Abstract
Exposure to Staphylococcal enterotoxin B (SEB) causes food poisoning, acute inflammatory lung injury, toxic shock syndrome, and often death. In this study, we investigated whether microRNA (miRNA) play a role in regulating SEB-driven inflammation in the lungs. Exposure to SEB caused immune cell infiltration, robust cytokine and chemokine production, compromised lung function, and 100% mortality in mice. We assessed miRNA and mRNA expression in lung infiltrating mononuclear cells following exposure to SEB and found 89 miRNA that were dysregulated (>2-fold) compared with vehicle controls. In silico analysis revealed that the miRNA exhibited biological functions pertaining to cell death and survival, cellular proliferation, and cell cycle progression. Through the use of q-RT PCR, we validated 9 specific miRNA (miR-155, miR-132, miR-31, miR-222, miR-20b, miR-34a, miR-192, miR-193*, and let-7e) and observed that they were predicted to bind the 3′-UTR of a number of genes that were either involved in the stringent regulation of inflammation (Smad3, Tgfb, Runx1, and Foxo3) or those that contributed to its exacerbation (Stat3, Ptgs2, Ccnd1, Ccne1, NfκB, and Tbx21). Further, by increasing or decreasing the levels of miR-132 (a miRNA highly induced by SEB), we noted the corresponding decrease or increase in the levels of its predicted target FOXO3. As a result of FOXO3 suppression by miR-132, we saw increase in Ifn-γ, Ccnd, and Ccne1. Taken together, our data support the role for miRNA in actively participating and orchestrating SEB-mediated inflammation in the lungs and provide several therapeutic targets for the treatment of SEB-driven toxicity via the modulation of miRNA.
Keywords: SEB, microRNA, inflammation, ALI, mortality
Staphylococcus aureus is a commonly occurring gram-positive pathogen implicated in a number of community and nosocomial infections ranging from skin infections, endocarditis, sepsis, and toxic shock (Lowy, 1998). Its pathogenicity can be attributed to a number of virulence factors such as polysaccharides, proteases, cell surface proteins, and in particular, its ability to secrete potent toxins such as Staphylococcal enterotoxin B (SEB) (Foster, 2004). Commonly referred to as a superantigen, SEB poses a threat as a biological weapon because it is effective at smaller quantities, is easily aerosolized, and disseminated. Consequently, the Center for Disease Control and Prevention has deemed SEB, a Category B select agent (Ulrich et al., 2001).
The consequences of SEB exposure are known to arise from an exaggerated immune response. Upon directly binding the non-polymorphic regions of the major histocompatibility complex class II (MHC II) on antigen presenting cells (APCs) and Vβ8 region of the T-cell receptor, SEB leads to the activation and clonal expansion of approximately 30–40% T cells (Kozono et al., 1995). Subsequently, a cytokine storm ensues leading to cellular infiltration, tissue damage, multiorgan failure, and death (Strandberg et al., 2010; Uchakina et al., 2013).
Although the impact of SEB exposure on key inflammatory signaling pathways such as the NFκB, MAPK has been extensively studied (Krakauer, 2013), we have only recently been made aware of the possible role of microRNA (miRNA) in SEB-mediated inflammation.
miRNA are a unique class of small (18–25 nt), single-stranded non-coding RNA molecules that have emerged as primary regulators of gene expression (Dai and Ahmed, 2011; Davidson-Moncada et al., 2010). Mammalian miRNA bind primarily to the 3′-UTR of their respective target mRNA causing mRNA instability and degradation and/or the disruption of translation (Davidson-Moncada et al., 2010). Although miRNAs are involved in the development and differentiation of cells, apoptosis, and hematopoiesis under normal conditions, inflammatory cues lead to their dysregulation, thereby enabling the targeting of key regulators of inflammation (Sonkoly et al., 2008). For example, in rheumatoid arthritis (RA), the overexpression of miR-146a in CD4+ T cells obtained from the synovial fluid is associated with increased expression of TNF-α (Li et al., 2010). Similarly, the inhibition of miR-126 in a house dust mite-induced model of asthma leads to the decrease in allergic inflammation (Mattes et al., 2009). Conversely, while high levels of miR-125b are required to maintain a naïve state of T cells, the activation of T cells leads to its downregulation and subsequent expression of its target gene, Ifng (Rossi et al., 2011).
Recently, our laboratory has demonstrated that intranasal (i.n.) exposure to SEB in C57BL/6 mice leads to the strong dysregulation of miRNA. miR-155 in particular promotes SEB-mediated acute inflammatory lung injury (Rao et al., 2014). In the current study, we administered SEB as a “Dual Hit” to C3H/HeJ mice which entails administering small quantities (5 and 2 μg) of SEB intranasally and intraperitoneally (i.p.), respectively, 2 h apart that results in the hyperactivation of the immune system and 100% mortality of mice. Upon investigating the effect of SEB exposure on the miRNA profile, we observed the aberrant expression of several miRNA that could potentially contribute to SEB-mediated inflammation. Further, applying bioinformatics tools and q-RT PCR, we established important links between the miRNA and their respective target genes. Using gain and loss of function experiments, we specifically demonstrate the possible role of miR-132 in mediating damage and death. Thus, our results provide further insights into the role of SEB-induced miRNA in contributing toward severe acute inflammatory lung injury and consequent mortality.
MATERIALS AND METHODS
Mice
Female C3H/HeJ mice (6–8 weeks) were obtained from the Jackson Laboratory. All mice were housed under pathogen-free conditions in the Animal Resource Facility, University of South Carolina, School of Medicine. All experiments using vertebrate animals were performed under protocols approved by the Institutional Animal Care and Use Committee at USC.
Administration of SEB
SEB was acquired from Toxin Technologies (Sarasota, Florida). SEB was administered as a “Dual Hit” as described previously (Huzella et al., 2009). The first dose of SEB was delivered by the i.n. route at a concentration of 5 μg/mouse in a 25-μl volume. Two hours after, the second dose of SEB was administered i.p. at a concentration of 2 μg/mouse in a 100-μl volume. Control mice were similarly given PBS (vehicle) as a dual dose. While mice were euthanized 72 h after SEB exposure in all experiments, survival of mice was monitored up to 5 days after exposure to SEB and any moribund mice were immediately euthanized.
Lung histopathology
72 h after exposure to either vehicle or SEB, lungs were excised and fixed in 10% formalin. Lung tissue was paraffin embedded and 5 μm serial sections were made. Subsequently, the sections were deparaffinized by dissolving in xylene, rehydrated in alcohol (100%, 95%, and 90%). The sections were stained with hematoxylin and eosin (H&E) and assessed with Nikon E600 light microscope. Images were taken at ×40 magnification.
Assessment of vascular leak
The percent of vascular leakage in the lungs was determined as described previously (Rieder et al., 2012; Saeed et al., 2012). Briefly, 72 h after vehicle or SEB exposure, mice were administered 1% Evans Blue dye in sterile PBS intravenously. Two hours later, the mice were euthanized and lungs perfused with heparinized PBS. The lungs were incubated in formamide at 37 °C for 24 h to extract the dye. The optical density (OD) of the supernatant was measured by a spectrophotometer at 620 nm. Percent increase in vascular leak was calculated using the following formula—(ODsample−ODcontrol/ODcontrol) × 100.
Preparation of lung infiltrating cells
72 h after exposure to vehicle or SEB, lungs were perfused with heparinized PBS and harvested. They were then homogenized using Stomacher 80 Biomaster blender from Seward (Davie, Florida) in 10 ml sterile PBS. Following washing with sterile PBS, the cells were separated by density gradient centrifugation at 500 × g for 30 min at 24 °C with the brake off. Mononuclear cell layer isolated was enumerated by the Trypan blue exclusion method using a hemocytometer.
Detection of cytokines
Cytokines in the broncheoalveolar lavage fluid (BALF) were obtained as described previously (Rao et al., 2014). Briefly, 72 h after vehicle or SEB exposure, mice were euthanized. The trachea was bound with a suture and the lung was excised as an intact unit along with the bound trachea. Sterile ice-cold PBS was injected through the trachea to collect the BALF. Cytokine detection was carried out using Bio-Plex Pro mouse cytokine 23-plex Assay from Biorad (Hercules, California).
