Abstract
Objective
The transcription factor PU.1 (encoded by Sfpi1) promotes myeloid differentiation but it is unclear what downstream genes are involved. MiRNAs are a class of small RNAs that regulate many cellular pathways including proliferation, survival and differentiation. The objective of this study was to identify miRNAs downstream of PU.1 that regulate hematopoietic development.
Materials and Methods
MiRNAs that change expression in a PU.1-inducible cell line were identified with microarrays. The promoter for a miRNA cluster upregulated by PU.1 induction was analyzed for PU.1 binding by electrophoretic mobility shift and chromatin immunoprecipitation assays. Retroviral transduction of hematopoietic progenitors was performed to evaluate the effect of miRNA expression on hematopoietic development in vitro and in vivo.
Results
We identified a miRNA cluster whose pri-transcript is regulated by PU.1. The pri-miRNA encodes three mature miRNAs: miR-23a, miR-27a, and miR-24-2. Each miRNA is more abundant in myeloid cells compared to lymphoid cells. When hematopoietic progenitors expressing the 23a cluster miRNAs were cultured in B cell promoting conditions we observed a dramatic decrease in B lymphopoiesis and an increase in myelopoiesis compared to control cultures. In vivo, hematopoietic progenitors expressing the miR-23a cluster generate reduced numbers of B cells compared to control cells.
Conclusions
The miR-23a cluster is a downstream target of PU.1 involved in antagonizing lymphoid cell fate acquisition. Although miRNAs have been identified downstream of PU.1 in mediating the development of monocytes and granulocytes, the 23a cluster is the first downstream miRNA target implicated in regulating the development of myeloid versus lymphoid cells.
Introduction
Transcription factors determine hematopoietic cell fate decisions through gene regulation. Gene targeting has demonstrated that factors such as GATA1, PU.1, and C/EBPα are essential for the development of specific blood lineages[1-4]. Since transcription is critical for programming development other mechanisms regulating gene expression may also be critical for determining blood cell fates. MicroRNAs (miRNAs) are a class of small (~22 nucleotides), non-coding regulatory RNAs, that regulate gene expression post-transcriptionally [5]. Unlike transcription factors that are often absolutely required for the expression of specific genes, miRNAs may fine-tune gene expression, and not act as on/ off switches [6, 7].
The ability of miRNAs to fine-tune gene expression may have dramatic effects on cellular differentiation. Others and we have shown that different concentrations of transcription factors direct distinct cell fate acquisition in several developmental systems[8-10]. Targeting the c-myb transcription factor with the miR-150 miRNA has developmental consequences in the hematopoietic system. C-myb expression in a Megakaryocyte-Erythrocyte Progenitor (MEP) affects development in a concentration-dependent manner with high c-myb directing erythroid differentiation and low c-myb directing megakaryocyte differentiation[11]. MiR-150 targets c-myb mRNA to promote megakaryocyte cell fate acquisition MEPs in vitro and in vivo[12, 13]. This study demonstrated that a miRNA could direct hematopoietic cell fate.
MiRNAs are also implicated in the development of macrophages and granulocytes. The first miRNA implicated in myeloid development was miR-223. Exogenous expression of miR-223 in the promyelocytic leukemia cell line NB4 induces granulocyte differentiation[14]. Other miRNAs implicated in myeloid development include miRNAs coded by the related miRNA clusters, miR-17-92 and miR-106a-92; and miR-21 [15, 16].
We have been interested in understanding the mechanism by which the transcription factor PU.1 regulates hematopoietic development[9]. Mice lacking PU.1 have defects in myelopoiesis[1, 3]. In addition high retroviral expression of PU.1 in multipotential progenitors directs myeloid cell development at the expense of B cell development[17]. We hypothesized that miRNAs are important downstream targets of PU.1 in promoting the myeloid lineage. In an attempt to identify miRNAs critical for myelopoiesis, miRNA profiling was performed with RNA isolated from a myeloid progenitor cell line expressing an inducible PU.1 protein [9, 18]. We focused analysis on three miRNAs that were upregulated by PU.1 activity, miRs-23a, -27a, and -24-2. These three miRNAs are clustered on a single pri-transcript and is referred to as the 23a cluster. The promoter for the cluster contains conserved binding sites for PU.1. We demonstrate that PU.1 binds to the promoter in vitro and in vivo. Similar to PU.1, ectopic expression of the cluster miRNAs in hematopoietic progenitors favors the development of myeloid cells at the expense of lymphoid cells in vitro. Bone marrow transplant assays demonstrate that the miR-23a cluster represses the development of B cells in vivo. Our data suggests that the miR-23a cluster is a downstream effecter of PU.1 in hematopoietic development.
Materials and Methods
RNA isolation and microarray analysis
RNA was isolated from PUER cells at days 0, 1, and 4 of treatment with 4-hydroxy-tamoxifen (OHT) using Trizol (Invitrogen). RNAs were used for microarray analysis by LC Biosciences (Houston, TX). Samples were enriched for small RNA, after which samples were labeled with Cy3 and Cy5 fluorescent dyes and hybridized to a μParaFlo microfluidics chip that held probes for 565 mature miRNA which represented the mouse miRNAs present in miRBase sequence database version 7.1 (Sanger Institute, Cambridge, U.K.; http://microrna.sanger.ac.uk.sequences). Analysis was performed in duplicate with independent RNA samples obtained from each of the timepoints.
