Abstract
Signaling mucins are evolutionarily conserved regulators of signal transduction pathways. The signaling mucin Msb2p regulates the Cdc42p-dependent mitogen-activated protein kinase (MAPK) pathway that controls filamentous growth in yeast. The cleavage and release of the glycosylated inhibitory domain of Msb2p is required for MAPK activation. We show here that proteolytic processing of Msb2p was induced by underglycosylation of its extracellular domain. Cleavage of underglycosylated Msb2p required the unfolded protein response (UPR), a quality control (QC) pathway that operates in the endoplasmic reticulum (ER). The UPR regulator Ire1p, which detects misfolded/underglycosylated proteins in the ER, controlled Msb2p cleavage by regulating transcriptional induction of Yps1p, the major protease that processes Msb2p. Accordingly, the UPR was required for differentiation to the filamentous cell type. Cleavage of Msb2p occurred in conditional trafficking mutants that trap secretory cargo in the endomembrane system. Processed Msb2p was delivered to the plasma membrane, and its turnover by the ubiquitin ligase Rsp5p and ESCRT attenuated the filamentous-growth pathway. We speculate that the QC pathways broadly regulate signaling glycoproteins and their cognate pathways by recognizing altered glycosylation patterns that can occur in response to extrinsic cues.
INTRODUCTION
Signaling mucins are evolutionarily conserved regulators of signal transduction pathways (1–4). Signaling mucins are composed of a highly glycosylated extracellular domain that contains a mucin homology domain (MHD), which is defined by tandem repeats rich in Ser/Thr/Pro residues. The extracellular domain is connected by a single-pass transmembrane (TM) alpha helix to a cytosolic signaling domain, which associates with a diverse array of proteins that regulate mitogen-activated protein kinase (MAPK) pathways, Akt, β-catenin, and other pathways (5–8). Signaling mucins are overexpressed in different cancers, where they contribute to cell proliferation and metastasis (6). They are diagnostic biomarkers for cancers (9) and targets for immunotherapies (10, 11). Therefore, the mechanisms by which signaling mucins and related glycoproteins are regulated is of intense interest.
In the budding yeast Saccharomyces cerevisiae, the mucin-like glycoprotein Msb2p regulates the MAPK pathway that controls filamentous growth, a cell differentiation behavior that occurs in response to nutrient limitation (12–14). The extracellular domain of Msb2p is extensively glycosylated. Msb2p is modified by N-linked and O-linked glycosylation and contains a canonical MHD that is itself highly glycosylated (15, 16). In a landmark study, Yang et al. identified Pmt4p as the major O-mannosyltransferase for Msb2p (17). Pmt4p is a member of an evolutionarily conserved protein mannosyl transferase (Pmt) gene family (2, 18). Msb2p also contains a cytosolic signaling domain. The cytosolic domain of Msb2p associates with the Rho GTPase Cdc42p (15), which is a ubiquitous regulator of cell polarity and signaling (19). Msb2p also associates with the tetraspan protein Sho1p (15, 20, 21) and functions with a third TM regulator, Opy2p (17, 22–26), to regulate the filamentous-growth pathway. Once activated, Cdc42p binds to the p21-activated kinase (PAK) Ste20p to regulate a canonical MAPK cascade (Ste11p [MAPK kinase kinase]→Ste7p [MAPK kinase]→Kss1p [MAPK]). The MAPK Kss1p controls the activity of transcription factors (Ste12p and Tec1p), which induce target genes that bring about differentiation to the filamentous cell type (14, 27).
An important challenge surrounding Msb2p and other mucin-like glycoproteins is to understand what these proteins might sense at the plasma membrane (PM). The filamentous-growth pathway is activated by glucose limitation (22, 28–30), yet whether or how Msb2p senses limiting glucose is not clear. Defects in protein glycosylation also activate the filamentous-growth pathway (17, 31, 32). It is known that a portion of the glycosylated extracellular domain of Msb2p plays an inhibitory role in MAPK signaling (100 to 950 amino acids [aa] [15]). Cleavage and release of the extracellular inhibitory domain of Msb2p by yapsin aspartyl-type proteases (33–36), particularly Yps1p, are required for filamentous-growth pathway activity (28).
Here, we describe a new connection between the processing of Msb2p and an internal quality control (QC) pathway that operates in the secretory pathway and is required for the activation of the filamentous-growth pathway. We show that glycosylation deficiency, which occurs in conditional protein glycosylation mutants, or growth in a nonpreferred carbon source (galactose) leads to underglycosylation of Msb2p. Underglycosylated Msb2p was efficiently processed by a mechanism involving the unfolded protein response (UPR), the major QC pathway that operates in the endoplasmic reticulum (ER) (37). The UPR regulator Ire1p, which detects misfolded and underglycosylated proteins in the ER (37–40), was required for proteolytic processing of Msb2p by regulating expression of YPS1, the major protease required for Msb2p cleavage. This activation mechanism connects the UPR to a cell differentiation response (filamentous growth) through a mucin-dependent ERK-type MAPK pathway. The regulatory mechanism described here may extend to other signaling glycoproteins whose glycosylation becomes altered in response to extracellular stimuli, leading to detection and modification by internal QC pathways.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains are listed in Table 1. Plasmids are listed in Table 2. Yeast strains were maintained in yeast extract and peptone medium containing 2% glucose (YEPD) or 2% galactose (GAL) unless otherwise indicated. Cells were grown at 30°C (49, 50). Temperature shift experiments were carried out at 37°C. For maintaining the selection of plasmids, cells were grown in synthetic medium. Gene disruptions were performed according to standard genetic techniques (51, 52), including the use of heterologous auxotrophic markers and antibiotic resistance markers for gene disruption and tagging (53). Epitope tagging with hemagglutinin (HA) or c-MYC epitopes (54) and GAL1 promoter and fluorescent tag (GFP) fusions (55) were carried out as described previously.
TABLE 1.
Strains used in the study
| Strain | Genotypea | Reference or source |
|---|---|---|
| PC244 | MATα ste4 FUS1-HIS3 ura3-52 pmi40-101b | 30 |
| PC313 | MATa ura3-52 | 12 |
| PC538 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 | 15 |
| PC544 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 bni1::KlURA3d | 41 |
| PC948 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 msb2::kanMX6 | 15 |
| PC999 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-HA(at aa 500) | 15 |
| PC1029 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 flo11::KlURA3 | 42 |
| PC1140 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ698-818-HA (ΔMHD) | 15 |
| PC1291 | SY1436 MATα ste4 FUS1-HIS3 ura3-52b | 15 |
| PC1516 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ100-818-HA | 15 |
| PC1523 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 ssk1::KlURA3 | 29 |
| PC1658 | NY13 MATa ura3-52c | 43 |
| PC1660 | NY61 MATa ura3-52 sec10-2c | 43 |
| PC1661 | NY64 MATa ura3-52 sec15-1c | 43 |
| PC1662 | NY402 MATa ura3-52 sec5-24c | 43 |
| PC1663 | NY410 MATa ura3-52 sec8-19c | 43 |
| PC1664 | NY412 MATa ura3-52 sec3-2c | 43 |
| PC2011 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-HA(at aa 500)-GFP(at aa 1306)::KanMX6g | 28 |
| PC2043 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 FLO11-HA | 42 |
| PC2094 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-GFP(at aa 1306)::KanMX6 | 28 |
| PC2212 | MATa ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100c | 33 |
| PC2213 | MATa ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 5ypsΔc | 33 |
| PC2138 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 cln1::KlURA3 | This study |
| PC2224 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ100-850-HA | 28 |
| PC2226 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ100-900-HA | 28 |
| PC2227 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ100-950-HA | 28 |
| PC2262 | MATa ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 yps1::LEU2c | 33 |
| PC2382 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 ste12::kanMX6 | 28 |
| PC2622 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 snf8::HYG | 44 |
| PC2963 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 FLO10-HA | 45 |
| PC3007 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 RAX2-HA | 45 |
| PC3008 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 SLG1-HA | 45 |
| PC3009 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 CTR1-HA | 45 |
| PC3063 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pep4::kanMX6 | Research Genetics |
| PC3156 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-HA(at aa 500)-GFP(at aa 1306)::KanMX6 pep4::KlURA3 | This study |
| PC3288 | SEY6210 MATα ura3-52 his3-200 trp1-901 lys2-801 suc2-9 leu2-3e | 46 |
| PC3289 | SEY6210.1 rsp5Δ::HIS3 pDsRED415-rsp5G753I rsp5-1e | 46 |
| PC3861 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 ste11::NAT | 22 |
| PC4994 | SEY6210 pik1::HIS3 pRS314pik1-83 (TRP1 CEN6 pik1-83)e | 47 |
| PC5659 | YPH500 MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 46 |
| PC5662 | RSY263 MATα ura3-52 leu2-3,112 sec12-4f | 48 |
| PC5732 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2pK1223R K1239R K1245R-HA(at aa 500)-GFP(at aa 1306)::KanMX | This study |
| PC5783 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 PGAL-Msb2p-HA(at aa 500)-GFP(at aa 1306)::KanMX6::NAT | This study |
| PC5834 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1209-1306-GFP::kanMX6 | This study |
| PC5836 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1223-1306-GFP::kanMX6 | This study |
| PC5838 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1239-1306-GFP::kanMX6 | This study |
| PC5840 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1245-1306-GFP::kanMX6 | This study |
| PC5842 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1290-1306-GFP::kanMX6 | This study |
| PC5844 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1291-1306-GFP::kanMX6 | This study |
| PC5846 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1294-1306-GFP::kanMX6 | This study |
| PC5848 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1298-1306-GFP::kanMX6 | This study |
| PC5850 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2pK1223R K1239R K1245R-HA(at aa 500)-GFP(at aa 1306)::KanMX::KlUra3 | This study |
| PC5916 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1304-1306::KlURA3 | This study |
| PC5918 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1223-1306::KlURA3 | This study |
| PC5919 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1239-1306::KlURA3 | This study |
| PC5920 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1245-1306::KlURA3 | This study |
| PC5921 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1290-1306::KlURA3 | This study |
| PC5922 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1291-1306::KlURA3 | This study |
| PC5923 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1298-1306::KlURA3 | This study |
| PC5924 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2Δ1301-1306::KlURA3 | This study |
| PC5951 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2L1301A Δ1304-1306::KlURA3 | This study |
| PC5952 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2G1302A Δ1304-1306::KlURA3 | This study |
| PC5953 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2W1303A Δ1304-1306::KlURA3 | This study |
| PC5956 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-GFP(at aa 1306)::KanMX6 | This study |
| PC5980 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 sec3::KlURA3 | This study |
| PC5987 | NY13 MATa ura3-52 yps1::KlURA3d pHA-MSB2-GFP | This study |
| PC5988 | NY412 MATa ura3-52 sec3-2 yps1::KlURA3 pHA-MSB2-GFPc | This study |
| PC5991 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 pmt4::NAT | This study |
| PC5997 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2S1300A Δ1304-1306::KlURA3 | This study |
| PC5998 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2N1299A Δ1304-1306::KlURA3 | This study |
| PC5999 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2S1292A Δ1304-1306::KlURA3 | This study |
| PC6000 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2R1293A Δ1304-1306::KlURA3 | This study |
| PC6001 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2P1294A Δ1304-1306::KlURA3 | This study |
| PC6002 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 MSB2I1295A Δ1304-1306::KlURA3 | This study |
| PC6032 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 ire1::KlURA3 | This study |
| PC6033 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 hrd1::KlURA3 | This study |
| PC6043 | SY1436 ste4 FUS1-HIS3 ire1::KlURA3b | This study |
| PC6044 | MATa ste4 FUS1-HIS3 ura3-52 pmi40-101 ire1::KlURA3b | This study |
| PC6048 | MATa ura3-52 ire1::NAT | This study |
| PC6320 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 lhs1::KlURA3 | This study |
| PC6321 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 Msb2p-HA(at aa 500) ire1::KlURA3 | This study |
| PC6322 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 hac1::KlURA3 | This study |
| PC6458 | MATα ste4 FUS1-HIS3 ura3-52 pmi40-101 ire1::KlURA3::FOA+::ura3 pUPRE-lacZb | This study |
| PC6459 | MATa ste4 ura3-52 PGAL-MSB2 | This study |
| PC6460 | MATa ura3-52 ire1::NAT PGAL-MSB2 | This study |
| PC6462 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 pmt4::NAT ire1::KlURA3 | This study |
| PC6463 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 ste12::kanMX6 ire1::KlURA3 | This study |
| PC6464 | MATa ste4 FUS1-lacZ FUS1-HIS3 ura3-52 pmt4::NAT ste12::KlURA3 | This study |
| PC6465 | MATa ura3-52 ire1::NAT ssk1::KlURA3 | This study |
| PC6467 | MATα ste4 FUS1-HIS3 ura3-52 pmi40-101 IRE1Δ673-1115::kanMX6b | This study |
All strains are in the Σ1278b background unless otherwise indicated.