In vitro assays
Splenocytes from naïve C3H/HeJ mice were harvested and cultured in complete RPMI (10% FBS, 10mM L-glutamine, 10 mM HEPES, 50μM β-mercaptoethanol, and 100 μg/ml penicillin). Cells were seeded at a density of 1 × 106 cells in a 96-well plate and stimulated with either PBS (vehicle) or SEB (1 μg/ml) for 24 h. The cells were then harvested to examine CD3, Vβ8, CD69, CD28, and CD25 percentages. For IFN-γ assessment, cell supernatants were isolated and assayed using mouse IFN-γ ELISA MAX kit (Biolegend, San Diego, California). Cellular proliferation was measured by similarly seeding and activating splenocytes for 48 h. In the last 12 h of incubation, 3[H]-Thymidine (2 μ Ci) was added to the cell culture. Cells were then collected using a harvester and thymidine incorporation was measured using a scintillation counter (Perkin Elmer).
Flow cytometry and antibodies
To determine the phenotypic characteristics of the lung infiltrating mononuclear cells and splenocytes obtained from in vitro cell culture assay above, cells were stained with the following fluorescent-conjugated antibodies—fluorescein isothiocyanate (FITC)-conjugated anti-CD8 (clone: 53-6.7), phycoerthyrin (PE)-conjugated anti-CD4 (clone: GK 1.5), FITC-conjugated anti-CD69 (clone: H1.2F3), PE-Cy5-conjugated anti-CD28 (clone 37.51), allophycocyanin (APC)-conjugated anti-CD3 (clone: 145.2 C11), PE-conjugated anti-Vβ8 (clone: KJ16-133.18), and PECy7-conjugated anti-CD25 (clone: PC61) from Biolegend. Stained cells were run and analyzed using Beckman Coulter 500 Flow Cytometer (Indianapolis, Indiana).
Total RNA extraction
Total RNA (including small RNA) was isolated from lung infiltrating mononuclear cells using the miRNAeasy kit from Qiagen (Valencia, California) according to manufacturer’s instructions. The purity and concentration of total RNA were confirmed spectrophotometrically by Nanodrop 2000c from Thermo Scientific (Wilmington, Delaware). The integrity of miRNA was further confirmed using Agilent 2100 BioAnalyzer (Agilent Tech, Palo Alto, California).
miRNA expression profiling
To profile the miRNA expression in the lung after SEB exposure, the Affymetrix GeneChip miRNA 3.0 array platform was used. The array identified 1111 mouse miRNA derived from the Sanger miRBase v17 (www.mirbase.org). Total RNA was labeled with Flash Tag Biotin HSR labeling kit from Affymetrix (Santa Clara, California) according to manufacturer’s instructions. Briefly, RNA spike control Oligos were added to the RNA and incubated with a Poly A Tailing master mix for 15 min. Next, the RNA was labeled with biotin using FlashTag Biotin HSR Ligation mix. For hybridization of the biotin-labeled samples to the array, a GeneChip Eukaryotic Hybridization Control kit comprising of bioB, bioC, bioD, and cre was used to create the array hybridization cocktail. Following incubation at 99 °C for 5 min, then 45 °C for 5 min, a small volume (100 μl) was injected into an array. The arrays were further incubated at 48 °C and 60 rpm for 16–8 h. Post-hybridization, the array was washed and stained with fluorescent-conjugated streptavidin using the Fluidics Station 450. The stained chip was scanned on a GeneChip Scanner (Affymetrix) to generate the data summarization, normalization, and quality control files. All miRNA microarray data were deposited into ArrayExpress database (www.ebi.ac.uk/arrayexpress/) under the ArrayExpress accession number E-MTAB-2641.
miRNA and mRNA target gene analysis
The fluorescent intensities obtained from the hybridization were log transformed, mean centered, and visualized as a heatmap. Hierarchical clustering was carried out using Ward’s method and similarity measurement was calculated using half square Euclidean distance. miRNA fold change differences between vehicle and SEB as obtained from the microarray was plotted as a fold change distribution plot. Further, only miRNA that showed >2-fold higher or lower values in SEB-exposed lung infiltrating cells compared with control were considered for analysis. Proportional Venn diagram was created using Venn Diagram Plotter (http://omics.pnl.gov/software/venn-diagram-plotter). To further assess the biological functions and visualize miRNA interactions with its respective target genes associated with the miRNA, we employed the use of the commercially available tool Ingenuity Systems, Ingenuity Pathway analysis (IPA) (Mountain View, California).
Real-time qPCR
Total RNA (miRNA and mRNA) obtained from lung infiltrating mononuclear cells or in vitro from splenocytes was converted to cDNA using miSript cDNA synthesis kit (Qiagen) according to manufacturer’s instructions. To validate and detect miRNA, qPCR was carried out using miScript primer assays (miR-132, miR-155, miR-31, miR-222, miR-20b, let-7e, miR-192, miR-193*, miR-34a, and control Snord96_a) that employ SYBR Green technology from Qiagen. Fold change was determined using the 2ΔΔCt method and expressed relative to control. For mRNA validation, SSO advanced SYBR green PCR kit from Biorad was used according to manufacturer’s instructions. β-Actin was used as the internal control. Primers (Table 1) were synthesized from integrated DNA technologies.
TABLE 1.
Real-Time qPCR Primer List
| Gene | Primer | Sequence 5′→3′ |
|---|---|---|
| Tbx21 | Tbx21-F | AAC CGC TTA TAT GTC CAC CCA |
| Tbx21-R | CTT GTT GTT GGT GAG CTT TAG C | |
| Ifn-γ | IFN-γ-F | GCG TCA TTG AAT CAC ACC TG |
| IFN-γ-R | GAG CTC ATT GAA TGC TTG GC | |
| Tgfb1 | Tgfb1-F | CCA CCT GCA AGA CCA TCG AC |
| Tgfb1-R | CTG GCG AGC CTT AGT TTG GAC | |
| Foxo3 | Foxo3-F | GCA AGC CGT GTA CTG TGG A |
| Foxo3-R | CGG GAG CGC GAT GTT ATC C | |
| Stat3 | Stat3-F | AAT ATA GCC GAT TCC TGC AGA G |
| Stat3-R | TGG CTT CTC AAG ATA CCT GCT C | |
| Nfκb | Nfκb-F | ATG GCA GAC GAT GAT CCC TAC |
| Nfκb-R | TGT TGA CAG TGG TAT TTC TGG TG | |
| Ccnd1 | Ccnd1-F | GCG TAC CCT GAC ACC AAT CTC |
| Ccnd1-R | ACT TGA AGT AAG ATA CGG AGG GC | |
| CcnE1 | CcnE1-F | GTG GCT CCG ACC TTT CAG TC |
| CcnE1-R | CAC AGT CTT GTC AAT CTT GGC A | |
| Cox2 | Cox2-F | TGA GCA ACT ATT CCA AAC CAG C |
| Cox2-R | GCA CGT AGT CTT CGA TCA CTA TC | |
| Smad3 | Smad3-F | AGG GGC TCC CTC ACG TTA TC |
| Smad3-R | CAT GGC CCG TAA TTC ATG GTG | |
| Runx1 | Runx1-F | GCA GGC AAC GAT GAA AAC TAC T |
| Runx1-R | GCA ACT TGT GGC GGA TTT GTA | |
| β-actin | β-actin-F | GGC TGT ATT CCC CTC CAT CG |
| β-actin–R | CCA GTT GGT AAC AAT GCC ATG T |
Transfection with miR-132 mimic and inhibitor
Splenocytes from naïve C3H/HeJ mice were harvested and cultured in 10 ml of complete RPMI. Cells were seeded at 2 × 105 cells per well in 24-well plates and transfected with synthetic miR-132 mimic (40nM) or mock transfected with Transfection reagent HiperFect from Qiagen for 24 h. For miR-132 inhibition, cells were activated with SEB (1 μg/ml) and transfected with either mock control or miR-132 inhibitor (100nM) also purchased from Qiagen. Twenty-four hours after transfection, total RNA was collected for q-RT PCR validation of miR-132, target gene Foxo3, and genes associated with cell cycle progression (Ccnd1, Ccne1) and inflammation (Ifn-γ).
Western blot
Splenocytes were mock transfected, transfected with miR-132 mimic, activated with SEB, and transfected with miR-132 synthetic inhibitor for 24 h. Protein extracts (>20 μg) were separated on a 10% SDS-polyacrylamide gel by electrophoresis (60 V for >2 h). Separated protein was transferred onto a nitrocellulose membrane. The membrane was probed with antibodies against FOXO3a (No. 2497S) from Cell signaling Technology and GAPDH (No. SC 25778) from Santa Cruz Biotechnology Inc.
Statistics
All statistical analyses were carried out using GraphPad Prism Software (San Diego). In all experiments, the number of mice used was 4–5 per group, unless otherwise specified. Results are expressed as means ± SEM. Student’s t test was used to compare 2 groups. A P value of less than 0.5 was considered statistically significant. Individual experiments were performed in triplicate and each experiment was performed independently at least 3 times to test reproducibility of results. Survival analysis was carried out using a log-rank test.