RNA isolation and Northern blot
Northern blots were prepared with RNA obtained from day 0, 2, 4 and 7 OHT-treated PUER cells. RNA was electrophoresed and transferred to Gene Screen Plus. Blots were probed with 32P-labeled oligonucleotides complimentary to miRNAs. Probes were hybridized to the membrane in hybridization buffer containing 7% SDS/ 0.2M Na2PO4 overnight at 35oC. The following day, membranes were washed in 2XSSPE/ 0.1% SDS and exposed to film.
Electrophoretic mobility shift assay (EMSA)
PU.1 binding sites in the mirn23a promoter were predicted using the Transcription Element Search Software (TESS) program (http://www.cbil.upenn.edu/cgi-bin/tess/tess). For EMSA experiments, nuclear extracts were prepared from 293T cells transfected with pcDNA3.1 or pcDNA3.1 PU.1. Nuclear protein was incubated with 32P 5′-end-labeled double-stranded oligonucleotides, +/− 50ng unlabeled competitor double stranded oligonucleotide. For supershifting experiments, 1 ul of anti-PU.1 antibody (sc-352X, Santa Cruz Biotechnology) was added to the reaction. Protein/ DNA complexes were separated by electrophoresis. Competitor oligonucleotides, WTMCSFR: tcgacctagctaaaaggggaagaagaggatcagc, MTMCSFR: tcgacctagctaaaagggatcggtatcggtaccgatcagc.
Chromatin immunoprecipitations (ChIP)
ChIP was performed using the ChIP-IT express kit (Active Motif) according to manufacturers instructions. Chromatin was incubated with either 2ug anti-PU.1, or anti-GATA1 (Santa Cruz Biotechnology, sc352, sc-265). The presence of the 23a promoter in immunoprecipitations was quantified by real-time PCR using Absolute Blue QPCR SYBR Green Mix (Thermo Scientific). The amplification primers were GATAAACGTGAGCCACCAAC and CCACCCCACACCACCTA.
Real-time PCR
RNA was isolated as described above from the indicated cell populations. Quantitative expression analysis was performed used miR-specific Taqman reagents (Applied Biosystems). Relative expression was calculated using the comparative 2ΔΔCt method. SnoRNA 202 expression was used to normalize miRNA expression across different RNA preparations. Results are represented as means +/− SEM of three independent experiments.
Retrovirus preparation
MSCV-EGFP and MSCV-EGFP miRNA expressing retroviral plasmids were cotransfected into 293T cells together with the retroviral packaging vector pCL-Eco (Imgenex) using Lipofectamine 2000 (Invitrogen). Forty-eight hours and 72h post-transfection retroviral supernatants were harvested and concentrated with Centricon Plus-70 filters (Millipore).
Retroviral infection and In vitro hematopoietic culture
The use of mice in these experiments was approved by the University of New Mexico LACUC (Protocol # 07UNM027). Bone marrow cells were isolated from femurs of 6-week old mice. Mature erythroid cells were removed by ammonium chloride lysis. Nucleated cells were lineage depleted with a MACS lineage cell separation kit according to manufacturers instructions (Miltenyi Biotec). Bone marrow was infected with retrovirus through 2 rounds of spinoculation. During infection cells cultured in IMDM supplemented with 10% defined FBS, Penicillin/Streptomycin, Glutamax, 2-mercaptoethanol (Invitrogen) 10ng/mL mIL-3, 20ng/mL mIL-6, mSCF 25ng/mL, mTPO 25ng/mL and 8ug/mL polybrene (Chemicon). Recombinant mouse cytokines obtained from R&D Systems, or Invitrogen. For myeloid conditions cells were cultured in IMDM an additional 4 days in the indicated cytokines. For evaluating B cell versus myeloid development, infected cells were co-cultured with OP9 cells in IMDM media containing 1ng/ml IL-7 and 5ng/ml Flt3L.
Sorting and cytocentrifugation of cells
For analysis of 23a cluster miRNA expression in primary cells, bone marrow was isolated from mouse femurs. Isolated cells were incubated with the following combination of antibodies: TERR119-FITC, CD11b-FITC, CD19-PE, and/or GR1-APC (EBioscience). Cells were then sorted on a MOFLO instrument in the UNM Cancer Center Flow Cytometry Shared Facility. Similarly cultured bone marrow cells infected with indicated retroviruses were sorted into GFP+CD11b+, and GFP+CD19+ cell populations after incubation with anti-CD19-PE, and anti-CD11b-APC (EBioscience). Progenitor populations were isolated as previously described[19]. For morphology evaluation isolated cells were cytocentrifuged onto glass slides, fixed and stained with HEMA 3 kit (Fisher). Photomicrographs of cytospins were taken with Axioskop Fluorescent microscope via a 40X objective and images analyzed with Slidebook software (UNM Cancer Center shared microscopy facility).
