ABSTRACT
The PhoPR two-component signal transduction system controls one of three responses activated by Bacillus subtilis to adapt to phosphate-limiting conditions (PHO response). The response involves the production of enzymes and transporters that scavenge for phosphate in the environment and assimilate it into the cell. However, in B. subtilis and some other Firmicutes bacteria, cell wall metabolism is also part of the PHO response due to the high phosphate content of the teichoic acids attached either to peptidoglycan (wall teichoic acid) or to the cytoplasmic membrane (lipoteichoic acid). Prompted by our observation that the phosphorylated WalR (WalR∼P) response regulator binds to more chromosomal loci than are revealed by transcriptome analysis, we established the PhoP∼P bindome in phosphate-limited cells. Here, we show that PhoP∼P binds to the chromosome at 25 loci: 12 are within the promoters of previously identified PhoPR regulon genes, while 13 are newly identified. We extend the role of PhoPR in cell wall metabolism showing that PhoP∼P binds to the promoters of four cell wall-associated operons (ggaAB, yqgS, wapA, and dacA), although none show PhoPR-dependent expression under the conditions of this study. We also show that positive autoregulation of phoPR expression and full induction of the PHO response upon phosphate limitation require PhoP∼P binding to the 3′ end of the phoPR operon.
IMPORTANCE The PhoPR two-component system controls one of three responses mounted by B. subtilis to adapt to phosphate limitation (PHO response). Here, establishment of the phosphorylated PhoP (PhoP∼P) bindome enhances our understanding of the PHO response in two important ways. First, PhoPR plays a more extensive role in adaptation to phosphate-limiting conditions than was deduced from transcriptome analyses. Among 13 newly identified binding sites, 4 are cell wall associated (ggaAB, yqgS, wapA, and dacA), revealing that PhoPR has an extended involvement in cell wall metabolism. Second, amplification of the PHO response must occur by a novel mechanism since positive autoregulation of phoPR expression requires PhoP∼P binding to the 3′ end of the operon.
INTRODUCTION
The PhoPR two-component signal transduction system (TCS) controls one of three responses activated by Bacillus subtilis to adapt to phosphate limiting conditions (PHO response). PhoPR is well characterized, with orthologous TCS being widely distributed among Gram-negative (e.g., PhoBR in Escherichia coli) and Gram-positive (e.g., PhoPR in B. subtilis and Staphylococcus aureus) bacteria. Although the PHO response is activated upon phosphate depletion in both E. coli and B. subtilis, the PhoR kinase senses a completely different signal in each bacterium. In E. coli the signal emanates from phosphate transport mediated by the PstSCAB2 phosphate transporter and the PhoU chaperone-like protein (1). It is proposed that formation of a PstSCAB2/PhoU/PhoR complex under conditions of phosphate excess (>4 μM) inhibits PhoR autokinase activity, thereby keeping the PHO response off (1). Inhibition is reduced or eliminated when phosphate levels fall below 4 μM thereby allowing the active PhoR to phosphorylate its cognate PhoB response regulator. In B. subtilis, the PhoR activation signal emanates from wall teichoic acid metabolism (WTA) (2). The PhoR kinase monitors the cellular level of the WTA synthetic intermediate undecaprenyl-P-P-1-GlcNAc-β(1-4)-ManNAc-(GroP)20 through an interaction with the intracellular PAS domain (2). This intermediate inhibits PhoR autokinase activity during growth under phosphate-replete conditions. When phosphate becomes limiting, PhoR kinase activity is initially triggered by an unknown mechanism that leads to a low-level PHO response. The response is then amplified by phosphorylated PhoP (PhoP∼P)-mediated reduction in the cellular level of the inhibitory WTA intermediate that is achieved by (i) initiating teichuronic acid synthesis through activation of the tuaA-tuaH (tuaA-H) operon expression and (ii) reducing WTA synthesis through repression of tagAB (2). The very different signals to which the orthologous PhoR kinases of E. coli and B. subtilis respond probably reflects the different levels of phosphate in the cell envelopes of these two bacteria (3, 4). It is interesting, however, that activation of both PhoR kinases occurs by a relief of inhibition mechanism (1, 2).
