ABSTRACT
Although Escherichia coli is a very small (1- to 2-μm) rod-shaped cell, here we describe an E. coli mutant that forms enormously long cells in rich media such as Luria broth, as long indeed as 750 μm. These extremely elongated (eel) cells are as long as the longest bacteria known and have no internal subdivisions. They are metabolically competent, elongate rapidly, synthesize DNA, and distribute cell contents along this length. They lack only the ability to divide. The concentration of the essential cell division protein FtsZ is reduced in these eel cells, and increasing this concentration restores division.
IMPORTANCE Escherichia coli is usually a very small bacterium, 1 to 2 μm long. We have isolated a mutant that forms enormously long cells, 700 times longer than the usual E. coli cell. E. coli filaments that form under other conditions usually die within a few hours, whereas our mutant is fully viable even when it reaches such lengths. This mutant provides a useful tool for the study of aspects of E. coli physiology that are difficult to investigate with small cells.
INTRODUCTION
We think of bacterial cells as small organisms. Cells in a growing Escherichia coli culture look like rigid rods 0.5 μm wide by 2 μm long. Each rod elongates to twice its original length by making new peptidoglycan in many disperse areas by using an enzyme complex including penicillin-binding protein 2 (PBP2). The organism then localizes peptidoglycan synthesis to midcell by using a different enzyme complex, based on PBP3. This changes the direction of cell wall synthesis; the cell wall invaginates from both sides, thus forming two identical daughter cells (1, 2, 3).
E. coli cells are small because their division controls are set accordingly (4). However, they can be much longer if a shift to PBP3 cannot be made but conditions still permit PBP2 function. This occurs under many circumstances. Mutants with conditional temperature-sensitive mutations in the gene coding for the essential cell division protein FtsZ have been isolated. These mutations result in the inhibition of division, accompanied by elongation into short-lived filaments, when cells are shifted from 37°C to 42°C (5).
Filaments also form when the SOS response is triggered under conditions of DNA damage (6) or when the cells are treated with the antibiotic aztreonam, which blocks division irreversibly by inhibiting the FtsI protein, involved in the formation of the septal ring (7). The filaments formed under these conditions, and others, are not viable and lyse within a few hours. As a result, it was largely believed that E. coli could not sustain a large cell size.
However, in this paper, we describe a mutant that forms very long viable (i.e., colony-forming) cells. This strain was derived from our earlier mutant carrying a deletion in metK, which codes for S-adenosylmethionine (SAM) synthetase; SAM is the major donor of methyl groups (8, 9). Our metK deletion mutant, MEW649, had to be exogenously provided with both SAM and methionine in order to grow, as well as with a SAM transporter, since E. coli does not natively express one (10).
The reason for the requirement for SAM was obvious, but the reason for the requirement for methionine was not. To clarify this, we isolated mutants of MEW649 that could grow without methionine. We found several mutants with mutations in the regulation of methionine biosynthesis, as one might expect. However, we found one derivative of MEW649, MNR2, that did not require methionine to grow and in which cell division was greatly altered. Much to our surprise, MNR2 formed the usual E. coli rods in minimal medium but could not divide regularly in a rich medium, such as Luria broth (LB). Indeed, the mutant formed extremely elongated (eel) cells, up to 700 μm long, in Luria broth without sodium chloride (LB no salt [LBNS]).
In this paper, we characterize this mutant and show that its metabolism supports rapid elongation at rates similar to those of its parent. Its principal defect is in division. This implies that most of the metabolic functions of E. coli are independent of cell division.
MATERIALS AND METHODS
General methods.
The E. coli K-12 strains and plasmids used in this study are listed in Table 1. The growth conditions and genetic methods have been described previously (11). Minimal medium (pH 7.2) was prepared with 0.2% ammonium sulfate, 0.1 M potassium phosphate, 0.05% isoleucine, and 0.05% valine. Both LB and LBNS contained 1% tryptone and 0.5% yeast extract, and LB also included 0.5% sodium chloride (12). LBMin had the components of minimal medium plus 1% tryptone and 0.5% yeast extract. All growth media contained 0.2% (wt/vol) glucose. Supplements were used at the following concentrations: methionine, 80 μg/ml; chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; tetracycline, 10 µg/ml; kanamycin, 50 μg/ml. For routine growth of MEW649 and MNR2, S-adenosylmethionine (SAM) was used at 100 μM as explained previously (10). In all experiments, the strains used were grown in minimal medium overnight, where they all formed small cells, and were then subcultured in the media indicated under the conditions specified.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype and/or relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| MEW1 | E. coli K-12 ilvA Δlac | 8 |
| MEW649 | MEW1 ΔmetK pSAMT; Chlr | 10 |
| SM10 | Donor strain for mini-Tn10 mutagenesis | 28 |
| MNR2 | MEW649 ybdMN::Tn10kan; filaments in rich medium; Chlr | This study |
| MNR2A | MNR2 pSAMT-A; Chls Ampr | This study |
| SNR | MNR2 metK+; Chlr | This study |
| EC448 | E. coli Δ(λattL-lom)::bla lacIq P208-ftsZ-gfp | 7 |
| MNR2 λftsZ-gfp | MNR2 Δ(λattL-lom)::bla lacIq P208-ftsZ-gfp | This study |
| CAG12149 | dsbG601::Tn10; Tetr | 29 |
| Plasmids | ||
| pLtet01 | pLtet01 promoter, p15A replicon, MCS-1; Chlr | 8 |
| pSAMT | pLtet01 carrying the sam gene from Rickettsia prowazekii; Chlr | 10 |
| pSAMT-A | Ampr derivative of pSAMT | This study |
| pSC189 | Suicide vector for mini-Tn10 mutagenesis | 30 |
| pftsZ | ASKA plasmid carrying ftsZ under the control of an IPTG-inducible promoter; Chlr | 16 |
| pftsW | ASKA plasmid carrying ftsW under the control of an IPTG-inducible promoter; Chlr | 16 |
Chlr, chloramphenicol resistant; Ampr, ampicillin resistant; Tetr, tetracycline resistant.