Bone marrow transplant assay
Female 6-7 week old BALB/c mice (Jackson Laboratories) were used as bone marrow donors and recipients. Donor mice were treated with 5mg of 5-fluorouracil (5-FU). 4 days post-treatment bone marrow was harvested and RBCs removed by hypotonic lysis. Nucleated bone marrow was spin-infected twice with the indicated viral supernatants. Cells were infected in media containing 6ng/ml rIL-3, 10ng/ml rIL-6, and 100ng/ml SCF. Transduced bone marrow cells were introduced into lethally irradiated (2 doses 450 rads) 8-week-old female recipients via tail vein injection. Recipients were sacrificed between 7 and 8 weeks transplant and single-cell suspensions were prepared from BM, and spleen. Contribution to hematopoietic lineages was examined with flow cytometry analyzing GFP and lineage specific cell surface protein expression
Results
Changes in miRNA expression as PUER cells differentiated into monocyte/macrophages
A PU.1−/− mouse myeloblast cell line expressing an estrogen receptor-PU.1 fusion protein (PUER) has been previously used as a model for monocyte differentiation in messenger RNA expression studies[20, 21]. Addition of the synthetic hormone 3-hydroxy tamoxifen (OHT) activates the PUER fusion protein and induces monocyte differentiation. We used this cell line to identify miRNAs that change expression during monocyte development. RNA was collected at days 0, 1 and 4 after addition of OHT. RNAs were hybridized to a miRNA microarray chip that contained probes representing 565 mouse miRNAs. In agreement with a previous human miRNA microarray study, we observed that miRNAs of the miR-17 cluster and the related 106a and 106b clusters (miRs -17, -18a, -20a, -92a, -92b, -93, and -106b) were downregulated during monocytic differentiation indicating that the PUER cells are a relevant model for examining monocyte miRNA expression (Fig 1 and Data not shown)[15].
Fig 1. Northern blots assaying expression of miRNAs during differentiation of PUER (monocytic) and 32D (granulocytic) differentiation.
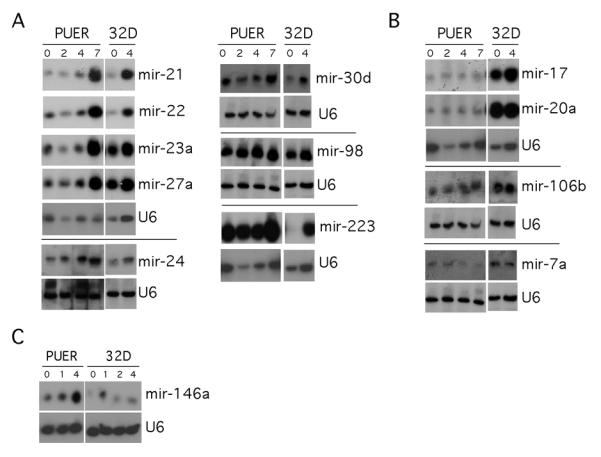
PUER cells were differentiated to monocytes with 100nM OHT. RNA was collected at days 0, 2, 4, and 7 post-treatment. 32Ds were differentiated to granulocytes with 10ng/ul G-CSF. RNA was collected at days 0 and 4 post-G-CSF treatment. Northern blots were prepared and probed for indicated miRNAs and U6 snRNA (small RNA loading control.) A) MiRNAs whose expression increased during monocytic and granulocytic development B) miRNAs whose expression preferentially increased during granulocytic development. C) MiRNAs whose expression preferentially increased during monocyte development.
Expression of several of the differentially expressed and/or highly expressed miRNAs was validated by northern blot with RNA obtained from PUER cells. In addition we examined the expression of these RNAs in the mouse 32Dcl3 cell line as they were differentiated to granulocytes with G-CSF. The majority of miRNAs that were analyzed were upregulated during both monocytic (PUER) and granulocytic (32Dcl3) differentiation (Fig 1A). Several miRNAs were shown to be upregulated only during granulocytic differentiation (Fig 1B). Of the miRNAs analyzed by northern blot, only miR-146a was observed to specifically increase abundance during monocyte differentiation (Fig 1C).
PU.1 associates with the 23a cluster promoter
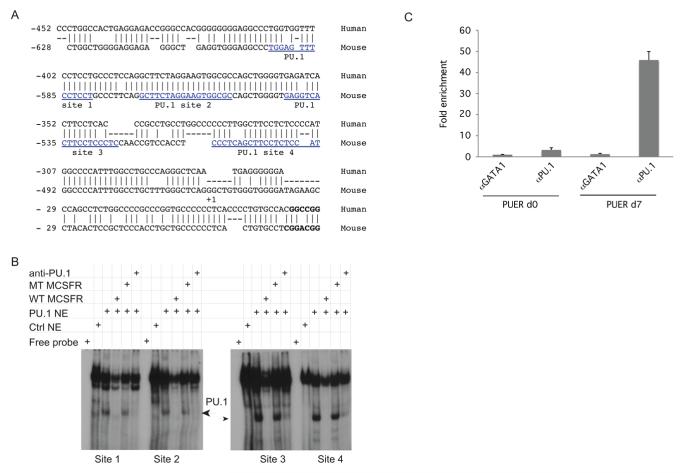
Of the miRNAs we identified as induced after OHT-treatment, miR-21 and miR-223 had promoters that were characterized as PU.1 regulated[22, 23]. We examined the promoters of other miRNAs that were upregulated in our screen for conserved PU.1 binding sites. We observed that the miR-23a cluster promoter contains 4 conserved PU.1 binding sites. Since this suggested that transcription of the 23a cluster gene is PU.1 regulated, we focused our attention on this miRNA cluster, which codes for the miR-23a, miR-27a, and miR-24-2 miRNAs (Fig 2A). To determine if PU.1 bound the promoter through these putative binding sites we performed electrophoretic mobility shift assays (EMSAs). Nuclear extracts from PU.1 expressing 293T cells were incubated with labeled probes representing each of the predicted PU.1 binding sites (Fig 2B). Presence of PU.1 in one of the retarded DNA complexes was shown by competition experiments with oligonucleotides from the MCSFR promoter containing wildtype (WT) and mutated (MT) PU.1 binding sites. The WT oligonucleotide could compete away a specific complex but the MT could not. Additionally PU.1 antibody ablated this DNA-protein complex. To determine if PU.1 interacted with the endogenous miR-23a promoter, we carried out chromatin immunoprecipitations (ChIPs) with untreated PUER cells or OHT-treated PUER cells. Analyzed by quantitative PCR there was over a 40-fold enrichment of the 23a cluster promoter in anti-PU.1 immunoprecipitates from d7 OHT treated PUER cells compared to GATA1 precipitates from d0 PUER cells (Fig 2C). We did not detect DNA upstream of the miR-23a promoter in immunoprecipitations with anti-PU.1 (Data not shown). These results indicated that PU.1 associates with the mirn23a (gene for the 23a cluster) promoter in myeloid cells.