Transcriptome and proteome analyses show that large numbers of genes are differentially expressed when E. coli and B. subtilis cells become phosphate limited, demonstrating that cellular adaptation to this condition requires significant physiological change (5–10). Moreover, the PhoBR and PhoPR regulons in E. coli and B. subtilis show several similarities. In E. coli activated PhoB∼P directly regulates expression of 31 genes in 9 transcription units, while in B. subtilis PhoP∼P directly regulates expression of 34 genes in 13 transcription units (1, 7, 9, 11). The genes of both regulons encode proteins involved in phosphorous assimilation (e.g., phosphate transporters and phosphodiesterases) and the regulators of their expression (i.e., PhoBR and PhoPR) (1, 7, 9, 11). However, due to the large phosphorous content of WTA and LTA, the B. subtilis regulon has a major cell wall component through PhoPR-mediated regulation of expression of teichoic (TagAB) and teichuronic (TuaA-H) acid synthetic enzymes (2, 4, 9). Thus, when B. subtilis cells become phosphate limited, PhoP∼P represses tagAB and activates tuaA-H expression (7, 9, 12, 13, 14). Teichuronic acid is a non-phosphate-containing anionic polymer that replaces teichoic acid in the wall of phosphate-limited cells. Reducing the teichoic acid content of cell walls therefore lowers the overall cellular requirement for phosphorous (15, 16). Moreover, scavenging enzymes probably liberate phosphate from the released WTA and recycle it for other cellular purposes.
The PhoPR regulon in B. subtilis was established by classical genetic and transcriptome analyses, identifying genes whose expression changed in a PhoPR-dependent manner upon the onset of phosphate limitation (7, 9, 11, 17). However, such approaches identify only genes whose transcription is regulated solely or primarily by PhoPR. A study of the closely related WalRK TCS shows that the gene cohort identified by transcriptome analysis is a subset of the chromosomal sites to which WalR∼P binds: only 7 genes were ascribed to the WalRK regulon by transcriptome analysis, whereas ChIP-chip studies show that WalK∼P binds to 22 chromosomal sites (18–21). Similarly, in E. coli, PhoB∼P binds to more chromosomal loci than were identified by transcriptomic and other studies (10). Such observations prompted us to establish the PhoP∼P bindome in B. subtilis cells by ChIP-chip analysis using high-density microarrays.
We report here that PhoP∼P binds to 25 chromosomal regions in phosphate-limited cells but does not bind to any locus in phosphate-replete cells. Thirteen of these loci are newly identified, with the remaining 12 sites being located within the promoters of previously identified PHO regulon genes. However, expression of genes at seven of the 13 newly identified binding sites is PhoPR independent. PhoP∼P binding is highly enriched throughout a 28-kb chromosomal region that encodes genes involved in cell envelope and nucleotide sugar metabolism. We identify a new peptide PipA (40 amino acids) that is highly induced in a PhoPR-dependent manner and localizes to the cytoplasm. Finally, we show that PhoP∼P binding to cognate sequences at the 3′ end of the operon is required for positive autoregulation of phoPR expression and for full induction of the PHO response upon phosphate limitation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in the present study are listed in Table 1. E. coli strains EC101 (repA+) (22) and TG-1 (23) were used for propagating plasmids. E. coli strains were grown in Luria-Bertani (LB) medium (23). B. subtilis strains were grown in LB medium, high-phosphate-defined medium (HPDM), or low-phosphate-defined medium (LPDM) (24). Media were supplemented when needed with antibiotics at the following concentrations: erythromycin at 150 μg/ml (for strain EC101), chloramphenicol at 5 or 10 μg/ml, MLS (macrolide-lincomycin-streptogramin B resistance; erythromycin at 1 μg/ml plus lincomycin at 25 μg/ml), and spectinomycin at 100 μg/ml. The optical density at 600 nm (OD600) was measured by using a UVmini-1240 UV-VIS spectrophotometer (Shimadzu Scientific Instruments).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| EC101 | JM101 with repA from pWV01 integrated into the chromosome (Kmr) | 22 |
| TG-1 | supE hsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB lacIq lacZΔM15) | 23 |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| AH024 | trpC2 phoPR::erm | 18 |
| LSB002 | trpC2 phoP::pLIS003 (PphoP phoP-FLAG) | 20 |
| LSB046 | trpC2 with clean deletion of 90 bp from 3′ end of phoR | Excision of pLIS009 |
| LSB088 | trpC2 in which PhoP boxes in 3′ phoR have been mutated to restriction sites | Excision of pLIS011 |
