Abstract
The gene encoding human hemojuvelin (HJV) is one of the genes that, when mutated, can cause juvenile hemochromatosis, an early-onset inherited disorder associated with iron overload. The 5′ untranslated region of the human HJV mRNA has two upstream open reading frames (uORFs), with 28 and 19 codons formed by two upstream AUGs (uAUGs) sharing the same in-frame stop codon. Here we show that these uORFs decrease the translational efficiency of the downstream main ORF in HeLa and HepG2 cells. Indeed, ribosomal access to the main AUG is conditioned by the strong uAUG context, which results in the first uORF being translated most frequently. The reach of the main ORF is then achieved by ribosomes that resume scanning after uORF translation. Furthermore, the amino acid sequences of the uORF-encoded peptides also reinforce the translational repression of the main ORF. Interestingly, when iron levels increase, translational repression is relieved specifically in hepatic cells. The upregulation of protein levels occurs along with phosphorylation of the eukaryotic initiation factor 2α. Nevertheless, our results support a model in which the increasing recognition of the main AUG is mediated by a tissue-specific factor that promotes uORF bypass. These results support a tight HJV translational regulation involved in iron homeostasis.
INTRODUCTION
Mammalian cells have the ability to change global and gene-specific translation in response to many different environmental stresses. Translation itself is regulated by a diverse set of mechanisms that act at the initiation step as well as during elongation and termination, and even after termination (1, 2). Translational regulation at the initiation step can be mediated via different cis-acting elements present in the 5′ untranslated region (5′ UTR), among them the upstream AUG codons (uAUGs) associated with upstream open reading frames (uORFs) (1–4). These uORFs are spread among different species and throughout the genome, but their prevalence has been difficult to calculate. The most recent studies estimate that about half of human transcripts contain at least one uORF (5), with these being conspicuously common in certain classes of genes, including oncogenes and genes involved in the control of cellular growth and differentiation (6, 7). A genetic and proteomic large-scale analysis revealed that under control conditions, uORFs reduce protein expression from the downstream main ORF by 30 to 80% (5); however, this repression can be alleviated in response to stress (4).
For a uORF to function as a translational regulatory element, its initiation codon (uAUG) must be recognized, at least at certain times, by the scanning 40S ribosomal subunit and associated initiation factors (8). One of the determinant factors of uAUG recognition by the translational machinery is the context of the start codon (9). According to Kozak's scanning model, the ideal context at the start AUG codon for higher eukaryotes is GCCRCCAUGG, where R can be G or A (10). The AnnAUGn and GnnAUGG contexts are considered to be strong enough to be recognized by the majority of scanning ribosomes (11). However, the ribosome might ignore the first AUG, scan right past it, and recognize the downstream AUG by leaky scanning (12). This mechanism depends not only on the AUG context but also on the proximity of the AUG codon to the cap, the length of the 5′ UTR, and the secondary structures of the transcript (13). Even after translation of the uORF, the main ORF can still be translated due to reinitiation (13, 14). Translation reinitiation occurs when the 40S subunit of the ribosome remains associated with the mRNA after uORF translation terminates, so it resumes scanning and reinitiates further downstream (13, 14). Translation reinitiation is thought to be an inefficient mechanism that depends on (i) the time required for the uORF translation, which is determined by the relative length of the uORF and the translation elongation rate; (ii) the translation initiation factors involved in the initiation event; and (iii) the length of the intercistronic region (15). A key factor for translation reinitiation is the reacquisition of a new ternary complex (eukaryotic translation initiation factor 2 [eIF2]–GTP–Met-tRNAi), which is essential for the recognition of a downstream AUG by the scanning 40S subunit (10, 15). As a result, eIF2α is one of the modulators of reinitiation efficiency (14, 15). In fact, the protein kinases that phosphorylate eIF2α are activated under stress conditions, resulting in global inhibition of translation (1). However, phosphorylation of eIF2α selectively promotes translational upregulation of mRNAs that contain uORFs by increasing uORF leaky scanning or translation reinitiation efficiency (4, 7, 16, 17).
During translation of a uORF, ribosomes may stall through direct interaction between the translating ribosome and the uORF-encoded peptide, at either the elongation or termination step (14). This ribosomal blockade might pose an obstacle to the scanning ribosome and thereby reduce the number of ribosomes that, by leaky scanning or reinitiation, gain access to the main AUG codon (13, 18). The uORFs characterized so far that act through this mechanism do not share any recognizable consensus sequence, suggesting that the different peptides interact with distinct sites in the translational machinery (13).
In adults, the hemojuvelin (HJV) gene is expressed mainly in the liver and in skeletal and cardiac muscles (19–21). HJV is a bone morphogenetic protein (BMP) coreceptor of BMP signaling (22). BMPs, which are members of the transforming growth factor beta (TGF-β) superfamily of cytokines, play an important role during development, being involved in proliferation, differentiation, and apoptosis (23). HJV acts through the BMP signaling pathway in order to modulate the expression of hepcidin, an iron homeostasis regulator (24). HJV binds to type I and type II BMP receptor complexes and induces their phosphorylation. Consequently, these activated complexes phosphorylate a subset of Smad proteins (Smads 1, 5, and 8) (24). These phosphorylated Smads have the ability to bind to Smad4, creating a complex that migrates into the nucleus, binding itself to specific DNA motifs and regulating gene transcription (25). Due to this process, HJV expression leads to an increase in hepcidin expression, demonstrating its fundamental role in systemic iron metabolism (22, 25). In fact, disruptions in the HJV gene may cause juvenile hemochromatosis, an early-onset hereditary hemochromatosis (26).
In this study, we demonstrate that the two uORFs (with 28 and 19 codons) present in the 5′ UTR of the human HJV mRNA have the ability to significantly decrease translational efficiency under normal conditions. Moreover, the C-terminal domain of the peptides encoded by these uORFs seems to be involved in the mechanism through which translational repression of the downstream main ORF occurs. However, this repression is significantly released in hepatic cells in response to an increase in iron. These results provide support for the understanding that HJV protein expression is tightly modulated at the translational level in response to iron overload.
MATERIALS AND METHODS
Plasmid constructs.
The expression vector pGL2-enhancer (Promega), encoding firefly luciferase (FLuc), was digested with BglII/HindIII to insert the human cytomegalovirus (HCMV) promoter sequence from the pcDNA3.1/hygro+ vector (Invitrogen), after its digestion with the same enzymes, creating the pGL2hCMV plasmid. The 5′ UTR of the human HJV mRNA was then cloned into the new pGL2hCMV vector, upstream of the firefly luciferase cistron, in such a way that the firefly luciferase AUG overlaps the main HJV AUG. For this purpose, the HJV 5′ UTR (325 nucleotides) was amplified by PCR, using cDNA from human liver, primer 1 with a HindIII linker, and primer 2, and the region between the HindIII and XbaI restriction sites was amplified from the pGL2hCMV plasmid by use of primers 3 and 4. The two obtained PCR products were then joined by a third amplification, using both amplification products obtained before as DNA templates, the forward primer from the first PCR, and the reverse primer from the second PCR (primers 1 and 4, respectively). The resulting DNA fragment was digested with HindIII/XbaI and then ligated to the pGL2hCMV plasmid previously digested with the same enzymes. The resulting construct was named the wild type (WT) (Fig. 1A), and its cloned sequence was confirmed by automated sequencing.