Fig. 2. PU.1 binds to conserved sequences in the 23a cluster promoter.
A) Alignment of conserved regions of the miR-23a cluster promoter from mouse and human. Numbering is relative to human start site. Putative PU.1 binding sites (AGGAA core sequence) are boxed in red. B) EMSA demonstrating PU.1 binds in vitro to each of the 4 conserved PU.1 sites (blue underlined sequence used as probes). Nuclear extracts (NE) prepared from mock (ctrl), or PU.1 transfected 293T cells. Controls include incubations with unlabeled wildtype and mutant competitor oligos as well as antibody to PU.1. C) Chromatin immunoprecipitation analysis. Sheared chromatin extracts were prepared from PUER cells untreated or treated with OHT for 7 days. Chromatin was immunoprecipitated with anti-PU.1 or as a negative control anti-GATA1 antibody. Quantitative SYBR green PCR was performed to determine whether the 23a promoter was present in the immunoprecipitates. Fold-enrichment of the 23a promoter above what was detected in GATA1 precipitates from untreated PUER cells is shown.
Mature 23a cluster miRNAs are predominantly expressed in myeloid cells
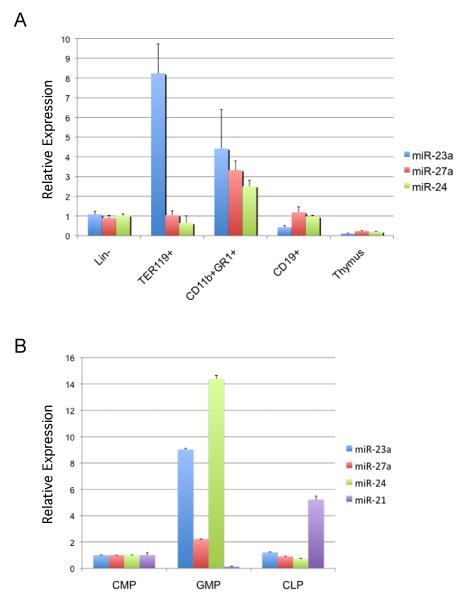
Regulation of the miR-23a cluster by PU.1 in the PUER cell line suggested that the 23a cluster is expressed preferentially in myeloid cells of the hematopoietic system. To confirm this hypothesis, we performed real-time reverse transcriptase PCR (RT-PCR) assays with RNA obtained from isolated bone marrow hematopoietic cells and thymocytes. We examined expression of 23a cluster miRNAs in Lin- bone marrow (pool of early hematopoietic progenitors), TERR119+ (erythroid), CD11b+GR1+ (myeloid), CD19+ (B cells), and thymocytes (T cells) by Taqman analysis (Fig 3A). Expression levels are shown relative to expression detected in the Lin- progenitors. All three miRNAs were more highly expressed in the CD11b+GR1+ myeloid cells compared to CD19+ B cells and thymocytes. This data is consistent with publicly available expression results for human hematopoietic cells available at microRNA.org (Fig S1B)[24]. Although miR-27a and miR-24 were also not as highly expressed in erythroid cells compared to myeloid cells we did observe high expression of miR-23a in TER119 erythroid cells. A similar expression analysis was carried out with murine hematopoietic cell lines representing multiple hematopoietic lineages (Fig S1A). Consistent with the results in primary cells we observed preferential expression of the 23a cluster miRNAs in myeloid cells compared to lymphoid cells. In contrast, there was low expression of miR-23a in the erythroid cell line examined. In mammals there exists a homologous cluster of miRNAs, which codes for miRs-23b, -27b, and -24-1. However data from human hematopoietic cells demonstrates that the 23a cluster miRNAs are preferentially expressed in blood cells (Fig S1B).
Fig. 3. Expression of miR-23a cluster members in primary bone marrow hematopoietic cells.
Taqman analysis (Applied Biosystems) of miR-23a, miR-27a, miR-24 expression in primary mouse hematopoietic populations. A) Lin- progenitors were prepared from bone marrow by depleting samples of cells expressing mature lineage markers. TER119+ erythroid cells, CD11b+GR1+ granulocytes and CD19+ B cells were isolated from bone marrow. For examination of miRNA expression in thymocytes, RNA was prepared from whole thymus. Expression was normalized to snoRNA 202 expression. Expression data is relative to Lin- cells. B) Bone marrow progenitors CMP, CLP, and GMP were isolated from mouse bone marrow. Expression of miRNAs is relative to expression in CMPs. MiR-21 expression was used as a control as its expression in these populations had been previously characterized[16].