| LSB396 | trpC2 ΔS1052 | LFH→168 followed by dif excision |
| LSB434 | trpC2 ΔamyE::pLIS065 (PS1052 gfp) | Linearized pLIS065→168 |
| LSB435 | trpC2 ΔthrC::pLIS066 (PxylA S1052) | Linearized pLIS066→168 |
| LSB448 | trpC2 pipA::pLIS071 (pipA′-gfp) | pLIS071→168 |
| LSB450 | trpC2 pipA::pLIS075 (PS1052 pipA-3×FLAG) | pLIS075→168 |
| Plasmids | ||
| pG+host4 | Ts derivative of pGK12 (Ermr) | 47 |
| pCA3XFLAG | Vector to tag proteins with 3×FLAG | 31 |
| pDG646 | Plasmid containing Ermr cassette | 27 |
| pDG782 | Plasmid containing Kmr cassette | 27 |
| pGFPamy | Vector for creation of GFP transcriptional fusions and insertion at the amyE locus | 28 |
| pBitter | Vector for inducible gene expression from the Pxyl promoter inserting at the thrC locus | This study |
| pDN1102 | pET21b containing phoP gene | 20 |
| pSKD14 | pET21d containing intracellular part of phoR gene | This study |
| pLIS009 | pG+host4 derivative containing 90-bp deletion in the 3′ UTR region of phoR (Ermr) | This study |
| pLIS011 | pG+host4 derivative containing mutated PhoP boxes in the 3′ UTR region of phoR (Ermr) | This study |
| pLIS065 | pGFPamy derivative containing PS1052 gfp transcriptional fusion | This study |
| pLIS066 | pBitter derivative for inducible (Pxyl) expression of S1052 (pipA) | This study |
| pLIS071 | pDG782 derivative containing a pipA-gfp translational fusion; 120-bp fragment consisting of the first half of pipA fused with gfp (Spcr) | This study |
| pLIS075 | pCA3XFLAG derivative containing 180-bp fragment of S1052, including pipA minus its stop codon | This study |
Ermr, erythromycin resistance; Spcr, spectinomycin resistance; Kmr, kanamycin resistance.
Strain and plasmid construction.
To create strains LSB046 (in which 90 bp from the 3′ of phoR between the stop codon and operon terminator are cleanly deleted) and LSB088 (in which the PhoP binding boxes are mutated to restriction sites), two fragments were amplified by using the primer pairs 3′phoR up fwd IF(B)/3′phoR up rev IF and 3′phoR do fwd IF/3′phoR do rev IF(H) (for LSB046) and the primer pairs 3′phoR up fwd IF(B)/3′phoR up rev MB and 3′phoR do fwd MB/3′phoR do rev IF(H) (for LSB088) from chDNA of B. subtilis 168 using Phusion polymerase. The fragments were fused by strand overlap extension PCR and cloned into BamHI-HindIII-digested pG+host4. The resulting plasmids, pLIS009 and pLIS011, were propagated in E. coli EC101. pLIS009 and pLIS011 were integrated into and excised from the B. subtilis chromosome as previously described (20). The desired excisants were verified by PCR screening and sequencing. In addition, the excisants containing the mutated PhoP boxes were identified by digestion of PCR-generated promoter fragments with EcoRI, PstI, NcoI, and XhoI.
Strain LSB396 was generated using adaptations of long flanking homology PCR (25) and a procedure for marker excision by Xer recombination (26). DNA fragments of ∼750 bp flanking the 5′ and 3′ of S1052 were PCR amplified using the primer pairs S1052 up fwd/S1052 up rev(dif) and S1052 do fwd(dif)/S1052 do rev using B. subtilis 168 chromosomal DNA as the template. These fragments were fused with an MLS resistance cassette flanked by dif sites amplified using the primers mls dif fwd and mls dif rev and plasmid pDGD646 DNA as the template (27). The resulting PCR product was used to transform B. subtilis 168. Transformants in which the S1052 locus is replaced with the antibiotic cassette were screened by direct colony-PCR, using the up-forward primer with a reverse primer that anneals inside the MLS resistance cassette. Selected integrants were grown in the absence of antibiotics for 24 h, plated on LB plates, and screened for antibiotic sensitivity, generated by excision of the antibiotic cassette. Deletion of S1052 was verified by diagnostic PCR and DNA sequencing.
Strain LSB434 was created by amplifying a 400-bp DNA fragment containing the promoter region of S1052 using the primer pair PpipA fwd/PpipA rev and cloning it into plasmid pGPFamy (28) using the ligation-independent cloning (LIC) procedure described by Botella et al. (29). The resulting pLIS065 plasmid was linearized using PstI and transformed into B. subtilis 168. Transformants were verified by diagnostic PCR and the lack of halos on 0.2% starch plates.
Plasmid pBitter was generated to place expression of genes under the control of the xylose inducible Pxyl promoter in single copy through integration into the chromosome by double crossover at the thrC locus. Primers OE254 and OE255 were used to amplify the xylose cassette containing the xylR gene and the xylA promoter from plasmid pSweet (30). The resulting PCR fragment was ligated into pDG1731 digested with BamHI and HindIII to create plasmid pBitter.