RESULTS
SEB Exposure Leads to Lung Inflammation and Acute Mortality
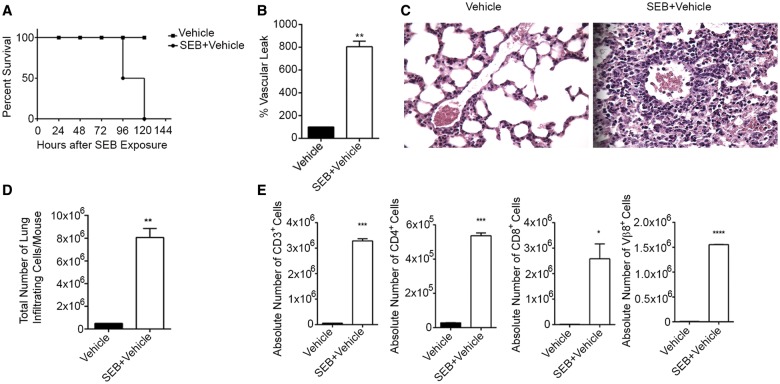
The “Dual Hit” model of SEB administration has been previously known to result in severe inflammation in the lungs and death of the mice (Huzella et al., 2009). In this study, we found that SEB administration resulted in 100% mortality in mice between 96 and 120 h (Fig. 1A) when compared with mice exposed to vehicle. Pulmonary damage was characterized by the disruption of lung integrity, vascular leak, and consequently, edema. To quantitate the extent of damage to the lungs after SEB exposure, mice were administered Evans Blue dye and vascular leak expressed as a percent increase over vehicle as described earlier (Rieder et al., 2012; Saeed et al., 2012). Our results revealed that SEB exposure had almost an 8-fold increase in vascular permeability compared with mice that were only exposed to vehicle (Fig. 1B). Further, a consequence of SEB exposure is the infiltration of immune cells around capillaries and bronchioles (Rao et al., 2014). Accordingly, histopathological examination of the lungs confirmed the presence of infiltrating immune cells after SEB exposure compared with vehicle alone (Fig. 1C). Further, upon enumeration of the total number of infiltrating mononuclear cells in the lung, we found a profound increase in the number of cells post-SEB exposure compared with vehicle (Fig. 1D). Because SEB exposure leads primarily to the clonal expansion of Vβ8+ T cells, we further assessed the immune subsets (CD4, CD8, and Vβ8) by staining with fluorescein-conjugated antibodies and analyzed using flow cytometry. As expected, we found significantly increased absolute cell counts in all 3 subsets after SEB exposure (Fig. 1E).
FIG. 1.
SEB exposure results in pulmonary inflammation and mortality of mice. C3H/HeJ mice were exposed to a “Dual Hit” of SEB and euthanized 72 h post-exposure. A, Survival curve of mice exposed to either vehicle or SEB. B, Measurement of vascular leak 24 h after exposure to the second dose of SEB. Mice were administered Evans Blue dye and following perfusion, lungs were placed in formamide. Absorbance was recorded at 620 nm and the percentage of vascular leak was calculated and graphically represented. C, Representative H&E (×40) staining of sections of the lung demonstrating immune cell infiltration. D, Total number of mononuclear cells infiltrating the lungs in vehicle or SEB exposed mice as determined by trypan blue exclusion method. E, Phenotypic characterization of mononuclear cells infiltrating the lung determined by staining cells with fluorescein-conjugated antibodies against CD4, CD8, and Vβ8 and conducting flow cytometric analysis. Absolute cell counts are represented graphically. Data are represented as mean ± SEM (n = 5) from 3 independent experiments. Statistical significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (when compared with vehicle).
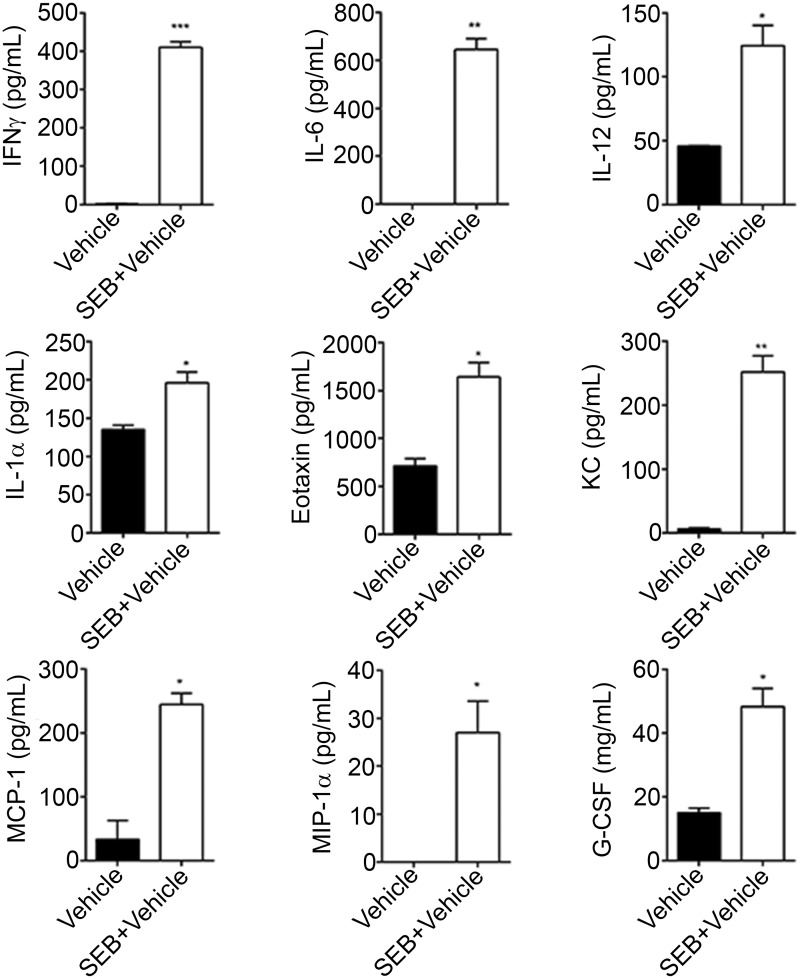
SEB, being a superantigen is known to result in the massive release of cytokines. Therefore, we obtained the BALF from mice that were exposed to vehicle or SEB and screened for several cytokines and chemokines using a Bioplex assay. Among the 23 cytokines and chemokines assayed as part of the Bioplex, IFN-γ, IL-6, IL-12, IL-1α, monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein-1 (MIP-1), granulocyte colony stimulating factor (G-CSF), eotaxin, and keratinocyte-derived chemokine (KC) (Fig. 2) were significantly increased in response to SEB when compared with vehicle.
FIG. 2.
Exaggerated expression of chemokines and cytokines after SEB exposure. Mice were exposed to either vehicle or SEB for 72 h and euthanized. The trachea was bound with a suture. Following the excision of the lung along with the trachea, 1 ml of ice-cold PBS was flushed through the trachea and collected as the broncheoalveolar lavage fluid (BALF). Cytokine and chemokine expression was analyzed using a Bioplex and the concentration quantified in pg/ml. Data are represented as mean ± SEM (n = 5) from 2 independent experiments. Statistical significance is indicated as follows *P < 0.05, **P < 0.01, ***P < 0.001 (when compared with vehicle).
SEB Exposure Leads to the Activation and Proliferation of Immune Cells in vitro
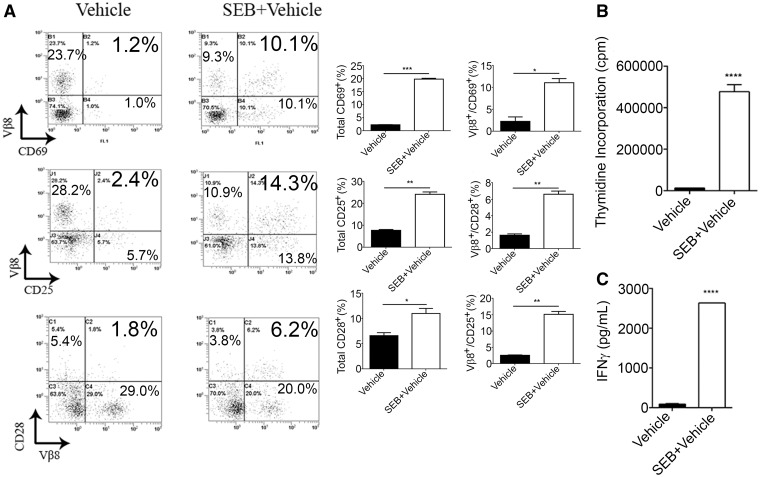
To determine whether SEB exposure can trigger activation of T cells, we exposed splenocytes to SEB in vitro for 24 h and assessed activation by flow cytometry. Compared with vehicle, SEB leads to increased frequencies of T-cell activation markers: CD3+Vβ8+CD69+, CD3+Vβ8+CD25+, and CD3+Vβ8+CD28+ (Fig. 3A). While we had earlier demonstrated that SEB administration leads to increased cell counts in the lung, we investigated whether activation of splenocytes with SEB could also lead to increased cellular proliferation. Indeed, activation with SEB for 48 h resulted in significantly higher thymidine counts when compared with vehicle (Fig. 3B). Cellular proliferation and activation, especially of T cells, is influenced by IFN-γ production. Consequently, we found exaggerated (>2500 pg) levels of IFN-γ (Fig. 3C), a hallmark cytokine associated with SEB exposure in the supernatants of cells activated with SEB. These data indicate that SEB triggers the activation and proliferation of cells, in vitro in addition to in vivo.