246-1-1 background.
W303 background.
KlURA3 refers to the Kluyveromyces lactis URA3 cassette.
Strains are in the SEY6210 background.
Strains are in the X2180-1A background.
Msb2p with HA attached at aa 500 and GFP attached at aa 1306 fused to KanMX6.
TABLE 2.
Plasmids used in the study
| Number | Plasmid | Description | Source or reference |
|---|---|---|---|
| PC1287 | λYES-MSB2 | AMP/CEN/URA3 | 15 |
| PC1456 | pHA-MSB2 | AMP/CEN/URA3 | 28 |
| PC1694 | pGFP-2-MSB2 | GFP at 246-539Δ; AMP/CEN/URA3 | This study |
| PC1696 | pGFP-1-MSB2 | GFP at 324-326Δ; AMP/CEN/URA3 | This study |
| PC2344 | pMBP-MSB2 | AMP/CEN/URA3 | This study |
| PC2417 | λYES-MSB2-2 extra repeats | AMP/CEN/URA3 | This study |
| PC2418 | λYES-MSB2-0 extra repeats | AMP/CEN/URA3 | This study |
| PC2419 | λYES-MSB2-1 extra repeats | AMP/CEN/URA3 | This study |
| PC2582 | pHA-MSB2-GFP | AMP/CEN/URA3 | This study |
| PC6469 | pGST-cLD IRE1 | E. coli expression | This study |
The plasmid containing the UPRE-lacZ reporter was provided by P. Walter (56). The plasmid containing the YPS1-lacZ reporter was provided by D. Krysan (33). A. Tartakoff (Case Western Reserve, Cleveland, OH) provided the sec12-4 and wild-type control strains. C. Burd (Yale University, New Haven, CT) provided the rsp5-1 strain (46). P. Novick (Yale University, New Haven, CT) provided exocytosis mutant strains. The plate-washing assay and agar invasion were examined as described previously (13). Biofilm and mat assays were performed on YEPD and YEP-GAL plates containing 0.3% agar (57). The single-cell invasive growth assay was performed as described previously (30).
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (15). Cells were grown in synthetic medium lacking uracil to maintain selection for the plasmids. Cells from a saturated culture were washed once in water and cultured in inducing medium (typically YEPD or YEP-GAL) until cells had reached mid-log-phase growth (∼5.5 h). At least two independent experiments were performed, and the average values were represented in Miller units. Error bars indicate standard deviations between trials.
qPCR analysis.
Quantitative PCR (qPCR) analysis was performed as described previously (58) using primers for YPS1 (forward, 5′-AACGTTACCGGGTTGTCTTT-3′; reverse, 5′-CGCTTGGAACAGAGGATGTAA-3′) and ACT1 (forward, 5′-GGCTTCTTTGACTACCTTCCAACA-3′; reverse, 5′-GATGGACCACTTTCGTCGTATTC-3′). The template cDNA was synthesized by an iScript cDNA synthesis kit (Bio-Rad, Carlsbad, CA) according to the manufacturer's suggested protocol. PCRs were run with iQ SYBR green Supermix (Bio-Rad, Carlsbad, CA). qPCR was performed for the following amplification cycles: initial denaturation for 8 min at 95°C, followed by 35 cycles of denaturation for 15 s at 95°C and annealing for 1 min at 60°C. The expression of genes was quantified using the 2−ΔΔCT method (141). The ACT1 gene (encoding actin) was used to normalize expression levels. Error bars indicate the SEM (standard errors of the means) from three independent trials.
Site-directed mutagenesis by in vivo recombination.
To generate Msb2p3KR and single-amino-acid substitutions (from residue 1285 to 1303) in the cytosolic domain of Msb2p, homologous recombination at the MSB2 locus in the genome was performed. Primers were designed with the desired nucleotide changes incorporated. pKlURA3 (PC5225) plasmid was used to generate a URA3 cassette with the incorporated point mutations in the flanking regions. The cassette was transformed into yeast strains and selected on synthetic medium lacking uracil and incubated for 4 to 5 days at 30°C. PCR analysis was performed for the verification of the integration of the cassette at the locus. Point mutations were confirmed by sequencing of PCR products flanking the region mutagenized.
To generate the Msb2p3KR-GFP strain, the designated K-to-R changes were incorporated in the forward primer. MSB2 was amplified from pHA-Msb2p-GFP with a reverse primer designed from the region downstream of the MSB2 open reading frame (ORF) containing the antibiotic resistance kanMX6 cassette. The amplified PCR product was transformed in the PC999 (HA-MSB2) strain. Transformants were selected on YEPD medium containing Geneticin for 4 to 5 days at 30°C. Positive isolates were confirmed by GFP fluorescence and sequencing.
Protein immunoblot analysis.
The glycosylation status of Msb2p was measured using a concanavalin A (ConA)-bound resin for glycoprotein isolation (89804; Pierce, Rockford, IL), and the manufacturer's protocol was followed. Immunoblotting was performed as described previously (15). Phosphorylated MAPKs were detected as described previously (59). In general, cell lysates were resolved by 10% SDS-PAGE analysis unless otherwise indicated. For immunoblot analysis, proteins were transferred to nitrocellulose membranes (Protran BA85; VWR International Inc., Bridgeport, NJ), which were incubated in blocking buffer (5% nonfat dry milk, 10 mM Tris-HCl [pH 8], 150 mM NaCl, and 0.05% Tween 20) for 16 h at 4°C.
Antibodies were used at the manufacturer's recommended concentrations. WesternBright quantum horseradish peroxidase (HRP) substrate (K-12042-D20; Advansta) was used to detect secondary antibodies. Antibodies to phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) (4370; Cell Signaling Technology) were used to detect phosphorylated Kss1p (1:3,000 dilution). Mouse monoclonal antibodies were used to detect green fluorescent protein (GFP) (clones 7.1 and 13.1; 11814460001; Roche Diagnostics). Antibodies to HA (11583816001; Roche) are commercially available. Secondary antibodies were goat anti-mouse IgG–HRP (170-6516; Bio-Rad) and goat anti-rabbit IgG–HRP (111-035-144; Jackson ImmunoResearch Laboratories, Inc.). Antibodies to ubiquitin were used (ab24686; Abcam). Protein concentration was measured using the bicinchoninic acid (BCA) kit (Thermo-Fisher, Waltham, MA) and by immunoblotting using antibodies against the Pgk1p protein (1:5,000 dilution; no. 459250, lot P0660; Life Technologies-Molecular Probes, Grand Island, NY). To measure protein turnover, the galactose-inducible promoter GAL1 fusions were constructed to drive expression of MSB2-GFP. Cells were grown in YEP-GAL medium for 4 h to induce expression, washed in water twice, and resuspended in YEPD medium. Cells were harvested over a time series, and cell extracts were examined by immunoblot analysis to measure protein levels.
Analysis of Msb2p cleavage in yapsin and conditional trafficking mutants.
All conditional alleles were compared to their isogenic parent strains. To investigate Msb2p cleavage and MAPK signaling in temperature-sensitive mutants, the following strains and conditions were used. All strains contained the pHA-Msb2p-GFP (PC2582) plasmid and were pregrown in SD-URA medium to maintain selection for the plasmid for 16 h and induced for 4 h at 37°C in prewarmed YEPD medium. For ER and Golgi trafficking mutants, wild-type (PC5659) and sec12-14 (PC5662) strains were examined. For phosphatidylinositol (PI) kinase mutants, wild-type (PC4990) and pik1-83 (PC4994) strains were examined. For exocyst mutants, wild-type (PC1658), sec3-2 (PC1664), sec5-24 (PC1662), sec8-19 (PC1663), sec10-2 (PC1660), sec15-1 (PC1661), yps1Δ (PC2262), sec3-2 yps1Δ (PC5988), and sec15-1 (PC1661) strains were examined. For yapsin mutants, wild-type (PC2212), yps1Δ (PC2262), and 5ypsΔ (PC2213) cells were induced in YEPD medium for 6 h at 30°C. In other experiments, wild-type (PC1291) and pmi40-101 (PC244) strains harboring pHA-Msb2p-GFP were grown with or without 50 mM mannose (Man) for 6 h in YEPD medium. To assess Msb2p levels in conditional RSP5 mutants, wild-type (PC3288) and rsp5-1 (PC3290) cells expressing either pGFP-Msb2p (PC1696) or pHA-Msb2p-GFP (PC2582) were grown in YEPD medium for 2 h at 37°C.
Protein localization and microscopy.
The localization of Msb2p was examined using plasmids pGFP-Msb2p (PC1696) and pMsb2p-GFP (PC2582). For some experiments, a dual-tagged functional fusion protein was used (pHA-Msb2p-GFP [PC2582]). For localization experiments involving sec mutants, cells were grown in SD-URA for 16 h at 30°C, shifted to 37°C for 4 h, and examined on a stage heated to 37°C.
Differential interference contrast (DIC) and fluorescence microscopy using fluorescein isothiocyanate (FITC) filter sets were performed using an Axioplan 2 fluorescence microscope (Zeiss) with a Plan-Apochromat 100×/1.4 (oil) objective (numeric aperture, 0.17). Digital images were obtained with the Axiocam MRm camera (Zeiss). Axiovision 4.4 software (Zeiss) was used for image acquisition and analysis. Cells were examined by oil immersion on glass slides (2947-75; Corning Inc., Corning, NY) with a glass coverslip (48366-227; VWR) using a 100× objective. Images were analyzed in Adobe Photoshop, where adjustments of brightness and contrast were made. A temperature control stage slide warmer (0115.000; PeCon GmbH, Germany) was used to maintain cells at 37°C for protein localization experiments in temperature-sensitive mutants.
In vitro pulldown assay.