Microscopy and cell length measurements.
All micrographs were taken with a Leica DMIRE2 microscope. Cells were fixed, and their lengths were measured as described previously (10, 13).
Slide cultures were prepared by coating glass slides with a thin layer of either minimal medium or LBNS in 2% agar. Diluted cultures were spotted onto the surface of the agar and were delicately topped with a coverslip, allowing the liquid (and therefore the cells) to spread evenly on the surface of the agar. The slides were placed in glass boxes with wet paper towels to maintain humidity.
Time lapse microphotography on slide cultures was carried out by spotting and spreading the liquid cultures on the slide as described above, placing the slides on the microscope stage, and photographing at 15-min intervals, a method that involved both a change to solid medium and a temperature change from 37°C to the temperature of the unheated room temperature microscope stage.
Construction of strain MNR2.
Strain MNR2 was constructed by mobilization of the mini-Tn10kan transposon from strain SM10 (14) into MEW649. The strains were grown in LB with appropriate antibiotics, and a 1:4 mixture of donor to recipient was incubated on LB plates with isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 8 h. The cells were then suspended in 10 mM MgSO4 and were plated on minimal medium with SAM, kanamycin, nalidixic acid (5 μg/ml), and anhydrotetracycline (100 ng/ml). Colonies selected in this way were referred to as methionine nonrequirers (MNR).
Assessment of viability.
To assess the viability of MNR2, cultures grown in minimal medium, LB, or LBNS for 6 h were placed in a mixture of ice and water to halt metabolism. A sample of each culture was taken for protein extraction and quantification as described previously (10). Another sample was serially diluted in cold LB, plated on minimal agar containing glucose and SAM, and incubated at 37°C. Then colonies were counted to estimate the number of CFU. Each of the three original cultures was assayed in triplicate.
Construction of MNR2 λftsZ-gfp.
Strain EC448 carries an ftsZ-gfp transcriptional fusion driven by an IPTG-inducible promoter (15). This cassette was transduced into MNR2 using phage P1, and transductants were selected on minimal agar with glucose, SAM, and ampicillin. The transductants were tested for the presence of the cassette by looking for FtsZ rings with an inducer added in minimal medium.
Construction of MNR2A/pftsZ and MNR2A/pftsW.
Plasmids pftsZ and pftsW (16) confer chloramphenicol resistance and therefore could not be transferred directly into MNR2, since pSAMT confers the same resistance. To replace pSAMT with pSAMT-A, which carries ampicillin resistance, pSAMT-A was transformed into MNR2, and transformants were selected on minimal medium with SAM and ampicillin. Ampicillin-resistant, chloramphenicol-sensitive clones were checked for the presence of pSAMT-A by restriction digestion, and their elongation in LBNS was verified. One such clone was termed MNR2A; pftsZ and pftsW were transformed into MNR2A, and transformants with chloramphenicol resistance were selected.
Western blotting.
Cell lysates were prepared by resuspending 1 ml of culture in sonication buffer (50 mM Tris [pH 8.0], 5% glycerol, 0.4 M NaCl) and then sonicating with a microtip for 3 cycles of 10 s each. The lysate was centrifuged to remove cell debris, and total protein in each cell-free lysate was quantified using the Bio-Rad protein assay. SDS-PAGE gels (12% acrylamide) were run by loading 2.5 μg of protein from each sample, and the resolved proteins were transferred to nitrocellulose membranes and were blocked in milk. The membrane was cut in half, and the halves were probed with either an anti-FtsZ or an anti-FtsI primary antibody, followed by an anti-rabbit secondary antibody (the antibodies were a kind gift from David Weiss). The membranes were then treated with the Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore) and were exposed to photographic film, which was then developed.
RESULTS
Isolation of an elongating mutant, MNR2.
In order to investigate the reason for the surprising methionine requirement caused by the deletion of the metK gene, we looked for gene knockout mutations that could relieve it. For this purpose, we created a library of MEW649 mutants carrying mini-Tn10kan insertions (17) at random locations on the chromosome (see Materials and Methods for details) and selected kanamycin-resistant, methionine-independent cells on minimal medium with SAM and kanamycin (i.e., without methionine).
Using this selection, we isolated many methionine-independent strains with mutations affecting the regulation of methionine biosynthesis but did not study them in detail. We also found one strain, MNR2 (i.e., MEW649 methionine nonrequiring), the subject of this paper, that had major defects in cell division.
In minimal medium (with or without methionine), MNR2 made rod-shaped cells similar to normal E. coli cells. However, when subcultured in LB, MNR2 produced biomass and elongated without division. When grown in LB with NaCl omitted (LBNS), the cells grew even longer and eventually became eel (extremely elongated) cells.
Sizes of MNR2 cells in various media.
