol. 2013;22:4055–4070. doi: 10.1111/mec.12373. [DOI] [PubMed] [Google Scholar]
- Dowle M., Hill N.J., Power M.L. Cryptosporidium from a free-ranging marsupial host: bandicoots in urban Australia. Vet. Parasitol. 2013;198:197–200. doi: 10.1016/j.vetpar.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Eldridge M.D.B., Close R.L. Brush-tailed rock-wallaby Petrogale penicillata. In: Strahan R., editor. The Mammals of Australia. Reed Books; Sydney: 2005. pp. 383–385. [Google Scholar]
- Elwin K., Hadfield S.J., Robinson G., Chalmers R.M. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol. Infect. 2012;140:673–683. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- Fall A., Thompson R.C.A., Hobbs R.P., Morgan-Ryan U.M. Morphology is not a reliable tool for delineating species within Cryptosporidium. J. Parasitol. 2003;89(2):399–402. doi: 10.1645/0022-3395(2003)089[0399:MINART]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Fayer R., Santín M., Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet. Parasitol. 2010;172(1–2):23–32. doi: 10.1016/j.vetpar.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Gatei W., Ashford R.W., Beeching N.J., Kang'ethe Kamwati S., Greensill J., Anthony Hart C. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 2002;8:204–206. doi: 10.3201/eid0802.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N.J., Deane E.M., Power M.L. Prevalence and genetic characterization of Cryptosporidium isolates from common brushtail possums (Trichosurus vulpecula) adapted to urban settings. Appl. Environ. Microbiol. 2008;74:5549–5555. doi: 10.1128/AEM.00809-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN . 2013. IUCN Red List of Threatened Species. Version 2013.2. [Google Scholar]
- Kock R.A., Woodford M.H., Rossiter P.B. Disease risks associated with the translocation of wildlife. Rev. Sci. Tech. 2010;29:329–350. doi: 10.20506/rst.29.2.1980. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leoni F., Amar C., Nichols G., Pedraza-Díaz S., McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 2006;55:703–707. doi: 10.1099/jmm.0.46251-0. [DOI] [PubMed] [Google Scholar]
- Menkhorst P., Hynes W.H. In: National Recovery Plan for the Brush-Tailed Rock-Wallaby Petrogale penicillata. Department of Sustainability and Environment, V., editor. 2010. East Melbourne. [Google Scholar]
- Moberg G.P. American Physiological Society; Bethesda, Maryland, USA: 1985. Animal Stress. [Google Scholar]
- Moir M.L., Vesk P.A., Brennan K.E.C., Poulin R., Hughes L., Keith D.A. Considering extinction of dependent species during translocation, ex situ conservation, and assisted migration of threatened hosts. Conserv. Biol. 2012;26:199–207. doi: 10.1111/j.1523-1739.2012.01826.x. [DOI] [PubMed] [Google Scholar]
- Morgan U.M., Constantine C.C., Forbes D.A., Thompson R.C.A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- Ng J., Yang R., Whiffin V., Cox P., Ryan U. Identification of zoonotic Cryptosporidium and Giardia genotypes infecting animals in Sydney's water catchments. Exp. Parasitol. 2011;128:138–144. doi: 10.1016/j.exppara.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Nolan M.J., Jex A.R., Koehler A.V., Haydon S.R., Stevens M.A., Gasser R.B. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 2013;47:1726–1740. doi: 10.1016/j.watres.2012.12.027. [DOI] [PubMed] [Google Scholar]
- O'Donoghue P.J. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- Power M.L. Biology of Cryptosporidium from marsupial hosts. Exp. Parasitol. 2010;124:40–44. doi: 10.1016/j.exppara.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Power M.L., Ryan U.M. A new species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) from eastern grey Kangaroos (Macropus giganteus) J. Parasitol. 2008;94:1114–1117. doi: 10.1645/GE-1508.1. [DOI] [PubMed] [Google Scholar]
- Power M.L., Shanker S.R., Sangster N.C., Veal D.A. Evaluation of a combined immunomagnetic separation/flow cytometry technique for epidemiological investigations of Cryptosporidium in domestic and Australian native animals. Vet. Parasitol. 2003;112:21–31. doi: 10.1016/s0304-4017(02)00414-4. [DOI] [PubMed] [Google Scholar]