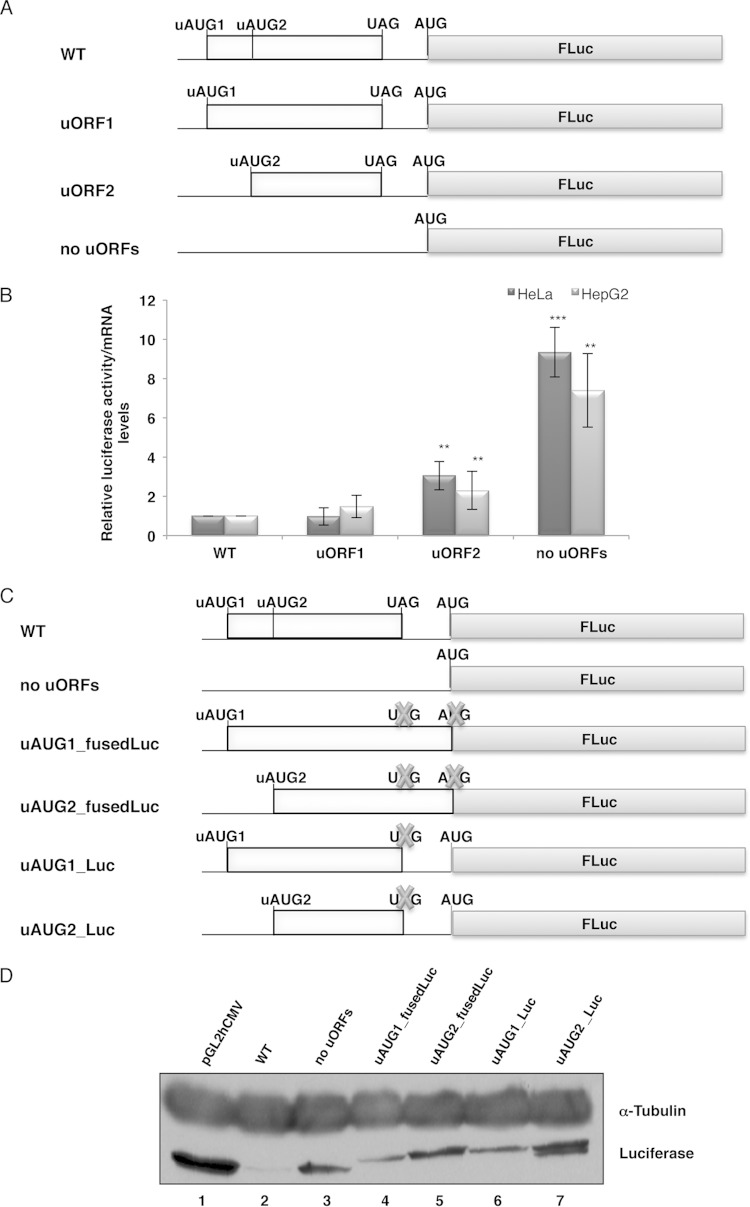
FIG 1.
The human HJV mRNA uORFs decrease the translational efficiency of the downstream main reporter ORF. (A) Schematic representation of reporter constructs. The human HJV 5′ UTR, encompassing the uORFs (light gray boxes) with intact initiation (uAUG) and termination (UAG) codons, was cloned into the empty vector (pGL2hCMV) upstream of the firefly luciferase gene coding region (FLuc) to create the wild-type construct (WT). The uAUGs were mutated individually (uORF1 and uORF2) or simultaneously (no uORFs). (B) The HJV 5′ UTR represses the activity of the downstream reporter coding sequence. HeLa and HepG2 cells were transiently cotransfected with each of the constructs shown in panel A and with the pRL-TK plasmid, encoding Renilla luciferase (RLuc). Cells were lysed 24 h later, and luciferase (Luc) activities and mRNA levels were measured by luminometry assays and RT-qPCR, respectively. The graph represents the data as translational efficiencies (relative luciferase activity/mRNA levels). Expression levels obtained from the WT construct were defined as 1. Average values and standard deviations for three independent experiments are shown. Statistical analysis was performed using Student's t test (unpaired, two-tailed). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Schematic representation of the constructs used for translation analysis. The WT and “no uORFs” constructs are the same as in panel A. The stop codon of each of the HJV uORFs was mutated in the uORF1 and uORF2 constructs, and each uORF was fused in frame with the downstream FLuc coding sequence, with (uAUG1_fusedLuc and uAUG2_fusedLuc) or without (uAUG1_Luc and uAUG2_Luc) mutation of the FLuc initiation (AUG) codon. Crosses represent the point mutations of the uORF stop codon and the FLuc initiation codon. (D) Translation initiation occurs at the HJV uORFs. Representative Western blot analyses show HeLa cell extracts transiently transfected with the constructs specified above the lanes. Immunoblotting was performed using an antiluciferase antibody to verify protein expression, and a human α-tubulin-specific antibody was used to control for variations in protein loading.
The WT construct was subjected to site-directed mutagenesis to obtain several mutant constructs, as follows. Site-directed mutagenesis was performed by using Pfu Turbo DNA polymerase (Invitrogen) as instructed by the manufacturer, mutagenic primers, and the WT plasmid, with the wild-type HJV 5′ UTR as the DNA template. PCR cycling was done as follows: after initial denaturation for 10 min at 95°C, PCR cycling parameters were 95°C for 1 min, 55°C for 1 min, and 72°C for 16 min, for a total of 11 cycles, with a final extension at 72°C for 10 min. Following DpnI (Fermentas) restriction digestion of the template, Escherichia coli DH5α bacteria were transformed with the mutagenesis reaction mixture, and transformants were selected on Luria-Bertani (LB) agar-ampicillin plates. The corresponding plasmid DNAs were purified from overnight cultures of single colonies by use of a Jetquick plasmid purification spin kit (Genomed) following the manufacturer's instructions. Confirmation of the correct cloned sequences containing the relevant mutations was carried out by automated sequencing. Using this strategy, the uORF1, uORF2, and “no uORFs” constructs (Fig. 1A), carrying a mutation(s) (ATG → TTG) in uAUG2, uAUG1, and both, respectively, were obtained with primers 5 to 8. The constructs uAUG1_fusedLuc, uAUG2_fusedLuc, uAUG1_Luc, and uAUG2_Luc (Fig. 1C) were obtained by using primers 9 and 10 in order to set the uORF and the luciferase cistron in frame by deletion of a T in the intercistronic sequence in the uORF1 and uORF2 constructs. Afterwards, the resulting constructs were subjected to a new site-directed mutagenesis in order to mutate the uORF stop codon by using primers 11 and 12. Finally, to obtain the constructs uAUG1_fusedLuc and uAUG2_fusedLuc (Fig. 1C), the ATG codon of the luciferase cistron was mutated to TTG by using primers 13 and 14. Next, the WT, uORF1, and uORF2 constructs were used as DNA templates in mutagenesis reaction mixtures, using primers 15 to 18, to obtain the “optimal uAUGs,” “optimal uAUG1,” and “optimal uAUG2” constructs, respectively (Fig. 2A). In these constructs, the uAUG1 flanking sequence was mutated from GAATCATGG to GAACCATGG, and the uAUG2 flanking sequence was mutated from GAGTAATGT to GAGCCATGG. The constructs named “mt stop uORFs,” “mt stop uORF1,” and “mt stop uORF2” (Fig. 2C) were obtained by mutating the uORF stop codon from TAG to AAG in the WT, uORF1, and uORF2 constructs, respectively, using primers 11 and 12. The “frameshift uORFs,” “frameshift uORF1,” and “frameshift uORF2” constructs (Fig. 3B) were also cloned by mutagenesis of the WT, uORF1, and uORF2 constructs, respectively, using primers 19 to 28. These primers can introduce two frameshifts: one frameshift is due to deletion of a T after uAUG1 (ATGGCTG → ATGGCG) and insertion of a C before uAUG2 (GATAGC → GATACGC), and the second frameshift is due to deletion of a T after uAUG2 (ATGTTT → ATGTT) and insertion of a C before the stop codon (TAGGTAG → TACGGTAG). Next, the frameshift uORFs, frameshift uORF1, and frameshift uORF2 constructs were used as templates to mutate the uORF stop codon from TAG to AAG by using primers 29 and 30, obtaining the “frameshift mt stop uORFs,” “frameshift mt stop uORF1,” and “frameshift mt stop uORF2” constructs (Fig. 3D). Finally, the “frameshift′2” to “frameshift′15” constructs (Fig. 4A) were sequentially derived, using the uORF2 construct as the template and primers 31 to 62, respectively, which shift the sequence until the codon mentioned in each construct designation.
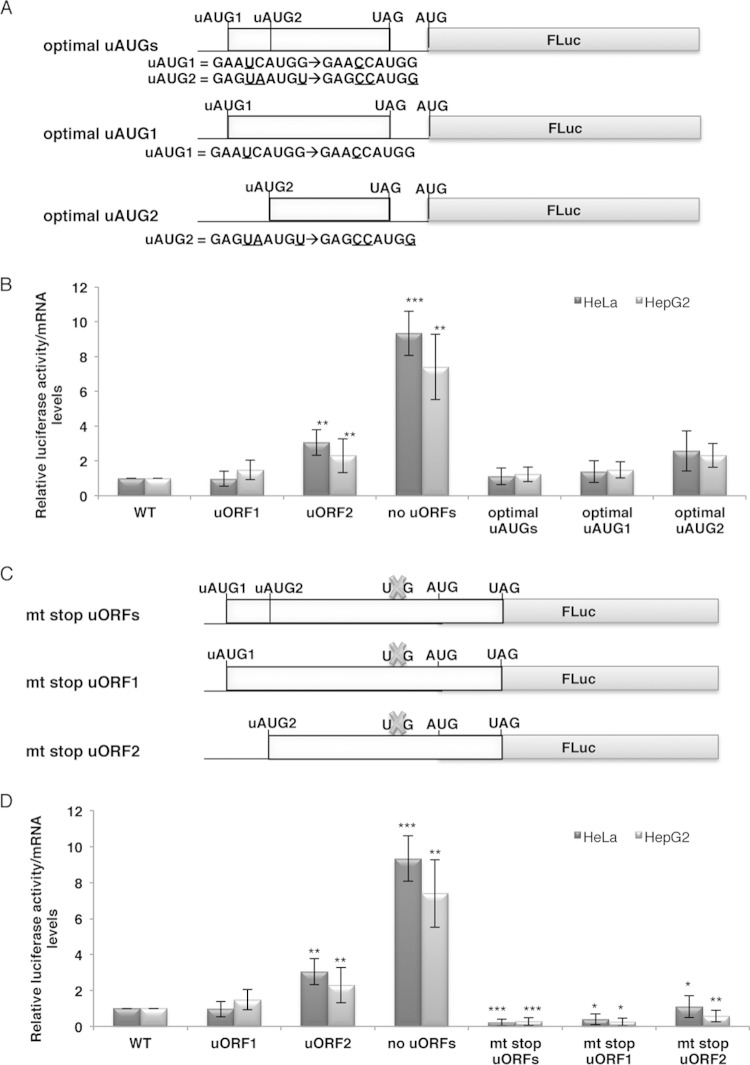
FIG 2.