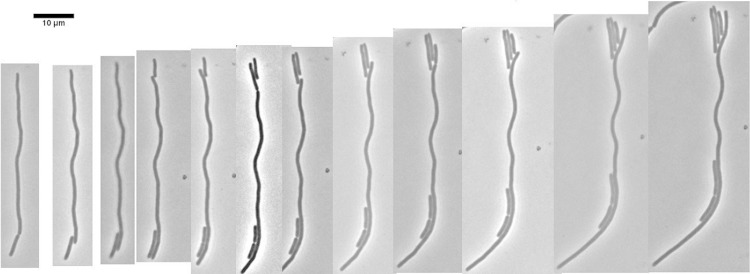
Strain MNR2 grows in phosphate-buffered minimal medium (11) with an average length of just over 4 μm (Table 2). These cells are the same length as cells of their parent strain, MEW649 (ΔmetK pSAMT), but longer than normal E. coli K-12 cells (2.7 μm) (10). When subcultured in either LB or LBNS liquid medium, MNR2 elongated rapidly, with the longest cell reaching 207 μm after 8 h in LBNS. These MNR2 eel cells could continue to elongate if transferred to LBNS slide cultures; the longest of these cells (Fig. 1) attained 750 μm, i.e., about 300 times the length of the same strain grown in glucose minimal medium (Fig. 1A) and 700 times the length of a normal E. coli K-12 cell (Fig. 1B).
TABLE 2.
Lengths of MNR2 cellsa
| Time (h) | Avg cell length (μm) (SD) in the following medium: |
Length of longest cell measured (μm) in the following medium: |
||||
|---|---|---|---|---|---|---|
| Minimal | LB | LBNS | Minimal | LB | LBNS | |
| 0 | 4.5 (1.9) | 3.8 (1) | 4.6 (1.8) | 13.5 | 6.3 | 9.6 |
| 2 | 4.4 (1.4) | 5.9 (4.9) | 6.8 (2) | 9.1 | 50 | 16.6 |
| 4 | 4.4 (1.5) | 9.5 (4.9) | 19.1 (12.9) | 13.5 | 32.5 | 59.3 |
| 6 | 3.6 (1) | 12.1 (8.5) | 22.4 (21.8) | 9.5 | 47.7 | 105.4 |
| 8 | 3.9 (1.1) | 15.5 (13.4) | 47.5 (41.4)b | 11.6 | 96.2 | 207.5 |
MNR2 cells were grown in minimal medium overnight, subcultured for 8 h in the indicated medium, fixed at the indicated time, and then photographed. The cell lengths were measured with the ObjectJ plugin of ImageJ.
Many of the cells measured were so long that they did not fit into the field at the magnification used (×1,000), so these values are an underestimate of the real mean length under this condition.
FIG 1.

Giant E. coli eel cell. A 24-h LBNS liquid culture of strain MNR2 was plated onto an LBNS slide and was incubated at 37°C for 24 h. Four adjacent overlapping fields were photographed, and the images were combined into a single image showing one intact cell and a second lysed cell. The insets show cells of strain MNR2 grown in minimal medium (A) and typical 1- to 2-μm E. coli K-12 (strain MEW1) cells grown in LBNS (B), both for 6 h. Bars, 10 μm.
We examined thousands of MNR2 eel cells in liquid LBNS without seeing visible cell wall constrictions or any indication of internal subdivision. The eel cells are so long that they sometimes lyse, as illustrated in a culture grown on an agar slide (Fig. 1). This lysis allows a very clear demonstration of the continuity of the cytoplasm. Thus, Fig. 2A shows a 280-μm cell, which had lost about half its cytoplasm, photographed in midlysis, demonstrating that the cytoplasm had no interruption for at least 130 μm. In a second such example, a lysed cell happened to fold in such a way as to leak its contents into a closed loop on the agar-coated slide, forming a pool of cytoplasm with visible, though unidentifiable, subcellular structures (Fig. 2B).
FIG 2.
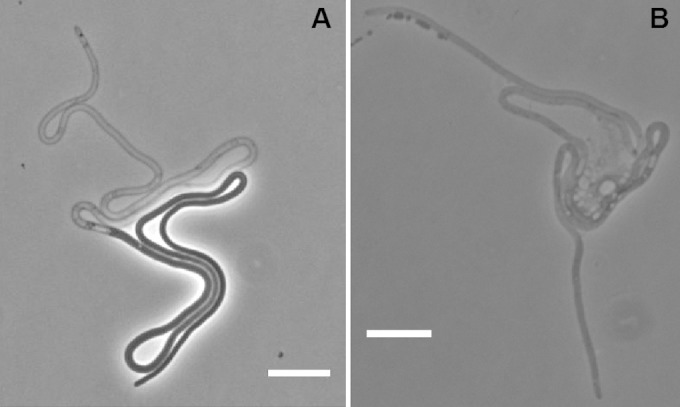
Lysis and continuous cytoplasm in eel cells. A 24-h LBNS culture of MNR2 was plated onto an LBNS slide and was incubated at 37°C for 16 h. Most cells either had not lysed or had lysed completely, but some were caught in midlysis, as seen in these images. Bars, 10 μm.
When subcultured in LB, MNR2 cells reached a mean length of 5.9 μm in 2 h and 15.5 μm in 8 h. Elongation was even faster in LBNS, the other classical formulation of LB (12), where MNR2 cells reached a mean length of 6.8 μm in 2 h. In 8 h, many cells were so long that they could not be photographed within a single frame at ×1,000 magnification. Thirty-eight percent were longer than 50 μm, and 18% were longer than 100 μm. While almost all cells in the MNR2 population elongated, none increased in width. Among several thousand cells measured, the diameter never increased, even when the cells reached lengths of >500 μm. Adding methionine (80 μg/ml) to LB or LBNS did not restore cell division, nor did it affect the shape or length of the cells in any of the media (data not shown).
Viability of long MNR2 cells.