The core endoplasmic reticulum (ER)-lumenal domain of Ire1p (cLD-Ire1; aa residues 111 to 411 [60]) was expressed and purified from the bacterial lysate by glutathione S-transferase (GST) fusion. Briefly, Bl21-DE3 bacterial cells containing the GST-cLD-Ire1p plasmid construct were grown at 37°C to an optical density at 600 nm (OD600) of 0.6. The expression of GST-cLD-Ire1p was induced by 0.5 mM isopropyl-â-d-thiogalactopyranoside (IPTG) at 22°C for 16 h. Cells were harvested and resuspended in the lysis buffer (1× phosphate-buffered saline [PBS], pH 7.4; 1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 mM dithiothreitol [DTT]; 1 mg/ml lysozyme) and further lysed by sonication. Clarified lysate was incubated with the glutathione-Sepharose 4B beads (GE 17-0756-01) at room temperature for 1 h. GST alone was used as a control. For the preparation of the yeast lysates, the pmi40-101 (PC244) mutant containing HA-Msb2p (PC2582) was grown in YEPD medium with or without mannose for 16 h to enrich for the underglycosylated form of Msb2p. The yeast cells were disrupted using a FastPrep-24 instrument (MP Biomedicals LLC, Solon, OH) in the lysis buffer (1× PBS, pH 7.4; 1 mM PMSF; 1 mM DTT; 1% NP-40). Precleared yeast lysates containing HA-Msb2p were divided equally and incubated with GST alone or GST-cLD-Ire1p immobilized onto glutathione beads at 4°C for at least an hour by end-to-end rotation. The beads were washed with 1× PBS, pH 7.4, 3 times. One hundred fifty microliters of the 1× SDS-PAGE dye was added to the beads and boiled for 10 min with frequent agitation.
Bioinformatics.
UB PRED (http://www.ubpred.org/) was used to identify candidate ubiquitinated lysines (61). NetOGlyc (http://www.cbs.dtu.dk/services/NetOGlyc/) was used to identify O-linked protein glycosylation sites for mucin glycoproteins (62). ImageJ analysis was used to quantitate band intensity for protein gels and immunoblots (http://imagej.nih.gov [63]) using the invert function and by subtraction of background signals. SGD was used as a resource for yeast gene annotation and analysis (http://www.yeastgenome.org). The crystal structure images of GFP (64) and MBP (65, 66) were visualized by PDB (http://www.rcsb.org/pdb/home/home.do). A chi-square test was used to determine statistical significance. A two-tailed unpaired Student's t test was used to generate P values.
RESULTS
Underglycosylated Msb2p is proteolytically processed at elevated levels.
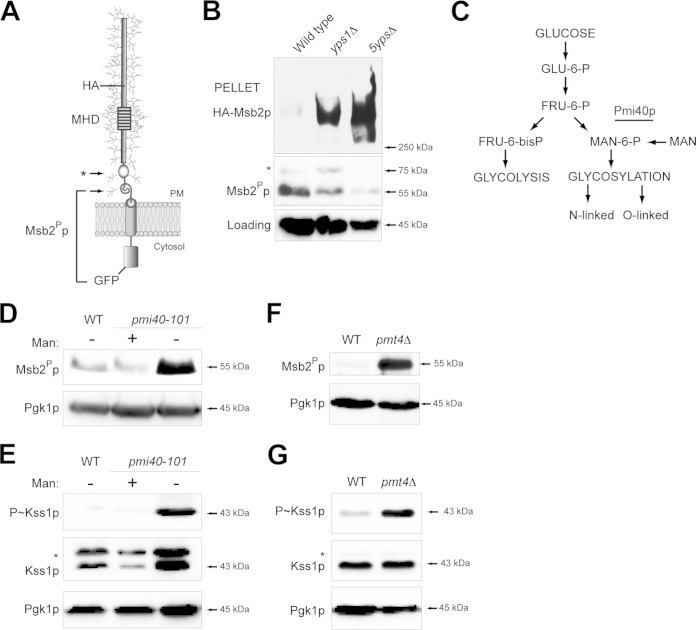
Msb2p is proteolytically processed in its extracellular domain by the aspartyl protease Yps1p (28). A portion of the extracellular domain (aa 100 to 900), which functions in an inhibitory capacity, is released from cells. The C-terminal portion of Msb2p, which includes the cytoplasmic signaling domain, remains associated with the plasma membrane (PM) (Fig. 1A; arrows refer to potential cleavage sites). A functional dual-tagged version of Msb2p, HA-Msb2p-GFP (HA epitope at aa 500 and GFP at the C-terminal aa 1306), was used to examine the proteolytic processing of Msb2p. The proteolytically processed C-terminal domain, referred to as Msb2Pp, migrated as a 55-kDa band by immunoblot analysis (Fig. 1B). Msb2Pp levels were reduced in the yps1Δ mutant (Fig. 1B). Msb2Pp levels were further reduced in a mutant lacking all five yapsins (Fig. 1B, 5ypsΔ mutant), consistent with the idea that multiple yapsins proteolytically process Msb2p. Consistent with this idea, the extracellular domain of Msb2p was not shed in yapsin mutants and accumulated in cell pellets (Fig. 1B, HA-Msb2p).
FIG 1.
Underglycosylated Msb2p is proteolytically processed at elevated levels. (A) The Msb2p protein is shown as a single-pass glycoprotein with the mucin homology domain (MHD). Cleavage sites corresponding to immunoblot data for cleaved Msb2p-GFP are indicated by arrows. Msb2Pp refers to the proteolytically processed form; the asterisk refers to a minor cleavage product. The positions of HA and GFP fusions are shown. (B) Cleavage of HA-Msb2p-GFP in the yps1Δ and 5ypsΔ mutants. The top blot was probed with anti-HA antibodies to show full-length Msb2p at >250 kDa (HA-Msb2p). The middle blot was probed with anti-GFP antibodies to show the proteolytically processed Msb2Pp-GFP fusion (Msb2Pp, 55 kDa; *, 75 kDa). Blots were probed with anti-Pgk1p antibody, which was used as a loading control for all experiments. (C) Pathway for the conversion of glucose into substrates for glycolysis and protein glycosylation. The Pmi40p enzyme is underlined. (D) Immunoblot of Msb2Pp in wild-type cells (WT) and the pmi40-101 mutant grown in YEPD (−Man) or YEPD plus 50 mM mannose (+Man) for 5.5 h. (E) Immunoblot of P∼Kss1p levels for the strains used in panel D. The asterisk refers to a background band seen under some conditions with the total Kss1p antibodies. (F) Msb2Pp levels in wild-type (WT) cells and the pmt4Δ mutant. Cells were grown in YEP-GAL for 6 h. (G) P∼Kss1p levels for the strains examined in panel F.
Msb2p is modified by N- and O-linked glycosylation (15, 16), which are posttranslational modifications that occur in the endoplasmic reticulum (ER) and Golgi apparatus (67, 68, and references therein). A high-throughput screening approach, called secretion profiling, uncovered mutants defective for N- and O-linked glycosylation that showed differential shedding of HA-Msb2p (69). The secretion profiling data suggested that changes in glycosylation of Msb2p correlate with its proteolytic processing. To investigate this possibility, the cleavage of Msb2p was examined in a conditional glycosylation mutant, pmi40-101 (30). Pmi40p converts fructose-6-P to mannose-6-P (70). Consequently, Pmi40p is involved in early steps of N- and O-linked glycosylation and related processes (71, 72). The glycosylation defect of the pmi40-101 mutant can be suppressed by growth of cells in media containing 50 mM mannose (Fig. 1C) (70, 73). Cleavage of Msb2p was elevated in the pmi40-101 mutant grown in media lacking mannose (Fig. 1D). Cleavage of Msb2p is required to activate the filamentous-growth pathway (28), which can be monitored by phosphorylation of the MAPK Kss1p (P∼Kss1p) (74–77). The filamentous-growth pathway was induced in the pmi40-101 mutant grown in media lacking mannose (Fig. 1E). Cleavage of Msb2p also was examined in cells lacking Pmt4p, the major O-glycosyltransferase that modifies Msb2p (17). The pmt4Δ mutant showed elevated processing (Fig. 1F) and an increase in MAPK activity (Fig. 1G). Msb2p is underglycosylated in protein glycosylation mutants, including pmi40-101 and pmt4Δ (15, 17, 78). Therefore, underglycosylation of Msb2p leads to elevated processing of the protein and activation of the filamentous-growth pathway.
UPR regulates cleavage of underglycosylated Msb2p and the filamentous-growth pathway.
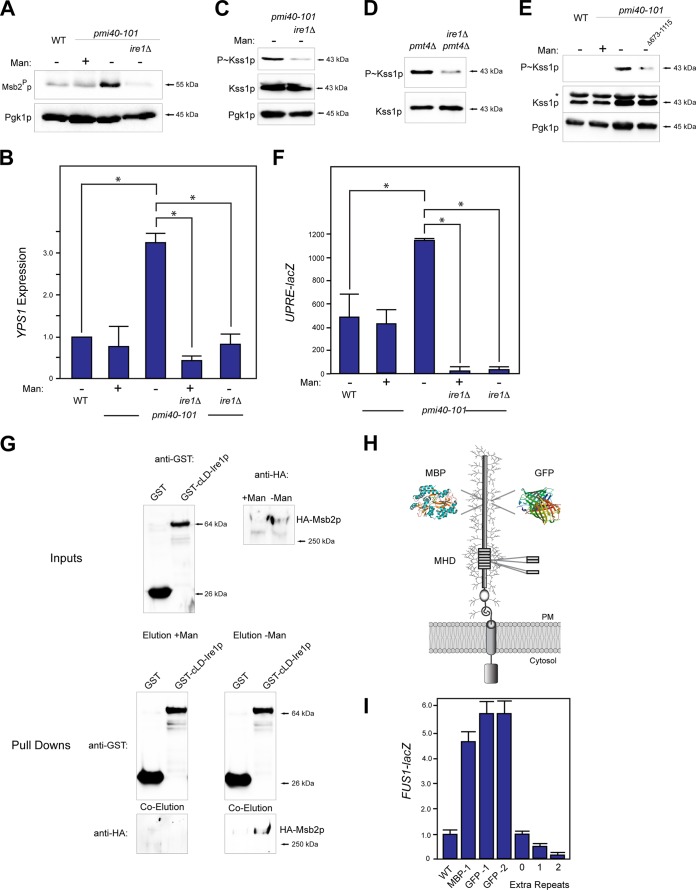
Underglycosylated proteins can become misfolded in the secretory pathway. QC pathways in the endomembrane system identify misfolded proteins and target them for proper folding or destruction. One of these pathways is the unfolded protein response (UPR) (37–40). A major regulator of the UPR is Ire1p, which recognizes misfolded proteins in the ER and induces a nuclear response (37, 39, 56, 60, 79). To test whether underglycosylated Msb2p is regulated by the UPR, proteolytic processing of Msb2p was examined in the pmi40-101 mutant lacking Ire1p. Cleavage of Msb2p was reduced in the pmi40-101 ire1Δ double mutant (Fig. 2A). Likewise, HA-Msb2p was shed at reduced levels in the ire1Δ mutant (see Fig. S1A in the supplemental material). Ire1p regulates the transcriptional induction of proteases, chaperones, and other enzymes (37) that include YPS1 (80, 81), which encodes the major protease for Msb2p. The expression of YPS1, assessed by qPCR and YPS1-lacZ analysis, was elevated in the pmi40-101 mutant (Fig. 2B). The elevated YPS1 expression seen in the pmi40-101 mutant was dependent on Ire1p (Fig. 2B). Therefore, the proteolytic processing of Msb2p in the pmi40-101 mutant results from UPR-dependent induction of YPS1 expression.
FIG 2.