The HJV main AUG is mostly recognized by translation reinitiation. (A) Schematic representation of reporter constructs. The WT, uORF1, and uORF2 constructs, carrying both HJV uORFs, uORF1, and uORF2 (light gray boxes), respectively, were used to mutate the uAUG sequence context and to improve it to an optimal Kozak context (as indicated below each uAUG), giving the optimal uAUGs, optimal uAUG1, and optimal uAUG2 constructs, respectively. (B) Preventing leaky scanning by improving the Kozak sequence context of the uAUGs does not affect luciferase protein expression levels. HeLa and HepG2 cells were transiently cotransfected with each of the constructs shown in panel A and with the pRL-TK plasmid, encoding Renilla luciferase (RLuc). The experimental procedure and analyses were identical to those described in the legend to Fig. 1B. For comparison, the chart also contains data imported from Fig. 1B. (C) Representation of reporter constructs. The mt stop uORFs, mt stop uORF1, and mt stop uORF2 constructs were obtained by mutating the uORF stop codon of the WT, uORF1, and uORF2 constructs, respectively. Crosses represent the point mutations of the uORF stop codon. (D) Preventing translation reinitiation by overlapping the HJV uORFs with the main firefly luciferase (FLuc) reporter ORF significantly decreases protein expression levels. Relative translation activities in HeLa and HepG2 cells for each of the constructs presented in panel C, as well as for the WT, uORF1, uORF2, and no uORFs constructs, were obtained as described for panel B. Data were analyzed as described in the legend to Fig. 1B. For comparison, the chart also contains data imported from Fig. 1B.
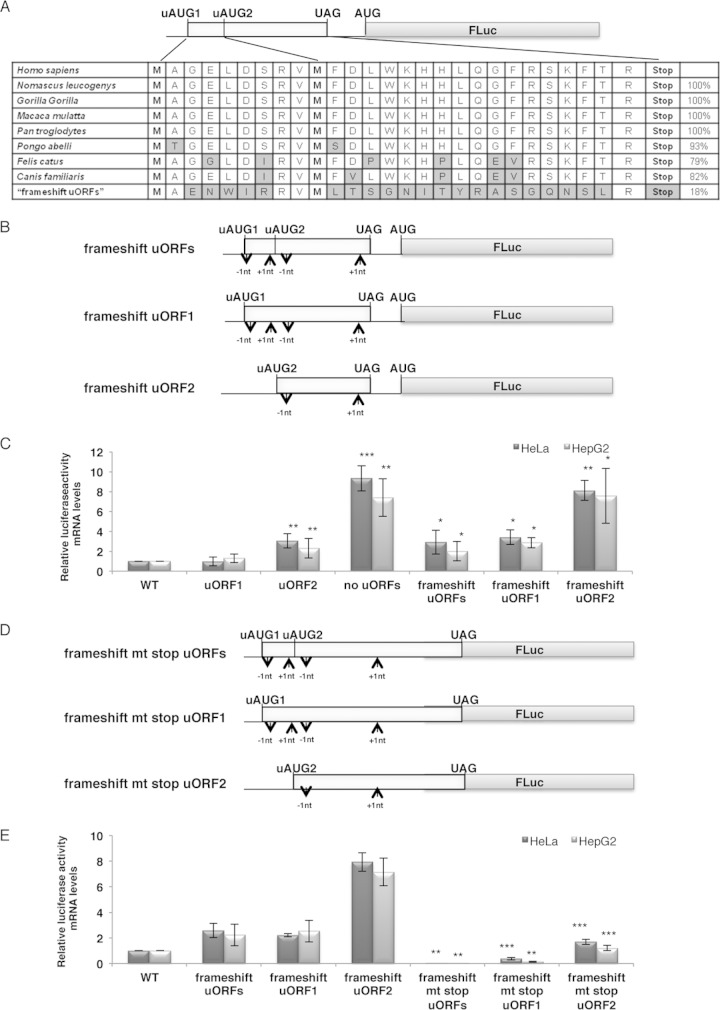
FIG 3.
The peptides encoded by the human HJV uORFs are conserved among different species and are competent to significantly inhibit downstream translation. (A) An alignment of the amino acid sequences of the peptides encoded by the human HJV uORFs and those of several mammalian species shows high degrees of similarity and the conservation of both uAUGs. Alignment of the peptide encoded by the frameshift uORFs construct is also shown. Percentages of similarity are indicated on the right. Amino acids are represented by letters; the nonconserved amino acids are indicated in gray boxes. (B) The frameshift uORFs, frameshift uORF1, and frameshift uORF2 constructs were obtained by mutagenesis of the WT, uORF1, and uORF2 constructs, respectively, by shifting the ORF without affecting the nucleotide sequence, as described in Materials and Methods. (C) Translational repression exerted by the HJV uORFs is peptide sequence dependent. Relative translation activities in HeLa and HepG2 cells are shown for each of the constructs presented in panel B. Relative translation activities were determined as described in the legend to Fig. 1B. For comparison, the chart also contains data imported from Fig. 1B. (D) Schematic representation of reporter constructs. The frameshift mt stop uORFs, frameshift mt stop uORF1, and frameshift mt stop uORF2 constructs were derived from the frameshift uORFs, frameshift uORF1, and frameshift uORF2 constructs, respectively, by mutation of the uORF stop codon. (E) The uORFs present in the frameshift constructs support very low levels of leaky scanning. Relative translation activities expressed in HeLa and HepG2 cells for each of the constructs presented in panel D, as well as for the WT, frameshift uORFs, frameshift uORF1, and frameshift uORF2 constructs, were obtained as described in the legend to Fig. 1B.
FIG 4.
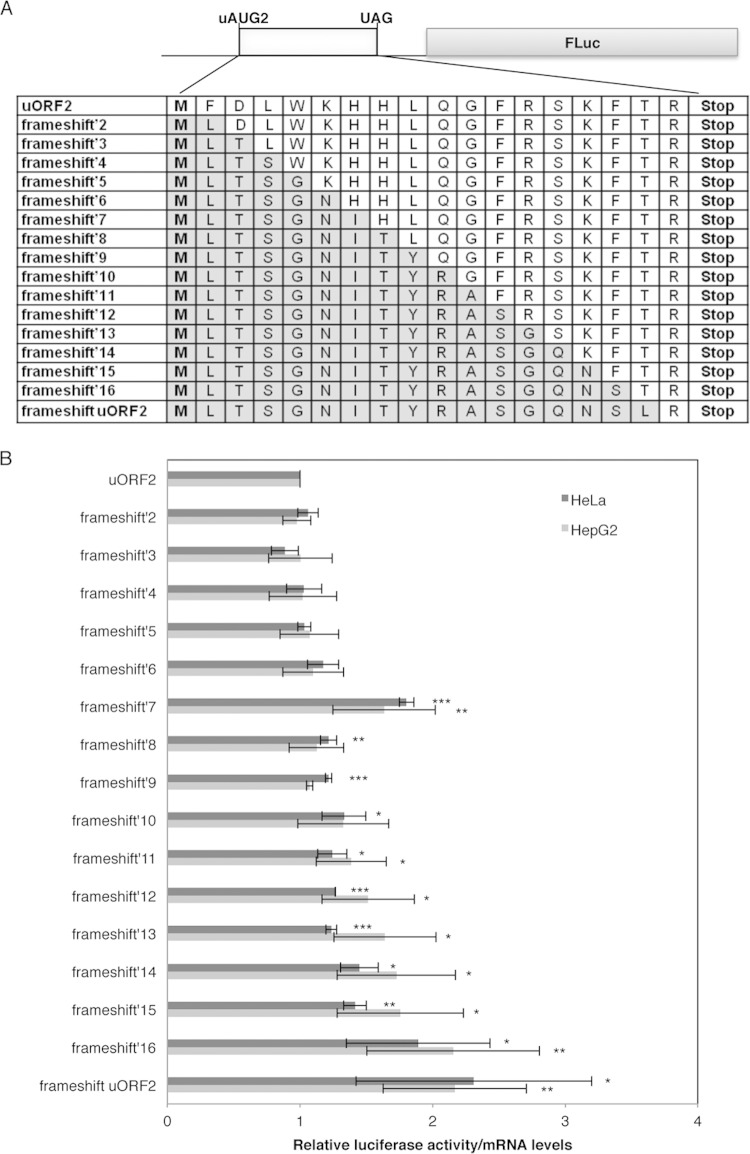
The carboxyl-terminal domain of the native HJV uORF2-encoded peptide is inhibitory to the downstream main ORF translation. (A) Representation of the uORF2 construct encoding the native peptide, with the amino acid sequence shown below. Also, the amino acid sequences of the frameshift′2 to frameshift′16 constructs are shown, as well as the amino acid sequence of the peptide encoded by the uORF present in the frameshift uORF2 construct. These mutants were obtained from the parental uORF2 construct, in which the amino acid sequence of the uORF2-encoded peptide was successively altered by consecutive frameshift mutations, using the same strategy as that for the frameshift uORF2 construct shown in Fig. 3B. Amino acids are represented by letters; the altered amino acids are indicated in gray. (B) Relative translation activities expressed in HeLa and HepG2 cells for each of the constructs presented in panel A were obtained as described in the legend to Fig. 1B.