Strain LSB435 was created by amplifying the S1052 region using the primer pair S1052 fwd(SphI)/S1052 rev(BamHI) and cloning it into SphI-BamHI-digested pBitter. The resulting pLIS066 plasmid was linearized using SmaI and transformed into B. subtilis 168. Transformants were verified as being MLS sensitive and threonine auxotrophs.
Strain LSB448, in which PipA is translationally fused with green fluorescent protein (GFP), was created by amplifying a 120-bp DNA fragment containing the sequence encoding the first 20 amino acids of PipA using the primer pair pipA-GFP fwd 2(E)/pipA-GFP rev 2 and a 716-bp fragment of gfpmut3 using the primer pair gfp fwd/gfp rev (BamHI). The fragments were fused by strand overlap extension PCR and cloned into EcoRI-BamHI-digested pDG782. The resulting plasmid, pLIS071, was transformed into B. subtilis 168. Transformants were selected on kanamycin (10 μg/ml) and verified by diagnostic PCR and sequencing.
Strain LSB450, in which PipA is C-terminally tagged with a 3×FLAG tag, was created by amplifying a 180-bp DNA fragment of S1052 containing the entire pipA open reading frame without the stop codon using the primer pair pipA fwd(H)/pipA rev(B) and cloning it into HindIII-BamHI-digested pCA3×FLAG (31). The resulting plasmid, pLIS075, was propagated in E. coli TG-1 and then transformed into B. subtilis 168. Transformants were selected on chloramphenicol (5 μg/ml), and correct insertion of the plasmid was verified by sequencing.
Chromatin immunoprecipitation using a tiled array and data analysis (ChIP-chip).
Strain LSB002 was grown in LPDM with the addition of 1 μg of erythromycin/ml plus 25 μg of lincomycin/ml (MLS) and 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C. Cells of strain 168 (mock precipitation) were grown in LPDM and 100 μM IPTG at 37°C. Samples were harvested at mid-exponential phase (200 ml of a culture at OD600 = 0.2) and at 2 h after the onset of phosphate starvation state (50 ml of a culture at T2). Cross-linking was performed by the addition of formaldehyde to a final concentration of 1%, followed by a 20-min incubation at room temperature with slow shaking. The cultures were then quenched with glycine (0.36 M final concentration) for 5 min at room temperature. Cells were collected by centrifugation at 4°C, and the pellets were washed twice with 50 ml of ice-cold buffer A (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) before being snap-frozen in a dry ice-ethanol bath.
Pellets were thawed at 37°C and resuspended in 0.5 ml of buffer B (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 0.1% Triton X-100) supplemented with 3 mg of lysozyme/ml and 0.1 mg of RNase A/ml (final concentrations) and incubated for 30 min at 37°C. The sample volume was adjusted to 1 ml with buffer B, and samples were placed on ice for 10 min. Genomic DNA was sheared by sonication (Diagenode Bioruptor Twin sonicator) into fragments between 0.2 and 1 kb. Sonicated samples were centrifuged for 10 min at 20,000 × g (4°C) to remove insoluble debris. Then, 50 μl of the cleared lysates was removed to serve as the input sample, to which 50 μl of 10× buffer C (10 mM Tris-HCl [pH 8], 10% sodium dodecyl sulfate [SDS], 10 mM EDTA) and 400 μl of buffer B were added, and the sample was stored on ice. The remaining cleared lysates were each added to 40 μl of anti-FLAG M2-agarose resin (Sigma-Aldrich, catalog no. A2220) equilibrated with ice-cold buffer B (according to the manufacturer's instructions), followed by incubation for 2 h at 4°C with mild agitation. Beads were then collected by centrifugation for 1 min at 5,000 × g (4°C) and washed three times with 0.5 ml of ice-cold buffer B. To reverse the cross-linking, the beads were transferred to new 1.5-ml Eppendorf tubes, resuspended in 0.5 ml of 1× buffer C, and incubated along with the input samples for ∼20 h at 65°C with shaking (950 rpm). Chromatin immunoprecipitation (ChIP) samples were collected by centrifugation for 1 min at 13,300 rpm (4°C). DNA from both ChIP and input samples was purified using a QIAquick PCR purification kit (Qiagen, 28106), eluted in 50 μl of H2O, and treated with 0.02 mg of RNase A (Roche)/ml for 30 min at 37°C. The RNase was eliminated by using the QIAquick PCR purification kit, and samples were eluted in 30 μl of H2O.
