Abstract
In biological processes, the balance between positive and negative inputs is critical for an effective physiological response and to prevent disease. A case in point is the germinal center (GC) reaction, wherein high mutational and proliferation rates are accompanied by an obligatory suppression of the DNA repair machinery. Understandably, when the GC reaction goes awry, loss of immune cells or lymphoid cancer ensues. Here, we detail the functional interactions that make microRNA 155 (miR-155) a key part of this process. Upon antigen exposure, miR-155−/− mature B cells displayed significantly higher double-strand DNA break (DSB) accumulation and p53 activation than their miR-155+/+ counterparts. Using B cell-specific knockdown strategies, we confirmed the role of the miR-155 target Aicda (activation-induced cytidine deaminase) in this process and, in combination with a gain-of-function model, unveiled a previously unappreciated role for Socs1 in directly modulating p53 activity and the DNA damage response in B lymphocytes. Thus, miR-155 controls the outcome of the GC reaction by modulating its initiation (Aicda) and termination (Socs1/p53 response), suggesting a mechanism to explain the quantitative defect in germinal center B cells found in mice lacking or overexpressing this miRNA.
INTRODUCTION
Somatic hypermutation (SHM) and class switch recombination (CSR) of the immunoglobulin genes are critical steps for the development of fully functional mature B cells. Several components of the cellular machinery that promotes SHM and CSR have been identified. Activation-induced cytidine deaminase (Aicda), an enzyme that deaminates cytosine to produce uracil in DNA, is thought to initiate and be essential for both SHM and CSR (1). Activation of uracil DNA glycosylase (UNG), ATM, histone H2AX, p53 binding protein 1 (53BP1), and the nonhomologous end joining protein Ku70/80, among others, also plays important roles in these processes (1, 2).
The physiological SHM and CSR involve DNA mutagenesis and double-strand DNA breaks (DSB). Thus, the cellular response to these injuries must be fine-tuned so as to neither excessively engage the repair checkpoints nor compromise the integrity of the rest of the genome (3). Transient transcription repression of multiple DNA repair genes by BCL6 and high-fidelity repair of nonimmunoglobulin genes account, at least in part, for a successful germinal center (GC) response (3–7). Furthermore, timely engagement of the p53 pathway protects against AID-dependent aberrant DNA damage and chromosomal translocations (8). However, less is known about the termination of these activities, which is a critical step to prevent loss of normal B lymphocytes.
MicroRNAs (miRNAs) are non-protein-coding small RNAs that regulate a vast array of physiological functions. miRNAs subtly downmodulate the expression of multiple proteins, thus functioning primarily as rheostats that match the cell needs seamlessly but effectively (9, 10). This unique property suggests that miRNAs may contribute to the control of SHM and CSR reactions. microRNA 155 (miR-155) plays an extensive role in immune cell biology; miR-155 knockout (KO) mice display a defective mature B cell development characterized by a decreased number of GC B cells, whereas an Eμ-miR-155 transgenic mouse model develops an oligoclonal proliferation which evolves to B cell lymphoma (11, 12). These observations suggest that miR-155 may regulate the B cell response to physiological DNA damage, i.e., the control of SHM and CSR, and mechanistically explain the phenotypes observed in these loss- and gain-of-function animal models. The discovery that miR-155 targets Aicda supports the notion that this miRNA controls the initiation of SHM and CSR (13, 14). However, in spite of these advancements, it remains mechanistically unexplained why following an antigen challenge, mice lacking miR-155 display an abnormally low number of GC B cells. We postulate that an exacerbated response to DNA damage accounts for this observation.
Here, we demonstrate that miR-155 plays a central role in modulating the extent of DSB and the amplitude of p53 activation and the DNA damage response (DDR) associated with the GC reaction; we used small interfering RNA (siRNA) approaches in untransformed mature B cells to link this hitherto-unreported observation to the known miR-155 targets Aicda and Socs1. Following immunization or after in vitro stimulation with lipopolysaccharide (LPS) and interleukin-4 (IL-4), mature B cells from miR-155−/− mice display a significantly higher accumulation of γH2AX at the DSB foci and heightened p53 activation, with attendant greater cell cycle arrest and apoptosis. Genetic suppression of p53 abrogated the excessive cell cycle arrest associated with miR-155 deficiency, whereas retrovirus-mediated ectopic expression of miR-155 rescued these B cells by dampening both γH2AX accumulation and p53 activity. These findings pointed to a role for miR-155 in controlling the DSB and p53 activation that accompany the GC reaction. We confirmed the participation of Aicda in this process by partially rescuing the excessive DSB and DDR via an siRNA-mediated knockdown. Further, we showed that the miR-155 target Socs1 directly binds to p53 and that an siRNA-mediated Socs1 knockdown significantly suppressed p53 activity in activated B cells. Additionally, using a complementary gain-of-function model, we showed that in B cells Socs1 facilitates p53 activation and localization to DSB foci. Together, these data suggest that miR-155 controls the outcome of the GC reaction at two levels, by modulating its initiation (Aicda targeting) and distally by influencing p53 response (at least in part via Socs1 targeting). These findings may help explain the aberrant loss or excess of GC B cells in miR-155 null or knock-in mice, respectively (12). Finally, our data add another layer, the functional suppression of p53 activity via Socs1 targeting, to the complex role played by miR-155 in tumor development and maintenance (11, 15–18).
MATERIALS AND METHODS
Mice and isolation of murine cells.
miR-155-deficient mice were obtained from Klaus Rajewsky's group. A local colony was maintained by breeding heterozygous mice. Sex matched, 8- to 15-week-old miR-155−/− and wild-type (WT) littermates were used in all assays reported. For a subset of experiments, mice were immunized with 100 μg of NP-CGG (4-hydroxy-3-nitrophenylacetyl chicken γ-globulin; Biosearch Technologies) on days 0 and 7 and were harvested on day 14. In naive or immunized mice, splenic mature B cells were isolated as we described previously (19), using the EasySep mouse B cell enrichment kit (StemCell Technologies); purity was confirmed by fluorescence-activated cell sorter (FACS)-based quantification of B220 or CD19 (>93%). These mature B cells were examined either immediately upon harvesting and purification or following 48 to 96 h of culture in RPMI 1640 supplemented with 20% fetal bovine serum (FBS), 100 μM β-mercaptoethanol, 10 mM HEPES, 2 μM l-glutamine, 0.1% penicillin-streptomycin, 20 μg/ml lipopolysaccharide (LPS), and 2.5 ng/ml murine interleukin-4 (IL-4). In addition, for a group of mice, thymus and bone marrow were also harvested under sterile conditions. In brief, thymocytes were cultured in a T cell medium containing RPMI 1640 supplemented with 20% FBS, anti-CD3 (1 μg/ml, UCTH1 clone; BD Biosciences), and anti-CD28 (2 μg/ml, CD28.2 clone; BD Biosciences) as previously described (20). Bone marrow was collected from the two legs (femur and tibia), and a mononuclear cell suspension was prepared after lysing red cells. Total cells were subsequently cultured in minimal essential medium (MEM) alpha supplemented with 10% FBS, 25 ng/ml stem cell factor (SCF), 20 ng/ml interleukin-6 (IL-6), 10 ng/ml interleukin-3 (IL-3), and 10 ng/ml thrombopoietin (TPO), as described previously (21). These T cells and hematopoietic cells, alongside with mature B cells not exposed to LPS–IL-4, were cultured for short time (2 h) or overnight and then irradiated (5 Gy, cesium 137) or treated with 30 μM Etoposide for 20 h, respectively. For the irradiated cohort, the cells were collected at 3 h postirradiation. All animal procedures were approved by the Institute Animal Care and Use Committee of the University of Texas Health Science Center San Antonio.
miR-155 transduction.