Ire1p regulates Msb2p cleavage and filamentous-growth pathway activity by regulating YPS1 expression in response to protein glycosylation deficiency. (A) Msb2p cleavage in wild-type cells and the pmi40-101 mutant grown with (YEPD + Man) or without (YEPD) mannose in combination with the ire1Δ mutant. (B) YPS1 expression was determined by qPCR and adjusted to ACT1 levels as a control. The indicated strains were grown in YEPD (−Man) or YEPD plus Man medium (+Man) for RNA preparation and qPCR analysis. The asterisk refers to a P value of <0.05. (C) P∼Kss1p levels in the pmi40-101 and pmi40-101 ire1Δ double mutant. (D) P∼Kss1p levels in the pmt4Δ and pmt4Δ ire1Δ double mutants grown in YEP-GAL. (E) P∼Kss1p levels for an Ire1p C-terminal truncation. (F) UPRE-lacZ activity was determined by β-galactosidase assays for the indicated strains and conditions. Experiments were performed in duplicate, and the average values are shown. Error bars represent the standard deviations between trials. The asterisk refers to a P value of <0.05. (G) In vitro pulldown of HA-Msb2p expressed in the pmi40-101 mutant in YEPD (with or without mannose) with the luminal domain of Ire1p, called cLD-Ire1p-GST. Input, pulldown, and coelutions are shown. (H) Msb2p with insertion of tandem repeats, MBP, or GFP shown. GFP-1 was inserted at residue 324, resulting in an in-frame deletion of aa 324 to 326. GFP-2 was inserted at residue 246 and resulted in a deletion of aa 246 to 539. MPB was inserted at residue 324 without deletion of amino acid residues. (I) MAPK activity was assessed by an Msb2p-dependent reporter (FUS1-lacZ, which in Σ1278b ste4 strains shows Msb2p dependence [15]). Differences are expressed as fold differences compared to those of wild-type cells. Error bars represent standard deviations between trials, which varied by less than 10%.
If Ire1p controls the processing of Msb2p, it also may regulate the activity of the filamentous-growth pathway. The ire1Δ mutant showed reduced P∼Kss1p levels in the pmi40-101 mutant grown in medium lacking mannose (Fig. 2C). Ire1p also was required for P∼Kss1p in the pmt4Δ mutant (Fig. 2D). Ire1p contains a cytosolic signaling domain (79) that deletion analysis showed was required for induction of the filamentous-growth pathway (Fig. 2E, Δ673-1115).
The pmi40-101 mutant exhibited elevated UPR activity, based on the activity of a transcriptional reporter (UPRE-lacZ [56]), which was measured by β-galactosidase assays (Fig. 2F). Induction of the UPRE-lacZ reporter in the pmi40-101 mutant was dependent on Ire1p (Fig. 2F). The pmi40-101 mutant also exhibited a growth defect on medium lacking mannose, which was exacerbated in cells lacking Ire1p (see Fig. S1B in the supplemental material). Altogether, the results show a role for the UPR in regulating cleavage of Msb2p and the filamentous-growth pathway during protein glycosylation deficiency.
The core ER luminal domain (cLD) of Ire1p recognizes the misglycosylated form of carboxypeptidase in the ER (60). Msb2p may be recognized by Ire1p or be modified as part of a general response to ER stress. To test whether Msb2p and Ire1p associate, the interaction between Msb2p and GST-cLD-Ire1p was examined by in vitro pulldown analysis. Underglycosylated HA-Msb2p in extracts prepared from the pmi40-101 mutant grown in medium lacking mannose (underglycosylated) associated with GST-cLD-Ire1p that was overexpressed and purified from Escherichia coli (Fig. 2G). HA-Msb2p in extracts prepared from the pmi40-101 mutant grown in medium containing mannose (fully glycosylated) did not associate with GST-cLD-Ire1p (Fig. 2G). Thus, it is possible that Ire1p interacts with the underglycosylated form of Msb2p.
We further hypothesized that altering the glycosylation of Msb2p in wild-type cells would impact the filamentous-growth pathway. To test this possibility, globular domains that are not normally glycosylated (e.g., maltose binding protein [MBP] and GFP) were inserted into the extracellular domain of Msb2p (Fig. 2H). The insertions resulted in induction of the filamentous-growth pathway based on a transcriptional reporter that, in filamentous strains lacking STE4, exhibits filamentous-growth pathway dependence (FUS1-lacZ) (Fig. 2I). The elevated signaling presumably resulted from reduced stability of the extracellular domain, which is reduced in GFP-Msb2p and MBP-Msb2p variants (see Fig. S1C in the supplemental material) with a concomitant increase in signaling (see Fig. S1D; shown for GFP-Msb2p). Increasing the glycosylation of Msb2p likewise might dampen the filamentous-growth pathway. The addition of tandem repeats to the MHD (Fig. 2H), which are highly glycosylated (15, 82), dampened the filamentous-growth pathway in a manner that corresponded with increasing repeat number (Fig. 2I). Thus, altering the Msb2p protein in a way that would be expected to impact its glycosylation profile alters the filamentous-growth pathway, which may occur by its recognition by Ire1p.
The UPR regulates Msb2p cleavage and the filamentous-growth pathway during invasive growth.
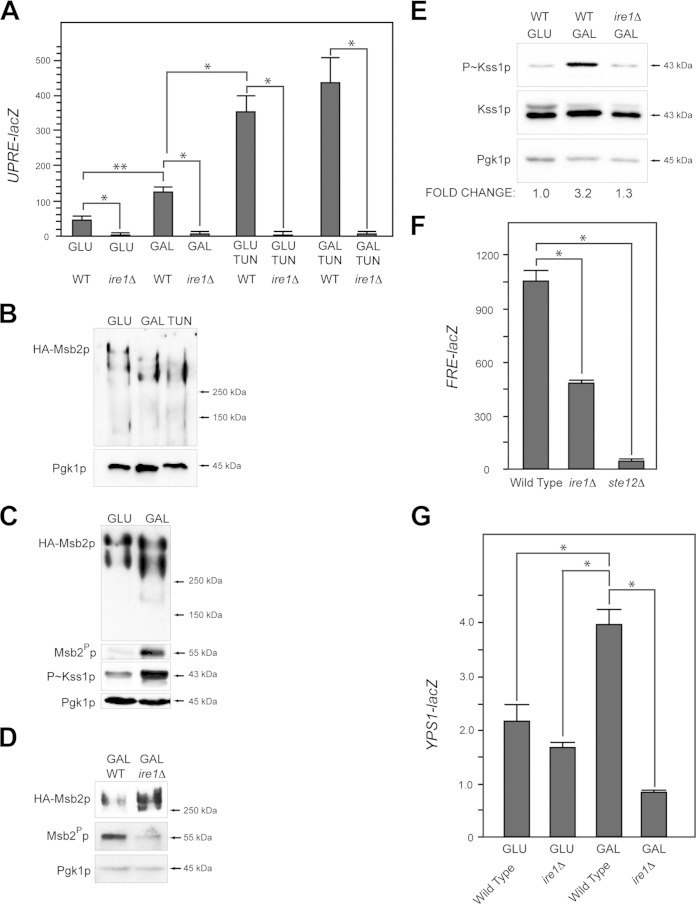
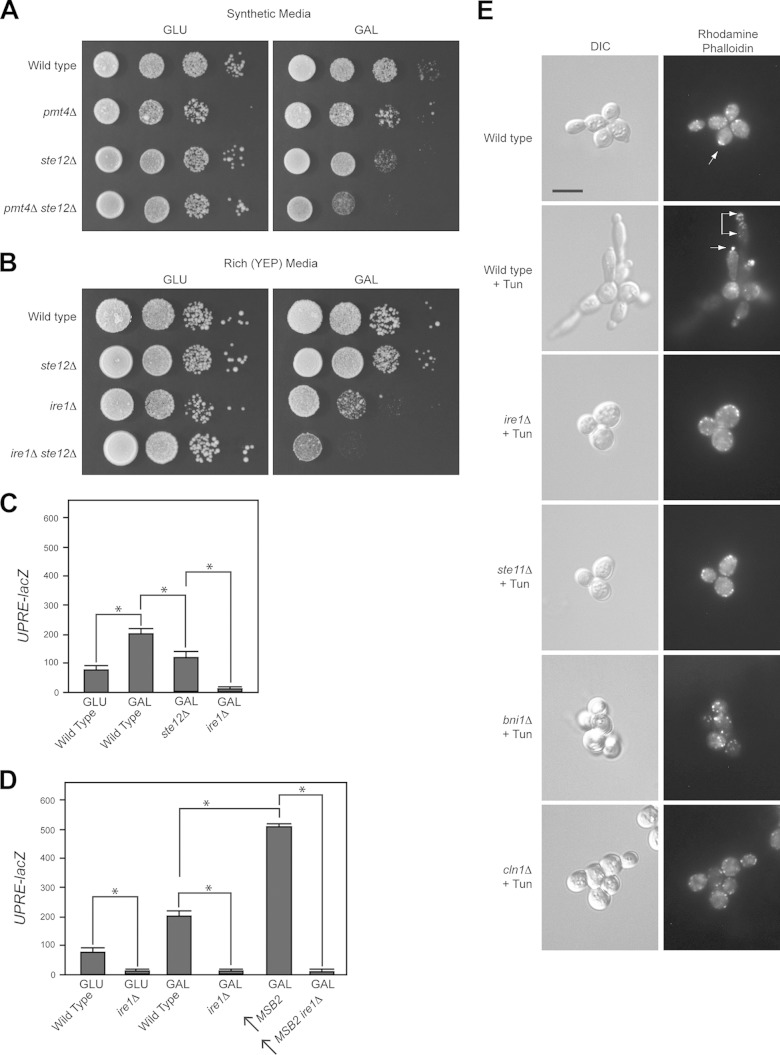
The filamentous-growth pathway is induced by defects in protein glycosylation and also by the limitation of nutrients like glucose (22, 28, 29). In mammals, glucose depletion activates the UPR (83–86). This may be because glucose is the precursor for glycolysis and oligosaccharide production (Fig. 1C); thus, limiting glucose may lead to a reduction in protein glycosylation. In yeast, the ire1Δ mutant showed a growth defect on poor carbon sources, including the nonpreferred carbon source galactose (see below) and other poor carbon sources (see Fig. S1E in the supplemental material). Growth in galactose modestly induced the UPRE-lacZ reporter (Fig. 3A) (58). Induction of the UPR by galactose required Ire1p (Fig. 3A). Tunicamycin (Tun) is an inhibitor of N-linked glycosylation (87), which leads to accumulation of misfolded proteins in the ER and activation of the UPR (37, 88, 89). Tunicamycin induced the UPRE-lacZ reporter to higher levels than galactose (Fig. 3A).
FIG 3.
Ire1p regulates cleavage of Msb2p and the filamentous-growth pathway during growth in galactose. (A) UPRE-lacZ activity for the indicated strains and conditions (TUN, 2.5 μg/ml tunicamycin). The experiment was performed in duplicate. Error bars represent the standard deviations between trials. *, P < 0.05; **, P < 0.09. (B) Immunoblot of elutions from ConA columns showing HA-Msb2p migration on low-percentage acrylamide gels (6% SDS-PAGE) under the indicated conditions. (C) Immunoblots of HA-Msb2p migration (top, anti-HA, 6% SDS-PAGE gel), Msb2p cleavage (anti-GFP immunoblot Msb2Pp), P∼Kss1p levels, and total protein levels (anti-Pgk1p) of extracts from cells grown in glucose (GLU, YEPD), galactose (GAL, YEP-GAL), or TUN (YEPD plus 2.5 μg/ml tunicamycin). (D) Immunoblots showing Msb2p cleavage in wild-type cells (WT) and the ire1Δ mutant. The anti-HA antibodies were used to evaluate HA-Msb2p, and GFP antibodies were used to evaluate Msb2Pp-GFP. (E) P∼Kss1p levels in wild-type cells (WT) and the ire1Δ mutant incubated in YEPD (GLU) and YEP-GAL (GAL) medium. Numbers refer to fold differences relative to the loading control determined by assessing band intensity by ImageJ. (F) FRE-lacZ expression in the wild type and ire1Δ and ste12Δ mutants grown in YEP-GAL for 12 h. The experiment was performed in duplicate. Error bars represent the standard deviations between experiments. *, P < 0.05. (G) YPS1-lacZ expression in the wild-type and ire1Δ strains in YEPD (GLU) and YEP-GAL (GAL) medium at 24 h. The experiment was performed in duplicate. Error bars represent the standard deviations between experiments. *, P < 0.05.