As far as we know, the entire population of strain MNR2 makes elongating cells. To see if these elongated cells were actually viable and able to form colonies at later times, we grew MNR2 in minimal medium, LB, and LBNS for 6 h and then determined the number of CFU as well as the total protein concentration per milliliter of culture (see Materials and Methods). As expected, the protein/CFU ratio was higher in LB than in minimal medium, and higher yet in LBNS, where the majority of the cells synthesize biomass but cannot divide (Table 3). That is, average cells in LBNS had 9 times more protein and were 10 times longer than average cells in minimal medium. If a significant number of LBNS cells were nonviable, the protein/CFU ratio would be much higher. These findings suggest that most of the long cells are alive and are capable of forming colonies once returned to minimal medium.
TABLE 3.
Ratio of total-protein content to number of cells in MNR2a
| Medium | Total protein concn (μg/ml) | CFU (millions) | Ratio of total protein to CFU |
|---|---|---|---|
| Minimal | 27 | 250 | 0.11 |
| LB | 95 | 750 | 0.13 |
| LBNS | 78 | 90 | 0.87 |
MNR2 cells were grown in minimal medium overnight and were subcultured for 6 h in the indicated medium. Then a sample was diluted and was plated on minimal agar to determine viable counts, while another sample was fixed for protein quantification.
Effect of medium composition on MNR2 elongation.
Whereas strain MNR2 elongates in LBNS, it does not elongate in minimal medium to which the components of LBNS have been added (LBMin). For example, MNR2 cells grown for 8 h in LBMin had a mean length of 5.4 μm, very similar to their length of 4 μm after 8 h in minimal medium but much shorter than eel cells grown in LBNS for 8 h (47.5 μm). We therefore made many unsuccessful attempts to inhibit division by adding organic chemicals, such as Casamino Acids, purines, thymidine, or methionine, and even 1% tryptone or 0.5% yeast extract, to minimal medium.
Elongation must depend on some difference between our minimal medium and our LB media other than their organic components. Since our minimal medium is made in 0.1 M phosphate buffer, it must have a high osmolality. We therefore measured the osmolalities of all our media. We found that minimal medium indeed had a high osmolality (332 mOsm/kg), as did LBMin (349 mOsm/kg). LB had a lower osmolality (256 mOsm/kg), while LBNS had a much lower osmolality (93 mOsm/kg). E. coli is reported to grow best in minimal medium with an osmolarity of about 300 to 500 mOsM, although it can tolerate osmolarities of 30 to 3,000 mOsM (18). We suggest that cell division in strain MNR2 requires a medium of high osmolality and that it is the low osmolality of LB and LBNS that delays the onset of division.
Occasional division of eel cells in LBNS.
MNR2 cells elongate so quickly that most of their basic metabolism must function efficiently. Staining with 4′,6-diamidino-2-phenylindole (DAPI) indicated that MNR2 cells could also segregate DNA, at least for as long as 8 h. However, the longer the cells, the more often we observed irregular DNA distribution and areas of continuous DAPI staining (data not shown).
This demonstrates that neither efficient metabolism nor DNA segregation requires cell division, as indicated, albeit less dramatically, by others (19). Indeed, although the eel cells do not have a mechanism to divide to form long daughter cells in enriched medium, they do produce occasional small rods, which can be seen clearly on LBNS agar slides. We made time lapse photographs of 50 individual eel cells on such slides, photographing each field at approximately 15-min intervals, and found most of the division at the ends, resulting in a great disparity in size between the division products. In Fig. 3, the original eel cell produced three small cells at one end and one at the other. All four cells then elongated. We suggest that the only point at which the eel cell can maintain its rigidity while placing a new septum is near its ends.
FIG 3.
Single-cell time lapse photographs showing division at ends. A 4-h culture of strain MNR2 in LBNS at 37°C was plated onto an LBNS slide, and a single cell, remaining at room temperature on the unheated microscope platform, was photographed every 15 min.
Response of eel cells to transfer to minimal medium.
The metabolism of eel cells is very efficient in LBNS, although they cannot divide and show little differentiation along their length other than occasional division at the ends. However, a culture consisting of undifferentiated eel cells grown in LBNS (Fig. 4A) and then plated onto minimal medium slides showed great differentiation along the length of the cells. The cells did not grow by horizontal extension as on LBNS. Instead, they grew by looping outward at many irregularly distributed sites along the eel cells (Fig. 4 B and C). These loops remained intact and grew larger for only a short time, after which constriction and cell division began, and small cells appeared. To the best of our knowledge, such loops in E. coli have not been reported previously.
FIG 4.
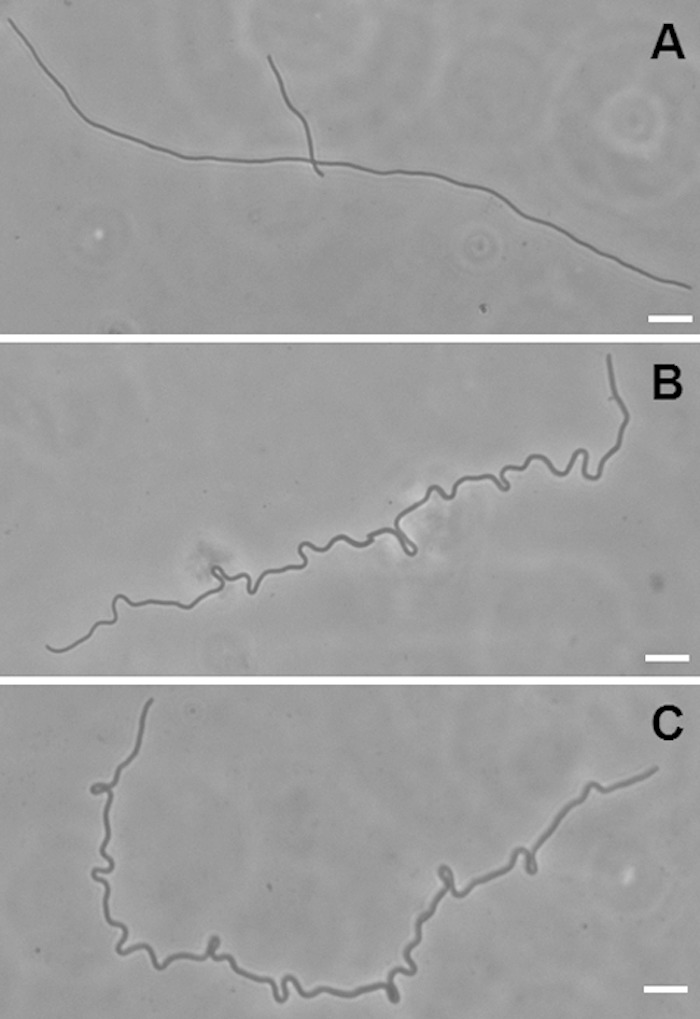
Loop formation in eel cells transferred to minimal medium. A 16-h culture of strain MNR2 in LBNS was plated onto minimal medium slides and was photographed immediately (A) or after 2.5 h at 37°C (B and C).