The generation of a murine stem cell virus (MSCV)–miR-155–enhanced green fluorescent protein (eGFP) bicistronic retrovirus construct and the virus production method were reported earlier (18). Purified splenic mature B cells from miR-155−/− mice (n = 4) were maintained in LPS- and IL-4-containing medium and transduced by spinoculation with MSCV or MSCV–miR-155 viruses every 24 h for a total of three rounds. The effectiveness of the transduction was confirmed by FACS (mean GFP-positive cells = 73%; range, 63% to 81%) and by measuring miR-155 expression with the stem-loop quantitative TaqMan real-time reverse transcription-PCR (RT-PCR) assay (MicroRNA assays; Applied Biosystems Inc.). Twenty-four hours after the last round of transduction, cells were collected for downstream assays.
Stable Socs1 expression in BaF3 cells.
A full-length murine Socs1 cDNA was PCR amplified, sequence verified, and cloned into the MSCV-eGFP vector. MSCV-Socs1-eGFP and empty MSCV-eGFP viruses were transduced in the murine pro-B cell line BaF3 and highly purified populations (>90% eGFP positive) obtained by FACS-based cell sorting. These stable B cell populations were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 2 μM l-glutamine, 0.1% penicillin-streptomycin, and 10 ng/ml murine interleukin-3 (IL-3).
siRNA-mediated downregulation of p53, Aicda, and Socs1 in murine B cells.
Purified mature B cells from miR-155−/− and WT littermates were transfected by electroporation with control siRNA oligonucleotides or a pool of sequences directed at p53, Aicda, or Socs1 (see Table S1 in the supplemental material). In brief, murine B cells cultured for 48 h were washed twice with cold phosphate-buffered saline (PBS) and resuspended into 400 μl of Opti-MEM medium containing 100 to 200 nM siRNA duplex. Electroporation was performed using a Bio-Rad Gene Pulser MX Cell (Bio-Rad Laboratories) with parameters set at 250 V, 975 μF, and ∞ resistance. The transfected cells were kept on ice for 10 min and then cultured for another 24 h or 48 h before the cells were harvested for downstream applications, including cell cycle analysis, γH2AX immunofluorescence (IF), Western blot analysis of total p53, phospho-p53, Aicda, and Soc1, and real-time RT-PCR-based quantification of p53 transcriptional targets, as well as p15, p16, and p57.
γH2AX and phospho-p53 IF.
Relevant single-cell suspensions were processed by cytospin onto polylysine slides and fixed with 4% paraformaldehyde solution at 4°C for 15 min, followed by a 5-min permeabilization step using 50 mM NaCl, 3 mM MgCl2, 200 mM sucrose, 10 mM HEPES, and 0.5% Triton X-100. Slides were blocked in 3% goat serum, 1% bovine serum albumin, and 0.1% Triton X-100 in Tris-buffered saline solution for 1 h at room temperature (or overnight at 4°C) and stained with anti-γH2AX-Ser139 (1:250) (2577; Cell Signaling Technology) overnight at 4°C (or for 1 h at room temperature) followed by Alexa Fluor 488-conjugated goat anti-rabbit antibody (1:500) (Invitrogen) for 30 min at room temperature. For BaF3 cells stably expressing empty MSCV or MSCV-Socs1 constructs, the immunofluorescence (IF) assays were performed in cultures grown in IL-3-free medium for 6 h, followed by ionizing radiation (IR) (5 Gy, cesium 137), with harvesting at 1 h postirradiation. Subsequently, the cells were processed as described above and stained with mouse anti-γH2AX-Ser139 (1:250) (05-636; EMD Millipore) and/or rabbit anti-phospho-p53–Ser15 (1:250) (9284; Cell Signaling Technology) overnight at 4°C, followed by incubation with secondary antibodies (Alexa Fluor 488–goat anti-rabbit IgG [1:500] and Texas Red–goat anti-mouse IgG [1:500], both from Invitrogen) for 1 h at room temperature. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and slides were mounted using Vectashield (Vector Laboratories). For single γH2AX staining, the fluorescence patterns were visualized by Nikon Eclipse 2000 fluorescence microscopy, and the data were analyzed and collected using NIS Elements software, as described previously (22). The quantification of γH2AX-positive signal was performed with the ImageJ software, with enumeration of foci in 50 nuclei/sample. Confocal imaging was performed using an inverted IX-81 Olympus microscope configured with an Olympus FV1000 confocal scanning system. The Uplanapo 60× oil objective (1.42 numerical aperture [NA]) was used for all data sets, and an additional electronic zoom of 2 was applied. The quantification of p-p53 and γH2AX colocalization signals was performed using the plug-in module provided for ImageJ, as previously described (23), with analysis of 100 cells/sample.
Western blots and immunoprecipitation.
Protein isolation, electrophoresis, and transfer to polyvinylidene difluoride (PVDF) membranes were performed as we previously described (24). Filters were immunoblotted with antibodies against p53 (sc-6243; Santa Cruz Biotechnology), phospho-p53 (Ser15, 9284; Cell Signaling Technology), p21 (F-5, sc-6246; Santa Cruz Biotechnology), phospho-STAT5–Tyr694 (4322; Cell Signaling Technology), Aicda (4975; Cell Signaling Technology), Socs1 (3950; Cell Signaling Technology), and β-actin (Sigma). In immunoprecipitation assays, 500 μg of protein from miR-155−/− or WT mature B cells was incubated with 1 μg of anti-53 antibody (sc-99; Santa Cruz Biotechnology) or mouse IgG control and protein A/G beads at 4°C overnight. Following washes and separation by electrophoresis, membranes were immunoblotted with anti-Socs1 or anti-p53 antibodies.
Real-time RT-PCR.
RNA isolation and cDNA generation were performed as we described previously (26). Real-time RT-PCR was used to quantify the expression of the cyclin-dependent kinase inhibitors (CDKi) p15 (Cdkn2b), p16 (Cdkn2a), p21 (Cdkn1a), and p57 (Cdkn1c) as well as the expression of the p53 transcriptional targets Cdc25c, Gadd45a, and Pcna. Expression was normalized to that of the housekeeping control TATA binding protein (TBP) gene, relative quantification was achieved by calculating the ΔΔCT, and expression was defined as 2−ΔΔCT, where either miR-155 WT B cells or those transduced with control vector or siRNA represented the baseline. All primers are listed in Table S2 in the supplemental material. Relative expression of miR-155 was defined by stem-loop quantitative TaqMan real-time reverse transcription-PCR (RT-PCR) assay (MicroRNA assays; Applied Biosystems Inc.). Small nucleolar (Sno) RNA 202 was used for normalizing the expression, as described previously (26).
Cell cycle and apoptosis analyses.
Cell cycle distribution and apoptosis were determined as we described before (27). In brief, B cells from WT or miR-155−/− mice (n = 6) were grown in the presence of LPS and IL-4 for 72 to 96 h, fixed with 70% ethanol at 4°C overnight, stained with propidium iodide, and analyzed by FACS. Annexin V staining (phycoerythrin [PE]-annexin V apoptosis detection kit I; BD Pharmingen) was used to determine the apoptosis rates of WT and miR-155−/− mouse B cells. An additional cohort of B cells from WT or miR-155−/− mice (n = 4) was investigated in the same manner following transfection with siRNA oligonucleotides directed at p53 or at a control innocuous sequence.
Cellular senescence assay.
Splenic mature B cells from WT or miR-155−/− mice were cultured in the presence of LPS and IL-4 for 72 h. Senescence-associated β-galactosidase (SA-β-gal) activity was determined using the senescence β-galactosidase staining kit (Cell Signaling Technology) following the general manufacturer's instructions and using 2.5 mg/ml of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 37°C for 16 to 24 h. Images were collected with a Nikon Digital Sight DS-Ri1 camera in a Nikon eclipse TE200 microscope (total magnification, ×400).
RESULTS
Heightened formation of γH2AX foci and p53 activation in miR-155−/− mature B cells.