FIG. 3.
Effect of SEB activation in vitro. Splenocytes seeded at a density of 1 × 106 cells were activated with 1 μg/ml of SEB. A, Twenty-four hours after activation, cells were harvested and triple stained in the following combinations: CD3+Vβ8+CD69+, CD3+Vβ8+CD25+, and CD3+Vβ8+CD28+. The representative dotplots shown are gated on CD3+ cells. B, Cell proliferation assay as measured by the incorporation of thymidine in splenocytes that were activated with SEB for 48 h. C, IFN-γ expression determined by ELISA. Samples were obtained from collecting the supernatants following SEB activation of splenocytes. Data are represented as mean ± SEM (n = 3) from 2 independent experiments. Statistical significance is indicated as follows: **P < 0.01, ***P < 0.001, ****P < 0.0001 (when compared with vehicle).
SEB Exposure Results in the Dysregulation of Several miRNA
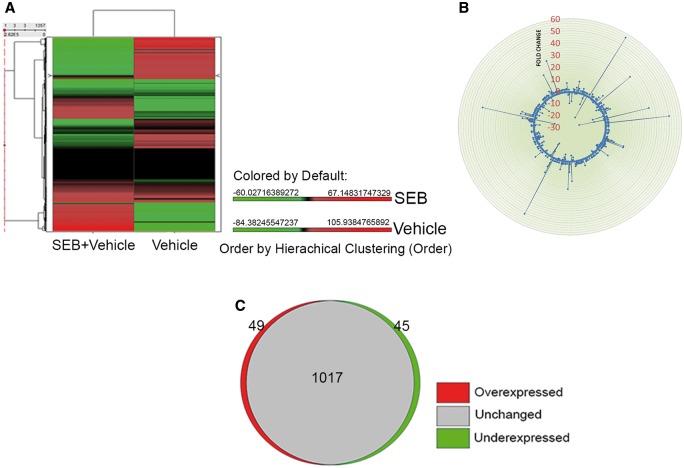
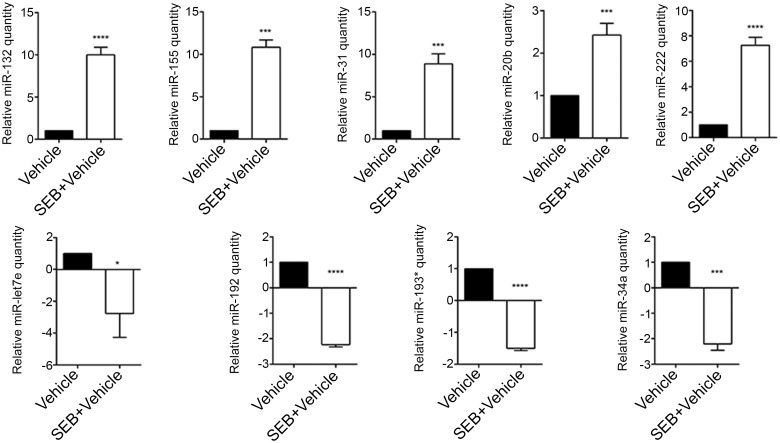
While miRNA are known to be induced during inflammation and play a critical role in its regulation, expression profile of miRNA during SEB-mediated lung inflammation and consequent death has not yet been investigated. Therefore, we extracted total RNA from the lung infiltrating mononuclear cells from mice that were either exposed to vehicle or SEB and performed miRNA microarray. The heatmap revealed a noticeable difference in fold change between vehicle and SEB-treated groups (Fig. 4A). To visualize the distribution of the miRNA, a radial fold change distribution plot was generated demonstrating that while most miRNA remained unchanged and were found around the circumference of the plot, a few miRNA were highly upregulated (up to 56-fold) or highly downregulated (up to 22-fold) when compared with vehicle (Fig. 4B). Among the miRNA assessed on the array, approximately 8.4% of the miRNA were either overexpressed or underexpressed greater than and equal to 2-fold in SEB exposed cells compared with vehicle (Fig. 4C). Next, IPA was employed to analyze on this fraction of dysregulated miR, which yielded a list of top upregulated (miR-132, miR-155, miR-31, miR-20b, and miR-222) and downregulated miRs (miR-192, miR-193*, let-7e, and miR-34a), whose fold change values as determined by the microarray, seed sequence, and miRbase accession numbers were tabulated (Table 2). Next, the miRNA expression levels in the lung infiltrating mononuclear cells were validated by q-RT PCR. These results corroborated the expression pattern observed with the microarray (Fig. 5).
FIG. 4.
SEB exposure leads to dysregulation of microRNA. Seventy-two hours after SEB exposure, lungs were perfused with heparinized PBS and were homogenized. After washing with sterile PBS, the mononuclear cell layer was isolated by density gradient centrifugation and total RNA was isolated. A, A heatmap depicting the differential expression of miRNA in the lungs of SEB exposed mice compared with vehicle. The accompanying color scape depicts mean expression values that are overexpressed (red) or underexpressed (green) above or below the mean, respectively. B, Fold change distribution plot of 1111 mouse-specific miRNA indicating several unchanged, upregulated, or downregulated miRNA. C, A proportional Venn diagram showing the number of miRNA that are overexpressed, underexpressed (<2-fold), or unchanged after SEB exposure.
TABLE 2.
List of Select Differentially Expressed (>2-fold) miRNA in Lung Infiltrating Mononuclear Cells after SEB Exposure Compared with Vehicle Control
| miRNA | Fold Change | Seed Sequence | Accession Number |
|---|---|---|---|
| Overexpressed | |||
| mmu-miR-132 | 56.7 | AACAGUC | MIMAT0000144 |
| mmu-miR-155 | 38.8 | UAAUGCU | MIMAT0000165 |
| mmu-miR-31 | 15.2 | GGCAAGA | MIMAT0000538 |
| mmu-miR-20b | 5.6 | AAAGUGC | MIMAT0003187 |
| mmu-miR-222 | 3.4 | GCUACAU | MIMAT0000670 |
| Underexpressed | |||
| mmu-miR-193* | −22.3 | GGGUCUU | MIMAT0004544 |
| mmu-miR-192 | −10.6 | UGACCUA | MIMAT0000517 |
| mmu-let-7e | −8.5 | GAGGUAG | MIMAT0000524 |
| mmu-miR-34a | −6.2 | GGCAGUG | MIMAT0000542 |
FIG. 5.
Experimental validation of Top overexpressed and underexpressed miRNA. Seventy-two hours after SEB exposure, lungs were perfused with heparinized PBS and were homogenized. After washing with sterile PBS, the mononuclear cell layer was isolated by density gradient centrifugation and total RNA was isolated. A, q-RT PCR validation of the IPA generated Top overexpressed miRNA. B, q-RT PCR validation of the IPA generated Top underexpressed miRNA. Snord96a was used as the small endogenous control and expression of miRNA was normalized to vehicle. Data are represented as mean ± SEM from replicate samples. Lung infiltrating mononuclear cells were pooled from 5 mice in each group. Statistical significance is indicated as follows: *P < 0.05, ***P < 0.001, ****P < 0.0001 (when compared with vehicle).