FtsZ levels and cell division in MNR2.
Clearly, strain MNR2 cells are metabolically competent and cannot divide in organic low-osmolality media. One of the earliest steps in E. coli cell division is the assembly of FtsZ into a ring at midcell. We therefore examined the possibility that the FtsZ ring would be less stable in strain MNR2 incubated at a low osmolality.
To visualize the distribution of FtsZ, we used an MNR2 derivative expressing IPTG-inducible ftsZ-gfp (λftsZ-gfp) in various media. In minimal medium, the distribution of green fluorescent protein (GFP)-labeled FtsZ was consistent with cells growing and dividing as small rods, 62% of which (91/145) formed rings, and constrictions were readily seen. However, FtsZ ring formation was greatly inhibited in MNR2 grown in LBNS: only 6% of eel cells (19/302) showed any trace of visible Z-rings, none of which coincided with a constriction.
The enormous decrease in FtsZ ring formation could be due either to an inability to assemble the ring or to decreased FtsZ availability due to less synthesis or lower stability. The Z-ring would not form, and the switch to PBP3 would not occur; instead, metabolism and elongation via PBP2 would continue. Indeed, depletion of FtsZ in an ftsZ-null mutant results in an 8-fold increase in the average cell length (20). Likewise, decreasing FtsZ levels below those of wild-type E. coli inhibits cell division and increases cell size (21).
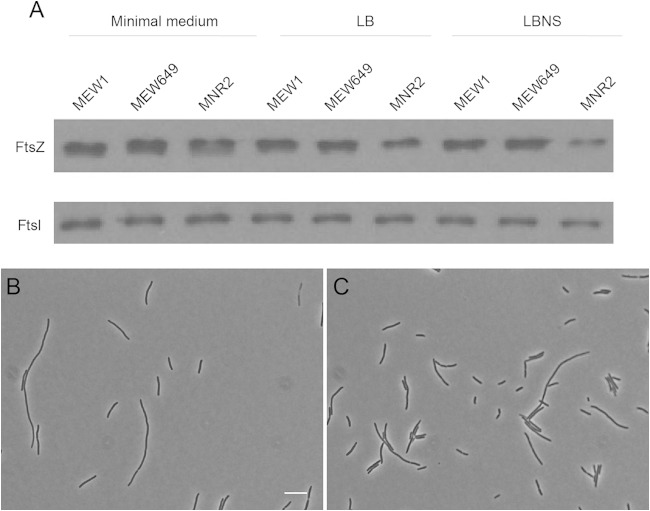
To test this hypothesis, we used Western blotting to compare the FtsZ levels of MEW1 (wild-type), MEW649 (ΔmetK), and MNR2 cells incubated in minimal medium, LB, and LBNS for 8 h. We found that the FtsZ contents of all three strains are similar when they grow in minimal medium as small rods but that in LBNS, and to a lesser extent also in LB, the FtsZ content of MNR2 is visibly lower than those of the other strains (Fig. 5A). The levels of FtsZ thus measured correlated with the osmolalities of the media; the lowest levels were seen in the medium with the lowest osmolality (LBNS).
FIG 5.
FtsZ levels in MNR2. (A) Strains MEW1, MEW649, and MNR2 were grown in minimal medium, LB, or LBNS, all supplemented with glucose. MEW649 and MNR2 were also supplemented with SAM, and MEW649 was supplemented with methionine. After 8 h, cells were lysed by sonication, and total protein was extracted and was analyzed by Western blotting (see Materials and Methods). (B and C) MNR2A/pftsZ was grown at 37°C in LBNS supplemented with glucose and SAM, either without IPTG (B) or with the addition of 10 μM IPTG (C). Cells were fixed after 8 h and were examined by phase-contrast microscopy. Bar, 10 μm. The mean lengths of the cells from all the photographs taken in this experiment are given in the text.
If the elongation of strain MNR2 in LBNS is due to decreased FtsZ levels, increasing cellular FtsZ levels should restore division in LBNS. To test this, we transformed MNR2 with pftsZ, which carries ftsZ under the control of an IPTG-inducible promoter (16). Even without any inducer, leaky expression from the plasmid resulted in dramatically shortened cells in LBNS (Fig. 5B), with an average cell length of 11 μm at 8 h, in contrast to >56 μm for MNR2 without the plasmid. Induction with 10 μM IPTG resulted in even shorter cells, with an average length of 7.3 μm at 8 h, as well as fewer eel cells and a visible increase in the number of short cells (Fig. 5C). In contrast, overexpression of a later cell division protein, FtsW, using the appropriate ASKA plasmid and a range of IPTG concentrations, had no effect on cell length (data not shown).