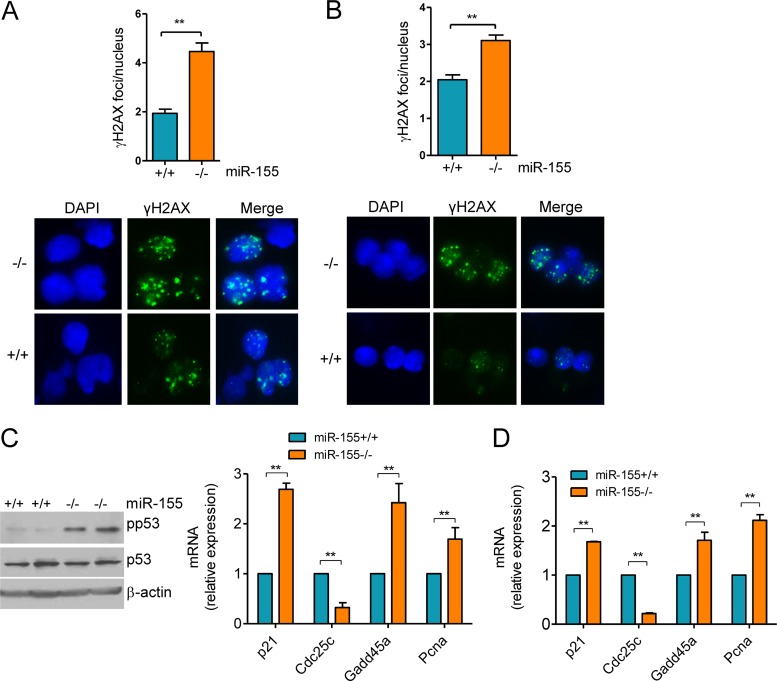
miR-155−/− mice display a significantly reduced fraction of GC B cells (12). A defect in cytokine production has been linked to this finding (12), but the full picture remains to be elucidated. miR-155 deficient B cells also develop an abnormally high degree of Aicda-dependent Myc-Igh translocation, suggesting that these cells may be exposed to excessive DNA damage (13). To explore the possibility that an exacerbated DDR contributes to the lower number of GC cells in miR-155−/− mice, we first quantified the extent of DSB. To that end, we used immunofluorescence (IF) to detect the phosphorylated (Ser-139) form of the histone variant H2AX (γH2AX), a highly specific and sensitive marker of double-strand DNA break and subsequent repair (28). Mature B cells were harvested from miR-155−/− and WT littermates (n = 18) and grown in medium containing LPS and IL-4 for 48 to 96 h, and γH2AX detection was performed by IF. Quantification of γH2AX foci (50 nuclei/mouse) showed a significantly higher DSB formation in miR-155−/− B cells than in WT controls (P < 0.01, two-tailed Student t test) (Fig. 1A). In parallel experiments and to demonstrate that in vitro exposure to LPS–IL-4 recapitulates the in vivo GC reaction, we immunized miR-155−/− and WT littermates (n = 6) with NP-CGG. Subsequently, mature B cells from these mice were isolated and processed directly for γH2AX focus detection; this analysis confirmed that an absence of miR-155 is associated with enhanced DSB focus formation (P < 0.01, two-tailed Student t test) (Fig. 1B). A physiological response to DNA damage is the engagement of repair checkpoints, in particular the p53 pathway. Nonetheless, this response is transiently repressed during the GC reaction to allow for the proper diversification of the immunoglobulin genes (3, 6). Thus, we examined whether loss of miR-155 interfered with p53 activation. The phosphorylation of p53's serine 18 (human Ser15), was markedly higher in mature B cells from miR-155−/− mice than in those from WT littermates (Fig. 1C). This serine residue is phosphorylated by ATM and ATR, and it is a marker of the induction of p53 by DNA damage (29). Concordantly, real-time RT-PCR-based quantification of four transcriptional targets of p53 (p21, Cdc25c, Gadd45a, and Pcna) demonstrated a significantly higher p53 activity in miR-155−/− than in WT B cells, irrespective of whether they derived from in vitro stimulation or immunized mice (P < 0.01, two-tailed Student t test) (Fig. 1C and D). We conclude that miR-155−/− B cells have an elevated rate of DNA double-strand breaks and activation of the p53 pathway, which may account for the diminished number of GC B cells in mice deficient for miR-155.
FIG 1.
Heightened formation of γH2AX foci and p53 activation in miR-155−/− mature B cells. (A) Mature B cells were harvested from miR-155 null and WT mice and grown in the presence of LPS–IL-4 for 96 h, and γH2AX was detected by IF. Quantification of γH2AX foci (50 nuclei counted per mouse) revealed a significantly higher DSB formation in miR-155 null than in WT control B cells (**, P ≤ 0.01, two-tailed Student t test). Data shown are mean ± standard deviation (SD) for the nine pairs of mice collected independently. A representative example of the γH2AX IF is also shown. (B) Mature B cells from NP-CGG-immunized mice were isolated and processed directly for γH2AX IF. Quantification of γH2AX foci confirmed the significantly more pronounced DSB formation in miR-155−/− B cells (P < 0.01, Student t test). Data shown are mean ± SD for the three pairs of mice collected independently. A representative example of the γH2AX IF is also shown. (C) Left, Western blot analysis of phospho-P53 (Ser18) levels in B cells from two pairs of miR-155+/+ and miR-155−/− mice shows a markedly higher p53 phosphorylation in cells lacking miR-155. Total p53 and B-actin immunoblotting confirmed that the changes noted are not related to distinct protein abundance. Right, real-time RT-PCR quantification of four p53 transcriptional targets (p21, Cdc25c, Gadd45a, and Pcna) defined a higher p53 activity in miR-155 null than in WT cells. (D) Similar real-time RT-PCR quantification was performed in B cells isolated from NP-CGG-immunized miR-155+/+ or miR-155−/− mice. In both instances, the expression of the p53 transcriptional targets was significantly influenced by miR-155 status (**, P ≤ 0.01, two-tailed Student t test). Note that Cdc25a expression is repressed by p53, whereas all other genes are induced. Data in panel C represent the mean ± standard error of the mean (SEM) of nine independent quantifications 18 mice); data in panel D are the mean ± SEM from three independent assays (6 mice). Data are displayed as relative expression, and all quantitative RT-PCRs were performed in triplicate.
Cell cycle arrest and apoptosis in miR-155−/− B cells.