Msb2p and other glycoproteins may be underglycosylated during growth in poor carbon sources. HA-Msb2p showed similar migration on SDS-PAGE from cells grown in galactose and cells treated with tunicamycin (Fig. 3B). Samples examined in Fig. 3B were eluted from a concanavalin A (ConA) column that binds oligosaccharides (90). Versions of Msb2p that lacked portions of its extracellular domain (see Fig. S1F in the supplemental material) and other glycoproteins (91) showed a similar pattern (see Fig. S1G), which indicates that many proteins are underglycosylated during growth in a nonpreferred carbon source.
The underglycosylation of Msb2p that occurs in galactose may induce its proteolytic processing and MAPK activation. Cleavage of Msb2p occurred at elevated levels in galactose (Fig. 3C), which corresponded to activation of the filamentous-growth pathway (Fig. 3C) (28). The UPR may regulate cleavage of underglycosylated Msb2p in this setting. Ire1p was required for Msb2p cleavage in galactose (Fig. 3D) and for activation of the filamentous-growth pathway based on the levels of P∼Kss1p (Fig. 3E) and the activity of a transcriptional reporter for the pathway (Fig. 3F) (92). Complete ablation of P∼Kss1p was not seen in the ire1Δ mutant. Quantitation of relative band intensities by ImageJ showed an ∼2.5-fold reduction in P∼Kss1p levels in the ire1Δ mutant across multiple trials. YPS1 expression was induced during growth in galactose in an Ire1p-dependent manner (Fig. 3G). The expression of yapsins and the cellular responses showed some differences depending on whether cells were experiencing a glycosylation defect or carbon source limitation, which indicates (as one might expect) that the two conditions elicit different cellular responses. The above-described results collectively show that the UPR regulates the cleavage of Msb2p and the filamentous-growth pathway during a nutritional response.
Ire1p regulates filamentous growth and biofilm/mat formation.
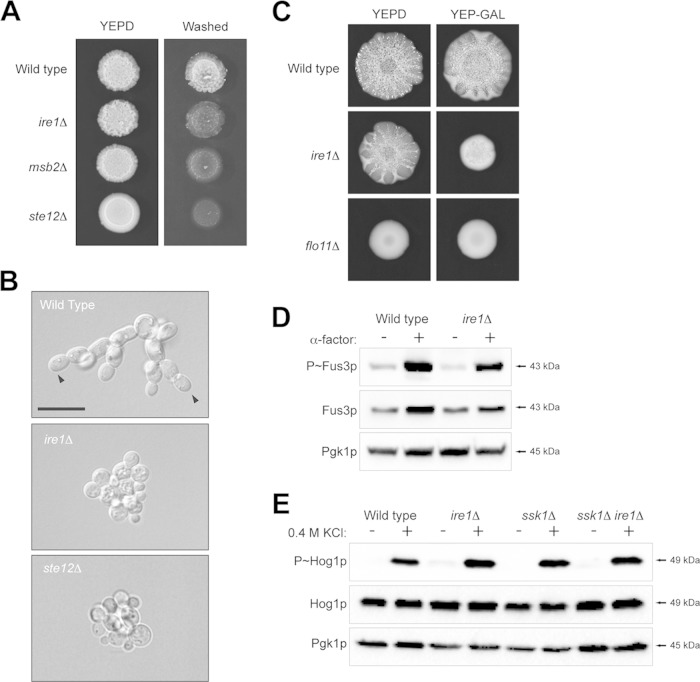
The fact that the UPR regulates the filamentous-growth pathway suggests a role for this pathway in regulating differentiation to the filamentous cell type. Filamentous growth in yeast involves changes in the cell cycle, cell adhesion, and cell polarity (93, 94). To evaluate the role of Ire1p in regulating filamentous growth, the ire1Δ mutant was compared to MAPK pathway mutants (msb2Δ and ste12Δ) in assays used to evaluate the filamentous-growth response. The plate-washing assay measures cell invasion into agar surfaces (13). The ire1Δ mutant was defective for invasive growth by the plate-washing assay (Fig. 4A). The invasive growth defect of the ire1Δ mutant resembled that of the msb2Δ mutant, whose defect was not as severe as that of the ste12Δ mutant, which is completely defective for MAPK signaling. The single-cell invasive growth assay allows for a quantitative measure of the changes in cell length and cell polarity (budding pattern) that accompany the filamentation response (30). The single-cell assay showed that the ire1Δ mutant was defective for cell elongation and distal-pole budding (arrowheads) to the same extent as the ste12Δ mutant (Fig. 4B). These phenotypes are controlled by the filamentous-growth pathway (94). Thus, Ire1p regulates filamentous growth in yeast.
FIG 4.
Role of Ire1p in regulating invasive growth, biofilm/mat formation, and other MAPK pathways that share components with the filamentous-growth pathway. (A) Wild-type, msb2Δ, ire1Δ, and ste12Δ strains were spotted onto YEPD medium. After 48 h, the plate was photographed, washed, and photographed again to reveal invaded cells. (B) Single-cell assay showing the growth after 16 h of strains on synthetic medium lacking glucose. Arrowheads refer to examples of distal-unipolar buds. Bar, 15 μm. (C) Biofilm/mat formation. Wild-type cells and ire1Δ and flo11Δ mutant cells were spotted on YEPD and YEP-GAL media (0.3% agar) for 3 days. (D) Phosphorylation of Fus3p in response to α-factor in wild-type cells and the ire1Δ mutant. (E) Phosphorylation of Hog1p in wild-type cells and the ire1Δ mutant exposed to 0.4 M KCl for 10 min in wild-type cells and an ssk1Δ background.
Yeast and other microbial species form biofilms/mats to facilitate spreading across surfaces (95, 96). Biofilm/mat formation was examined on YEPD and YEP-GAL media. On YEPD, the ire1Δ mutant was modestly defective for the expansion of biofilms/mats based on visual inspection of colonies (Fig. 4C). Biofilm/mat formation in yeast requires the cell adhesion flocculin Flo11p (57). The flo11Δ mutant showed a complete defect in biofilm/mat formation (Fig. 4C). On YEP-GAL, the ire1Δ mutant showed a full defect in biofilm/mat formation (Fig. 4C), which may result (at least in part) from its growth defect under this condition.
Other regulators of the UPR and other QC pathways were also evaluated. Hac1p is the major transcription factor of the UPR that is activated by Ire1p during ER stress (56, 97). The hac1Δ mutant exhibited the same phenotype as the ire1Δ mutant by the plate-washing assay (see Fig. S2A in the supplemental material). The ire1Δ and hac1Δ mutants showed an equivalent defect in filamentous-growth pathway activity as assessed by a growth reporter (see Fig. S2B). Lhs1p is a target of the UPR that functions as an exchange factor for the ER chaperone Hsp70p (98–100). The lhs1Δ mutant showed a defect in invasive growth (see Fig. S2A), although not to the same extent as the ire1Δ and hac1Δ mutants. The ER-associated degradation pathway (ERAD) also regulates the response to protein misfolding in the ER by removing proteins from the ER for degradation in the proteosome (101). A major component of ERAD is Hrd1p (102). The hrd1Δ mutant was not defective for invasive growth (see Fig. S2C), which indicates that ERAD is not a major regulator of filamentous growth.
The filamentous-growth MAPK pathway is composed of proteins that also regulate other MAPK pathways (14, 27, 103). Ire1p was tested for roles in regulating the mating and HOG pathways. The ire1Δ mutant was not defective for mating based on the phosphorylation of the MAPK Fus3p in response to pheromone (Fig. 4D). The HOG pathway is composed of two branches, the Sho1p branch and the Sln1p branch (104, 105). The ire1Δ mutant and the ire1Δ ssk1Δ double mutant (that lacks the Sln1p branch) were not defective for HOG pathway activation in response to osmotic stress based on phosphorylation of the MAPK Hog1p (Fig. 4E). Therefore, the UPR does not play a major role in regulating the mating pathway. Activation of the HOG pathway does occur in response to galactose, which also requires Ire1p (58). The results shown here bring to light a new role for the UPR in regulating fungal behavioral responses through the regulation of a differentiation (ERK-type) MAPK pathway.
Role of the filamentous-growth pathway in regulating the response to low nutrient availability.
We have previously shown that the filamentous-growth pathway contributes to the viability of the pmi40-101 mutant (32). The filamentous-growth pathway was tested for a broader role in the response to low nutrient levels. The filamentous-growth pathway contributed to viability of the pmt4Δ mutant, where the pmt4Δ ste12Δ double mutant was more defective for growth than the single mutants (pmt4Δ and ste12Δ mutants) alone (Fig. 5A). This is indicative of a genetic interaction between the two pathways (106). The filamentous-growth pathway also contributed to viability of the ire1Δ mutant (Fig. 5B). Thus, the filamentous-growth pathway and the UPR share a function that is important for survival on poor-carbon-source media.
FIG 5.
Roles of UPR in regulating polarity and growth in poor carbon sources. (A) Serial dilutions were spotted onto synthetic medium containing glucose or galactose. The ste12Δ mutant showed a modest growth defect on synthetic media with galactose. (B) Serial dilutions were spotted onto YEPD and YEP-GAL media. (C) UPRE-lacZ activity of the indicated strains in YEPD (GLU) and YEP-GAL (GAL). The experiment was performed in duplicate; error bars show standard deviations between strains. *, P < 0.05. (D) UPRE-lacZ activity was performed as described for panel C. The arrow refers to Msb2p overexpressed by the pGAL1 promoter. (E) Rhodamine-phalloidin staining of wild-type cells and the indicated mutants grown to saturation in YEPD medium. Tun, tunicamycin. Bar, 5 μm.
What role might the filamentous-growth pathway play? To begin to address this question, the activity of the UPR was compared in cells lacking or overproducing filamentous-growth pathway components. Cells lacking an intact filamentous-growth pathway showed reduced UPRE-lacZ activity (Fig. 5C, ste12Δ). Cells overexpressing Msb2p, which activates the filamentous-growth pathway (15), showed an increase in UPRE-lacZ activity (Fig. 5D). Both of these changes were dependent on Ire1p (Fig. 5C and D). These results demonstrate that the filamentous-growth pathway promotes the activity of the UPR during growth in galactose. Moreover, the data suggest that positive feedback occurs between the two pathways: the UPR regulates the activity of the filamentous-growth MAPK pathway, and the MAPK pathway acts upstream or in parallel with the UPR by regulating UPR activity. The filamentous-growth pathway was not found to show responses to every UPR stressor (e.g., DTT [H. Adhikari and P. J. Cullen, unpublished results]), so its role in promoting the UPR response may be specific for the conditions tested here.