Since these reduced levels could be due to a mutation in ftsZ or one of its promoters, we sequenced the entire ftsQAZ region in MEW1 and MNR2 but found no mutations in any base pair in the ftsQAZ region relative to the NCBI database sequences (NC_000913.2). This may suggest that the mutation affects a regulator(s) of FtsZ synthesis or stability.
Effect of MetK deficiency on elongation.
MNR2 carries two known mutations: the ΔmetK deletion, inherited from its parent, MEW649, and a mini-Tn10kan insertion. To see if restoring a functional SAM synthetase would allow strain MNR2 to divide normally in LBNS, we transduced wild-type metK from MEW1 to MNR2 and selected SAM-nonrequiring (SNR) derivatives on minimal medium with SAM omitted, but with methionine added as a precaution. The transductants did not require an exogenous supply of either SAM or methionine. Using PCR with the same primers we have used in the past (8), we confirmed the presence of the wild-type metK gene in a typical SNR transductant (Fig. 6A). We then measured the lengths of SNR cells 8 h after subculture and found that SNR made longer cells in LBNS than in minimal medium. Although these LBNS cells were much shorter than our MNR2 eel cells, they were still significantly longer than MEW1 and MEW649 cells (Fig. 6B). When we looked at the distribution of lengths, we found that it was very even in minimal medium but that in LBNS, the spread was much larger and the histogram was skewed toward higher lengths (Fig. 6C). Five percent of the cells were too long to be counted mechanically and included in the histogram; they had an average length of 67 μm, and the longest was 106 μm long. Clearly, restoration of a functional metK gene results in shorter cells. However, the 2-phase histogram suggests that the cell metabolism is precariously balanced and that some cells still elongate. Moreover, addition of SAM to cultures in either minimal medium or LBNS did not change the mean length or the distribution. Thus, while eel cell formation is strongly influenced by metK deletion, factors other than the concentration of SAM in the cell must also be involved.
FIG 6.
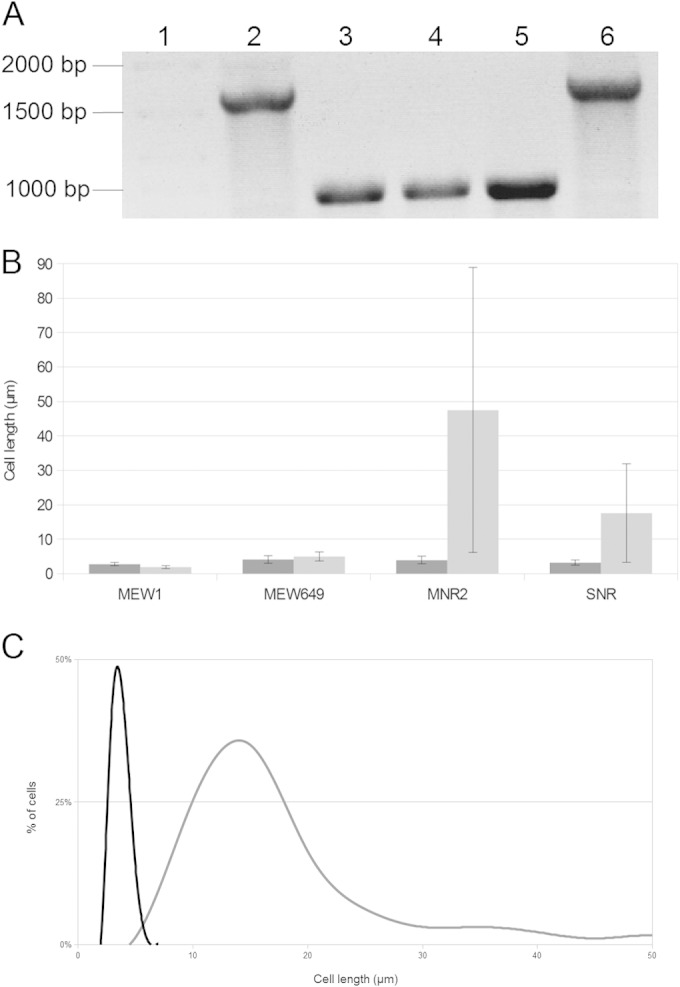
Effect of introducing wild-type metK on the length of MNR2 cells. (A) PCR amplification of the metK gene, showing the restoration of the wild-type gene in SNR transductants. Lanes: 1, DNA ladder; 2, strain MEW1; 3 and 4, strain MEW649; 5, strain MNR2; 6, strain SNR. (B) Mean lengths of cells of strains MEW1, MEW649, MNR2, and SNR grown in minimal medium (dark shading) or LBNS (light shading) for 8 h. Error bars indicate standard deviations. (C) Distribution of lengths of SNR cells grown in either minimal medium (black curve) or LBNS (gray curve) for 8 h. The histogram shows the relative number of cells (as a percentage of all cells) with the indicated length in each culture. Cell length was measured using ImageJ with the ObjectJ plugin.
Characterization of the mini-Tn10kan insert in strain MNR2.
Since we isolated strain MNR2 by insertion mutagenesis, one would expect that the loss of function of the gene with the insert would contribute to elongation. Using inverse PCR (22, 23), we located the mini-Tn10kan insertion in the overlap region of two unannotated genes, ybdN and ybdM, and sequenced the DNA on both sides of the insertion to confirm this. We also used Southern blotting to confirm that this was the only insertion in the chromosome of MNR2 (data not shown). Nonetheless, the mini-Tn10kan insert was not the cause of the differences between strains MEW649 and MNR2. We showed that inserting the mini-Tn10kan transposon from MNR2 into MEW649 did not induce elongation and that restoring wild-type ybdNM by transduction linked to dsbG::Tn10tet did not prevent it.