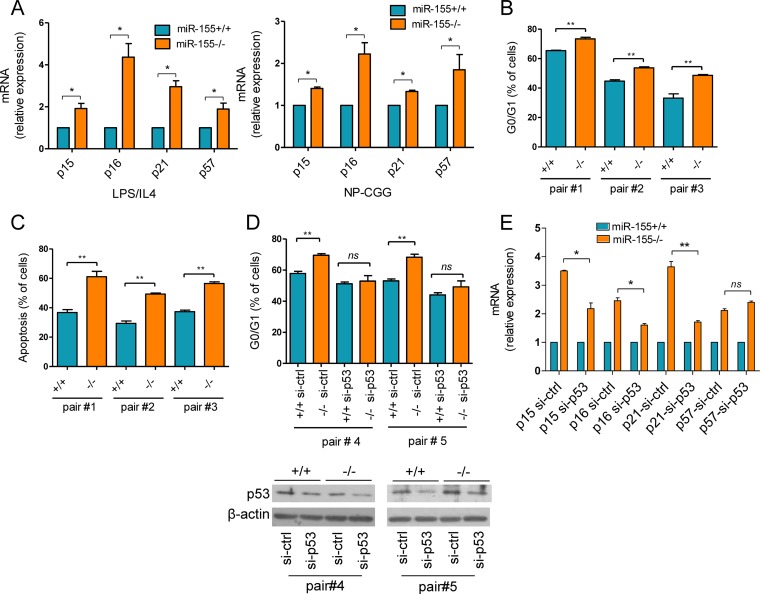
In response to DNA injury, p53 induces apoptosis, transient cell cycle arrest, or senescence (29). The excessive engagement of the p53 pathway that we identified in miR-155−/− mice suggests that the reported loss of the GC B cells in this model may be associated with a p53-regulated apoptosis or cell cycle arrest. To address this issue, we first confirmed that miR-155−/− mice developed fewer GC cells than WT littermates (CD95+/PNAhigh) (see Fig. S1 in the supplemental material). Next, we defined the cell cycle profile, measured the apoptosis rate, and quantified the expression of the cyclin-dependent kinase inhibitor (CDKi) genes (p15, p16, p21, and p57) in mature B cells from miR-155−/− and WT controls (n = 6 to 18). Following in vitro exposure to LPS–IL-4 or in vivo immunization, we detected a significantly higher expression of all CDKi genes examined in miR-155−/− cells than in WT cells (P < 0.05, two-tailed Student t test) (Fig. 2A). Concordantly, after 72 h in culture, we detected a modest, but significant, higher percentage of G0/G1-arrested cells in the miR-155−/− that in WT controls (P < 0.01, two-tailed Student t test) (Fig. 2B). These nonimmortalized mature B cells became apoptotic after 4 days in culture, again at a significantly higher rate in miR-155−/− cells than in WT controls (P < 0.01, two-tailed Student t test) (Fig. 2C). Although p53 activation preferentially induces apoptosis in murine B cells, the higher expression of CDKi genes and cell cycle arrest suggested that premature senescence may be associated with miR-155 deficiency in mature B cells. Unfortunately, the quantification of β-galactosidase activity, an important senescence marker, is compromised in our model because the targeting construct used to generate the miR-155−/− mice includes a LacZ reporter (encoding β-galactosidase) under the control of the BIC gene (the primary miR-155 transcript) promoter (12). Indeed, we found that mature B cells from the miR-155−/− mice readily stain for β-galactosidase upon LPS exposure in vitro (data not shown), which further supports the concept that miR-155 expression is induced during, and important for, the GC reaction. Finally, to ascertain the contribution of p53 to these processes, we isolated mature B cells from a cohort of miR-155−/− and WT mice (n = 4) and used electroporation to transfect them with control or p53-directed siRNA oligonucleotides. A significantly higher percentage of G0/G1-arrested cells was readily detected in siRNA control-transfected miR-155−/− cells (P < 0.01, two-tailed Student t test), whereas this difference was abrogated upon p53 knockdown (Fig. 2D). Next, we examined the consequences of suppressing p53 in these cells for p16, p15, p21 and p57 expression. Inhibiting the p53 expression significantly restricted the upregulation of p15, p16, and p21 (but not p57) that is associated with in vitro activation of miR-155 KO B cells (Fig. 2E). Unfortunately, following electroporation of siRNA oligonucleotides we could not capture the difference in apoptosis rate between miR-155 null and WT cells. We attributed this unexpected result to the effects of electroporation, which by inflicting broad cell damage may have impaired our ability to detect subtle differences in apoptosis rate. Thus, we cannot determine at the moment if p53 also mediates the effects of miR-155 on the apoptosis of mature B cells. We conclude that the aberrant p53 activation found in miR-155−/− B cells elicits a spectrum of exacerbated responses, including heightened cell cycle arrest and apoptosis, which may contribute to the loss of GC B cells in mice lacking miR-155.
FIG 2.
Excessive cell cycle arrest and apoptosis in miR-155−/− B cells. (A) Real-time RT-PCR quantification of the cyclin-dependent kinase inhibitor genes p15, p16, p21, and p57 was performed in miR-155 null or WT B cells following exposure to LPS–IL-4 (left panel) or NP-CGG immunization (right panel). We detected a significantly higher expression of all CDKi genes in miR-155−/− than in miR-155+/+ cells. Data in the left and right panels represent the mean ± SEM for nine and three independent quantifications (18 and 6 mice), respectively. Data are displayed as relative expression; all quantitative RT-PCRs were performed in triplicate. (B) Propidium iodide (PI) staining was used to determine the cell cycle profiles of mature B cells from three pairs of miR-155+/+ and miR-155−/− littermates exposed to LPS–IL-4 for 72 h. The absence of miR-155 was significantly associated with a higher percentage of G0/G1-arrested cells. (C) Annexin V staining was used to quantify apoptosis in three pairs of miR-155+/+ and miR-155−/− littermates exposed to LPS–IL-4 for 96 h. A significantly higher fraction of miR-155 null than WT cells were positive for annexin V. Data shown in panels B and C are mean ± SD of independent measurements performed in triplicate. (D) Mature B cells from two pairs of miR-155+/+ and miR-155−/− littermates were grown in medium containing LPS–IL-4 for 48 h and electroporated with control (ctrl) or p53-specific siRNA oligonucleotides, and the cell cycle profile was determined 24 h later. Knockdown of p53 abrogated the excessive G0/G1 arrest found in miR-155−/− B cells (compare si-ctrl versus si-p53-transfected cells). Western blots (bottom) confirmed the effective KD of p53 in this model. (E) Real-time RT-PCR of p15, p16, p21, and p57 in miR-155+/+ or miR-155−/− B cells following si-ctrl or si-p53 transfection. The higher levels of p15, p16, and p21 in miR-155−/− cells was significantly limited by the KD of p53. Data shown are the mean ± SD for the cells shown in panel D, displayed as relative expression normalized by the values obtained with miR-155+/+ cells. *, P < 0.05; **, P ≤ 0.01 (two-tailed Student t test).
miR-155 reconstitution rescues the aberrant DSB formation and p53 activation in mature B cells.
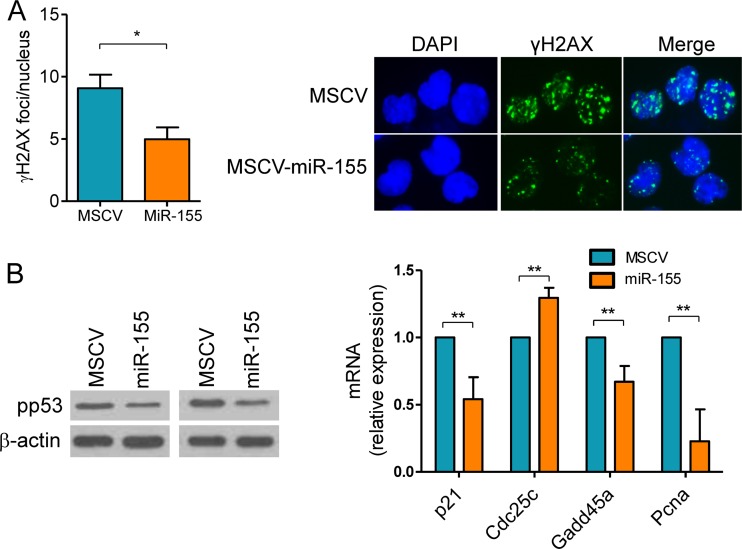
To confirm the cell-autonomous role of miR-155 in modulating the DSB accumulation and DDR associated with the GC reaction, we rescued its expression in miR-155−/− B cells. In these assays, mature B cells from miR-155−/− mice (n = 4) were transduced with an MSCV–miR-155 retrovirus or an empty MCSV control. The effectiveness of this process was confirmed by FACS (GFP > 75%) and stem-loop real-time quantification of miR-155 (see Fig. S2 in the supplemental material). In these assays, cells transduced with MSCV–miR-155 formed significantly fewer DSB foci (P < 0.05, two-tailed Student t test), as determined by γH2AX IF, than their MSCV control counterparts and showed significantly lower p53 activity, as defined by its phosphorylation levels and transcriptional target expression (P < 0.01, two-tailed Student t test) (Fig. 3A and B). Thus, expression of miR-155 limits the extension of DNA damage and p53 activation associated with the GC reaction in a cell-autonomous manner.