Specific progenitor populations were examined for expression of individual miR-23a cluster members to determine if the cluster members were expressed early in development and whether the increased expression in myeloid cells compared to lymphoid cells was an early or late event. RNA was prepared from Common myeloid Progenitors (CMPs), Granulocyte-Monocyte Progenitors (GMPs) and Common Lymphoid Progenitors (CLPs). MiR-21 was used as a control as its expression was previously characterized in hematopoietic progenitors[16]. Consistent with what was observed in the mature cells, expression of miRs-23a, -27a, and -24 was higher in GMPs compared to CLPs or the CMP (Fig 3B). Expression of the cluster correlates with the development of the monocyte/ granulocyte lineages. MiR-21 expression in these populations was consistent with previous observations[16]. The expression analysis demonstrates that miRs-23a, -27a, and -24 are more highly expressed in myeloid progenitors and their offspring than lymphoid progenitors and mature B cells.
Expression of the miR-23a cluster does not affect monocyte versus granulocyte differentiation
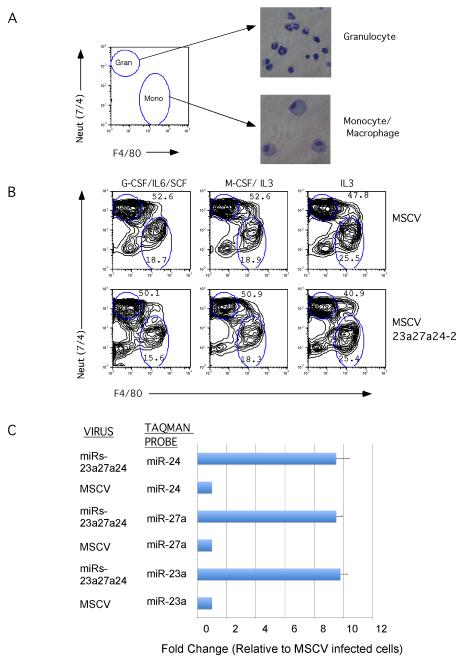
We previously observed that distinct PU.1 concentrations determine whether a myeloid progenitor becomes a monocyte (high PU.1) or granulocyte (low PU.1)[9]. To investigate whether the miR-23a cluster mediates this effect downstream of PU.1, we cloned the 23a cluster pri-transcript into the MSCV-EGFP retroviral plasmid. We infected Lin- mouse bone marrow progenitors with either MSCV or MSCV-23a cluster retrovirus (Retroviral expression in blood cells of mature miRNAs was confirmed by Taqman analysis, Fig 4C). Cells were cultured for 4 days under three different cytokine conditions that promote myeloid development. Analyzing the differentiation of the cells by expression of the cell surface markers F4/80 (monocytic) and Neut (granulocytic) we did not observe any differences in development between MSCV and 23a cluster infected hematopoietic cells (Fig 4A,B). This result demonstrates that the cluster does not affect commitment of myeloid progenitors to monocyte and granulocytes.
Fig 4. Expression of the miR-23a cluster in hematopoietic progenitors does not affect differentiation into monocytes and granulocytes.
Lin- cells were infected with MSCV or MSCV-23a27a24 retrovirus. Infected cells were then cultured 4 days in IMDM media containing the indicated cytokines A) Monocyte and granulocyte differentiation was evaluated by cell surface expression of F4/80 and Neut (7/4) on infected cells (GFP+). Neut+ fraction of cells contains granulocytes and the F4/80+ fraction contains monocytes B) No differences were detected in monocyte versus granulocyte differentiation between MSCV and miRNA expressing cells under any of the cytokine conditions tested. Percentages of cells in the monocyte and granulocyte gates are shown. C) MPRO cells (murine promyelocytic cell line) were infected with indicated retroviral plasmids. GFP+ (infected) cells were isolated, expanded, and lysed for RNA isolation. Taqman analysis was performed to quantify expression of the miR23a miRNAs. Expression is relative to MPRO cells infected with empty MSCV virus. Demonstrates that the cluster virus generates all three mature miRNAs.
The miR-23a cluster promotes myeloid development over lymphoid development
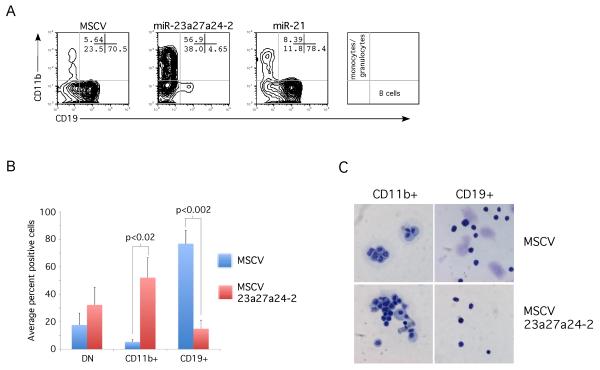
We next tested whether the miR-23a cluster mediates the ability of PU.1 to promote myeloid development at the expense of B cell development. We used a similar strategy as above except that infected hematopoietic progenitors were co-cultured on OP9 stromal cells with IL-7 and Flt3L, which promotes pro-B cell growth[25]. Cells were cultured with the indicated viruses and then co-cultured with OP9 cells for 12 days. Myeloid versus B cell differentiation was evaluated by the cell surface expression of CD11b (myeloid) and CD19 (B cell). As expected MSCV infected cells predominantly generated CD19+ B cells (Fig 5A). Between 50% and 70% of MSCV infected cells were CD19+, whereas only approximately 5% to 15% of MSCV-infected cells were CD11b+ cells (Fig 5B). When cells were infected with a 23a cluster expressing, differentiation was dramatically shifted to myeloid development. Over 50% of the cluster infected cells were CD11b+, with less than 20% of the cells positive for CD19 (Fig 5B). The effect is not due to a global defect in miRNA processing that may occur due to overexpression of a pri-miRNA. Ectopic expression of miR-21 did not change the balance between myeloid and lymphoid development (Fig 5A). Sorting GFP+CD11b+ and GFP+CD19+ cells and examining the morphology of the cells by histochemical staining confirmed the use of CD11b and CD19 as faithful markers of differentiation (Fig 5C).