Equal concentrations of ChIP DNA and input samples were amplified by using a GenomePlex complete whole-genome amplification (WGA) kit (Sigma-Aldrich, catalog no. WGA2-50RXN) according to the manufacturer's instructions, purified using the QIAquick PCR purification kit, and eluted in 30 μl of H2O. Ten microliters of each of the purified amplified samples was subjected to a second round of amplification using the WGA kit in accordance with the manufacturer's instruction, purified using the QIAquick PCR purification kit, and eluted in 30 μl of H2O. Next, 3.5 to 4 μg of twice-amplified ChIP DNA and input samples (of both tagged and mock precipitation experiments) at a concentration of ∼250 ng/μl (A260/A280 ≥1.7 and A260/A230 ≥ 1.6) from three separate ChIP experiments were submitted to NimbleGen. ChIP DNA was labeled with Cy5 dye, input DNA was labeled with Cy3 dye, and the samples were applied to the BaSysBio B. subtilis T2 385K array (32).
The ChIP-chip data were processed with Ringo, a Bioconductor package for the analysis of ChIP-chip data from the NimbleGen platform (33). After the data were read into the tool, images were generated for quality assessment. The data were normalized by using a Tukey-biweight scaling procedure. This method is recommended by NimbleGen to scale ChIP-chip readouts so that the data are centered on zero. Peaks were then smoothed by computing medians in a running window approach. The window size was set to 700 bp to correlate with the average fragment size. During this step, the values from three replicates were combined. The smoothed control data were then subtracted from the smoothed sample data to remove background noise. After a noise threshold was calculated for each antibody on the smoothed and background-subtracted data, the significant peaks were called with a probability threshold of 99.9999%.
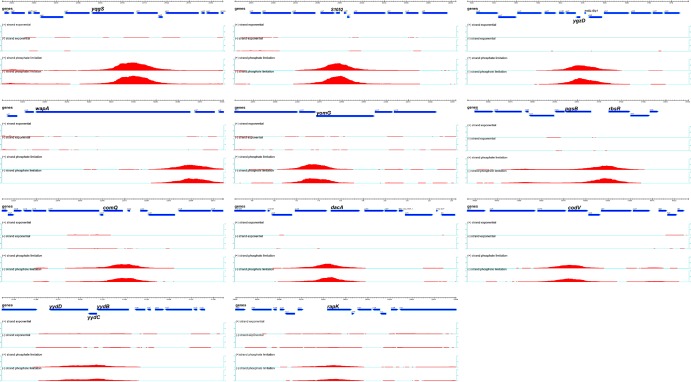
Only peaks with a significant signal on more than 30 contiguous tiles were considered. The score assigned to a peak is the sum of the log ratios (signals) for the probes within the peak; the Max (log2) value is the highest measure of binding signal within the peak, and the size of the peak is the length of DNA covered by the probes that have a signal above the background threshold. It is important to visualize binding peak morphology (Fig. 1; see also Fig. S1 in the supplemental material), since a particular score could represent either a narrow peak with a very high signal or a broad peak with a low signal.
FIG 1.
PhoP∼P binding to new chromosomal loci in vivo. Binding of PhoP∼P to chromosomal DNA was determined by ChIP-chip analysis using a tiled array. Chromosomal loci are named in each panel. The genetic organization and direction of transcription of genes in the region are represented by blue arrows with the dimensions shown above in kilobases. The intensity (peak height) and extent (peak length) of PhoP∼P binding is represented by the red peak whose position corresponds to that region of the chromosomal locus (blue arrows). The fold enrichment of PhoP∼P binding is shown on the vertical axes located on the left and right of each panel. The (+)/(−) strand exponential shows PhoP∼P binding on the plus (+) and minus (−) DNA strands at this locus in cells growing exponentially in LPDM; the (+)/(−) strand phosphate limitation shows PhoP∼P binding on the plus (+) and minus (−) DNA strands at this locus in phosphate-limited cells.
RNA extraction and RT-qPCR.