Functional Analysis of the Dysregulated miRNA
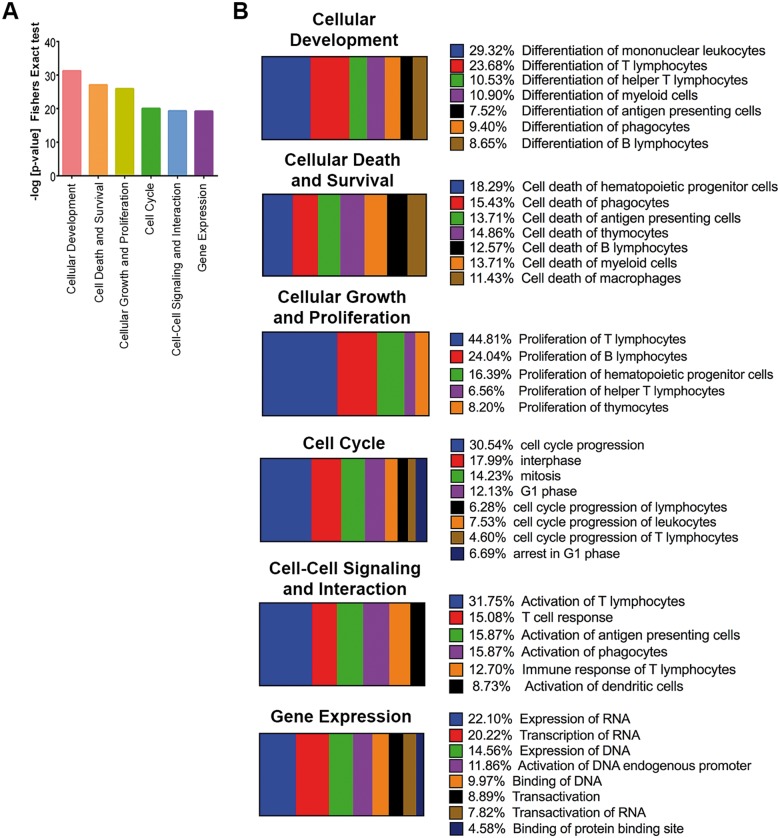
To examine the possible biological functions associated with the over and underexpressed validated miRNA, a combined set of the highly predicted and experimentally observed target genes of these SEB-induced miRNA were subjected to IPA core analysis. Statistically significant (Fishers’ exact test) enrichment of genes into functional annotation categories were generated and plotted as a bar graph. We observed that cellular development, cell death and survival, cellular growth and proliferation, cell cycle, cell-cell signaling and interaction, and gene expression were highly (P < 0.001) enriched functions (Fig. 6A), suggesting that these functions are most likely to be regulated by SEB-induced miRNA. A further dissection of these highly predicted or experimentally observed genes revealed that a majority were involved in very specific functions. For example, almost half of the genes linked with cellular development category were specifically involved in the differentiation of mononuclear leukocytes and T lymphocytes. Further, a majority (>45%) of the molecules associated with cellular growth and proliferation were linked specifically to propagation of T lymphocytes. Similarly, cell-cell signaling and interactions predominantly comprised the specific activation of T lymphocytes and T-cell responses (Fig. 6B), characteristic features of SEB-induced inflammation. Thus, our results suggested that the miRNA expression profile and its associated biological functions, as indicated in silico, correlated with experimentally observed lung inflammation accompanying SEB exposure.
FIG. 6.
In silico analysis of SEB-deregulated miRNA. Ingenuity pathway analysis (IPA) was used to analyze a combined set of the highly predicted and experimentally observed miRNA target genes for miRNA that were over or underexpressed (<2-fold). A, Bar graph highlighting the enrichment of target genes into functional annotation categories with the enrichment P-values plotted on the Y-axis. B, Horizontal slice plot depicting the percentage of the miRNA-associated molecules that can be attributed to very specific biological functions.
Predicted mRNA Targets of the Dysregulated miR after SEB Exposure
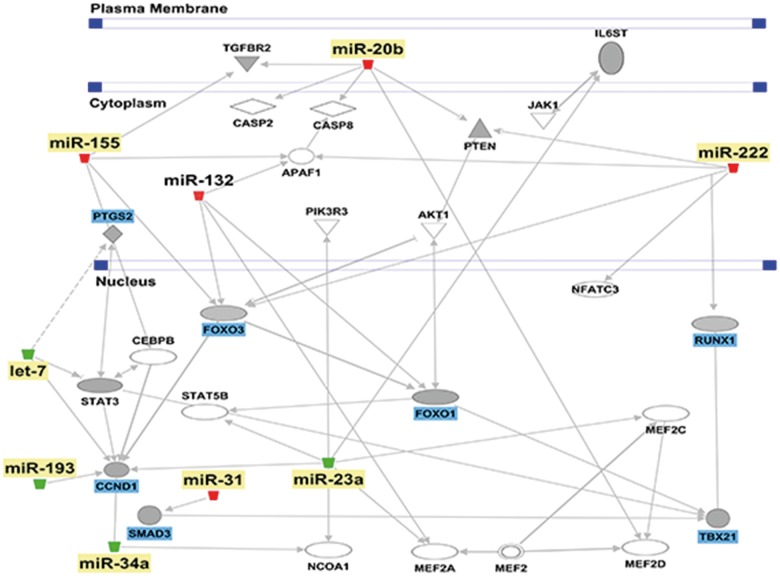
Next, we identified possible miR targets by employing IPA. Highly predicted, experimentally observed as well as moderately predicted targets of the validated miRNA were filtered using the miRNA target filter tool. Only those molecules that were associated with the inflammatory process were considered for analysis. Among these target genes, we found that the upregulated miRNA were predicted or experimentally observed to target transcription factors and genes involved in the regulation of inflammation. For example, miR-132 and miR-155 targeted B-cell CLL/lymphoma 10 (BCL10), an inducer of apoptosis. Similarly, miR-20b and miR-222 target the suppressor of cytokine signaling (Socs) genes, which are negative regulators of several proinflammatory cytokines (Table 3). In contrast, the downregulated miRNA such as let-7e, miR-34a, miR-192, and miR-193* were predicted to target proinflammatory molecules such as those involved in cellular proliferation, activation, and cytokine production (Table 3). IPA pathway designer tool was also used to visualize the miRNA-mRNA interactions (Fig. 7) which provided further insight into how the over and underexpressed miRNA target key molecules and collectively act to regulate inflammation.
TABLE 3.
List of Predicted or Experimentally Observed miRNA Target Genes Associated with Inflammation
| miRNA | Target Genes |
|---|---|
| miR-132 ↑ | Foxo1, Foxo3, Tgfb2, Bcl10, Smad2, Smad5 |
| miR-155 ↑ | Bcl10, Foxo3, Socs1, Smad2 |
| miR-20b ↑ | Runx1, Mcl1, Pten, Socs6, Socs7, Tgfbr2 |
| miR-222 ↑ | Foxo3, Gata4, Pten, Socs1, Socs3 |
| miR-31 ↑ | Foxp3, Smad3, Rorc |
| let7e ↓ | Ccnd1, Ccne1, Cd80, Cd86, Cdkn1a, Il6, Il6r, Stat3 |
| miR-192 ↓ | Nfat5, Cxcr5, Traf5, Traf6, Zeb1, Ccne1 |
| miR-193* ↓ | Ccnd1, Ets1, Esr1, Il12rg, Cd247, Ptgs2 |
| miR-34a ↓ | Ccnd1, Bcl2, Il6r, Il2rb, Map2k1, Pik3r2 |
Up arrow indicates Upregulated miRNA and Down Arrow indicates Downregulated miRNA.
FIG. 7.
In silico analysis of the predicted mRNA targets. IPA generated network highlighting the interaction between validated miRNA (yellow) and their respective target genes (blue).