Fig 5. The 23a cluster promotes myeloid development of hematopoietic progenitors.
A) Lin- mouse hematopoietic progenitors were infected with empty viral vector, 23a cluster virus, or miR-21 virus. Cells were cultured on OP9 cells 12 days in presence of IL-7 and Flt3L. Differentiation analyzed by FACs by gating on GFP+ population and staining with fluorescently tagged antibodies to CD11b (myeloid), and CD19 (pro-B cells). Percentage of CD11b+, CD19+ and CD11b-CD19− cells is shown in top panel. . Lower panel denotes the actual number of events (cells) that were observed to be double negative, CD11b+, or CD19+. B) Average expression of CD19 and CD11b plotted from 4 independent OP9 co-culture experiments. Differences in the generation of myeloid and B cells were determined to be statistically significant by unpaired student-t-test. C) Cytospins of sorted GFP+CD11b+, and GFP+CD19+ cells from both MSCV and MSCV-23a27a24 cultures. Morphology indicates that CD11b, and CD19 expression is differentiating between myeloid and B cells.
MiR-24 is necessary and sufficient for the myeloid promoting activity of the cluster
To determine which of the cluster miRNAs was most responsible for the myeloid promoting activity we generated MSCV viruses that expressed miR-23a alone, miR-27a alone and miR-24 alone. Lin- hematopoietic progenitors were infected and cultured as described above. Neither miR-23a nor miR-27a alone was able to significantly increase the production of CD11b+ myeloid cells (Fig 6A). However the miR-24 alone virus increased the production of CD11b+ cells and decreased production of CD19+ B cells. This indicated that miR-24 expression was necessary and sufficient to induce in vitro myeloid differentiation from hematopoietic progenitors.
Fig 6. MiR-24 is necessary and sufficient to promote myeloid development of hematopoietic progenitors cultured on OP9 cells.
To determine if any single miRNA in the cluster was more responsible for the myeloid inducing activity we performed OP9 co-culture assays with cells infected with MSCV, 23a cluster, 23a only, 27a only, and 24 only expressing retroviruses. A) Infected Lin- progenitors were cultured on OP9 cells in the presence of Flt3L and IL-7 for 12 days. Differentiation was evaluated by cell surface expression of CD11b and CD19. (Cell percentages shown) B) Taqman analysis of RNA isolated from MPRO cells infected with MSCV viruses expressing a single cluster miRNA. Demonstrates that the indicated viruses produce the expected miRNAs in hematopoietic cells.
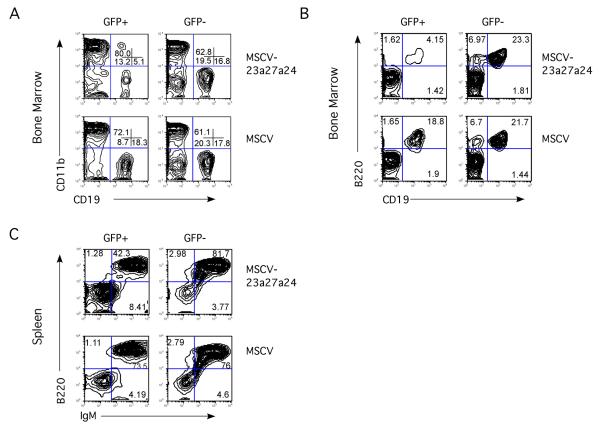
The 23a cluster inhibits B cell development in vivo
To determine whether the 23a cluster has the same activity in vivo, we performed bone marrow transplants. Stem/progenitor-cell enriched bone marrow cells from donor mice were transduced with either miR-23a cluster retrovirus or control virus, and then transplanted into lethally irradiated syngeneic recipient mice. As with in vitro experiments both viruses expressed a GFP marker. Hematopoiesis was analyzed by flow cytometry between 6 and 7 weeks post-injection. This allowed for sufficient time to assay short-term reconstitution of the lymphoid and myeloid compartments[26]. We did not observe a consistent increase in myeloid cells produced by miR-23a infected bone marrow cells in vivo. Consistent with the in vitro results, there was a reproducible 3-fold decrease in CD19+ B cells in the bone marrow generated from miR-23a expressing cells compared to GFP only expressing controls (Fig 7A, B). In addition we observe on average a 2-fold reduction in splenic B220+ B cells comparing miR-23a-infected cells to control cells. These results demonstrate that the cluster is able to inhibit B cell development in vivo as well as in vitro.
Fig 7. Expression of the 23a cluster in hematopoietic progenitor inhibits B cell development in vivo.