We also found that treatment of cells grown to saturation with Tun caused cells to be longer in appearance (Fig. 5E). Actin staining with the dye rhodamine-phalloidin confirmed that cells exhibited enhanced polarized growth under this condition (Fig. 5E, arrows refer to the polarized actin cytoskeleton). The filamentous-growth pathway regulates cell polarity (94) and the cell cycle (107), which contribute to changes in cell length. The elongated morphology induced by Tun was dependent on Ire1p (Fig. 5E, ire1Δ) and the filamentous-growth pathway (Fig. 5E, ste11Δ). The filamentous-growth pathway regulates cell polarity through the formin Bni1p (108, 109). Bni1p was required for the elongated morphology induced by Tun (Fig. 5E). The cyclin Cln1p is a transcriptional target of the filamentous-growth pathway (107) and also was required (Fig. 5E). Thus, in response to an established inducer of the UPR, the filamentous-growth pathway controls a morphogenetic response that involves changes in the cell cycle and cell polarity.
Msb2p can be proteolytically processed in the secretory pathway.
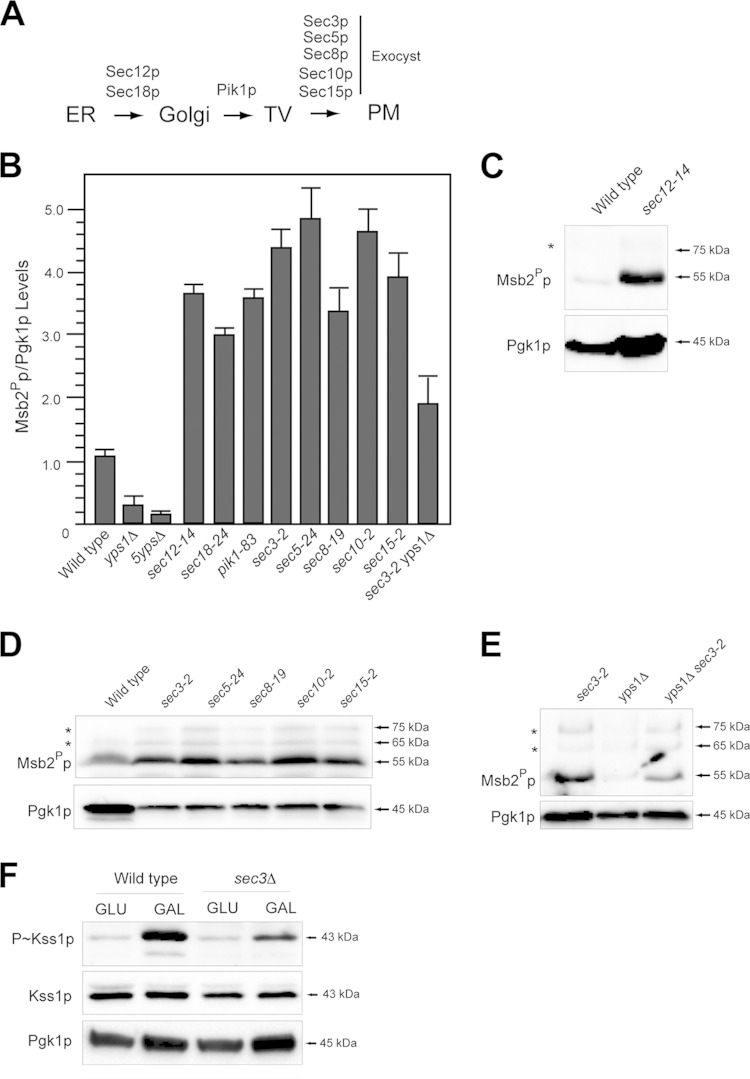
Protein glycosylation occurs in the ER and Golgi compartments (37). The UPR functions in the ER (60). Underglycosylated Msb2p may be proteolytically processed in the endomembrane system. To test this possibility, protein trafficking was arrested at specific points along the secretory pathway with conditional (temperature-sensitive; ts) mutants (Fig. 6A), and cleavage of Msb2p was evaluated. Cleavage of Msb2p occurred in most trafficking mutants tested. These included the sec12-14 mutant (Fig. 6B and C), the PI kinase mutant pik1-83 (Fig. 6B) (47, 110–112), and exocytosis mutants (Fig. 6B and D) (113–117). In these mutants, Msb2p was cleaved more efficiently than in wild-type cells, possibly due to cotrapping Yps1p and Msb2p in the secretory pathway or because the turnover of Msb2Pp was inhibited (see below). Yps1p functions at the PM (118) and has been reported to function in the Golgi apparatus (34, 80, 119). Proteolytic processing of Msb2p in the sec3-2 mutant required Yps1p (Fig. 6B and E). Residual cleavage of Msb2p in the sec3-2 yps1Δ double mutant may be mediated by other yapsins (28, 33).
FIG 6.
Proteolytic processing of Msb2p in protein-trafficking mutants. (A) Proteins that regulate trafficking in the secretory pathway. Mutants were examined at the nonpermissive temperature (4 h of growth in YEPD at 37°C) to arrest protein trafficking at different points along the secretory pathway. TV, transit vesicles. (B) Quantitation of Msb2Pp adjusted to total protein levels for the indicated mutants. The Msb2Pp/Pgk1p ratio for wild-type cells was set to 1 and compared to those of other mutants, assessed by band intensity by immunoblot analysis and analyzed by ImageJ. Intensities varied less than 10% between trials. (C) Msb2p cleavage in the sec12-14 mutant. Asterisks refer to minor cleavage products for panels C to E. (D) Msb2p cleavage in exocyst mutants. (E) Msb2p cleavage in sec3-2 yps1Δ mutant alongside control strains. (F) P∼Kss1p levels in wild-type cells and the sec3Δ mutant incubated for 4 h in YEPD (GLU) or YEP-GAL (GAL).
The fact that Msb2p can be processed in the secretory pathway suggested the possibility that it activates the filamentous-growth pathway from internal compartments. The sec3Δ mutant showed reduced activation of the filamentous-growth pathway by galactose (Fig. 6F). The sec3Δ and other exocyst mutants showed a full growth defect at 37°C (see Fig. S3A in the supplemental material). In the exocyst mutant, Msb2p accumulated in a punctate pattern (see Fig. S3B and C) and was not shed (see Fig. S3D and E). Thus, Msb2p must be delivered to the PM for full activation of the filamentous-growth pathway.
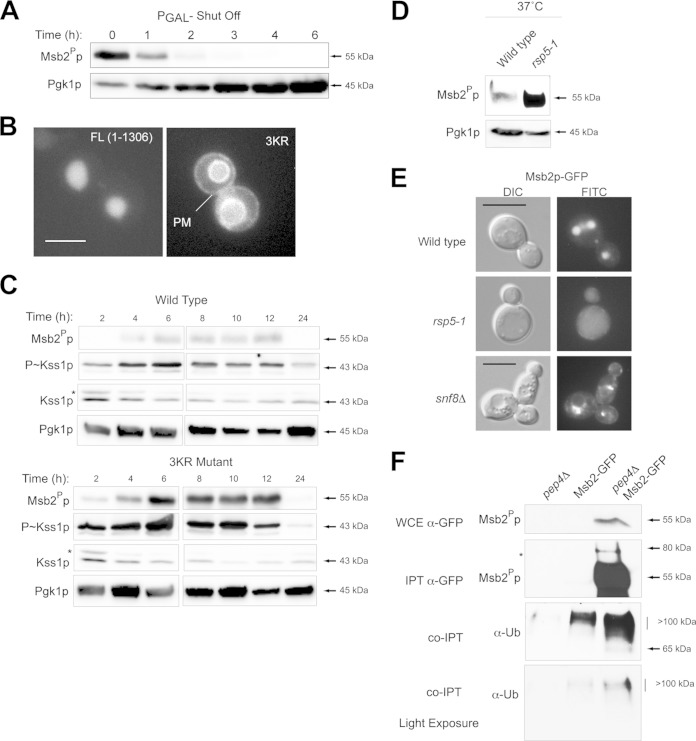
Fate of proteolytically processed Msb2p.
What is the fate of proteolytically processed Msb2Pp? The proteolytically processed form of Msb2p is required for filamentous-growth MAPK pathway activity. Promoter shutoff experiments showed that Msb2Pp was turned over in ∼20 min (Fig. 7A). This contrasts with the stability of the extracellular domain, which has a half-life of ∼7 days (78). To define how Msb2Pp is turned over, deletion analysis of the cytoplasmic domain was performed, which identified separate regions that control MAPK signaling and turnover (see Fig. S4 in the supplemental material). The turnover domain of Msb2p contained three lysines, which are commonly modified by ubiquitin attachment to direct proteins for turnover (120, 121). Site-directed mutagenesis was used to change the lysines to arginines to construct Msb2pK1223R K1239R K1245R or Msb2p3KR. Compared to wild-type Msb2p-GFP (full length [FL]; aa 1 to 1306), which is found mainly in the vacuole, Msb2p3KR-GFP was at the PM (Fig. 7B) and the vacuolar membrane. Msb2p3KR-GFP was also present at higher levels in the cell (Fig. 7C, Msb2Pp) and caused an increase in filamentous-growth pathway activity (Fig. 7C, P∼Kss1p). These results show that Msb2p is delivered to the PM. Turnover of Msb2p from the PM attenuates the filamentous-growth pathway.
FIG 7.
Roles for Rsp5p and ESCRT in regulating the turnover of Msb2p. (A) Promoter shutoff showing Msb2Pp levels at the indicated time points. (B) Localization of full length (FL; aa 1 to 1306) and Msb23KRp-GFP (3KR). Bar, 5 μm. (C) Role of lysines in the turnover domain of Msb2p in impacting the stability of the protein and MAPK activity. (Top) Msb2p-GFP and Msb23KRp-GFP levels over a culture-growth cycle at the indicated time points. (Middle) P∼Kss1p activity. (Bottom) Pgk1p levels. Proteins also were examined side by side on separate blots to directly compare protein levels. (D) Msb2Pp-GFP levels in wild-type cells and in mutants harboring temperature-sensitive rsp5 alleles. Strains were grown at the nonpermissive temperature (37°C) and evaluated by immunoblot analysis. (E) The localization of Msb2p-GFP in wild-type cells, the rsp5-1 mutant (at 37°C), and the snf8Δ (ESCRT) mutant. (F) Msb2p-GFP was immunoprecipitated (IPT) from wild-type cells and the pep4Δ mutant, and extracts were probed using anti-GFP and anti-UB antibodies. Stabilization of Msb2p in the pep4Δ mutant resulted in higher levels of the ubiquitin-modified forms of the protein. Bottom panel, lighter exposure. Ub, ubiquitin; WCE, whole-cell extract.
Rsp5p is an essential HECT-type E3 ubiquitin ligase that regulates the turnover of most, if not all, PM proteins in yeast (122). Msb2Pp was stabilized in cells containing temperature-sensitive alleles of RSP5 (Fig. 7D). In the rsp5-1 mutant, Msb2p was not delivered to the vacuole (Fig. 7E), which indicates that, like for many proteins (123, 124), Rsp5p is required for the sorting and turnover of endocytosed Msb2p. Immunoprecipitation of Msb2p-GFP pulled down ubiquitin-modified forms of the protein (Fig. 7F). The ESCRT complex is responsible for trafficking ubiquitinated cargoes from the PM to the endosome (125, 126). Msb2p accumulated in the multivesicular body (MVB; or prevacuolar compartment) in ESCRT mutants (Fig. 7E). Thus, Rsp5p and ESCRT regulate the turnover of Msb2Pp.