Cell culture and plasmid transfection.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (Gibco). HepG2 cells were grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum. Transient transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen), following the manufacturer's instructions, in 35-mm plates. For gene constructs cloned into the pGL2hCMV plasmid, 1,500 ng of the test DNA construct was cotransfected with 500 ng of the pRL-TK plasmid, which encodes Renilla luciferase (RLuc), as a control for luminescence assays, and the cells were harvested after 24 h. Treatment with 20 μM holotransferrin (Sigma-Aldrich), which mimics iron overload conditions, was performed 2 h after transfection, and cells were harvested 24 h later. To activate eIF2α kinases and induce eIF2α phosphorylation, cells taken 4 h after transfection were treated with 1 μM thapsigargin (Enzo-Life Technologies) for 24 h.
Luminometry assay.
Lysis was performed in all cell lines by use of passive lysis buffer (Promega), and luminescence was measured in a Lucy 2 luminometer (Anthos Labtec) with a dual-luciferase reporter assay system (Promega) according to the manufacturer's standard protocol. One microgram of extract was assayed for firefly and Renilla luciferase activities. Ratios show the units of firefly luciferase (FLuc) activity normalized to those of Renilla luciferase (RLuc) activity, and each value was derived from three independent experiments.
RNA isolation.
Total RNA from transfected cells was prepared using a Nucleospin RNA extraction II kit (Macherey-Nagel) according to the manual provided by the manufacturer. After this step, all RNA samples were treated with RNase-free DNase I (Ambion) and purified by phenol-chloroform extraction.
Reverse transcription-quantitative PCR (RT-qPCR).
Synthesis of cDNA was carried out using 1 μg of total RNA and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed in an ABI Prism 7000 sequence detection system, using Sybr green master mix (Applied Biosystems). Specific primers for the firefly luciferase (primers 63 and 64) and Renilla luciferase (primers 65 and 66) genes were designed using ABI Primer Express software (6). Hypoxanthine phosphoribosyltransferase 1 (Hprt1) (primers 67 and 68)- and transferrin receptor 1 (TfR1) (primers 69 and 70)-specific primers were designed by Crespo et al. (27). Quantification was performed using the relative standard curve method (ΔΔCT; Applied Biosystems). The following cycling parameters were used: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 61°C. Technical triplicates from three or four independent experiments were assessed in all cases.
SDS-PAGE and Western blotting.
Protein lysates were resolved by 10% SDS-PAGE according to standard protocols and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were probed using mouse monoclonal anti-α-tubulin (Sigma) at a 1:10,000 dilution, mouse antiluciferase (Invitrogen) at a 1:200 dilution, rabbit polyclonal anti-eIF2α (Cell Signaling) at a 1:500 dilution, and rabbit polyclonal anti-phospho-Ser52 eIF2α (Invitrogen) at a 1:500 dilution. Detection was carried out using secondary peroxidase-conjugated anti-mouse IgG (Bio-Rad) and anti-rabbit IgG (Bio-Rad) antibodies, followed by chemiluminescence.
Statistical analysis.
Results are expressed as means ± standard deviations. Student's t test was used for estimation of statistical significance (unpaired, two-tailed). Significance for statistical analysis was defined as having a P value of <0.05.
RESULTS
The human HJV uORFs inhibit translation of the downstream main ORF.
The human HJV mRNA (NM_213653) presents a 5′ UTR with 325 nucleotides. Within this sequence, there are two AUGs located upstream of the main ORF, which in turn encodes the functional HJV protein. These two uAUGs are in frame with the same stop codon, located upstream of the main AUG, and thus create two overlapped uORFs which are 52 nucleotides upstream of the main AUG (Fig. 1A). The first uORF (uORF1) has 28 codons, and the second one (uORF2) has 19 codons. Both uAUG1 (GAAUCAUGG) and the main AUG (UAGGUAUGG) have very good sequence contexts, showing Kozak matches of G/A at position −3 and G at position +4 of the A(+1)UG codon; in contrast, uAUG2 (GAGUAAUGU) lacks a suitable base at position +4, despite having an adequate base at position −3. Thus, at least uAUG1 is found within a sequence context that might allow initiation of translation by ribosomes. This observation led us to investigate the effects of the HJV uORFs in modulating the translation of the downstream coding sequence.
In order to assess whether HJV uORFs can regulate translation of the downstream ORF, we designed a reporter construct containing a DNA fragment with 325 bp corresponding to the intact human HJV 5′ leader sequence fused upstream of the firefly luciferase cistron (WT construct) (Fig. 1A). Additionally, to determine the importance of each of the HJV uAUGs in regulating translation, we constructed a series of plasmids (derived from the WT construct) containing a point mutation in each uAUG (AUG → UUG) or both, creating the uORF1, uORF2, and “no uORFs” constructs (Fig. 1A). These constructs were transiently transfected into human hepatoma HepG2 cells. These cells were chosen because they correspond to liver cells, where endogenous HJV is expressed (19, 28). Cellular extracts were prepared and assayed for luciferase activity, and total RNA was isolated to quantify the luciferase mRNA levels by RT-qPCR. The FLuc activity of each construct was normalized to the activity units from RLuc expressed from the cotransfected pRL-TK plasmid. The FLuc mRNA level of each construct was also normalized to the coexpressed RLuc mRNA level. The relative luciferase activity was then normalized to the corresponding relative mRNA level and compared to that of the WT control, arbitrarily defined as 1 (Fig. 1B).
The results show that the intact HJV 5′ UTR present in the wild-type (WT) transcript induced a 7-fold repression (P = 0.004) of relative luciferase activity compared to that from the “no uORFs” construct without uORFs (Fig. 1B). In addition, compared with the WT control, mutation of the first uAUG (uORF2 construct) led to an increase in relative luciferase activity of 2.3-fold (P = 0.004), whereas mutation of the second uAUG (uORF1 construct) did not significantly affect relative luciferase activity (1.5-fold increase; P = 0.092) (Fig. 1B). Taken together, these results indicate that the intact HJV uORFs repress translation. However, uORF1 is a stronger repressor than uORF2, with uORF1 being as repressive as both uORFs in the WT transcript (Fig. 1B).
To test the generality of these data, we analyzed the relative luciferase activities of the same reporter constructs expressed in HeLa cells. Similar results were observed in these cells (Fig. 1B), suggesting that translation is strongly repressed by the HJV mRNA 5′ UTR and that this translational regulation might be a general phenomenon observed in different cell lines.