Strains 168, AH024 and LSB088 were inoculated from HPDM overnight cultures into LPDM medium. Cultures were grown at 37°C with shaking at 220 rpm in an orbital shaker (New Brunswick, Edison, NJ). Samples were harvested at designated times and centrifuged for 2 to 3 min at 4°C, and cell pellets were snap-frozen in a dry ice-ethanol bath. Pellets were either stored at −70°C or processed immediately. Total RNA was prepared using the GenElute mammalian total RNA miniprep kit (Sigma catalog no. RTN70-1KT) according to the manufacturer's instructions, except that the cells were first broken using a Fast-Prep shaker (Bio 101), as previously described (34), and the addition of an on-column DNase treatment step, using 10 U of RQ1 DNase (Promega) for 15 min at 37°C. cDNA synthesis was conducted using the Transcriptor high-fidelity cDNA synthesis kit (Roche) using 1 μg of DNase-treated total RNA and a random hexamer primer according to the manufacturer's instructions. Parallel reaction mixtures lacking reverse transcriptase were carried out and used as the templates to ensure successful DNA removal. Reverse transcription-quantitative PCR (RT-qPCR) analysis of transcript levels was conducted using a LightCycler 480 SYBR green I Master kit (Roche) on a LightCycler 480 instrument (Roche). Reactions were set up in duplicate and crossing points (Cp) were determined using the second derivative maximum method of the LightCycler 480 software (v1.5.0). The level of 16S rRNA was used as a reference to normalize samples and relative expression ratios were calculated using the 2−ΔΔCT method (35). qPCR primers were designed using PrimerExpress 3.0 software, and a melting-curve cycle was included for every primer set to check the specificity of each amplification.
Gel mobility shift DNA binding assays.
His6-tagged PhoP and PhoR′ were purified from strain BL21(DE3) as previously described (36). PhoP was cloned in NdeI/XhoI-digested pET21b after amplification with the primer pair PHOP5PET/PHOP3PET to generate plasmid pDN1102. The intracellular region of PhoR was cloned in NcoI/XhoI-digested pET21d after amplification with PhoRF(NcoI)/PhoRR(XhoI) to generate plasmid pSKD14. DNA fragments spanning the 3′ untranslated region (UTR) region of phoR (329 bp) were amplified from either B. subtilis 168 (3′phoR) or LSB088 (3′phoR*) chromosomal DNA by PCR using Phusion polymerase (Fermantas) and the polA EMSA fwd(biotinylated)/polA EMSA rev primer pair. The biotinylated fragments were gel purified according to standard procedures. Binding assays were performed as follows. Increasing amounts of PhoP were incubated with 1 μM PhoR′ in the presence of 1 mM ATP, 10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 5% glycerol, 1 mM EDTA, 4 mM dithiothreitol, 0.5 μg of sheared herring sperm DNA, and 4 μg of bovine serum albumin (BSA). The phosphorylation reaction took place for 15 min at room temperature, 2 ng of biotinylated probe was added, and the reaction mixture was incubated for 10 min at room temperature. The reactions were loaded on 6% 1× Tris-acetate-EDTA–polyacrylamide gels, and electrophoresis took place at 4°C. Bands were detected after electrotransfer to BioDyne membrane (Pall) using a Phototope-Star detection kit (NEB) according to the manufacturer's instructions.
Western blot analysis.
Strains 168, AH024 (ΔphoPR), LSB088 (mutated 3′phoR), and LSB450 (PipA-3×FLAG) were grown in LPDM at 37°C. Samples were harvested by centrifugation at 7,100 × g for 3 min at 4°C. The cell pellets were resuspended to an OD600 of ∼0.04/μl in lysis buffer (10 mM Tris [pH 8], 0.5 mM EDTA, 100 μg of lysozyme/ml, 10 μg of DNase/ml, Calbiochem protease inhibitor cocktail set III), followed by incubation at 37°C for 30 min. An equal volume of 2× SDS-PAGE (100 mM Tris [pH 6.8], 4% SDS, 20% glycerol) loading buffer, together with 0.025% (final concentration) β-mercaptoethanol, was added to the lysates. The samples were thoroughly vortexed and then boiled for 10 min. The protein concentration was determined (with aliquots of the samples prior to addition of β-mercaptoethanol) using a bicinchoninic acid protein assay kit (Novagen) with a standard curve generated using BSA. Samples (20 μg of total protein) were separated on either 10% (for PhoP/PhoR) or 15% (for PipA-3×FLAG) SDS-PAGE Tris-glycine polyacrylamide gels and transferred to a polyvinylidene difluoride membrane (Roche) by electroblotting. To detect PhoP and PhoR, the membranes were incubated with a 1:2,500 dilution (PhoP) or a 1:1,000 dilution (PhoR) of primary polyclonal antibody raised against purified PhoP and PhoR proteins in New Zealand White rabbits according to the standard procedure used by Dundee Laboratories (Dundee, Scotland). The PhoR antibody was affinity purified using PhoR antigen coupled to CnBr-activated Sepharose as previously described (36). The membranes were then incubated with a 1:30,000 dilution of goat anti-rabbit antibody–horseradish peroxidase conjugate (Sigma-Aldrich). To detect PipA-3×FLAG, the membranes were incubated with a 1:1,000 dilution of primary monoclonal anti-FLAG M2 antibody produced in mouse (Sigma-Aldrich, catalog no. F1804), followed by incubation with a 1:30,000 dilution of goat anti-mouse antibody–horseradish peroxidase conjugate (Sigma-Aldrich). Protein-antibody complexes were detected with a WesternBright Sirius Western blotting detection kit (Advansta, catalog no. K-12043-D10).