We conclude that eel cell formation is independent of changes in ybdNM and that MNR2 must carry some as yet unidentified mutation(s) other than the ybdNM::Tn10 insertion.
DISCUSSION
In this study, we show that elongation and synthesis of protoplasm in E. coli are independent of septum formation and cell division. We describe a mutant of E. coli K-12, strain MNR2, that divides to form small rods in glucose minimal medium but forms extremely elongated (eel) cells, up to at least 750 μm and without septa, in rich media such as Luria broth without salt. This is comparable to the lengths of two of the longest bacteria known, Thiomargarita namibiensis (750 μm) and Epulopiscium fishelsoni (700 μm).
One of the remarkable features of E. coli (and other bacteria) is the efficiency and accuracy of its metabolism, which allows it to produce new cells rapidly, with no excess metabolites going to waste and with very few mutants. The division time varies with the growth medium, but in any given medium, it is closely controlled. As shown elegantly recently by the laboratory of Suckjoon Jun, this control relies on sensing the extent of volume change and results in the maintenance of cell size (4). Nonetheless, we see here that E. coli can maintain a functional metabolism in the total absence of cell division.
Strain MNR2 grown in LBNS has a functional metabolism, allowing it to produce biomass and distribute its contents inside the cell. The eel cells are viable, as judged by their ability to produce colonies on minimal medium plates. However, they have no superstructure to protect them and no specialized division system; they could not sustain themselves indefinitely in LBNS or outside the laboratory. Epulopiscium fishelsoni is a symbiont living in the surgeonfish gut in close metabolic coordination with its host and is much wider and more structured than MNR2 (24). Thiomargarita namibiensis survives as a free-living organism as a chain of large cocci with a viscous protective coat (25). Both have elaborate division systems to produce daughter cells.
The eel cells can in fact divide. As they elongate, particularly in LB but also in LBNS, they occasionally split off small rods, usually at the ends, and both the rod and the elongated parent continue to elongate. When the eel cells are transferred to minimal medium, it becomes obvious that they are not homogeneous with respect to division in minimal medium. They first make loops, suggesting that they are growing in many scattered spots, and many of these later produce viable rods and eventually microcolonies. In other eel cells, and sometimes in other areas within the same cell, loops do not form, and that part of the cell is unchanged over the same time frame.
In the interval prior to the activation of septum formation and invagination, these loops may form as a response to the mechanical difficulties caused by the rapid elongation of a long cell adhering to a solid agar surface. While the force required to push apart two E. coli cells of normal size is probably not negligible, the force required to push two lengths of an eel cell apart is immensely greater. Insertion of new peptidoglycan by horizontal expansion in the localized area where cell division occurs would require pushing the entire cell on either side, a process that would be hindered by the weight of the cell and its adherence to the agar. This resistance may be at least partly relieved by the outward turn.
We made a considerable effort to determine the reason why MNR2 division is inhibited in LB and LBNS. We have tentatively ascribed this inhibition to the low osmolality of these media and suggest that this, or some other feature of the medium, interferes with the production of a functional FtsZ ring. We examined eel cells grown with FtsZ-GFP, and these indeed had very few Z-rings. We therefore determined the amount of FtsZ by Western blotting and found that LBNS-grown MNR2 cells were considerably depleted in FtsZ. Since increasing FtsZ levels in MNR2 with a plasmid-carried inducible ftsZ gene reduced the production of long cells, it seems likely that the mutant really is deficient in FtsZ. However, we cannot explain why this is so, since we found that the sequences of the ftsQAZ genes and their regulatory region contained no mutation.
We showed in earlier studies that cells that could not deaminate l-serine formed long filaments (26). Cell division in these filaments was blocked shortly after Z-ring formation, and we suggested that alterations in serine degradation might also alter the one-carbon pool and consequently alter a metabolic trigger for cell division (13). We suggest here that cell division in MNR2 is blocked at a point even earlier than Z-ring assembly. We have not been able to elucidate the underlying mechanism or the precise factor(s) involved, but there is a precedent in E. coli for the inhibition of cell division in response to nutritional status. An interesting model attributes this blockage to the glucosyltransferase OpgH, a nutrient-dependent regulator of E. coli size (i.e., division). E. coli cells are longer in rich medium than in minimal medium. OpgH regulates this change in length by binding to FtsZ, blocking Z-ring assembly and allowing cells to grow longer (27). We have suggested previously that a chemical modification, e.g., a methylation, may be needed before and/or in the early stages of FtsZ ring formation (13). This could involve the modification of a regulator similar to OpgH, but one that directly reduces the levels of FtsZ instead of sequestering the molecules.
We also attempted to determine the nature of the mutation that produced the MNR2 phenotype. The parent of MNR2 carries a deletion in metK causing a requirement for SAM, which is also required by MNR2. This, of course, does not cause the elongation phenotype. However, the restoration of wild-type metK in strain SNR greatly reduced elongation, suggesting that a deficiency of SAM synthetase is involved in the production of eel cells.
We isolated strain MNR2 using a mini-Tn10kan transposon and located the mutation in the overlap region of the ybdN and ybdM genes. This insertion, however, had nothing to do with the phenotype. Transferring it to the parent (MEW649) did not induce elongation, and replacing it with the nonmutated genes in strain MNR2 did not prevent elongation.