DISCUSSION
A prevailing view of mucin-type receptors is that they function at the PM to sense an (as yet poorly defined) external cue. Here, we describe an internal activation mechanism for signaling mucins. We show that proteolytic processing of a signaling mucin in the secretory pathway is controlled by the UPR, a major QC pathway that operates in the ER. This new view of signaling mucin regulation inversely links the glycosylation of mucins to their activity. Our findings also bring to light a new role for the UPR in regulating an ERK-type differentiation pathway. Furthermore, we provide evidence for an ERK-type pathway in contributing to the response to ER stress in protein glycosylation mutants and during growth on poor carbon sources. Given that signaling mucins, ERK-type MAPK pathways, and the UPR are evolutionarily conserved throughout eukaryotes, these findings may broadly apply to many systems.
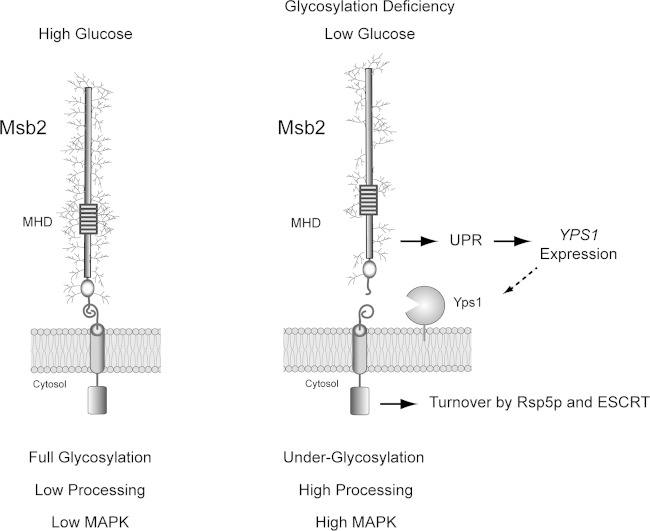
We specifically show that underglycosylation of the yeast signaling mucin Msb2p induces its proteolytic processing by a mechanism that involves the UPR (Fig. 8). In the model, we propose that under high-nutrient conditions, Msb2p is highly glycosylated and poorly cleaved. As a result, the activity of the filamentous-growth MAPK pathway is low (Fig. 8). Two established triggers of the filamentous-growth pathway (carbon source depletion and glycosylation deficiency) lead to underglycosylation of Msb2p, which is recognized by the UPR regulator Ire1p. Ire1p controls the expression of Yps1p, which is the major protease that cleaves Msb2p (Fig. 8). In this way, Msb2p may transmit signals about glycosylation status and nutrition by the extent of glycosylation of its extracellular domain.
FIG 8.
Role of the UPR in regulating Msb2p cleavage and activation of the filamentous-growth pathway. (Left) During growth in glucose-replete conditions, Msb2p is fully glycosylated. As a result, Msb2p is not efficiently processed and MAPK activity is low. (Right) During growth in poor carbon sources (like galactose) or in cells experiencing a protein glycosylation deficiency, Msb2p becomes underglycosylated. Underglycosylated Msb2p is recognized by Ire1p, a major regulator of the UPR. Ire1p regulates expression of YPS1 (dashed arrow), resulting in elevated cleavage of Msb2p and activation of the filamentous-growth pathway. The proteolytically processed form of Msb2p is turned over by Rsp5p and ESCRT to attenuate the filamentous-growth pathway.
There are several possible ways in which glycosylation may be tied to its processing. The most direct possibility is that elevated levels of Yps1p lead to elevated Msb2p processing. A related possibility is that the loss of specific glycosyl modifications in Msb2p's cleavage domain increase accessibility by Yps1p. We do not favor this possibility, because altered glycosylation in regions of the protein far from the cleavage domain potently activate the filamentous-growth pathway. Another related possibility is that underglycosylated Msb2p is trapped in the secretory pathway, which increases Yps1p-dependent proteolysis. Msb2p functions with other PM proteins, including Sho1p and Opy2p. Opy2p also is glycosylated, but the glycosylation sites of Opy2p are not thought to play a role in the regulation of the filamentous-growth pathway (17).
The UPR previously has been tied to the regulation of filamentous growth in a different way. Schroder and colleagues (127, 128) showed that the UPR plays an inhibitory role in filamentous growth in cells responding to nitrogen limitation. Thus, the UPR may play different roles in regulating filamentous growth depending on the specific nutrient pool that is limited.
An interesting issue raised by the study is whether other glycoproteins are regulated by QC pathways in the endomembrane system. There are well-established connections between stress response and protein- and lipid-linked oligosaccharide metabolism (71, 129). Stress- and nutrient-dependent changes in oligosaccharide levels can trigger the UPR (83–86). Given that highly specific changes in glycosylation regulate diverse proteins like Notch (130), dystroglycan (131–133), amyloid precursor protein (134, 135), and MUC1 (136), one can speculate that UPR regulates these proteins under some conditions. For example, in cancer cells, the global glycosylation of proteins is impacted (137, 138), which includes MUC1, a major activator of the proliferative RAS-MEK-ERK MAPK pathway (9, 139, 140). Aberrantly glycosylated MUC1 and other proteins may be recognized by QC pathways in this setting, which may impact signaling pathway outputs. It will be interesting to explore the roles of the UPR and other QC pathways in regulating the extent of glycoprotein receptor function.
Supplementary Material
ACKNOWLEDGMENTS
Thanks go to Scott Emr (Cornell University, Ithaca, NY), Peter Novick (Yale Univeristy, New Haven, CT), Jeremy Thorner (UC Berkeley), Chris Burd (Yale University, New Haven, CT), Damian Krysan (University of Rochester), Alan Tarkakoff (Case Western Reserve University, Cleveland, OH), Steve Free (SUNY—Buffalo), Wei Guo (UPENN), Jason MacGurn (Vanderbilt), and Michael Yu (SUNY—Buffalo) for providing strains, reagents, and/or suggestions. Thanks go to Seth Nickerson, Geno Stolfa, and Brandon Zawacki for help with experiments.
P.J.C. is supported by a grant from the U.S. Public Health Service (GM098629).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01501-14.
REFERENCES
- 1.Kufe DW. 2009. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian E, Ten Hagen KG. 2009. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J 26:325–334. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bafna S, Kaur S, Batra SK. 2010. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen PJ. 2011. Post-translational regulation of signaling mucins. Curr Opin Struct Biol 21:590–596. doi: 10.1016/j.sbi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharbanda A, Rajabi H, Jin C, Tchaicha JH, Kikuchi E, Wong KK, Kufe DW. 2014. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res 20:5423–5434. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kufe DW. 2013. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene 32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Caffrey TC, Steele MM, Mohr A, Singh PK, Radhakrishnan P, Kelly DL, Wen Y, Hollingsworth MA. 2014. MUC1 regulates cyclin D1 gene expression through p120 catenin and beta-catenin. Oncogenesis 3:e107. doi: 10.1038/oncsis.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Wells A, Padilla MT, Kato K, Kim KC, Lin Y. 2014. A signaling pathway consisting of miR-551b, catalase and MUC1 contributes to acquired apoptosis resistance and chemoresistance. Carcinogenesis 35:2457–2466. doi: 10.1093/carcin/bgu159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Elssen CH, Frings PW, Bot FJ, Van de Vijver KK, Huls MB, Meek B, Hupperets P, Germeraad WT, Bos GM. 2010. Expression of aberrantly glycosylated Mucin-1 in ovarian cancer. Histopathology 57:597–606. doi: 10.1111/j.1365-2559.2010.03667.x. [DOI] [PubMed] [Google Scholar]
- 10.Blixt O, Bueti D, Burford B, Allen D, Julien S, Hollingsworth M, Gammerman A, Fentiman I, Taylor-Papadimitriou J, Burchell JM. 2011. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res 13:R25. doi: 10.1186/bcr2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitler BG, Menzl I, Huerta CL, Sands B, Knowlton W, Chang A, Schroeder JA. 2009. Intracellular MUC1 peptides inhibit cancer progression. Clin Cancer Res 15:100–109. doi: 10.1158/1078-0432.CCR-08-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Styles CA, Fink GR. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 13.Roberts RL, Fink GR. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 14.Bardwell L. 2006. Mechanisms of MAPK signalling specificity. Biochem Soc Trans 34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen PJ, Sabbagh W Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous-growth pathway in yeast. Genes Dev 18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lien EC, Nagiec MJ, Dohlman HG. 2013. Proper protein glycosylation promotes mitogen-activated protein kinase signal fidelity. Biochemistry 52:115–124. doi: 10.1021/bi3009483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang HY, Tatebayashi K, Yamamoto K, Saito H. 2009. Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. EMBO J 28:1380–1391. doi: 10.1038/emboj.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentzsch M, Tanner W. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J 15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 19.Bi E, Park HO. 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Rourke SM, Herskowitz I. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunanithi S, Cullen PJ. 2012. The filamentous growth MAPK pathway responds to glucose starvation through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics 192:869–887. doi: 10.1534/genetics.112.142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, Tatebayashi K, Tanaka K, Saito H. 2010. Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol Cell 40:87–98. doi: 10.1016/j.molcel.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. 2006. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J 25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truckses DM, Bloomekatz JE, Thorner J. 2006. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol Cell Biol 26:912–928. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito H. 2010. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol 13:677–683. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitoniak A, Birkaya B, Dionne HM, Vadaie N, Cullen PJ. 2009. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol Biol Cell 20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen PJ, Sprague GF Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci U S A 97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee BN, Elion EA. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A 96:12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155:1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell 4:1364–1374. doi: 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egel-Mitani M, Flygenring HP, Hansen MT. 1990. A novel aspartyl protease allowing KEX2-independent MF alpha propheromone processing in yeast. Yeast 6:127–137. doi: 10.1002/yea.320060206. [DOI] [PubMed] [Google Scholar]
- 35.Komano H, Fuller RS. 1995. Shared functions in vivo of a glycosyl-phosphatidylinositol-linked aspartyl protease, Mkc7, and the proprotein processing protease Kex2 in yeast. Proc Natl Acad Sci U S A 92:10752–10756. doi: 10.1073/pnas.92.23.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen V, Cawley NX, Brandt J, Egel-Mitani M, Loh YP. 1999. Identification and characterization of Saccharomyces cerevisiae yapsin 3, a new member of the yapsin family of aspartic proteases encoded by the YPS3 gene. Biochem J 339(Part 2):407–411. doi: 10.1042/0264-6021:3390407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox JS, Shamu CE, Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206. doi: 10.1016/0092-8674(93)90648-A. [DOI] [PubMed] [Google Scholar]
- 38.Ng DT, Spear ED, Walter P. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol 150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox JS, Chapman RE, Walter P. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cullen PJ, Sprague GF Jr. 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell 13:2990–3004. doi: 10.1091/mbc.E02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karunanithi S, Vadaie N, Chavel CA, Birkaya B, Joshi J, Grell L, Cullen PJ. 2010. Shedding of the Mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr Biol 20:1389–1395. doi: 10.1016/j.cub.2010.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finger FP, Novick P. 2000. Synthetic interactions of the post-Golgi sec mutations of Saccharomyces cerevisiae. Genetics 156:943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavel CA, Caccamise LM, Li B, Cullen PJ. 2014. Global regulation of a differentiation MAPK pathway in yeast. Genetics 198:1309–1328. doi: 10.1534/genetics.114.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkaya B, Maddi A, Joshi J, Free SJ, Cullen PJ. 2009. Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth. Eukaryot Cell 8:1118–1133. doi: 10.1128/EC.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, Burd CG. 2008. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol Biol Cell 19:4694–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Audhya A, Foti M, Emr SD. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell 11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaiser CA, Schekman R. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61:723–733. doi: 10.1016/0092-8674(90)90483-U. [DOI] [PubMed] [Google Scholar]
- 49.Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 51.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldstein AL, Pan X, McCusker JH. 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15:507–511. doi:. [DOI] [PubMed] [Google Scholar]
- 53.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- 54.Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 55.Gillen KM, Pausch M, Dohlman HG. 1998. N-terminal domain of Gpa1 (G protein alpha) subunit) is sufficient for plasma membrane targeting in yeast Saccharomyces cerevisiae. J Cell Sci 111(Part 21):3235–3244. [DOI] [PubMed] [Google Scholar]