Immunoprecipitation.
Immunoprecipitation experiments were conducted twice. Strains 168 and LSB450 were grown in LPDM (with the addition of 5 μg of chloramphenicol/ml for LSB450) at 37°C. Samples were harvested 30 min after the onset of a phosphate starvation state (T+0.5). Cross-linking was performed by the addition of formaldehyde to a final concentration of 0.6%, followed by a 20-min incubation at 37°C with slow shaking. The cultures were then quenched with glycine (0.125 M final concentration) for 5 min at room temperature. The cells were collected by centrifugation at 4°C, and the pellets were washed twice with 50 ml of ice-cold buffer A (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) before being snap-frozen in a dry ice-ethanol bath. Pellets were thawed then resuspended in 12 ml of ice-cold immunoprecipitation buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) containing 120 μl of Calbiochem protease inhibitor cocktail set III. The cells were then disrupted by using a French press. Samples were centrifuged for 10 min at 8,000 rpm (4°C) to remove insoluble debris. The clarified lysates were then added to 100 μl of anti-FLAG M2-agarose resin (Sigma-Aldrich catalog no. A2220) equilibrated with ice-cold 1× wash buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA) according to the manufacturer's instructions and incubated overnight at 4°C with mild agitation. Beads were then collected by centrifugation for 1 min at 5,000 rpm (4°C), transferred to 2-ml Eppendorf tubes, and washed three times with 1 ml of immunoprecipitation buffer. FLAG-tagged protein was eluted from the beads by incubation with 200 μl of FLAG elution buffer (1× wash buffer containing 150 ng of 3×FLAG peptide [Sigma-Aldrich catalog no. F4799]/μl) for 30 min at 4°C, followed by centrifugation for 1 min at 5,000 × g. Residual FLAG-tagged protein was eluted by washing the beads two times with 200 μl of 1× wash buffer at 25°C. The protein concentration in the eluted samples was determined by using a bicinchoninic acid protein assay kit (Novagen) with a standard curve generated using BSA. The presence of PipA-FLAG in the samples was determined by Western blot analysis as described above. Eluant proteins were trichloroacetic acid (TCA) precipitated using a 1/100 volume of 100% TCA (1 h on ice and then overnight at −20°C), followed by five washes with acetone, and then air dried.
LTQ-Orbitrap Velos mass spectrometry.
The immunoprecipitates from PipA-FLAG and untagged PipA-expressing B. subtilis wild-type strains were separated by SDS–15% PAGE and in-gel tryptic digested as described by Chi et al. (37). Tryptic peptides were subjected to a reversed-phase column chromatography and mass spectrometry (MS) and tandem MS (MS/MS) data were acquired with the LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source as described by Chi et al. (37). PipA-FLAG interaction partners were identified by searching all MS/MS spectra in “dta” format against a B. subtilis target decoy protein sequence database (38) that was extracted from UniprotKB release 12.7 (39) using Sorcerer-SEQUEST (SEQUEST, v2.7, rev.11; Thermo Electron including Scaffold 4.0; Proteome Software, Inc., Portland, OR). The SEQUEST search was carried out with following parameters: a parent ion mass tolerance of 10 ppm and a fragment ion mass tolerance of 1.00 Da. Up to two tryptic miscleavages were allowed. Methionine oxidation (15.994915 Da) and cysteine carbamidomethylation (57.021464 Da) were set as variable modifications. Proteins were identified by at least two peptides, applying a stringent SEQUEST filter. SEQUEST identifications required at least ΔCn scores (the normalized score difference between the currently selected total identified peptide spectra [PSM] and the highest-scoring PSM for that spectrum) of >0.10 and XCorr scores of greater than 2.2, 3.3, and 3.75 for doubly, triply, and quadruply charged peptides. Protein quantification for the PipA-interaction partners was performed using the Scaffold proteome software and spectral counting from two biological replicates each for the PipA-FLAG versus untagged PipA-expressing B. subtilis wild-type strains.
Microarray data accession number.
Microarray data have been submitted to the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information and assigned the accession number GSE65273.
RESULTS
PhoP∼P binds to 25 chromosomal regions.