Marrow from 5-FU treated mice was infected with 23a cluster virus, and control MSCV virus. Cells were transplanted into irradiated mice and hematopoiesis examined by FACs between 6 and 7 weeks post-transplant. GFP positive and negative fractions were examined for hematopoietic development by cell surface markers. A) Bone marrow was examined for myeloid versus B lymphoid development by expression of CD11b, and CD19. B) Bone marrow B cell development was specifically examined by expression of CD19 and B220. C) Contribution to splenic B cells was examined by expression of B220 and IgM. Representative data is shown. Seven mice transplanted with MSCV-infected bone marrow and 9 mice transplanted with miR-23a cluster-infected bone marrow were examined.
Discussion
Our study demonstrates that the miR-23a cluster is a potent inhibitor of B lymphopoiesis both in vitro and in vivo. We also observed the cluster promoting myeloid development in vitro, which is similar to what occurs when cultured progenitor cells are transduced with a PU.1 retrovirus[17]. Induction of the cluster miRNAs in the PUER cells and association of PU.1 with the mirn23a promoter suggest that 23a cluster is a target of PU.1. Previously it has been shown that PU.1 positively regulates the expression of miRs-21, -223, -342, and -424[22, 23, 27, 28]. Regulation of all these miRNAs has been reported to affect monocyte and/or granulocyte differentiation[14, 16, 27, 28]. Of these, only miR-21 and miR-424 expression has been shown to affect the development of primary hematopoietic cells[16, 28]. However these miRNAs enhance monocyte differentiation approximately two-fold, which is relatively modest compared to the effects we observe with the miR-23a cluster.
None of the three mature miRNAs generated from the 23a cluster have been shown previously to affect lymphoid development, however two cluster members have been implicated in the differentiation of other blood lineages. Exogenous miR-24 expression in human CD34+ hematopoietic progenitor cells inhibits activin A induced erythroid development[29]. MiR-27a has been implicated in influencing megakaryocytic differentiation of K562 cells [30] and granulocytic differentiation of 32D cells[31]. Our data clearly demonstrates that the miR-23a cluster miRNAs affect the development of primary hematopoietic cells both in vitro and in vivo.
We are currently investigating the mechanism by which these effects are mediated. The cluster could be blocking the ability of hematopoietic progenitors to commit to the lymphoid lineages. Alternatively the cluster may modify cellular proliferation and/ or survival of hematopoietic cells. As shown in Table 1, validated miR-24 targets include the pro-apoptotic proteins APAF1, and caspase 9, as well as cell cycle inhibitor INK4A. Downregulation of caspase 9 and/or APAF9 in myeloid cells may be allowing them to survive in our OP9 cultures. In vivo the bone marrow environment is favorable to myeloid growth so proapoptotic genes may not be induced under these conditions. If the increase in myeloid percentage observed in culture were due to silencing the expression of pro-apoptotic mRNAs induced in the myeloid cytokine poor cultures this would explain why an expansion of myeloid cells is not observed in vivo. Alternatively or in cooperation with decreased apoptosis, reduced expression of INK4A potentially could also lead to a higher number of myeloid cells relative to B cells by increasing cell division.
Table 1.
Validated miR-24 Target Genes
| Target Gene | Reference |
|---|---|
| DHFR | Mishra, P.J. et al. PNAS 2007 |
| ALK4 | Wang, Q. et al. Blood 2008 |
| p16 (INK4a) | Lal, A. et al. PLOS One 2008 |
| H2AX | Lal, A. et al. Nat. Struct. Mol. Bio. 2009 |
| Caspase 9 | Walker , J.C. et al. Genes Dev. 2009 |
| APAF1 | Walker , J.C. et al. Genes Dev. 2009 |
| myc | Lal, A. et al. Mol. Cell 2009 |
| E2F2 | Lal, A. et al. Mol. Cell 2009 |
| MKP-7 | Zaidi et al. Cancer Res. 2009 |
| HNF4α | Takagi, S. JBC 2009 |
| Trb3 | Chan, M.C. EMBO J. 2009 |
| AE1 | Wu, J. Oncogene 2010 |
The inhibition of B cell development may be mediated though distinct targets. Of the known miR-24 targets, H2AX and c-myc are potential candidates for mediating the effects on lymphopoiesis that we observe. Knockout of H2AX results in a decrease in both T and B cells without a specific block in development, which is not due to defects in antigen receptor rearrangements[32, 33]. Recently c-myc has been identified as a miR-24 target[34]. Myc is induced by IL-7 signaling in pro-B cells and is required for B cell differentiation[35, 36]. Lack of c-myc has pleiotropic effects on hematopoietic development that appear more severe than the effects of miR-24[37]. However miR-24’s effect on hematopoiesis could be due to a knockdown of myc expression, which could result in a phenotype distinct from elimination of myc. Single miRNAs are able to regulate the expression of multiple mRNAs so the inhibition of B cell development and enhancement of in vitro myeloid growth may require the downregulation of multiple target mRNAs.
In summary we have identified the 23a miRNA cluster as a downstream target of the myeloid transcription factor, PU.1. The functions of 23a cluster miRNAs have not been well characterized in normal hematopoietic cells. Here we have shown in primary mouse blood cells, that the cluster potently inhibits B lymphopoiesis both in vitro and in vivo. Identification of hematopoietic 23a cluster targets promises to reveal novel proteins regulating the growth and/or differentiation of lymphoid and myeloid cells.