FIG 3.
miR-155 reconstitution rescues the aberrant DSB formation and p53 activation in B cells. Mature B cells were isolated from four miR-155−/− mice and successfully transduced with a retrovirus expressing this miRNA or an empty control (MSCV). (A) Cells transduced with MSCV-miR-155 formed significantly fewer DSB foci (*, P < 0.05, Student t test), as determined by γH2AX IF, than their control counterparts. Data shown are mean ± SD from the four independent assays; 50 nuclei were counted per cell type in each assay. A representative example of the γH2AX IF is also shown. (B) Left panel, Western blot analysis of phospho-p53 (Ser18) levels in miR-155−/− B cells transduced with empty MSCV or MSCV–miR-155 shows a decrease in p53 phosphorylation following miR-155 expression. Right panel, real-time RT-PCR quantification of p53 transcriptional targets confirms the significant suppression of p53 activity upon miR-155 reconstitution (**, P ≤ 0.01, Student t test). Data shown are the mean ± SD from three independent assays (6 mice) performed in triplicate and are displayed as relative expression.
miR-155 regulation of DNA damage is cell and context specific.
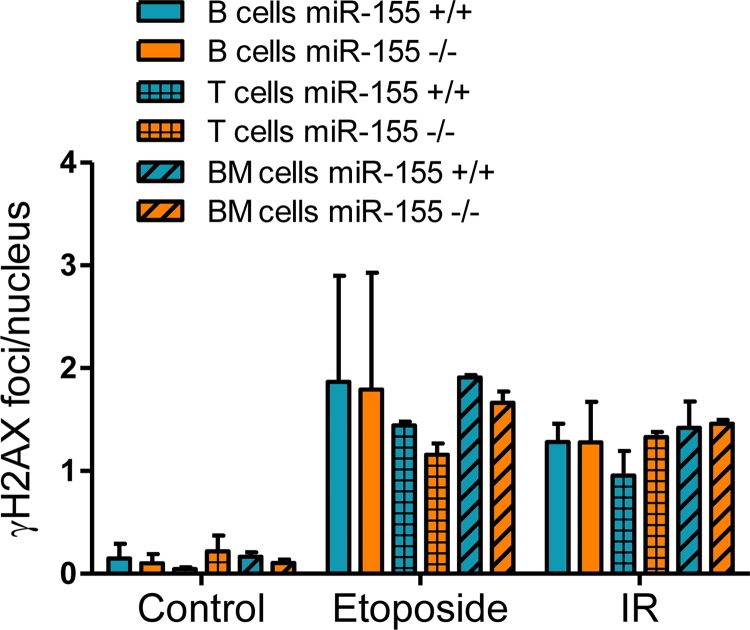
In mature B cells, miR-155 regulates the extent of DSB and p53 activation (Fig. 1 and 3). Here, we examined if this regulatory control extended to other genotoxic stimuli and cell types. To that end, splenic mature B cells, thymocytes, or bone marrow cells were isolated from miR-155−/− and WT littermates (n = 4) and exposed to etoposide (30 μM for 20 h) or IR (5 Gy, with cell collection after 3 h). Using γH2AX IF, we found a marked induction of DSB in all cell types exposed to etoposide or IR (Fig. 4). Likewise, in comparison to control (vehicle-exposed or nonirradiated cells), there was a significant modulation of p53 target gene expression following etoposide or IR treatment (see Fig. S3 in the supplemental material). However, irrespective of the cell type or genotoxic stimuli, there was no significant difference in the number of γH2AX foci between WT and miR-155−/− cells or p53 transcriptional target expression. As a control, the mature B cells were also exposed to LPS–IL-4, and we readily detected the expected significantly higher DSB accumulation in the miR-155−/− model (see Fig. S3 in the supplemental material). We conclude that in this model, the control of DSB extent and p53 activation by miR-155 may be restricted to B cells undergoing the GC reaction.
FIG 4.
miR-155 regulation of DNA damage is cell and context specific. Mature B cells, thymocytes, and bone marrow cells were harvested from miR-155 null or WT mice. Exposure of these cell to ionizing irradiation (IR) (5 Gy, with collection after 3 h) and etoposide (30 μM for 20 h) induced robust γH2AX accumulation at the DSB foci in all three cell populations (compare control [vehicle exposed, nonirradiated] versus etoposide- or IR-treated cells). However, irrespective of the cell type or genotoxic stimuli, there was no significant difference in the number γH2AX foci between WT and miR-155−/− cells. Data shown are mean ± SD for foci per nucleus. For each assay, approximately 50 nuclei were counted per mouse. p53 activity data are shown in Fig. S3 in the supplemental material.
Aicda overexpression contributes to the excessive DSB and DDR in miR-155-deficient mature B cells.
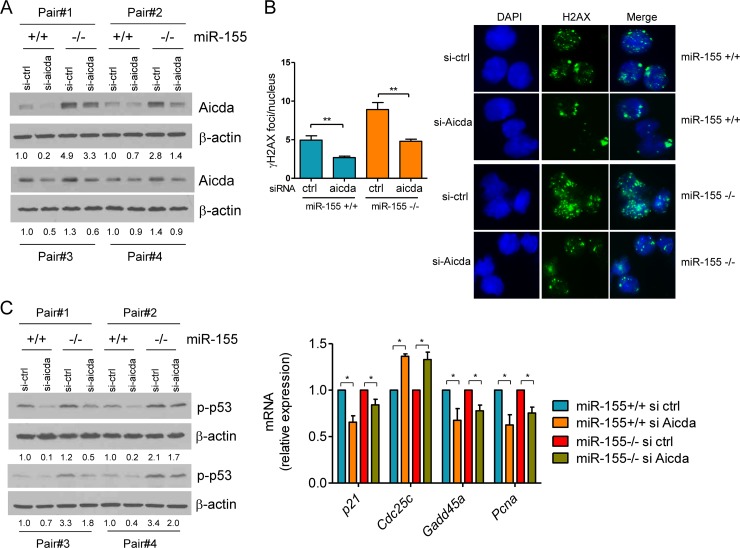
We showed that the regulation of DNA double-strand breaks by miR-155 is predominant in mature B cells undergoing the GC reaction (Fig. 1, 3, and 4). These data suggested a potential role for Aicda, a known miR-155 target (13, 14) and a key regulator of CSR, a B cell-specific process that is associated with double-strand DNA breaks (1, 2). To test this concept, we used an RNAi strategy to isolate the contribution of Aicda to the enhanced DSB accumulation and DDR found in miR-155−/− B cells. We transfected a pool of Aicda-directed or control siRNA oligonucleotides in mature B cells from miR-155−/− or WT littermates (n = 8) and measured DSB and p53 activity. Aicda expression was consistently higher in miR-155−/− cells (Fig. 5A), as expected for an miR-155 target. Importantly, Aicda expression was effectively suppressed by the siRNAs (Fig. 5A), resulting in a significant decrease in the formation of γH2AX foci (Fig. 5B) (P < 0.01, two-tailed Student t test). Notably, knockdown of Aicda corrected the excessive DNA breakage that typifies miR-155−/− B cells, as characterized by the now indistinguishable accumulation of DSB foci in WT B cells transfected with a control siRNA and miR-155−/− cells transfected with Aicda siRNAs (Fig. 5B). To determine whether this limited DSB formation decreased p53 activation, we examined its phosphorylation levels and the expression of its transcriptional targets. As we showed above, phosphorylation of p53 was higher in miR-155−/− cells, but it was effectively suppressed by the knockdown of Aicda (Fig. 5C). The decrease in p53 phosphorylation levels was accompanied by a significant change in the expression of its transcriptional targets (P < 0.05, two-tailed Student t test) (Fig. 5C). We conclude that the elevated p53 activity of miR-155−/− B cells is at least in part secondary to the unchecked Aicda levels. Thus, the targeting of Aicda by miR-155 is a key cellular safeguard to limit the extent of the DNA damage and p53 activation associated with the process of antibody diversification within the germinal center.
FIG 5.