For reasons outlined by Ghigo and colleagues (14), mini-Tn10 selection was not a wise choice of method for mutagenesis. Among other problems, it involved the use of nalidixic acid, which is itself a mutagen. We assume, therefore, that strain MNR2 must have at least one more mutation responsible for eel cell formation. Nonetheless, the strain as we studied it was genetically stable and useful for investigating the physiology of long E. coli cells.
ACKNOWLEDGMENTS
We are grateful for NSERC grant 6050, which financed this work, and to Concordia University Graduate Studies for financial support for Z. W. El-Hajj.
We are also grateful to the National BioResource Project and to the Coli Genetic Stock Centre, both of which supplied us with many strains. We have been greatly aided by D. Weiss, who provided us with the antibodies and the λftsZ-gfp construct used in this study, by A. Victor, who helped with the cell length measurements, and by M. Peters and W. Zerges, who greatly assisted with Western blotting. We also thank J. Dhaliwal, M. Jaramillo Garcia, C. van Oostende, J. A. Kornblatt, A. Krus, C. Woldringh, D. Allcock, K. Ardat, K. Mathee, S. Booker, and V. Mathur for valuable discussions.
E.B.N. dedicates this paper to Harold Newman, Harold Amos, V. Mathur, and B. Anhang, who over many years strongly supported her attempts to be a scientist.
REFERENCES
- 1.Donachie WD, Begg KJ, Vicente M. 1976. Cell length, cell growth and cell division. Nature 264:328–333. doi: 10.1038/264328a0. [DOI] [PubMed] [Google Scholar]
- 2.den Blaauwen T. 2013. Prokaryotic cell division: flexible and diverse. Curr Opin Microbiol 16:738–744. doi: 10.1016/j.mib.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Cava F, Kuru E, Brun YV, de Pedro MA. 2013. Modes of cell wall growth differentiation in rod-shaped bacteria. Curr Opin Microbiol 16:731–737. doi: 10.1016/j.mib.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. 2015. Cell-size control and homeostasis in bacteria. Curr Biol 25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricard M, Hirota Y. 1973. Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J Bacteriol 116:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones CA, Holland IB. 1984. Inactivation of essential division genes, ftsA, ftsZ, suppresses mutations at sfiB, a locus mediating division inhibition during the SOS response in E. coli. EMBO J 3:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Arends SJ, Weiss DS, Newman EB. 2005. A deficiency in S-adenosylmethionine synthetase interrupts assembly of the septal ring in Escherichia coli K-12. Mol Microbiol 58:791–799. doi: 10.1111/j.1365-2958.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Newman EB. 2002. Studies on the role of the metK gene product of Escherichia coli K-12. Mol Microbiol 43:1651–1656. doi: 10.1046/j.1365-2958.2002.02856.x. [DOI] [PubMed] [Google Scholar]
- 9.Greene RC. 1996. Biosynthesis of methionine, p 542–560. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 10.El-Hajj ZW, Reyes-Lamothe R, Newman EB. 2013. Cell division, one-carbon metabolism and methionine synthesis in a metK-deficient Escherichia coli mutant, and a role for MmuM. Microbiology 159:2036–2048. doi: 10.1099/mic.0.069682-0. [DOI] [PubMed] [Google Scholar]
- 11.Newman EB, Miller B, Colebrook LD, Walker C. 1985. A mutation in Escherichia coli K-12 results in a requirement for thiamine and a decrease in l-serine deaminase activity. J Bacteriol 161:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinella D, D'Ari R. 1994. Thermoinducible filamentation in Escherichia coli due to an altered RNA polymerase beta subunit is suppressed by high levels of ppGpp. J Bacteriol 176:966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, El-Hajj ZW, Newman E. 2010. Deficiency in l-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J Bacteriol 192:5515–5525. doi: 10.1128/JB.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrières L, Hémery G, Nham T, Guérout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 17.Kleckner N, Bender J, Gottesman S. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol 204:139–180. doi: 10.1016/0076-6879(91)04009-D. [DOI] [PubMed] [Google Scholar]
- 18.Cayley S, Record MT Jr. 2004. Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein-DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J Mol Recognit 17:488–496. doi: 10.1002/jmr.695. [DOI] [PubMed] [Google Scholar]
- 19.Bernander R, Nordström K. 1990. Chromosome replication does not trigger cell division in E. coli. Cell 60:365–374. doi: 10.1016/0092-8674(90)90588-6. [DOI] [PubMed] [Google Scholar]
- 20.Dai K, Lutkenhaus J. 1991. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol 173:3500–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrido T, Sánchez M, Palacios P, Aldea M, Vicente M. 1993. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J 12:3957–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchetina E, Newman EB. 1995. Identification of Lrp-regulated genes by inverse PCR and sequencing: regulation of two mal operons of Escherichia coli by leucine-responsive regulatory protein. J Bacteriol 177:2679–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X. 2008. Reduced intracellular SAM can increase the expression of met gene under the SAM-MetJ mechanism in Escherichia coli. M.Sc. thesis Concordia University, Montreal, Quebec, Canada. [Google Scholar]
- 24.Angert ER, Clements KD, Pace NR. 1993. The largest bacterium. Nature 362:239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- 25.Schulz HN, Brinkhoff T, Ferdelman TG, Marine MH, Teske A, Jorgensen BB. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493–495. doi: 10.1126/science.284.5413.493. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Newman E. 2008. Deficiency in l-serine deaminase results in abnormal growth and cell division of Escherichia coli K-12. Mol Microbiol 69:870–881. doi: 10.1111/j.1365-2958.2008.06315.x. [DOI] [PubMed] [Google Scholar]
- 27.Hill NS, Buske PJ, Shi Y, Levin PA. 2013. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet 9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. doi: 10.1016/S0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]