- 56.Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds TB, Fink GR. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 58.Adhikari H, Cullen PJ. 2014. Metabolic respiration induces AMPK- and Ire1p-dependent activation of the p38-type HOG MAPK pathway. PLoS Genet 10:e1004734. doi: 10.1371/journal.pgen.1004734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee MJ, Dohlman HG. 2008. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol 18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gardner BM, Walter P. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. 2010. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Julenius K, Molgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 63.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 65.Laganowsky A, Zhao M, Soriaga AB, Sawaya MR, Cascio D, Yeates TO. 2011. An approach to crystallizing proteins by metal-mediated synthetic symmetrization. Protein Sci 20:1876–1890. doi: 10.1002/pro.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodseth LE, Martineau P, Duplay P, Hofnung M, Quiocho FA. 1990. Crystallization of genetically engineered active maltose-binding proteins, including an immunogenic viral epitope insertion. J Mol Biol 213:607–611. doi: 10.1016/S0022-2836(05)80246-3. [DOI] [PubMed] [Google Scholar]
- 67.Ecker M, Mrsa V, Hagen I, Deutzmann R, Strahl S, Tanner W. 2003. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep 4:628–632. doi: 10.1038/sj.embor.embor864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiro RG. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R–56R. doi: 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- 69.Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. 2010. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet 6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Payton MA, Rheinnecker M, Klig LS, DeTiani M, Bowden E. 1991. A novel Saccharomyces cerevisiae secretory mutant possesses a thermolabile phosphomannose isomerase. J Bacteriol 173:2006–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lehrman MA. 2006. Stimulation of N-linked glycosylation and lipid-linked oligosaccharide synthesis by stress responses in metazoan cells. Crit Rev Biochem Mol Biol 41:51–75. doi: 10.1080/10409230500542575. [DOI] [PubMed] [Google Scholar]
- 72.Fabre E, Hurtaux T, Fradin C. 2014. Mannosylation of fungal glycoconjugates in the Golgi apparatus. Curr Opin Microbiol 20:103–110. doi: 10.1016/j.mib.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Smith DJ, Proudfoot A, Friedli L, Klig LS, Paravicini G, Payton MA. 1992. PMI40, an intron-containing gene required for early steps in yeast mannosylation. Mol Cell Biol 12:2924–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Courchesne WE, Kunisawa R, Thorner J. 1989. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell 58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 75.Ma D, Cook JG, Thorner J. 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell 6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cook JG, Bardwell L, Thorner J. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 77.Madhani HD, Styles CA, Fink GR. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673–684. doi: 10.1016/S0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 78.Meem MH, Cullen PJ. 2012. The impact of protein glycosylation on Flo11-dependent adherence in Saccharomyces cerevisiae. FEMS Yeast Res 12:809–818. doi: 10.1111/j.1567-1364.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P. 2011. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol 193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller KA, DiDone L, Krysan DJ. 2010. Extracellular secretion of overexpressed glycosylphosphatidylinositol-linked cell wall protein Utr2/Crh2p as a novel protein quality control mechanism in Saccharomyces cerevisiae. Eukaryot Cell 9:1669–1679. doi: 10.1128/EC.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. 2006. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells 11:59–69. doi: 10.1111/j.1365-2443.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 82.Silverman HS, Sutton-Smith M, McDermott K, Heal P, Leir SH, Morris HR, Hollingsworth MA, Dell A, Harris A. 2003. The contribution of tandem repeat number to the O-glycosylation of mucins. Glycobiology 13:265–277. doi: 10.1093/glycob/cwg028. [DOI] [PubMed] [Google Scholar]
- 83.Doerrler WT, Lehrman MA. 1999. Regulation of the dolichol pathway in human fibroblasts by the endoplasmic reticulum unfolded protein response. Proc Natl Acad Sci U S A 96:13050–13055. doi: 10.1073/pnas.96.23.13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peluso RW, Lamb RA, Choppin PW. 1978. Infection with paramyxoviruses stimulates synthesis of cellular polypeptides that are also stimulated in cells transformed by Rous sarcoma virus or deprived of glucose. Proc Natl Acad Sci U S A 75:6120–6124. doi: 10.1073/pnas.75.12.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pouyssegur J, Shiu RP, Pastan I. 1977. Induction of two transformation-sensitive membrane polypeptides in normal fibroblasts by a block in glycoprotein synthesis or glucose deprivation. Cell 11:941–947. doi: 10.1016/0092-8674(77)90305-1. [DOI] [PubMed] [Google Scholar]
- 86.Shiu RP, Pouyssegur J, Pastan I. 1977. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci U S A 74:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heifetz A, Keenan RW, Elbein AD. 1979. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry 18:2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 88.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 89.Patil C, Walter P. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13:349–355. doi: 10.1016/S0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 90.Cummings RD, Etzler ME. 2009. Antibodies and lectins in glycan analysis. In Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (ed), Essentials of glycobiology, 2nd ed. CSHL Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 91.Kung LA, Tao SC, Qian J, Smith MG, Snyder M, Zhu H. 2009. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol Syst Biol 5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Madhani HD, Fink GR. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 93.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090. doi: 10.1016/0092-8674(92)90079-R. [DOI] [PubMed] [Google Scholar]
- 94.Cullen PJ, Sprague GF Jr. 2012. The regulation of filamentous growth in yeast. Genetics 190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Verstrepen KJ, Klis FM. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol 60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 96.Nobile CJ, Mitchell AP. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol 8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 97.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. 1996. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 98.Hale SJ, Lovell SC, de Keyzer J, Stirling CJ. 2010. Interactions between Kar2p and its nucleotide exchange factors Sil1p and Lhs1p are mechanistically distinct. J Biol Chem 285:21600–21606. doi: 10.1074/jbc.M110.111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ. 2009. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem 284:31564–31571. doi: 10.1074/jbc.M109.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tyson JR, Stirling CJ. 2000. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J 19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD. 1988. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell 54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 102.Kanehara K, Xie W, Ng DT. 2010. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol 188:707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schwartz MA, Madhani HD. 2004. Principles of map kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 104.Maeda T, Wurgler-Murphy SM, Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 105.Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 106.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, Vander Sluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. 2010. The genetic landscape of a cell. Science 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Madhani HD, Galitski T, Lander ES, Fink GR. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci U S A 96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 109.Goehring AS, Mitchell DA, Tong AH, Keniry ME, Boone C, Sprague GF Jr. 2003. Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol Biol Cell 14:1501–1516. doi: 10.1091/mbc.E02-06-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Flanagan CA, Schnieders EA, Emerick AW, Kunisawa R, Admon A, Thorner J. 1993. Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262:1444–1448. doi: 10.1126/science.8248783. [DOI] [PubMed] [Google Scholar]
- 111.Walch-Solimena C, Novick P. 1999. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol 1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 112.Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. 1999. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem 274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- 113.TerBush DR, Maurice T, Roth D, Novick P. 1996. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 114.Bowser R, Muller H, Govindan B, Novick P. 1992. Sec8p and Sec15p are components of a plasma membrane-associated 19.5S particle that may function downstream of Sec4p to control exocytosis. J Cell Biol 118:1041–1056. doi: 10.1083/jcb.118.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.TerBush DR, Novick P. 1995. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol 130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Julius D, Schekman R, Thorner J. 1984. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell 36:309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- 117.Novick P, Ferro S, Schekman R. 1981. Order of events in the yeast secretory pathway. Cell 25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 118.Gagnon-Arsenault I, Parise L, Tremblay J, Bourbonnais Y. 2008. Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol Microbiol 69:982–993. doi: 10.1111/j.1365-2958.2008.06339.x. [DOI] [PubMed] [Google Scholar]
- 119.Sievi E, Suntio T, Makarow M. 2001. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic 2:896–907. doi: 10.1034/j.1600-0854.2001.21205.x. [DOI] [PubMed] [Google Scholar]
- 120.Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 121.Hicke L, Dunn R. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 122.Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. 2004. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell 15:468–480. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.MacDonald C, Stringer DK, Piper RC. 2012. Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic 13:586–598. doi: 10.1111/j.1600-0854.2011.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Risinger AL, Kaiser CA. 2008. Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking. Mol Biol Cell 19:2962–2972. doi: 10.1091/mbc.E07-06-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hurley JH, Emr SD. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct 35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Teis D, Saksena S, Emr SD. 2009. SnapShot: the ESCRT machinery. Cell 137:182–182.e1. doi: 10.1016/j.cell.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 127.Schroder M, Chang JS, Kaufman RJ. 2000. The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Genes Dev 14:2962–2975. doi: 10.1101/gad.852300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schroder M, Clark R, Liu CY, Kaufman RJ. 2004. The unfolded protein response represses differentiation through the RPD3-SIN3 histone deacetylase. EMBO J 23:2281–2292. doi: 10.1038/sj.emboj.7600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao N, Shang J, Huynh D, Manthati VL, Arias C, Harding HP, Kaufman RJ, Mohr I, Ron D, Falck JR, Lehrman MA. 2011. Mannose-6-phosphate regulates destruction of lipid-linked oligosaccharides. Mol Biol Cell 22:2994–3009. doi: 10.1091/mbc.E11-04-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. 2000. Fringe is a glycosyltransferase that modifies Notch. Nature 406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 131.Takeuchi H, Haltiwanger RS. 2014. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun 453:235–242. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wells L. 2013. The o-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J Biol Chem 288:6930–6935. doi: 10.1074/jbc.R112.438978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281–291. doi: 10.1016/S0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 134.Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. 1997. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc Natl Acad Sci U S A 94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Skovronsky DM, Moore DB, Milla ME, Doms RW, Lee VM. 2000. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J Biol Chem 275:2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- 136.Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. 2005. The MUC1 SEA module is a self-cleaving domain. J Biol Chem 280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 137.Bones J, Byrne JC, O'Donoghue N, McManus C, Scaife C, Boissin H, Nastase A, Rudd PM. 2011. Glycomic and glycoproteomic analysis of serum from patients with stomach cancer reveals potential markers arising from host defense response mechanisms. J Proteome Res 10:1246–1265. doi: 10.1021/pr101036b. [DOI] [PubMed] [Google Scholar]
- 138.Satomaa T, Heiskanen A, Leonardsson I, Angstrom J, Olonen A, Blomqvist M, Salovuori N, Haglund C, Teneberg S, Natunen J, Carpen O, Saarinen J. 2009. Analysis of the human cancer glycome identifies a novel group of tumor-associated N-acetylglucosamine glycan antigens. Cancer Res 69:5811–5819. doi: 10.1158/0008-5472.CAN-08-0289. [DOI] [PubMed] [Google Scholar]
- 139.Saeland E, Belo AI, Mongera S, van Die I, Meijer GA, van Kooyk Y. 2012. Differential glycosylation of MUC1 and CEACAM5 between normal mucosa and tumour tissue of colon cancer patients. Int J Cancer 131:117–128. doi: 10.1002/ijc.26354. [DOI] [PubMed] [Google Scholar]
- 140.Xu Y, Zhang L, Hu G. 2009. Potential application of alternatively glycosylated serum MUC1 and MUC5AC in gastric cancer diagnosis. Biologicals 37:18–25. doi: 10.1016/j.biologicals.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 141.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.