To confirm that each of the uAUGs of the HJV uORFs can in fact be recognized by the ribosome, several constructs were cloned, in which each of the HJV uORFs was fused in frame with the FLuc ORF. This was achieved by site-directed mutagenesis of the WT construct to introduce a mutation at the uORF stop codon (UAG → AAG) and a deletion of one nucleotide in the intercistronic region, with or without mutating the FLuc main AUG (AUG → UUG), creating the uAUG1_fusedLuc, uAUG2_fusedLuc, uAUG1_Luc, and uAUG2_Luc constructs (see Materials and Methods and Fig. 1C). These constructs, as well as the WT and “no uORFs” control constructs and the parental pGL2hCMV vector, were transiently transfected into HeLa cells. Twenty-four hours later, cell extracts were purified and analyzed by Western blotting using a specific antibody that recognizes firefly luciferase, with α-tubulin as a loading control (Fig. 1D). The results show that luciferase protein expression was observed from the WT and “no uORFs” constructs and from the pGL2hCMV control vector (Fig. 1D, lanes 2 and 3 versus lane 1). However, FLuc expression from the WT transcript seemed to be much lower than that from the “no uORFs” construct, in accordance with the previous data (Fig. 1B) showing that HJV uORFs repress translation. In addition, lanes 4 and 5 of Fig. 1D show expression of a protein with a molecular weight slightly higher than that of the native FLuc protein seen in lanes 1, 2, and 3. These data demonstrate that a uORF-luciferase fusion protein is expressed from the uAUG1_fusedLuc and uAUG2_fusedLuc constructs and thus provide evidence that translation can be initiated at each of the uAUGs. The same uORF-luciferase fusion protein was also expressed from the uAUG1_Luc and uAUG2_Luc constructs (Fig. 1D, lanes 6 and 7). However, uAUG2_Luc mRNA produced an additional luciferase protein product with the molecular weight of native FLuc, which indicates that translation was also initiated at the main AUG. Thus, some of the scanning ribosomes skipped the first AUG codon (i.e., uAUG2) of the uAUG2_Luc mRNA and directed translation from the downstream main AUG codon, showing that ribosomal leaky scanning of HJV uAUG2 can occur. These results are in accordance with the observation that HJV uAUG2 is in a weaker Kozak sequence context (29) than that of uAUG1. Furthermore, these data also demonstrate that human HJV uORF1 and, to a lesser extent, uORF2 are indeed efficiently recognized and translated by the ribosome and thus are functional, creating strong barriers to scanning ribosomes.
The HJV main AUG codon is mostly recognized by translation reinitiation.
Our results indicate that HJV uORF1 is a strong barrier to scanning ribosomes, which corroborates well with its very good uAUG Kozak consensus sequence, while uORF2 is less repressive, as it presents a uAUG codon in a weaker context. These results suggest that, most of the time, uAUG1 is recognized by the scanning ribosome, and the ribosome may resume scanning after translation of the inhibitory uORF1 and reinitiate translation at the main AUG. Alternatively, if the ribosome bypasses uAUG1, it may recognize uAUG2 and may reinitiate at the main AUG, or it may skip the uAUG2 codon and continue scanning until the main AUG. To test these mechanisms and to better confirm the mechanism through which the main AUG is indeed recognized by the ribosome, we first considered testing whether the main AUG is recognized by translation reinitiation. In order to test this mechanism, the sequence contexts of both uAUGs were mutated to an optimal Kozak context (“optimal uAUGs” construct) (Fig. 2A). In parallel, the sequence context of each uAUG was individually mutated to an optimal Kozak context, while the other uORF was inactivated by mutation of the corresponding uAUG (AUG → UUG) (“optimal uAUG1” and “optimal uAUG2” constructs) (Fig. 2A). After transient transfection of HeLa and HepG2 cells with each of these constructs, cells were lysed and protein and total RNA were isolated. Relative luciferase activity from each construct was analyzed as described above. Results were normalized to the control (the WT construct) and compared with those of the corresponding original construct (Fig. 2B). The results show that the optimal uAUGs construct was expressed at the same levels as the WT construct in both HeLa (P = 0.543) and HepG2 (P = 0.414) cells (Fig. 2B). Also, the constructs with only one uAUG in an optimal Kozak sequence context (optimal uAUG1 and optimal uAUG2 constructs) (Fig. 2A) were expressed at levels comparable to those of the uORF1 and uORF2 constructs in both HeLa (P = 0.168 for optimal uAUG1 versus uORF1 and P = 0.439 for optimal uAUG2 versus uORF2) and HepG2 (P = 0.999 for optimal uAUG1 versus uORF1 and P = 0.960 for optimal uAUG2 versus uORF2) cells (Fig. 2B). These data indicate that the HJV uAUGs, or at least uAUG1, are recognized by the ribosome with a high frequency, with the HJV uORFs, or at least uORF1, being efficiently translated, and thus translation of the downstream main ORF seems to occur mainly by reinitiation.
Next, we addressed whether the HJV main ORF could be translated, at least sometimes, by the leaky scanning mechanism. In this mechanism, the scanning ribosome would bypass and scan through the inhibitory uORFs and initiate translation at the downstream main AUG (12). To test for the occurrence of this mechanism, we mutated the stop codon of the HJV uORFs to a sense codon (UAG → AAG), resulting in extended uORFs that overlap, by 23 out-of-frame nucleotides, the coding region of the downstream luciferase reporter ORF; these overlapped uORFs (“mt stop uORFs” construct) (Fig. 2C), if recognized by the ribosome, would inhibit translation reinitiation. Additionally, each of the uORFs was overlapped individually with the main ORF by the stop codon mutation, while the other uORF was inactivated by mutation of the corresponding AUG (AUG → UUG) (“mt stop uORF1” and “mt stop uORF2” constructs) (Fig. 2C). Relative luciferase activities from these constructs were analyzed as described above. The results showed significant decreases of expression levels, to approximately null levels, between the mt stop uORF1 and uORF1 constructs for both cell lines (P = 0.017 for HeLa cells and P = 0.006 for HepG2 cells). Comparison of relative luciferase levels of the mt stop uORF2 construct and the uORF2 construct also revealed significant reductions in protein expression in both cell lines (4-fold reduction [P = 0.003] for HepG2 cells and 3-fold reduction [P = 0.005] for HeLa cells), but protein expression did not reach levels as low as those with the mt stop uORF1 construct (Fig. 2D). Thus, these data indicate that the native uORF2 allows more ribosomal leaky scanning than uORF1, in accordance with the fact that the uAUG2 Kozak sequence context is weaker than that of uAUG1. In addition, comparison of the relative luciferase levels of the mt stop uORFs construct and the WT construct showed a significant reduction in the expression of the mt stop uORFs construct in both HeLa (P = 1.27 × 10−5) and HepG2 (P = 6.13 × 10−5) cells, reaching approximately null levels (Fig. 2D). These data indicate that HJV uORFs, and more specifically uORF1, are efficiently recognized by the ribosome. The facts that when translation reinitiation is inhibited by the overlapped uORFs or overlapped uORF1, translation of the main ORF is strongly inhibited, and that null levels of luciferase activity are obtained in both cases confirm that translation of the HJV main ORF occurs largely by reinitiation after translation of uORF1. These results support a low level of ribosomal leaky scanning of uORF1.
The peptides encoded by the human HJV uORFs are conserved among different species and are competent to inhibit downstream translation.
Alignment of the human HJV mRNA 5′ UTR nucleotide sequence with those of Nomascus leucogenys, Gorilla gorilla, Macaca mulatta, Pan troglodytes, Pongo abelli, Felis catus, and Canis familiaris shows high degrees of similarity and the conservation of both uAUGs. In addition, an alignment of the amino acid sequences among these species also reveals high degrees of similarity (Fig. 3A). This amino acid conservation among species may reflect an important evolutionary selection pressure, revealing their role in translation regulation of HJV protein expression. This might be a consequence of the conserved nucleotide sequence of the mRNA or reflect an important evolutionary selection pressure on the peptide sequence. To address the question of whether the mRNA or the peptide sequence is important for inhibition of downstream translation, a nucleotide was deleted (−T) downstream of uAUG1 (ATGGCTG → ATGGCG) and one was inserted (+C) upstream of uAUG2 (GATAGC → GATACGC); a nucleotide was also deleted (−T) downstream of uAUG2 (ATGTTT → ATGTT) and a nucleotide inserted (+C) upstream of the uORFs stop codon (TAGGTAG → TACGGTAG) (“frameshift uORFs” construct) (Fig. 3B). The resulting frameshifts change the peptide sequence of each uORF, without affecting the position of uAUG2. Additionally, each of the uORFs was frameshifted individually, while the other uORF was inactivated by mutation of the corresponding AUG (AUG → UUG) (“frameshift uORF1” and “frameshift uORF2” constructs) (Fig. 3B). In all these constructs, the context sequence of the native uAUGs was maintained. After transient transfection of these constructs into HeLa and HepG2 cells, their relative luciferase activities were analyzed as described above (Fig. 3C). The results showed that the mutations of the frameshift uORFs construct significantly affected its relative luciferase activity levels in both HeLa (3-fold increase; P = 0.020) and HepG2 (2-fold increase; P = 0.011) cells (Fig. 3C). Parallel results were observed between the frameshift uORF1 and uORF1 constructs in both cell lines (Fig. 3C). Lastly, when we compared relative luciferase activity levels of the frameshift uORF2 construct and the uORF2 construct, we also observed an increase of about 3-fold (P = 0.003 for HeLa cells and P = 0.028 for HepG2 cells), but frameshift uORF2 expression reached levels similar to those expressed in the “no uORFs” mRNA in both cell lines (Fig. 3C). Thus, there was a derepression of translation when the peptide sequence encoded by each of the HJV uORFs was modified; however, the final effect of uORF2 frameshift was more pronounced, as it induced a complete translational derepression (Fig. 3C). These results indicate that synthesis of native peptides encoded by HJV uORFs mediates repression of main ORF translation; however, the native uORF2-encoded peptide exerts a more evident effect.