Aicda overexpression contributes to the excessive DSB and p53 activity in miR-155-deficient mature B cells. (A) Western blot analysis of mature B cells from miR-155 null or WT mice (four pairs of mice, n = 8) shows higher expression of Aicda in cells lacking the miRNA (for each pair of mice, compare si-ctrl in miR-155+/+ versus miR-155−/− mice). The display also demonstrates the effective downmodulation of Aicda by the siRNA-based strategy (compare si-ctrl to si-aicda lanes). (B) Quantification of γH2AX by IF demonstrates that downregulation of Aicda significantly limits DSB formation (**, P ≤ 0.01, two-tailed Student t test). Data shown are mean ± SD of γH2AX foci (50 nuclei scored/mouse) obtained from four pairs of miR-155+/+ and miR-155−/− littermates. A representative example of the γH2AX IF is also shown. (C) Left, Western blot analysis of p-p53 (Ser18) demonstrates its suppression following the knockdown of Aicda (displayed in panel A). Right, real-time RT-PCR quantification of p53 transcriptional targets confirms the significant suppression of p53 activity upon Aicda knockdown (*, P < 0.05, Student t test). Data shown are the mean ± SD from four independent assays (8 mice) performed in triplicate and are displayed as relative expression (si-ctrl versus si-Aicda). Densitometric quantification is shown below the Western blots (A and C); data are normalized by β-actin and displayed relative to the values found in miR-155+/+ B cells transfected with the si-ctrl oligonucleotides.
miR-155 regulates the extent of p53 activity in mature B cells by targeting Socs1.
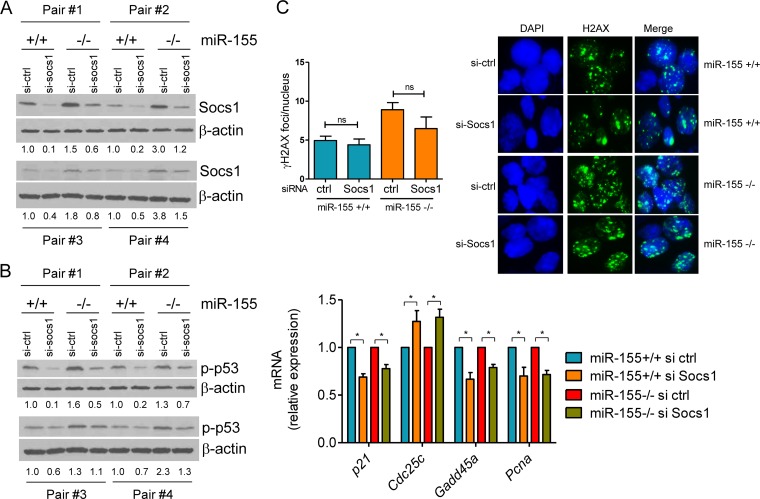
The lower number of GC B cells in miR-155−/− mice suggests that not only the Aicda-mediated DNA double-strand breaks is aberrant in this context but perhaps also the resolution of these DNA lesions. p53 is a central regulator of cell fate following DNA injury (29). Thus, an abnormally active p53 pathway, above and beyond its excessive induction secondary to Aicda deregulation, may play a role in the loss of GC B cells found in the miR-155−/− mice. To explore this possibility and absent any evidence that miR-155 could directly target p53, we focused on well-characterized miR-155 targets that are both relevant to immune cell biology and may regulate p53 activity. Socs1, an miR-155 target (30), is known primarily as a negative regulator of cytokine signaling (31). However, a less well defined function of Socs1 is to bind to p53 and promote its phosphorylation, transcriptional activity, and DNA binding. This unexpected role for Socs1 has been recently characterized in epithelial cells and in response to STAT5 signaling (25). However, it remains to be determined whether Socs1 modulates p53 function in response to other stimuli or in B cells. Here, we first showed that p53 and Socs1 also form a complex in murine mature B cells exposed to LPS–IL-4 (see Fig. S4 in the supplemental material). These data suggest that the expected higher expression of Socs1 in the absence of miR-155 may contribute to the differential p53 engagement that we found in miR-155−/− and WT B cells (Fig. 1, 3, and 5). To address this possibility, we used an siRNA strategy. We transfected a pool of Socs1-directed or control siRNA oligonucleotides in mature B cells from miR-155−/− or WT littermates (n = 8) and measured p53 phosphorylation and modulation of its transcriptional targets. Socs1 expression was consistently higher in miR-155−/− cells (Fig. 6A), as expected for an miR-155 target gene. Importantly, Socs1 expression was effectively suppressed by siRNA oligonucleotides (Fig. 6A). Phosphorylation of p53 was also higher in miR-155−/− cells, but it was effectively suppressed by the knockdown of Socs1 (Fig. 6B). The decrease in p53 phosphorylation levels was accompanied by a significant change in the expression of its transcriptional targets (P < 0.05, two-tailed Student t test) (Fig. 6B). These data confirmed that Socs1 regulates p53 activity in B cells undergoing the GC reaction. However, the reported localization of Socs1 to DNA breaks following STAT5 signaling (25) raised the possibility that in addition to regulating p53 phosphorylation/activation, Socs1 could play a broader role in controlling the extent of DNA damage. To shed light on this possibility, we investigated whether the knockdown of Socs1 modified the differential accumulation of phosphorylated H2AX in the DSB foci of miR-155−/− or WT B cells. In contrast to the effects of the siRNA-mediated Aicda downregulation (Fig. 5B), suppression of Socs1 did not significantly limit the abundance of DSB foci (Fig. 6C). We conclude that in mature B cells Socs1 positively regulates p53 activity and that its unchecked expression in an miR-155−/− context contributes to the excessive engagement of p53 but not to the aberrant DSB accumulation.
FIG 6.
miR-155 regulates the extent of p53 activity in mature B cells by targeting Socs1. (A) Western blot analysis of Socs1 expression demonstrates its higher expression in mature B cells from miR-155 null than from WT mice (for each pair of mice, compare si-ctrl in miR-155+/+ versus miR-155−/− mice) and their effective suppression by an siRNA-based strategy (for each mouse, compare si-ctrl to si-Socs1). (B) Left, Western blot analysis shows that Socs1 knockdown suppresses p53 phosphorylation (Ser18); right, real-time RT-PCR quantification of p53 transcriptional targets confirms the significant suppression of p53 activity upon Socs1 downregulation (*, P < 0.05, Student t test). Data shown are the mean ± SD from four independent assays (8 mice) performed in triplicate and are displayed as relative expression (si-ctrl/si-Socs1). Densitometric quantification is shown below the Western blots (A and B); data are normalized by β-actin and displayed relative to the values found in miR-155+/+ B cells transfected with the si-ctrl oligonucleotides. (C) Quantification of γH2AX by IF demonstrates that downregulation of Socs1 does not change the accumulation of DSB foci (ns, P = 0.21, Student t test). Data shown are mean ± SD of γH2AX foci (50 nuclei scored/sample) obtained from four pairs of miR-155+/+ and miR-155−/− littermates. A representative example of the γH2AX IF is also shown.
Stable expression of Socs1 in BaF3 cells enhances p53 phosphorylation, transcriptional activity, and its colocalization with γH2AX at the DSB foci.