Experimental Validation of mRNA Target Genes
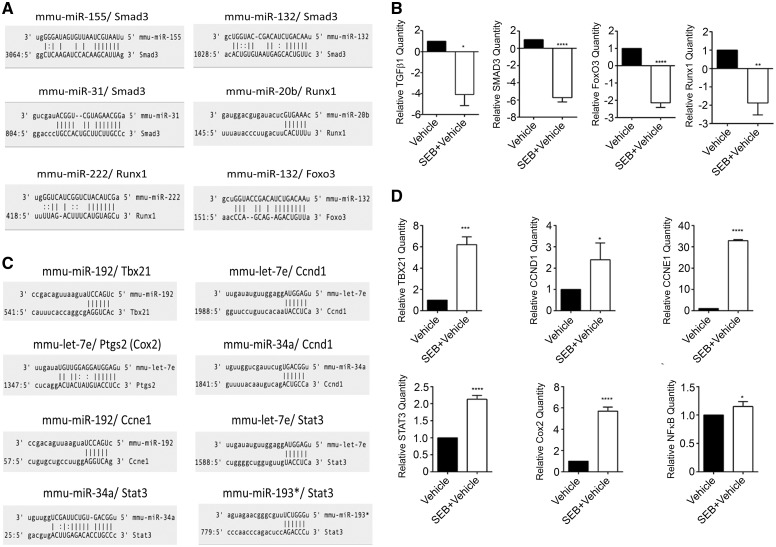
To validate and confirm some of the predicted target genes, q-RT PCR was carried out using mRNA from lung infiltrating mononuclear cells following exposure to vehicle or SEB. Interestingly, upon using microRNA.org (www.microRNA.org) miRNA-mRNA alignment tool, we observed that a few of the upregulated miRNA aligned to the 3′-UTR of some of these target genes. For example, miR-155, miR-132, miR-31 bind Smad3, while miR-20b and miR-222 aligned to the 3′-UTR of Runx1. miR-132, which was highly induced following SEB exposure, aligned with the 3′-UTR of Foxo3, with the potential to target it (Fig. 8A). We observed significant downregulation of Smad family member 3 (Smad3), transforming growth factor, beta 1 (Tgfβ1), forkhead box O3 (Foxo3), and runt-related transcription factor 1 (Runx1) (Fig. 8B), molecules that have been experimentally shown to regulate cellular proliferation, cell cycle progression, and cytokine production when induced. Further, the microRNA.org tool suggested that a few underexpressed validated miRNA, ie, miR-192, miR-34a, and let-7e aligned to the 3′-UTR of these genes (Fig. 8C) while several genes associated with the proinflammatory pathway such as T-box 21 (Tbx21), signal transducer, and activation of transcription 3 (Stat3), prostaglandin-endoperoxidase synthase 2 (Ptgs2), nuclear factor kappa (Nfκb), and cell cycle progression genes such as Cyclin D1(Ccnd1) and Cyclin E1 (Ccne1) were induced upon SEB exposure (Fig. 8D). Altogether, our results indicated that SEB causes alterations in the expression profiles of a significant number of miRNA, which in turn may either allow for the expression of proinflammatory genes or lead to the degradation of anti-inflammatory molecules, thereby collectively contributing to severe lung inflammation.
FIG. 8.
Experimental validation of miRNA target genes. A, Schematic illustration of SEB-induced overexpressed miRNA targeting the 3′-UTR of potential target genes. B, q-RT PCR validation of SEB-induced miRNA mRNA targets. C, Schematic illustration of SEB-induced underexpressed miRNA that target the 3′-UTR of potential target genes. D, q-RT PCR validation of SEB-induced miRNA mRNA targets. Data are represented as mean ± SEM from replicate samples. Lung infiltrating mononuclear cells were pooled from 5 mice in each group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (when compared with vehicle).
miR-132 Targets Foxo3, an Inhibitor of Cellular Proliferation and Cell Cycle Progression
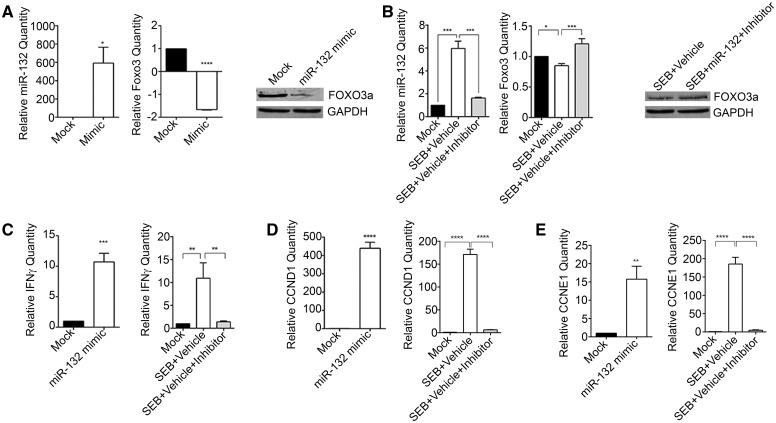
miRNA microarray analysis demonstrated that miR-132 was the most highly induced after SEB exposure (Table 2). Therefore, we rationalized that miR-132 may play a prominent role in mediating inflammation and damage in the current model. Additionally, among the many miR-132 predicted target genes, transcriptional factor Foxo3 was especially interesting not only because of its observed role in inhibiting cellular proliferation and curbing cell cycle progression, but also due to the fact that miR-132 aligned to the 3′-UTR of Foxo3. To confirm the binding of miR-132 to Foxo3, we transfected splenocytes with a miR-132 synthetic mimic. While the levels of miR-132 were highly increased with the mimic, mRNA and protein levels of FOXO3 inversely correlated with its expression (Fig. 9A). Similarly, SEB activation of splenocytes lead to the expected increase in miR-132, but the inhibition of miR-132 resulted in the derepression of FOXO3 (Fig. 9B). These results suggested that miR-132 strongly targets FOXO3. Since FOXO3 plays a role in the inhibition of cell cycle progression and inflammatory cytokine progression, we analyzed levels of Ifn-γ, Ccnd1, and Ccne1 after transfection with miR-132 mimic and inhibitor. We observed that the overexpression of miR-132 corresponded positively with the expression of Ifn-γ, Ccnd1, and Ccne1 (Fig. 9C), while its inhibition decreased their levels significantly (Fig. 9D) suggesting that targeting of FOXO3 by miR-132 prevents regulation of inflammatory cytokine production and cell cycle progression.
FIG. 9.
miR-132 targets Foxo3 and leads to increased IFN-γ, CCND1, and CCNE1 levels. A, Splenocytes obtained from mice were transfected either with miR-132 mimic (Mimic) or Mock transfection control (Mock) for 24 h. miR-132 and Foxo3 levels were determined by RT PCR and Western blot. B, Splenocytes were activated with (1 μg/ml) and for 24 h. Cells were then transfected with 100nM miR-132 inhibitor (Inhibitor) or Mock transfection control (Mock) for another 24 h. Foxo3 levels were determined via RT PCR and Western blot. For miRNA normalization, Snord96_a was used as internal control. For mRNA, β-actin was used as the internal control. C, Levels of Ifn-γ after transfection with miR-132 mimic and miR-132 inhibitor were assessed by RT PCR. D, Expression of cell cycle progression genes Ccnd1 and Ccne1 by RT PCR after transfection with miR-132 mimic and inhibitor. Data are represented as mean ± SEM from replicate samples. Statistical significance is indicated as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
DISCUSSION
The recent discovery of miRNA has propelled our understanding of gene regulation. These non-coding, evolutionarily conserved RNA molecules function primarily as repressors of gene expression and are estimated to regulate approximately 30% of all human genes (Lewis et al., 2005). While the role of miRNA in mammalian cells was first brought to the forefront in cancer (Calin et al., 2002), their importance in a number of immune cell activation and functions has since been stressed (Baltimore et al., 2008). With regards to inflammation, several changes in the transcriptional repertoire are accompanied with perturbation in miRNA (O’Connell et al., 2012). As a result, it is conceivable that miRNA control several aspects of the inflammatory process.
In the current study, we have profiled the miRNA expression patterns following exposure to SEB. Using in silico analysis and q-RT PCR approaches, we identified several miRNA that were altered in response to SEB. Importantly, these miRNA were found to be closely associated with key genes either involved in the control of inflammation or those that contributed directly to SEB-mediated inflammatory processes. By showcasing the effects of specific over and underexpressed miRNA, we provide evidence that the outcome of SEB exposure such as severe lung inflammation and death of mice can be explained, at least in part, to the intricate miRNA alterations and miRNA-mRNA interactions.
Following the unconventional binding of SEB to the MHC class II on APCs and subsequent activation of Vβ8 region of the T-cell receptor, the immune system gets hyperactivated due to the presence of large proportions (>30%) of Vβ+ T cells. As a consequence, such T cells get clonally expanded resulting in a cytokine storm (Rajagopalan et al., 2006). Consistent with previous reports, we also report an SEB-mediated increase in T cells, specifically Vβ8+ subsets both in lung infiltrating cells in vivo and in vitro. Furthermore, the assessment of cytokines in the BALF revealed an expected increase in IFN-γ, IL-6, IL-12, and IL-1α, cytokines typically produced in response to SEB (Faulkner et al., 2005; Miethke et al., 1993) and chemokines—MCP-1, MIP-1, G-CSF, KC, Eotaxin, and MCP-1, significant for their role in the induction of inflammatory cell migration to the lungs (Liu et al., 2009).
Although several murine models have been developed to study the toxic effects of SEB (Krakauer et al. 2010), very few models exhibit mortality in the absence of the use of additional potentiating agents such as D-gal or LPS to mimic the consequence of exposure to the toxin in humans (Miethke et al., 1992; Savransky et al., 2003). The dual administration of SEB while addressing this concern has previously been reported to result in severe lung inflammation and the subsequent death of mice (Huzella et al., 2009). In line with this previous observation, we found dual exposure to SEB resulted in extensive inflammation of the lung characterized by immune cell infiltration, accumulation of cytokine and chemokines in the BALF, compromised lung function, and eventually the mortality of mice.