Since the frameshift uORF2 construct was expressed at levels similar to those of the “no uORFs” construct (Fig. 3C), we hypothesized that this result may have been due to the fact that the uORFs present in the frameshift constructs were not recognized and translated by the ribosome, despite their unaltered uAUG contexts. To control for this possibility, we took advantage of knowing that the overlap of the HJV mRNA uORFs with the downstream main ORF inhibits translation reinitiation of the main ORF, as previously seen in Fig. 2, which confirms their recognition by the ribosome. Therefore, we mutated the stop codon of the previous frameshift constructs to a sense codon (UAG → AAG), resulting again in extended uORFs that overlap the coding region of the downstream luciferase reporter ORF by 23 out-of-frame nucleotides (frameshift mt stop uORFs, frameshift mt stop uORF1, and frameshift mt stop uORF2 constructs) (Fig. 3D). We then expressed and analyzed these constructs in HeLa and HepG2 cells as described above (Fig. 3E). The results were normalized to those of the WT construct and compared to those of the corresponding parental frameshift construct. The data show that the uORFs that overlapped with the reporter main ORF strongly inhibit protein expression; indeed, the “frameshift mt stop uORFs” construct was expressed at null levels in both cell lines (P = 0.001 for HeLa cells and P = 0.004 for HepG2 cells) (Fig. 3E). Parallel results were obtained with the “frameshift mt stop uORF1” construct (P = 3.17 × 10−5 for HeLa cells and P = 0.001 for HepG2 cells) (Fig. 3E). In comparing expression levels of the “frameshift mt stop uORF2” and frameshift uORF2 constructs, we noted that expression of the “frameshift mt stop uORF2” construct significantly decreased (5-fold decrease [P = 0.0001] for HeLa cells and 6-fold decrease [P = 0.0002] for HepG2 cells) but did not reach null levels, again providing evidence for a ribosomal leaky scanning capacity of uORF2 (Fig. 3E). Nevertheless, this set of data also proves that the HJV uORFs (mainly uORF1) are indeed efficiently recognized and translated by the ribosome, with uORF2 being recognized less competently than uORF1. In addition, a comparison of results from Fig. 2D and 3E demonstrates that uORF sequence frameshifting does not affect the competence of uORFs being recognized by the ribosome. Also, the data from Fig. 3 show that each of the native HJV uORF-encoded peptides is able to repress translation of the downstream main ORF.
The carboxyl terminus of the native HJV uORF2-encoded peptide is inhibitory to the downstream main ORF translation.
To better understand the peptide sequence requirements for HJV uORF-mediated translation repression, and knowing that uORF2 shares the same encoded peptide sequence with uORF1, we performed a systematic frameshift mutagenesis analysis of uORF2 to determine which amino acid residues are important for translational repression. Therefore, we cloned several mutants of the parental uORF2 construct in which the amino acid sequence of the uORF2-encoded peptide was successively altered by frameshift mutations, as previously performed to obtain the frameshift uORF2 construct (Fig. 4A). The effect of each mutation on reporter protein levels was compared with protein expression levels from the uORF2 construct (Fig. 4B). The results showed significant increases of the protein levels from the “frameshift′7” through frameshift uORF2 constructs expressed in HeLa cells (Fig. 4B). Similar results were observed in HepG2 cells (Fig. 4B). These results indicated that the peptides with altered amino acids at positions 7 to 16 had less ability to repress translation than that of the native peptides (Fig. 4B). Moreover, the increase in reporter protein levels was more substantial (about 2-fold increase) when the consecutive alterations of amino acids reached codons 16 and 17 in the frameshift′16 and frameshift uORF2 constructs, which are codons contiguous to the uORF stop codon. These results indicate that the C-terminal region of the peptide encoded by the native HJV uORF2 has the ability to repress translation of the downstream main ORF.
HJV uORF-mediated translational regulation responds to iron overload in hepatic cells.
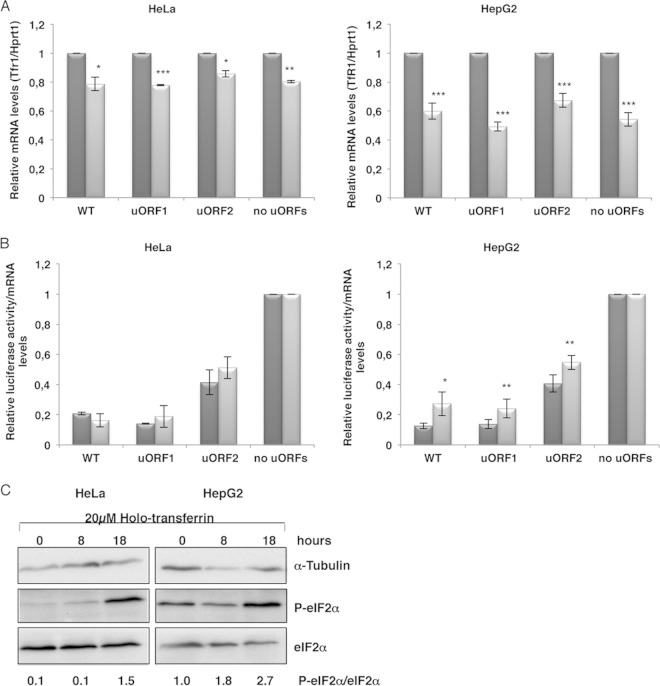
As mentioned before, HJV is responsible for the transcriptional activation of hepcidin that is involved in regulating iron homeostasis. When iron levels increase, HJV signaling is activated in order to increase the hepcidin levels, which triggers a decrease of dietary iron absorption (24, 30). Due to this HJV response to iron overload, we next examined whether the stress of iron overload affects HJV translational efficiency through its uORFs. To examine the influence of iron overload on the HJV uORF-mediated translational control, HepG2 and HeLa cells were transiently transfected with the WT, uORF1, uORF2, and “no uORFs” constructs and then incubated in medium with 20 μM holotransferrin to mimic iron overload. Eighteen hours after exposure, cells were lysed and protein and RNA were extracted and analyzed (Fig. 5). Since it has been shown that the transferrin receptor 1 (TfR1) mRNA is downregulated in cases of iron overload, which leads to a decrease of iron uptake by cells (30), the TfR1 mRNA levels were monitored by RT-qPCR to control the iron overload condition (Fig. 5A). This analysis showed a significant decrease in the relative TfR1 mRNA levels induced by holotransferrin treatment in both cell lines, as expected; however, the difference was more significant in HepG2 cells (Fig. 5A). Under normal and iron overload conditions, the relative luciferase activities of all constructs were evaluated as described above, and the results were normalized to those of the “no uORFs” construct under each condition (Fig. 5B). The results showed that in HeLa cells, iron overload had no significant effect on relative luciferase activity from either construct (Fig. 5B). However, in HepG2 cells, the relative luciferase activity from the WT construct was significantly increased (2-fold; P = 0.01) after 18 h of iron overload (Fig. 5B). Furthermore, this significant increase of protein levels between treated and untreated cells was also observed with the uORF1 construct (P = 0.008). With the uORF2 construct, the expression increased to a lower level (1.4-fold increase; P = 0.003). These results show that the HJV uORFs direct a translational derepression in response to iron overload, particularly in HepG2 cells.
FIG 5.
HJV uORF-mediated translational regulation responds to iron overload in hepatic cells. The WT, uORF1, uORF2, and “no uORFs” constructs were separately cotransfected with a plasmid encoding Renilla luciferase (pRL-TK) in HeLa and HepG2 cells. Two hours after transfection, cells were left untreated (dark gray bars) or treated (light gray bars) for 18 h with 20 μM holotransferrin. (A) To control the iron overload conditions, TfR1 mRNA levels were monitored by RT-qPCR and normalized to those of the housekeeping Hprt1 (hypoxanthine phosphoribosyltransferase 1) mRNA. (B) Untreated (dark gray bars) or treated (light gray bars) transfected cells were lysed, and relative luciferase activities were quantified as described in the legend to Fig. 1B. (C) Representative Western blot analyses of HeLa and HepG2 cell extracts left untreated (0 h) or treated with 20 μM holotransferrin for 8 or 18 h. Immunoblotting was performed using human eIF2α- and human phosphorylated eIF2α-specific antibodies to control for the state of eIF2α phosphorylation, and using a human α-tubulin-specific antibody to control for variations in protein loading.