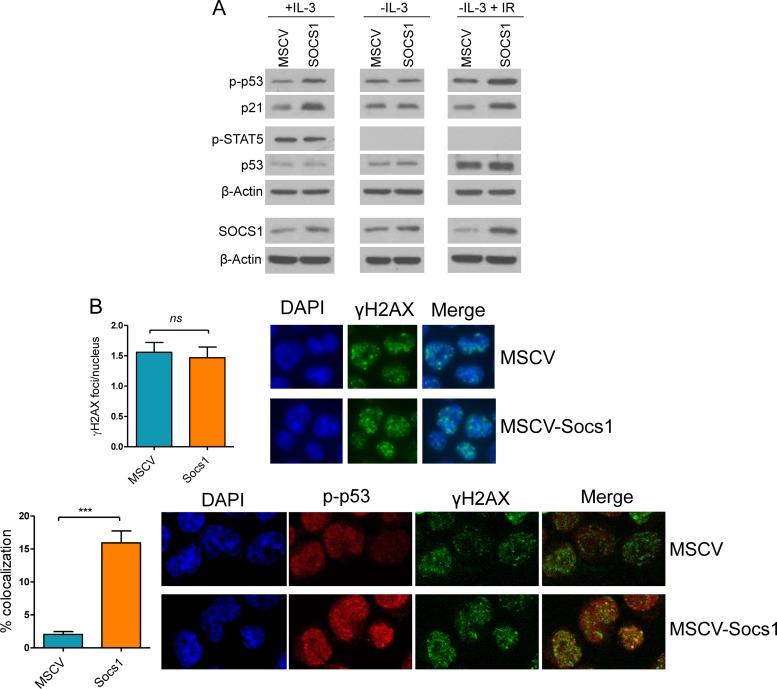
Using an siRNA approach, we showed that Socs1 influences p53 phosphorylation and activity in untransformed mature B cells (Fig. 6). To expand on these observations and better characterize the interplay between Socs1 and p53 in B cells, we generated a complementary model of stable Socs1 expression in the pro-B cell line BaF3. In agreement with the results obtained with the loss-of-function assays (Fig. 6), upon subtle elevation of Socs1 levels, p53 phosphorylation (Ser18) and activity levels (defined by the induction of the p53 target, p21) were markedly increased (Fig. 7A, left panel). In epithelial cells, the interplay between Socs1 and p53 has been shown to be dependent on cytokine signals and STAT5 activation (25). To examine the contribution of this signaling module to Socs1-mediated regulation of p53 activity in B lymphocytes, we took advantage of dependence of BaF3 cells on IL-3. Upon brief withdrawal (6 h) of IL-3 from the culture medium, we detected the expected loss of STAT5 phosphorylation at tyrosine 694 (an obligatory modification for STAT5 activation), which was accompanied by abrogation of the Socs1-mediated differential p53 phosphorylation/activation (Fig. 7A, middle panel). Thus, in B cells cytokine signaling appears to influence the Socs1/p53 cross talk. To define whether Socs1 could also influence p53 activity outside the constraints of cytokine signaling, we used a classical approach to induce p53 activation, ionizing irradiation. In these assays, BaF3-Socs1 cells and their isogenic controls expressing an empty vector were grown for 6 h in IL-3-deprived medium, subjected to IR (5 Gy), and harvested 1 h later for further characterization. Using Western blotting, we detected higher phospho-p53 levels and expression of its transcriptional target p21 in Socs1-expressing cells (Fig. 7A, right panel), similar to the effects associated with IL-3 exposure (Fig. 7A, left panel). Next, we used IF to further examine the influence of Socs1 on p53 in B cells subjected to IR. Here, while the extent of IR-induced DSB (γH2AX staining) was indistinguishable between BaF3 cells stably expressing Socs1 and their empty-MSCV isogenic controls, the former showed a significantly higher rate of phospho-p53/γH2AX colocalization (Fig. 7B), suggesting a role for Socs1 in facilitating p53 positioning and activation at the DSB foci. As expected, given our Western blot data (Fig. 7A), the phospho-p53 levels were higher in cells ectopically expressing Socs1. Collectively, these data demonstrate that Socs1 positively regulates p53 activity in B cells and show that this interplay is active in both cytokine-dependent and -independent environments. These findings also highlight a mechanism by which Socs1 may function as a tumor suppressor, and they substantiate the concept that elevated Socs1 levels, at least in part by enhancing p53 function, may contribute to the loss of GC B cells in miR-155 KO mice.
FIG 7.
Socs1 modulates p53 phosphorylation, activity, and subcellular localization. (A) Left panel, Western blot analysis of BaF3 cells cultured in the presence of IL-3 demonstrates higher p53 phosphorylation (Ser18) and p21 expression (a surrogate measure of p53 activity) upon stable expression of Socs1. Middle panel, Western blot analysis demonstrates that withdrawal of IL-3 for 6 h abrogates the Socs1-mediated increase in p53 phosphorylation/activity. Right panel, BaF3 cells were cultured in the absence of IL-3 for 6 h and subjected to IR (5 Gy). Western blot analysis demonstrates higher p53 phosphorylation and p21 expression in BaF3 cells stably expressing Socs1 than in MSCV-only isogenic controls. Phospho-STAT5 expression confirms the effectiveness of IL-3 signaling (compare left and middle/right panels). For each panel, p53 Western blots indicate that Socs1 primarily influences phosphorylation and not total protein abundance. The Western blot at the bottom of each panel confirms the higher expression of Socs1 upon its stable expression in BaF3 cells. (B) Top, quantification of γH2AX by IF demonstrates that stable expression of Socs1 in BaF3 cells does not increase the formation of DSB foci following IR. Data shown are mean ± SD of foci per nucleus (150 nuclei counted in each cell type). A representative example of the γH2AX IF is also shown. Bottom, following IR, BaF3 cells stably expressing Socs1 display a significantly higher colocalization of p-p53 and γH2AX signals than the isogenic control cells. The quantification of p-p53 and γH2AX signal colocalization was performed using the plug-in module provided for ImageJ, with analysis of 128 and 114 cells for MSCV and MSCV-Socs1, respectively. The data shown are mean ± SD of the percentage of colocalizing signals in each cell type (***, P ≤ 0.001, two-tailed Student t test). All assays for this figure were confirmed in two or more biological replicates.
DISCUSSION
In this report, we defined a role for miR-155 in controlling the extent of the double-strand DNA break and DNA damage response that is germane to the GC reaction. These findings may explain the aberrant loss or excess of GC B cells found in miR-155 null or knock-in mice, respectively (12). Mechanistically, we mapped this miR-155-mediated fine-tuning to its known targets, Aicda and Socs1. Thus, at the initiation point, miR-155 limits the magnitude and duration of SHM and CSR by suppressing Aicda expression. In a concerted fashion, miR-155 inhibition of Socs1 limits p53 engagement, contributing to the downregulation of repair checkpoints needed for a successful GC response. We propose that the absence of this dual targeting plays a significant, cell-autonomous role in the loss of GC B cells that typifies the miR-155 knockout mice (12). In this model, upon antigen exposure, overexpression of Aicda induces excessive double-strand DNA breaks, and this event, coupled with the high expression of Socs1, aberrantly engages p53, resulting in excessive cell death. Notably, the observation that genetic suppression of p53 abrogated the excessive cell cycle arrest that characterizes miR-155−/− activated B cells supported a main role of p53 in this process. Nonetheless, it is possible that in addition to the excessive p53 activation, other regulators of DNA damage/DNA repair have an impact on some of our observations. Indeed, a link between miR-155 and DNA repair, via the direct targeting of RAD51, has been shown earlier in a breast cancer cell line model (32).
Our data also unveiled a hitherto-unappreciated, Socs1-dependent interplay between miR-155 and the p53 pathway in B cells. Ectopically expressed Socs1 in epithelial cells had been shown earlier to bind to p53 and, in the context of STAT5 signaling, to influence its activity (25). Here, we showed that endogenous Socs1 complexes with p53 in mature B cells and positively influences its phosphorylation and transcriptional regulator activity. Accordingly, siRNA-mediated downregulation of Socs1 curtailed p53 activity and partially restored the excessive engagement of p53 found in miR-155−/− B cells. Further, stable ectopic expression of Socs1 in a murine pro-B cell line elevated p53 phosphorylation and activity and significantly increased its colocalization with γH2AX at the DSB foci. Interestingly, and contrary to the data obtained with epithelial cells, we found that in B lymphocytes the influence of Socs1 on p53 activity was broader and could also be detected in the absence of cytokine signaling, that is, following IR-induced DNA damage. The mechanism by which Socs1 appears to facilitate p53 positioning at the DSB foci is also of interest. We showed that in B cells Socs1 complexes with p53, while others showed that Socs1 also associates with ATM (25), probably positively influencing its phosphorylation at the site of DNA breaks. These data suggest that cells expressing high levels of Socs1 may be primed for the recruitment and activation of p53 at the site of DNA damage, thus offering a view on how Socs1 may suppress tumor development. Future studies on the kinetics of p53 localization and activation in distinct Socs1 contexts will allow a thorough examination of this concept. We suggest that this Socs1-p53 cross talk may also influence T cell biology. In particular, the reported decrease in Treg cell numbers and proliferative potential in an miR-155 null Socs1 overexpression setting (30) may in part be associated with heightened p53 activation and the attending aberrant cell cycle arrest, senescence, and/or apoptosis that we uncovered in B cells.