Supplementary Material
Fig 1S. Relative expression mir-23 cluster miRNAs in mouse hematopoietic cell lines human hematopoietic cells. A) Taqman analysis (Applied Biosystems) of miR-23a, miR-27a, miR-24 and miR-23b expression in cell lines representing erythroid, myeloid and lymphoid lineages. RNA was prepared from the indicated cell lines. G1E cells are an erythroblast cell line expressing an estrogen (E2) inducible GATA1 protein. Addition of E2 induced differentiation to early erythrocytes. MPROs are a promyelocyte cell line that undergoes granulocytic differentiation upon all trans retinoic acid (ATRA) addition. PUER cells are a bipotential myeloid cell line expressing an OHT-inducible PU.1 protein. 100nM dose of OHT promotes monocyte development. A20 and EL4 are B and T lymphoma cell lines respectively. B) Expression of miR-23a, -23b, -27a, -27b, and 24 in isolated human hematopoietic populations. Arbitrary expression units shown on y-axis. Data obtained from www.microrna.org[24]. 23a cluster mature miRNAs preferentially expressed in hematopoietic cells.
Acknowledgements
We would like to thank Brandy Comyford and Shannon Fitzpatrick for their technical assistance, and Ed Bedrick for statistics consultation. The MSCVEGFP vector was generously provided by Rodney DeKoter (University of Western Ontario). G1E-ER4 cells were a generous gift from Mitch Weiss (University of Pennsylvania). 32Dcl3 cells were provided by Allan Friedman (Johns Hopkins).
Support and Financial Disclosure Declaration This work was supported by an American Cancer Society Research Scholar Grant (RSG-06-170-01-LIB, R.D.), an American Society of Hematology Junior Faculty Scholar Grant (R.D.), and a grant from the dedicated health research funds of the University of New Mexico School of Medicine (R.D.). J.M. was partially supported by an Institutional Research Training Award from the American Society of Hematology.
Financial conflicts: Kimi Y. Kong,, none; Kristin S. Owens, none; Jason H. Rogers, none; Jason Mullenix, none; Chinavenmeni S. Velu, none; H. Leighton Grimes, none; and Richard Dahl, none.
References
- [1].McKercher SR, Torbett BE, Anderson KL, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. Embo J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- [2].Pevny L, Lin CS, D’Agati V, Simon MC, Orkin SH, Costantini F. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 1995;121:163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- [3].Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- [4].Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [6].Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- [7].Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- [8].Dahl R, Simon MC. The importance of PU.1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis. 2003;31:229–233. doi: 10.1016/s1079-9796(03)00152-9. [DOI] [PubMed] [Google Scholar]
- [9].Dahl R, Walsh JC, Lancki D, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- [10].Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. Embo J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- [13].Lu J, Guo S, Ebert BL, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- [15].Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- [16].Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- [18].Walsh JC, DeKoter RP, Lee HJ, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- [19].Horman SR, Velu CS, Chaubey A, et al. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- [21].Weigelt K, Lichtinger M, Rehli M, Langmann T. Transcriptomic profiling identifies a PU.1 regulatory network in macrophages. Biochem Biophys Res Commun. 2009;380:308–312. doi: 10.1016/j.bbrc.2009.01.067. [DOI] [PubMed] [Google Scholar]
- [22].Fujita S, Ito T, Mizutani T, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- [23].Fukao T, Fukuda Y, Kiga K, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- [24].Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, Zuniga-Pflucker JC. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc Natl Acad Sci U S A. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Allman D, Sambandam A, Kim S, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- [27].De Marchis ML, Ballarino M, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009 doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- [28].Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Q, Huang Z, Xue H, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- [30].Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Feng J, Iwama A, Satake M, Kohu K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. Br J Haematol. 2009;145:412–423. doi: 10.1111/j.1365-2141.2009.07632.x. [DOI] [PubMed] [Google Scholar]
- [32].Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lal A, Pan Y, Navarro F, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lal A, Navarro F, Maher CA, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Habib T, Park H, Tsang M, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Morrow MA, Lee G, Gillis S, Yancopoulos GD, Alt FW. Interleukin-7 induces N-myc and c-myc expression in normal precursor B lymphocytes. Genes Dev. 1992;6:61–70. doi: 10.1101/gad.6.1.61. [DOI] [PubMed] [Google Scholar]
- [37].He C, Hu H, Braren R, et al. c-myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development. 2008;135:2467–2477. doi: 10.1242/dev.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig 1S. Relative expression mir-23 cluster miRNAs in mouse hematopoietic cell lines human hematopoietic cells. A) Taqman analysis (Applied Biosystems) of miR-23a, miR-27a, miR-24 and miR-23b expression in cell lines representing erythroid, myeloid and lymphoid lineages. RNA was prepared from the indicated cell lines. G1E cells are an erythroblast cell line expressing an estrogen (E2) inducible GATA1 protein. Addition of E2 induced differentiation to early erythrocytes. MPROs are a promyelocyte cell line that undergoes granulocytic differentiation upon all trans retinoic acid (ATRA) addition. PUER cells are a bipotential myeloid cell line expressing an OHT-inducible PU.1 protein. 100nM dose of OHT promotes monocyte development. A20 and EL4 are B and T lymphoma cell lines respectively. B) Expression of miR-23a, -23b, -27a, -27b, and 24 in isolated human hematopoietic populations. Arbitrary expression units shown on y-axis. Data obtained from www.microrna.org[24]. 23a cluster mature miRNAs preferentially expressed in hematopoietic cells.