Phosphorylation of eIF2α is a rapid consequence of many cellular stresses, reducing the availability of competent initiation complexes. Indeed, when eIF2α is phosphorylated, the ternary complex becomes scarce and global translation is compromised (1). Despite eIF2α phosphorylation, the presence of uORFs can promote the increased expression of certain stress-related mRNAs (7). Based on these data, we next asked whether cellular holotransferrin treatment is accompanied by eIF2α phosphorylation. To test this hypothesis, eIF2α phosphorylation was examined by immunoblotting using an anti-phospho-eIF2α antibody in HeLa and HepG2 cells left untreated or treated with 20 μM holotransferrin for 8 or 18 h. Detection of α-tubulin was used to control the amount of loaded protein (Fig. 5C). The results showed that the extent of eIF2α phosphorylation was increased by holotransferrin treatment for 18 h, in both cell lines (Fig. 5C). Despite the increase of eIF2α phosphorylation in both cell lines (Fig. 5C), our results showed that a significant translational derepression occurred only in HepG2 cells (Fig. 5B). Thus, we concluded that if eIF2α phosphorylation is involved in the mechanism through which translational derepression occurs, it is not the only factor promoting it; instead, a tissue-specific factor might also be involved.
HJV uORF-mediated translational regulation responds to eIF2α phosphorylation in hepatic cells but not in HeLa cells.
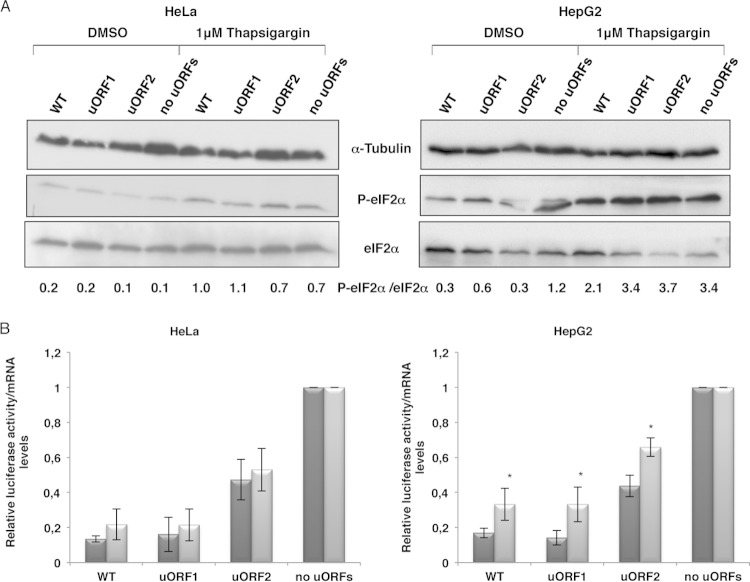
Based on the previous data, we next tested whether the treatment of HeLa and HepG2 cells with the potent endoplasmic reticulum stress agent thapsigargin, which directly activates eIF2α kinases without another signaling pathway (31, 32), would induce translational derepression of the reporter luciferase ORF. Thus, cells were transiently transfected with the WT, uORF1, uORF2, and “no uORFs” constructs and then left untreated or treated with 1 μM thapsigargin. After 18 h of exposure, cells were analyzed as described previously, and results were normalized to those of the “no uORFs” construct under each condition. The extent of eIF2α phosphorylation in untreated and thapsigargin-treated cells was examined by immunoblotting as described above. Figure 6A shows that the extent of eIF2α phosphorylation was increased by thapsigargin treatment in both cell lines. Figure 6B shows that phosphorylation of eIF2α effectively induced a significant increase (2-fold) of luciferase translation in the WT mRNA (P = 0.020), specifically in treated compared to untreated HepG2 cells (Fig. 6B). In addition, reporter translation of the uORF1 (P = 0.025) and uORF2 (P = 0.012) mRNAs also increased significantly (2- and 1.5-fold, respectively) (Fig. 6B). In contrast, treatment of HeLa cells induced irrelevant increases in reporter activity from the WT (P = 0.183), uORF1 (P = 0.524), and ORF2 (P = 0.586) constructs. From these findings, it is evident that eIF2α phosphorylation, although occurring in both cell lines, promotes a significant increase in reporter translation, especially in hepatic HepG2 cells. Therefore, these results uncover an additional tissue-specific factor involved in the mechanism through which HJV uORF-mediated translational derepression occurs in liver cells in response to iron overload.
FIG 6.
HJV uORF-mediated translational regulation responds to eIF2α phosphorylation in hepatic cells. The WT, uORF1, uORF2, and “no uORFs” constructs were separately cotransfected with a plasmid encoding Renilla luciferase (pRL-TK) into HeLa and HepG2 cells. Six hours after transfection, cells were left untreated (dimethyl sulfoxide [DMSO]) or treated with 1 μM thapsigargin for 24 h. (A) Representative Western blot analyses of HeLa and HepG2 cell extracts left untreated or treated as indicated. Immunoblotting was performed using human eIF2α- and human phosphorylated eIF2α-specific antibodies to control for eIF2α phosphorylation conditions, and a human α-tubulin-specific antibody was used to control for variations in protein loading. (B) Relative luciferase activities were quantified as described in the legend to Fig. 1B.
DISCUSSION
With the aim of investigating the functions of the two overlapping and in-frame uORFs (uORF1 and uORF2) present in the 5′ UTR of the human HJV mRNA, here we show that both uORFs are functional, preventing the main ORF from being translated with full competence, in both HeLa and HepG2 cells (Fig. 1). However, uORF1 is the major translational repressor of the main ORF. Note that the effect of uORF1 alone was similar to that obtained when both uORFs were present (Fig. 1). This high level of competence was expected, since uAUG1 lies in a very good AUG sequence context, having suitable nucleotides in the −3 and +4 positions. The better context of uAUG1 leads to the recognition of uORF1 by the ribosome in the vast majority of the rounds of translation, strongly reducing the production of the functional protein. On the other hand, uORF2 represses translation with less efficiency due to its weaker AUG sequence context (it lacks the appropriate nucleotide at position +4). It may function as a backup for those few rounds of translation in which the ribosome bypasses uAUG1, creating an additional barrier to the ribosome. This potential fail-safe mechanism might help to secure translational repression of the main ORF. Apparently, a similar fail-safe system has been described previously for the yeast GCN4 transcript (33).
Aiming to test the mechanism through which translation initiation occurs in the main ORF of the HJV mRNA we showed (Fig. 2) that reinitiation is the major mechanism in both HeLa and HepG2 cells. Since reinitiation depends on the length of the intercistronic region, on the secondary structures of the transcript, and on the uORF length (12–15, 34), our results illustrate that the human HJV mRNA intercistronic region has favorable features for reinitiation; also, the uORF properties (such as length and translational rate) seem to be appropriate for reinitiation to occur. In contrast, the results revealed a very low leaky scanning capacity in both cell lines (Fig. 2B). This low level of leaky scanning can be justified by the strong contexts of the uAUGs, especially uAUG1, which is recognized by the scanning ribosome most of the time.
To understand whether the HJV uORF-encoded peptides play a role in translational repression, we analyzed a set of constructs in which frameshift mutations were introduced into the uORFs without changing the nucleotide sequence (Fig. 3). This analysis revealed that the HJV uORF-encoded peptides, together or individually (uORF1 or uORF2), are involved in the mechanism by which translation of the downstream main ORF is repressed (Fig. 3). Also, we observed that the C-terminal region of the uORF-encoded peptides is involved in repressing translation of the downstream main ORF (Fig. 4). This effect may consist of ribosome stalling due to the ability of the peptide sequence to act in a cis manner to impede ribosomal movement during either elongation or termination, although future experiments are needed to confirm such an occurrence. Such a mechanism has already been described for the transcripts encoding the fungal arginine attenuator peptide (35) and, in humans, the CCAAT/enhancer-binding protein homologous protein (CHOP) (36). Nevertheless, we cannot exclude the possibility that this effect occurs in trans, as more recently shown for a human transcript encoding a microsomal epoxide hydrolase (EPHX1) (37).
In the present study, our data show that both HJV uORFs, when translated, are able to repress translation through the same mechanisms: translation reinitiation and uORF-encoded peptide-dependent repression. Indeed, the conservation of the HJV uORF-encoded peptide sequences among different species supports this hypothesis. Nevertheless, we cannot rule out the alternative hypothesis, in which the uORF1- and uORF2-encoded peptides may act differently. Length and/or the secondary structure of the encoded peptide may affect its function. It is possible that the two encoded peptides interact differently with the ribosome, depending on their lengths and/or secondary structures. For example, by being shorter, the uORF2-encoded peptide might be more effective at interacting with the translational machinery and thus might stall the ribosome with a higher efficiency.