The miR-155–Aicda–Socs1 interplay described here illustrates the relevance of this miRNA in controlling the GC B cell development. However, these data also highlight the complex role of miR-155 in a malignant B cell context. Taken in isolation, the miR-155-mediated suppression of Aicda, a central mediator of DSB and oncogenic chromosomal translocations in B cells, confers a tumor suppressor character to this miRNA (13). The transcriptional repression of miR-155 by BCL6 gives support to this observation (16). Conversely, the targeting of Socs1 and the consequent functional downmodulation of p53 highlight the potential oncogenic behavior of miR-155. Previous reports of putative inactivating Socs1 mutations, as well as p53 loss, in GC-derived lymphomas strengthens this concept (33, 34). The lymphomagenic role of miR-155 is also supported by earlier work from our group and others linking this miRNA to tumor suppressor genes and pathways (17–19, 35) and by the development of B cell malignancies in Eμ-miR-155 mice (11).
How can we reconcile these distinct roles of miR-155 in malignant B cell biology? The differences in biological models notwithstanding, we suggest that the genetic and cellular context is important. Thus, in a p53-deficient background, miR-155 may function as a tumor suppressor gene, for its loss, and associated overexpression of Aicda, will result in excessive accumulation of double-strand DNA breaks in the absence of a functional DNA repair machine. Interestingly, BCL6 suppresses both miR-155 (16) and p53 (6), and thus such a scenario can be envisioned in GC-derived BCL6-translocated lymphomas. In addition, recent evidence suggests that miR-155 is important to an effective T cell-mediated antitumor response (15) and that in its absence tumors may escape immune surveillance. Conversely, we submit that aberrant overexpression of miR-155 in Aicda-independent B cell developmental stages (e.g., pre- or post-GC cells) or in other cell types in which miR-155 is known to target genes and pathways via canonical and noncanonical binding sites (36) will reveal the oncogenic properties of this miRNA through the direct or indirect downmodulation of the transforming growth factor β (TGF-β), JAK-STAT, SHIP1, RB, p53, and other tumor-suppressive nodes (17–19, 35–37).
In summary, in this work, we showed that miR-155, by targeting Aicda and Socs1, controls the extents of double-strand DNA breaks and the DNA damage response in normal mature B cells. The interplay between Socs1 and p53 in B cells is extensive and culminates with activation of the latter. Thus, the suppression of Socs1 defines miR-155 as a negative regulator of the p53 pathway, a finding of potential relevance to normal B cell development and possibly to miR-155-associated malignancies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Klaus Rajewsky for providing the miR-155 knockout mice and Patricia Dahia for comments and suggestions throughout the execution of this project.
This work was supported by a grant from the National Cancer Institute (R01-CA138747) (R.C.T.A), a Young Investigator Award from the Voelcker Fund (R.C.T.A), a Veterans Administration Merit Award (I01-BX001882), and a National Cancer Institute Cancer Center Support Grant (P30 CA054174).
D.J., H.B., A.-P.L., L.W., and M.O. performed experiments and contributed to their design and analysis, and R.C.T.A. conceived the project, performed experiments, designed the experimental strategy, supervised the study and analysis, and wrote the manuscript. All authors read the manuscript and agreed with its contents.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01446-14.
REFERENCES
- 1.Keim C, Kazadi D, Rothschild G, Basu U. 2013. Regulation of AID, the B-cell genome mutator. Genes Dev 27:1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z, Pone EJ, Al-Qahtani A, Park SR, Zan H, Casali P. 2007. Regulation of aicda expression and AID activity: relevance to somatic hypermutation and class switch DNA recombination. Crit Rev Immunol 27:367–397. doi: 10.1615/CritRevImmunol.v27.i4.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 4.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol 6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 5.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. 2007. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol 8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 6.Phan RT, Dalla-Favera R. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 7.Frankenberger S, Davari K, Fischer-Burkart S, Bottcher K, Tomi NS, Zimber-Strobl U, Jungnickel B. 2014. Checkpoint kinase 1 negatively regulates somatic hypermutation. Nucleic Acids Res 42:3666–3674. doi: 10.1093/nar/gkt1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A 103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O'Connell RM. 2012. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep 2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, Dalla-Favera R. 2012. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med 209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. 2010. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A 107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Aguiar RC. 2014. MicroRNA-155 controls RB phosphorylation in normal and malignant B lymphocytes via the noncanonical TGF-beta1/SMAD5 signaling module. Blood 123:86–93. doi: 10.1182/blood-2013-07-515254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkuri KR, Herzenberg LA, Niemi AK, Cowan T. 2007. Importance of culturing primary lymphocytes at physiological oxygen levels. Proc Natl Acad Sci U S A 104:4547–4552. doi: 10.1073/pnas.0611732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 22.Qin Y, Deng Y, Ricketts CJ, Srikantan S, Wang E, Maher ER, Dahia PL. 2014. The tumor susceptibility gene TMEM127 is mutated in renal cell carcinomas and modulates endolysosomal function. Hum Mol Genet 23:2428–2439. doi: 10.1093/hmg/ddt638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 24.Bouamar H, Abbas S, Lin AP, Wang L, Jiang D, Holder KN, Kinney MC, Hunicke-Smith S, Aguiar RC. 2013. A capture-sequencing strategy identifies IRF8, EBF1, and APRIL as novel IGH fusion partners in B-cell lymphoma. Blood 122:726–733. doi: 10.1182/blood-2013-04-495804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabrese V, Mallette FA, Deschenes-Simard X, Ramanathan S, Gagnon J, Moores A, Ilangumaran S, Ferbeyre G. 2009. SOCS1 links cytokine signaling to p53 and senescence. Mol Cell 36:754–767. doi: 10.1016/j.molcel.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 26.Ortega M, Bhatnagar H, Lin AP, Wang L, Aster JC, Sill H, Aguiar RC. 14 October 2014. A microRNA-mediated regulatory loop modulates NOTCH and MYC oncogenic signals in B- and T-cell malignancies. Leukemia doi: 10.1038/leu.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. 2012. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci U S A 109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinner A, Wu W, Staudt C, Iliakis G. 2008. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pant V, Quintas-Cardama A, Lozano G. 2012. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 120:5118–5127. doi: 10.1182/blood-2012-05-356014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davey GM, Heath WR, Starr R. 2006. SOCS1: a potent and multifaceted regulator of cytokines and cell-mediated inflammation. Tissue Antigens 67:1–9. doi: 10.1111/j.1399-0039.2005.00532.x. [DOI] [PubMed] [Google Scholar]
- 32.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, Carasi S, Palmieri D, Costinean S, Shapiro CL, Huebner K, Croce CM. 2014. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A 111:4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mottok A, Renne C, Seifert M, Oppermann E, Bechstein W, Hansmann ML, Kuppers R, Brauninger A. 2009. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood 114:4503–4506. doi: 10.1182/blood-2009-06-225839. [DOI] [PubMed] [Google Scholar]
- 34.Young KH, Leroy K, Moller MB, Colleoni GW, Sanchez-Beato M, Kerbauy FR, Haioun C, Eickhoff JC, Young AH, Gaulard P, Piris MA, Oberley TD, Rehrauer WM, Kahl BS, Malter JS, Campo E, Delabie J, Gascoyne RD, Rosenwald A, Rimsza L, Huang J, Braziel RM, Jaffe ES, Wilson WH, Staudt LM, Vose JM, Chan WC, Weisenburger DD, Greiner TC. 2008. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 112:3088–3098. doi: 10.1182/blood-2008-01-129783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. 2009. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A 106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. 2010. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.