Previously, we studied the effect of a single high dose (50 μg) administration of SEB intranasally in C57BL/6 mice and studied lung inflammation (Rao et al., 2014). In this model, although mice exhibit symptoms of acute inflammatory lung injury, there is no mortality seen in mice exposed to SEB. Nonetheless, we demonstrated the predominant role of miR-155 in mediating acute inflammatory lung injury in this model (Rao et al., 2014). The current “Dual-Hit” model, because of smaller quantities of SEB and high mortality, is more relevant to SEB exposure in humans. It is for this reason that we aimed to highlight the miRNA profile after acute SEB exposure in this model. While we also observe the induction of miR-155 in the current model, we see a similar upregulation of other miRNA such as miR-31, miR-20b, miR-222, and miR-132 and the significant downregulation of miRNA such as let-7e, miR-34a, miR-192, and miR-193*. Interestingly, the expression of these miRNA has been associated with a variety of inflammatory conditions suggesting that they may collectively act to orchestrate the severe inflammation observed after SEB exposure. For example, miR-31 is overexpressed in psoriasis skin and mediates its proinflammatory role by targeting STK40, a negative regulator of NFκB (Xu et al., 2013). Similarly, miR-155 expression correlates positively with that of NFκB activation in Heliobacter pylori infection and in a murine model of chronic alcohol consumption and inflammation (Bala et al., 2011; Xiao et al., 2009). Given that SEB exposure is known to activate the NFκB signaling pathway (Kissner et al., 2011) and cause the expression of proinflammatory cytokines, as indicated by our results, it is possible that specific SEB-induced miRNA may function via the activation of NFkB.
Related to the expression of NFkB is the SEB-induced activation of the STAT family of proteins, especially STAT3 (Plaza et al., 2004). Activated by cytokines such as IL-6, the STAT3 molecule, along with NFκB leads to cellular proliferation, cell survival, cytokine production, and immune cell activation (Yu et al., 2009), key inflammatory pathways we observed experimentally in this study. In addition to finding an increase in Stat3 mRNA levels after SEB exposure, we demonstrate that miR-let7e, miR-34a, and miR-193* that are predicted to target STAT3 are significantly downregulated, thereby facilitating its expression.
Previous reports indicated that the overexpression of miR-34a and miR-193* leads to cell cycle arrest and repression of cellular proliferation by the targeting of CCND1 (Chen et al. 2010; Sun et al., 2008). Similarly, while miR-192 is predicted to target CCNE1, let-7e has been experimentally validated to directly degrade it in hepatocellular carcinoma cells (Zhang et al., 2014). Because these miRNA were specifically downregulated upon SEB activation, we observed the unrestrained expression of Ccnd1 and Ccne1 suggesting that the significant downregulation of these miRNA may contribute to cell cycle progression and cellular proliferation.
Consistent with previous studies (Busbee et al., 2014; Green et al., 1992), we have demonstrated that SEB exposure predominantly affects T cells, leading to their activation (as indicated by assessment of activation markers) or their recruitment into the lung in large numbers. Interestingly, our in silico analysis of dysregulated miRNA and its target genes primarily highlighted a number of T-cell-specific functions, such as those that promote T-cell activation and proliferation. Of interest, Runx1, a transcriptional factor that drives various aspects of T-cell activation and differentiation was highlighted by our analysis. Functionally, Runx1 levels remain high in naïve CD4+ T cells but the activation of T cells turns off its expression (Wong et al., 2012a,b). Moreover, Runx1 deficiency leads to T-cell proliferation and the induction of IL-2 (Wong et al., 2012a,b). We observed that the SEB-induced downregulation of Runx1 corresponded inversely with the induction of miR-20b and miR-222, miRNA that are specifically predicted to bind the 3′-UTR of Runx1, suggesting that these miRNA may contribute to SEB-mediated T-cell proliferation and activation.
Comparable to the role of Runx1 in activated CD4+ T cells, the impairment of the TGFβ/SMAD3 pathway also leads to the activation and proliferation of T cells (Delisle et al., 2013). Furthermore, TGFβ-mediated cellular apoptosis and cell cycle arrest are known to occur through the activation of SMAD3 (Ten Dijke et al., 2002). Our data demonstrated a strong downregulation of Smad3 and Tgfβ1 mRNA levels. Interestingly, 3 of the most highly overexpressed miRNA, ie, miR-132, miR-155, and miR-31 were predicted to align to the 3′-UTR of Smad3, strongly suggesting a role of these miRNA in this pathway.
It is striking that the microarray results revealed a very high expression of miR-132, which was validated by q-RT PCR. Although expressed in high levels in the brain with the capacity to influence neuronal differentiation and synaptic plasticity (Lungu et al., 2013; Scott et al., 2012), a few studies have explored its role in inflammation. For example, it was shown that human monocytes exposed to LPS lead to the increase in miR-132 (Taganov et al., 2006). Furthermore, miR-132 was overexpressed in peripheral blood mononuclear cells in patients with RA (Pauley et al., 2008). In a mouse model of multiple sclerosis, mice deficient in miR-132 were more resistant to the development of experimental autoimmune encephalomyelitis and had lower frequencies of Th1 and Th17 cells, suggesting its role in promoting inflammation (Nakahama et al., 2013). In direct contrast to this study however, miR-132 was demonstrated to have an anti-inflammatory role, whereby the targeting of acetylcholinesterase by miR-132 promoted brain-body-resolution of inflammation (Hanieh and Alzahrani, 2013). Our studies both in silico and experimentally, however, revealed that miR-132 could potentially contribute to SEB-induced inflammatory process. In fact, miR-132 was predicted to moderately target Bcl10 and Tgfb2, but was highly predicted to target Smad2, Smad5, and the Foxo set of transcriptional factors, especially Foxo3. It is known that FOXO3 overexpression leads to apoptosis and that one of its primary functions is to regulate unprecedented cellular proliferation (Hedrick et al., 2012). Moreover, the absence of FOXO3 leads to hyperactivation of T cells, autoimmunity, and lymphoadenopathy (Lin et al., 2004). Prompted by the importance of FOXO3 in controlling inflammation, we pursued gain and loss of function experiments with synthetic miRNA mimic and inhibitors and found an inverse relationship between miR-132 and Foxo3. Indeed, upon assessing the expression of IFN-γ, the hallmark cytokine of SEB-mediated inflammation and cell cycle progression genes, our data revealed that the inhibition of miR-132 leads to the decrease in proinflammatory genes. Taken together, the data suggested that the targeting of Foxo3 by the highly expressed miR-132 could offer an explanation for uninhibited cellular proliferation, the expression of cell cycle progression genes, and the production of a number of inflammatory cytokines in response to SEB.
In summary, we have effectively profiled several miRNA following exposure to the superantigen, SEB. Our data revealed that SEB significantly altered the expression of miRNA predicted to have proinflammatory roles. With the consequence of SEB exposure being extensive lung inflammation and acute mortality of mice it is likely that several miRNA are acting in concert to enhance the severity of the disease. Additionally, since miRNA have several cotargets, its impact would be expected to be widespread and involve the simultaneous activation and crosstalk of a number of inflammatory signaling pathways as highlighted in the study. To the best of our knowledge we are the first to report the differential regulation of miRNA in response to SEB in a murine model of acute inflammatory lung injury. While it would be interesting to identify a principal miRNA, the inhibition of which rescues mice from SEB-induced toxicity our results nevertheless point toward the potential therapeutic modulation of miRNA to mitigate SEB-induced toxicity.
FUNDING
National Institutes of Health grants (P01AT003961, R01AT006888, R01ES019313, R01MH094755, P20 GM103641) and the VA Merit Award BX001357.