Despite prior studies showing that HJV liver membrane protein expression itself does not appear to be regulated by iron in mice and rats (38), our data have shown that in response to iron overload, translational repression mediated by the human HJV uORFs is significantly relieved in hepatic HepG2 cells (Fig. 5 and 6). This might be due to the differences in the uORF-encoded peptides observed between humans, mice, and rats, which may reflect a fine-tuning regulatory mechanism that appeared during mammalian evolution. In addition, we observed that each HJV uORF responds equally to iron overload in hepatic cells, which supports the above-mentioned fail-safe function of uORF2. We also observed that the upregulation of protein levels occurs concomitantly with eIF2α phosphorylation (Fig. 5). Although the finding that iron overload induces eIF2α phosphorylation seems to be very interesting, to our knowledge, nothing is known about eIF2α phosphorylation by holotransferrin or by other sources of iron; indeed, it may result from an indirect mechanism. Nevertheless, the consequent phosphorylation of eIF2α may mediate the HJV uORF ribosomal bypass, as previously shown in other systems, such as the mammalian ATF4 mRNA (39–41). However, we show here that translational upregulation in response to the abnormal stimulus of iron overload, although involving eIF2α phosphorylation, needs an additional hepatic tissue-specific regulator(s), since phosphorylation of eIF2α occurs in both cell lines tested but protein upregulation is seen only in HepG2 cells (Fig. 5 and 6). Note that our data show that HJV uORF-mediated translational regulation in response to iron overload occurs in hepatic cells, which corresponds to a tissue where HJV expression occurs in adults (19, 21). Nevertheless, we cannot rule out the hypothesis that this translational regulation also occurs in myocytes, which likewise express HJV.
Our results support a model in which the increasing recognition of the HJV main AUG, observed in hepatic cells under iron overload conditions, is mediated by a tissue-specific factor that, together with phosphorylated eIF2α and lowered levels of eIF2-GTP, increases the time required for scanning ribosomes to scan through the inhibitory uORFs and instead translate the main HJV coding region. We do not yet completely understand the biochemical basis for the potential ribosomal bypass of the uORFs in the HJV mRNA in response to iron overload in hepatic cells. Additional contributors to this bypass may be the eIF2α phosphorylation-mediated expression regulation of other critical translation factors or tissue-specific regulators that would then facilitate the bypass of the HJV uORFs. A noteworthy report was recently published, showing for the first time how specific proteins can function as selective regulators of translation reinitiation in transcripts containing uORFs with strong Kozak sequences (42).
In conclusion, our results show that under physiological conditions, translation of the HJV uORFs serves as a strong barrier that inhibits translation of the downstream main ORF. Under conditions of iron overload, enhanced eIF2α phosphorylation in combination with a liver-specific factor(s) significantly increases ribosomal bypass of the uORFs, and translation of the downstream main ORF occurs with a higher efficiency. Overall, the current results report that the HJV uORFs have the ability to modulate the levels of functional protein according to the cellular iron levels.
ACKNOWLEDGMENTS
This research was partially supported by the Fundação para a Ciência e a Tecnologia (grants UID/MULTI/04046/2013 to BioISI and SFRH/BD/63581/2009 to C.B.).
The pRL-TK plasmid was a kind gift of Margarida Gama-Carvalho. Automated sequencing was performed at the Unidade de Tecnologia e Inovação (Departamento de Genética Humana, Instituto Nacional de Saúde Doutor Ricardo Jorge). Also, we thank Luka Clarke for English editing of the manuscript.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, Willis AEE. 2012. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci 13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa C, Peixeiro I, Romão L. 2013. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somers J, Pöyry T, Willis AE. 2013. A perspective on mammalian upstream open reading frame function. Int J Biochem Cell Biol 45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A 106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wethmar K, Smink JJ, Leutz A. 2010. Upstream open reading frames: molecular switches in (patho)physiology. Bioessays 32:885–893. doi: 10.1002/bies.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa C, Romão L. 2014. Translation of the human erythropoietin transcript is regulated by an upstream open reading frame in response to hypoxia. RNA 20:594–608. doi: 10.1261/rna.040915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández G, Altmann M, Lasko P. 2010. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem Sci 35:63–73. doi: 10.1016/j.tibs.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Churbanov A, Rogozin IB, Babenko VN, Ali H, Koonin EV. 2005. Evolutionary conservation suggests a regulatory function of AUG triplets in 5′-UTRs of eukaryotic genes. Nucleic Acids Res 33:5512–5520. doi: 10.1093/nar/gki847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kochetov AV. 2008. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays 30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 12.Kozak M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol 115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris DR, Geballe AP. 2000. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20:8635–8642. doi: 10.1128/MCB.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachs MS, Geballe AP. 2006. Downstream control of upstream open reading frames. Genes Dev 20:915–921. doi: 10.1101/gad.1427006. [DOI] [PubMed] [Google Scholar]
- 15.Pöyry T, Kaminski A, Jackson RJ. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palam LR, Baird TD, Wek RC. 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y. 2008. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem 283:2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 18.Geballe AP, Morris DR. 1994. Initiation codons within 5′-leaders of mRNAs as regulators of translation. Trends Biochem Sci 19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 19.Papanikolaou G, Samuels ME, Ludwig EH, Macdonald MLE, Franchini PL, Dubé M, Andres L, Macfarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. 2004. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet 36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 20.Malyszko J. 2009. Hemojuvelin: the hepcidin story continues. Kidney Blood Press Res 32:71–76. doi: 10.1159/000208988. [DOI] [PubMed] [Google Scholar]
- 21.Martinez AR, Niemla O, Parkkila S. 2004. Hepatic and extrahepatic expression of the new iron regulatory protein hemojuvelin. Haematologica 89:1441–1445. [PubMed] [Google Scholar]
- 22.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. 2006. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 23.Nohe A. 2004. Signal transduction of bone morphogenetic protein receptors. Cell Signal 16:291–299. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Anderson GJ, Frazer DM. 2006. Iron metabolism meets signal transduction. Nat Genet 38:503–504. doi: 10.1038/ng0506-503. [DOI] [PubMed] [Google Scholar]
- 25.Anderson GJ, Darshan D, Wilkins SJ, Frazer DM. 2007. Regulation of systemic iron homeostasis: how the body responds to changes in iron demand. Biometals 20:665–674. doi: 10.1007/s10534-006-9030-2. [DOI] [PubMed] [Google Scholar]
- 26.De Gobbi M, Roetto A, Piperno A, Mariani R, Alberti F, Papanikolaou G, Politou M, Lockitch G, Girelli D, Fargion S, Cox TM, Gasparini P, Cazzola M, Camaschella C. 2002. Natural history of juvenile haemochromatosis. Br J Haematol 117:973–979. doi: 10.1046/j.1365-2141.2002.03509.x. [DOI] [PubMed] [Google Scholar]
- 27.Crespo AC, Silva B, Marques L, Marcelino E, Maruta C, Costa S, Timóteo A, Vilares A, Couto FS, Faustino P, Correia AP, Verdelho A, Porto G, Guerreiro M, Herrero A, Costa C, de Mendonça A, Costa L, Martins M. 2014. Genetic and biochemical markers in patients with Alzheimer's disease support a concerted systemic iron homeostasis dysregulation. Neurobiol Aging 35:777–785. doi: 10.1016/j.neurobiolaging.2013.10.078. [DOI] [PubMed] [Google Scholar]
- 28.Niederkofler V, Salie R, Sigrist M, Arber S. 2004. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci 24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Pantopoulos K. 2011. Regulation of cellular iron metabolism. Biochem J 434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7:1153–1163. doi: 10.1016/S1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 32.Koumenis C, Naczki C, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG, Koritzinsky M. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol Cell Biol 22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunišová S, Valášek LS. 2014. Fail-safe mechanism of GCN4 translational control—uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res 42:5880–5893. doi: 10.1093/nar/gku204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer HA, Thomas AAM. 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem J 367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei J, Wu C, Sachs MS. 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol Cell Biol 32:2396–2406. doi: 10.1128/MCB.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jousse C, Bruhat A, Carraro V, Urano F, Ferrara M, Ron D, Fafournoux P. 2001. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res 29:4341–4351. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen HL, Yang X, Omiecinski CJ. 2013. Expression of a novel mRNA transcript for human microsomal epoxide hydrolase (EPHX1) is regulated by short open reading frames within its 5′-untranslated region. RNA 19:752–766. doi: 10.1261/rna.037036.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krijt J, Frýdlová J, Kukačková L, Fujikura Y, Přikryl P, Vokurka M, Nečas E. 2012. Effect of iron overload and iron deficiency on liver hemojuvelin protein. PLoS One 7:e37391. doi: 10.1371/journal.pone.0037391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ron DWP. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 40.Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Küchler K, Stoecklin G, Duncan KE, Teleman AA. 2014. DENR–MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]