- Power M.L., Slade M.B., Sangster N.C., Veal D.A. Genetic characterisation of Cryptosporidium from a wild population of eastern grey kangaroos Macropus giganteus inhabiting a water catchment. Infect. Genet. Evol. 2004;4:59–67. doi: 10.1016/j.meegid.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Power M.L., Cheung-Kwok-Sang C., Slade M., Williamson S. Cryptosporidium fayeri: diversity within the gp60 locus of isolates from different marsupial hosts. Exp. Parasitol. 2009;121:219–223. doi: 10.1016/j.exppara.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Reed C., Sturbaum G.D., Hoover P.J., Sterling C.R. Cryptosporidium parvum mixed genotypes detected by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 2002;68(1):427–429. doi: 10.1128/AEM.68.1.427-429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G., Wright S., Elwin K., Hadfield S.J., Katzer F., Bartley P.M. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int. J. Parasitol. 2010;40:1539–1548. doi: 10.1016/j.ijpara.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Ryan U.M., Power M.L. Cryptosporidium species in Australian wildlife and domestic animals. Parasitology. 2012;139:1673–1688. doi: 10.1017/S0031182012001151. [DOI] [PubMed] [Google Scholar]
- Ryan U.M., Power M.L., Xiao L. Cryptosporidium fayeri n. sp. (Apicomplexa: Cryptosporidiidae) from the red kangaroo (Macropus rufus) J. Eukaryot. Microbiol. 2008;55:22–26. doi: 10.1111/j.1550-7408.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- Ryan U.M., Fayer R., Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141(13):1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Short J. The Australian Animal Welfare Strategy. Australian Government Department of Agriculture, Fisheries and Forestry; Canberra, ACT: 2009. The characteristics and success of vertebrate translocations within Australia. [Google Scholar]
- Spencer P.B., Odorico D.M., Jones S.J., Marsh H.D., Miller D.J. Highly variable microsatellites in isolated colonies of the rock-wallaby (Petrogale assimilis) Mol. Ecol. 1995;4:523–525. doi: 10.1111/j.1365-294x.1995.tb00250.x. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Beser J., Axen C., Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J. Clin. Microbiol. 2014;52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Lal A.A., Xiao L. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 2002;88:388–394. doi: 10.1645/0022-3395(2002)088[0388:MPAERO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Tanriverdi S., Arslan M.O., Akiyoshi D.E., Tzipori S., Widmer G. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol. 2003;130(1):13–22. doi: 10.1016/s0166-6851(03)00138-5. [DOI] [PubMed] [Google Scholar]
- Waldron L.S., Ferrari B.C., Power M.L. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Exp. Parasitol. 2009;122:124–127. doi: 10.1016/j.exppara.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Waldron L.S., Cnheung-Kwok-Sang C., Power M.L. Wildlife-associated Cryptosporidium fayeri in human, Australia. Emerg. Infect. Dis. 2010;16:2006–2007. doi: 10.3201/eid1612.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron L.S., Dimeski B., Beggs P.J., Ferrari B.C., Power M.L. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl. Environ. Microbiol. 2011;77:7757–7765. doi: 10.1128/AEM.00615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren K.S., Swan R.A., Morgan-Ryan U.M., Friend J.A., Elliot A. Cryptosporidium muris infection in bilbies (Macrotis lagotis) Aust. Vet. J. 2003;81:739–741. doi: 10.1111/j.1751-0813.2003.tb14602.x. [DOI] [PubMed] [Google Scholar]
- Wultsch C., Waits L.P., Kelly M.J. Noninvasive individual and species identification of jaguars (Panthera onca), pumas (Puma concolor) and ocelots (Leopardus pardalis) in Belize, Central America using cross-species microsatellites and faecal DNA. Mol. Ecol. Resour. 2014;14:1171–1182. doi: 10.1111/1755-0998.12266. [DOI] [PubMed] [Google Scholar]
- Xiao L. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 2002;185:1846–1848. doi: 10.1086/340841. [DOI] [PubMed] [Google Scholar]
- Xiao L., Bern C., Limor J., Sulaiman I., Roberts J., Lal A.A. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 2001;183:492. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- Xiao L., Fayer R., Ryan U., Upton S.J. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L.H., Escalante L., Yang C.F., Sulaiman I., Escalante A.A., Montali R.J. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Fenwick S., Potter A., Ng J., Ryan U. Identification of novel Cryptosporidium genotypes in kangaroos from Western Australia. Vet. Parasitol. 2011;179:22–27. doi: 10.1016/j.vetpar.2011.02.011. [DOI] [PubMed] [Google Scholar]