REFERENCES
- Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. (2011). Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 286, 1436–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D., Boldin M. P., O’Connell R. M., Rao D. S., Taganov K. D. (2008). MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9, 839–845. [DOI] [PubMed] [Google Scholar]
- Busbee P. B., Nagarkatti M., Nagarkatti P. S. (2014). Natural indoles, indole-3-carbinol and 3,3′-diindolymethane, inhibit T cell activation by staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol. Appl. Pharmacol. 274, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Feilotter H. E., Pare G. C., Zhang X., Pemberton J. G., Garady C., Lai D., Yang X., Tron V. A. (2010). MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am. J. Pathol. 176, 2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R., Ahmed S. A. (2011). MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 157, 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson-Moncada J., Papavasiliou F. N., Tam W. (2010). MicroRNAs of the immune system: roles in inflammation and cancer. Ann. N. Y. Acad. Sci. 1183, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle J. S., Giroux M., Boucher G., Landry J. R., Hardy M. P., Lemieux S., Jones R. G., Wilhelm B. T., Perreault C. (2013). The TGF-beta-Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes Immun. 14, 115–126. [DOI] [PubMed] [Google Scholar]
- Faulkner L., Cooper A., Fantino C., Altmann D. M., Sriskandan S. (2005). The mechanism of superantigen-mediated toxic shock: not a simple Th1 cytokine storm. J. Immunol. 175, 6870–6877. [DOI] [PubMed] [Google Scholar]
- Foster T. J. (2004). The Staphylococcus aureus “superbug”. J. Clin. Invest. 114, 1693–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. M., Turka L. A., June C. H., Thompson C. B. (1992). CD28 and staphylococcal enterotoxins synergize to induce MHC-independent T-cell proliferation. Cell Immunol. 145, 11–20. [DOI] [PubMed] [Google Scholar]
- Hanieh H., Alzahrani A. (2013). A microRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur. J. Immunol. 43, 2771–2782. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Hess Michelini R., Doedens A. L., Goldrath A. W., Stone E. L. (2012). FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12, 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzella L. M., Buckley M. J., Alves D. A., Stiles B. G., Krakauer T. (2009). Central roles for IL-2 and MCP-1 following intranasal exposure to SEB: a new mouse model. Res. Vet. Sci. 86, 241–247. [DOI] [PubMed] [Google Scholar]
- Kissner T. L., Ruthel G., Alam S., Ulrich R. G., Fernandez S., Saikh K. U. (2011). Activation of MyD88 signaling upon staphylococcal enterotoxin binding to MHC class II molecules. PLoS One 6, e15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozono H., Parker D., White J., Marrack P., Kappler J. (1995). Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity 3, 187–196. [DOI] [PubMed] [Google Scholar]
- Krakauer T. (2013). Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins (Basel) 5, 1629–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer T., Buckley M., Fisher D. (2010). Murine models of staphylococcal enterotoxin B-induced toxic shock. Mil. Med. 175, 917–922. [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Li J., Wan Y., Guo Q., Zou L., Zhang J., Fang Y., Zhang J., Zhang J., Fu X., Liu H., Lu L., Wu Y. (2010). Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res. Ther. 12, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Hron J. D., Peng S. L. (2004). Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity 21, 203–213. [DOI] [PubMed] [Google Scholar]
- Liu D., Zienkiewicz J., DiGiandomenico A., Hawiger J. (2009). Suppression of acute lung inflammation by intracellular peptide delivery of a nuclear import inhibitor. Mol. Ther. 17, 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. [DOI] [PubMed] [Google Scholar]
- Lungu G., Stoica G., Ambrus A. (2013). MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci. Lett. 553, 153–158. [DOI] [PubMed] [Google Scholar]
- Mattes J., Collison A., Plank M., Phipps S., Foster P. S. (2009). Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. U.S.A. 106, 18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke T., Wahl C., Heeg K., Echtenacher B., Krammer P. H., Wagner H. (1992). T cell-mediated lethal shock triggered in mice by the superantigen staphylococcal enterotoxin B: critical role of tumor necrosis factor. J. Exp. Med. 175, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke T., Wahl C., Regele D., Gaus H., Heeg K., Wagner H. (1993). Superantigen mediated shock: a cytokine release syndrome. Immunobiology 189, 270–284. [DOI] [PubMed] [Google Scholar]
- Nakahama T., Hanieh H., Nguyen N. T., Chinen I., Ripley B., Millrine D., Lee S., Nyati K. K., Dubey P. K., Chowdhury K., et al. (2013). Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 110, 11964–11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R. M., Rao D. S., Baltimore D. (2012). microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312. [DOI] [PubMed] [Google Scholar]
- Pauley K. M., Satoh M., Chan A. L., Bubb M. R., Reeves W. H., Chan E. K. (2008). Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res. Ther. 10, R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza R., Vidal S., Rodriguez-Sanchez J. L., Juarez C. (2004). Implication of STAT1 and STAT3 transcription factors in the response to superantigens. Cytokine 25, 1–10. [DOI] [PubMed] [Google Scholar]
- Rajagopalan G., Sen M. M., Singh M., Murali N. S., Nath K. A., Iijima K., Kita H., Leontovich A. A., Gopinathan U., Patel R., David C. S. (2006). Intranasal exposure to staphylococcal enterotoxin B elicits an acute systemic inflammatory response. Shock 25, 647–656. [DOI] [PubMed] [Google Scholar]
- Rao R., Nagarkatti P., Nagarkatti M. (2014). Staphylococcal enterotoxin B (SEB)-induced microRNA-155 targets suppressor of cytokine signaling-1 (SOCS1) to promote acute inflammatory lung injury. Infect. Immun. 82, 2971–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. A., Nagarkatti P., Nagarkatti M. (2012). Multiple anti-inflammatory pathways triggered by resveratrol lead to amelioration of staphylococcal enterotoxin B-induced lung injury. Br. J. Pharmacol. 167, 1244–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi R. L., Rossetti G., Wenandy L., Curti S., Ripamonti A., Bonnal R. J., Birolo R. S., Moro M., Crosti M. C., Gruarin P., et al. (2011). Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12, 796–803. [DOI] [PubMed] [Google Scholar]
- Saeed A. I., Rieder S. A., Price R. L., Barker J., Nagarkatti P., Nagarkatti M. (2012). Acute lung injury induced by Staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc. Microanal. 18, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savransky V., Rostapshov V., Pinelis D., Polotsky Y., Korolev S., Komisar J., Fegeding K. (2003). Murine lethal toxic shock caused by intranasal administration of staphylococcal enterotoxin B. Toxicol. Pathol. 31, 373–378. [DOI] [PubMed] [Google Scholar]
- Scott H. L., Tamagnini F., Narduzzo K. E., Howarth J. L., Lee Y. B., Wong L. F., Brown M. W., Warburton E. C., Bashir Z. I., Uney J. B. (2012). MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur. J. Neurosci. 36, 2941–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E., Stahle M., Pivarcsi A. (2008). MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18, 131–140. [DOI] [PubMed] [Google Scholar]
- Strandberg K. L., Rotschafer J. H., Vetter S. M., Buonpane R. A., Kranz D. M., Schlievert P. M. (2010). Staphylococcal superantigens cause lethal pulmonary disease in rabbits. J. Infect. Dis. 202, 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. (2008). Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 582, 1564–1568. [DOI] [PubMed] [Google Scholar]
- Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Dijke P., Goumans M. J., Itoh F., Itoh S. (2002). Regulation of cell proliferation by Smad proteins. J. Cell Physiol. 191, 1–16. [DOI] [PubMed] [Google Scholar]
- Uchakina O. N., Castillejo C. M., Bridges C. C., McKallip R. J. (2013). The role of hyaluronic acid in SEB-induced acute lung inflammation. Clin. Immunol. 146, 56–69. [DOI] [PubMed] [Google Scholar]
- Ulrich R. G., Sidell S., Taylor T. J., Wilhelmsen C. L., Franz D. R. (2001). Textbook of Military Medicine: Medical Aspects of Chemical and Biological Warfare . Office of the Surgeon General, Department of the Army, USA. [Google Scholar]
- Wong W. F., Kohu K., Chiba T., Sato T., Satake M. (2012a). Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology 132, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W. F., Kohu K., Nakamura A., Ebina M., Kikuchi T., Tazawa R., Tanaka K., Kon S., Funaki T., Sugahara-Tobinai A., et al. (2012b). Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J. Immunol. 188, 5408–5420. [DOI] [PubMed] [Google Scholar]
- Xiao B., Liu Z., Li B. S., Tang B., Li W., Guo G., Shi Y., Wang F., Wu Y., Tong W. D., et al. (2009). Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 200, 916–925. [DOI] [PubMed] [Google Scholar]
- Xu N., Meisgen F., Butler L. M., Han G., Wang X. J., Soderberg-Naucler C., Stahle M., Pivarcsi A., Sonkoly E. (2013). MicroRNA-31 is overexpressed in psoriasis and modulates inflammatory cytokine and chemokine production in keratinocytes via targeting serine/threonine kinase 40. J. Immunol. 190, 678–688. [DOI] [PubMed] [Google Scholar]
- Yu H., Pardoll D., Jove R. (2009). STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hu S., Zhang X., Wang L., Zhang X., Yan B., Zhao J., Yang A., Zhang R. (2014). MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 443, 1078–1084. [DOI] [PubMed] [Google Scholar]