The chromosomal regions to which activated PhoP∼P binds in phosphate-limited cells were identified by a ChIP on chip analysis using high-density microarrays. The profile of PhoP∼P binding to chromosomal DNA was established in cells growing under phosphate-replete (PhoP is not activated) and phosphate-limited (PhoP is activated by phosphorylation [PhoP∼P]) conditions (9). Our results are presented in graphic form to show the location and extent of PhoP∼P binding at newly identified (Fig. 1 and 2) and previously identified (Fig. 2; see Fig. S1 in the supplemental material) loci and tabulated according to PhoP∼P binding score (Table 2) (see reference 21). PhoP binds to 25 regions of the B. subtilis chromosome in phosphate-limited cells but does not bind detectably to any chromosomal region in cells growing exponentially under phosphate-replete conditions (Fig. 1 and Table 2; see Fig. S1 in the supplemental material). The threshold of biologically relevant PhoP∼P binding was arbitrarily set at yurI, a gene previously identified as a member of the PhoPR regulon by transcriptome analysis (Table 2) (7, 9). The specificity of PhoP∼P binding at these 25 chromosomal loci is supported by the following control experiments: (i) the PHO response is induced normally in cells with FLAG-tagged PhoP (data not shown), (ii) binding is not observed at any chromosomal locus when PhoP is not FLAG-tagged, (iii) binding of PhoP∼P to chromosomal DNA is observed in phosphate-limited cells but not in phosphate-replete cells, and (iv) binding of PhoP∼P is observed within the promoters of most of the previously identified PHO regulon genes (see genes marked with asterisks in Table 2; see also Fig. S1 in the supplemental material).
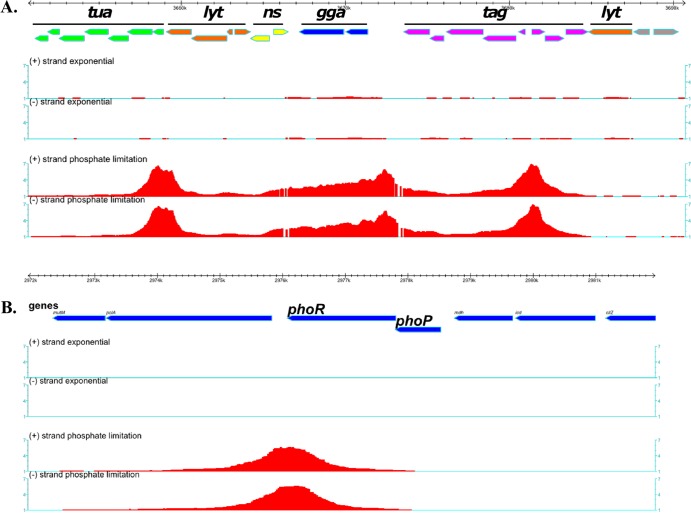
FIG 2.
PhoP binding to new targets in vivo. Binding of PhoP∼P to chromosomal DNA was determined by ChIP on chip analysis using a tiled array. The symbols and organization are as described for Fig. 1. (A) PhoP∼P binds to an ∼28-kb region that is enriched in genes associated with cell wall metabolism and especially anionic polymers. Green arrows, the tuaA-H operon; blue arrows, ggaAB; purple arrows, tagABC and tagDEFGH; orange arrows, lytR, lytABC, and lytD; yellow arrows, mnaA and gtaB. (B) PhoP∼P binds to DNA at the 3′ end of the phoPR operon with no detectable binding at the promoter that is located between mdh and phoP.
TABLE 2.
PhoP∼P binding to chromosomal DNA in vivo determined by genome-wide ChIP-on-chip analysis
| Gene(s)a | Scoreb | Max (log2)c | Size (bp)d | Regulation | Function(s) | Fold changee in: |
|
|---|---|---|---|---|---|---|---|
| WT | ΔphoPR strain | ||||||
| ggaA | 995.8 | 2.55 | 11,414 | Poly(glucosyl N-acetylgalactosamine 1-phosphate) glucosyltransferase, biosynthesis of teichoic acid | –3.9 | –2.8 | |
| tagA* | 632.7 | 2.79 | 8,030 | UDP-N-acetyl-d-mannosamine transferase, biosynthesis of teichoic acid | –5.4 | 2.1 | |
| tuaA* | 484.3 | 2.72 | 5,206 | Lipid carrier sugar transferase, biosynthesis of teichuronic acid | 112.3 | 1.1 | |
| yqgS | 333.6 | 2.39 | 3,529 | Minor lipoteichoic acid synthase | –1.9 | –1.2 | |
| pstS* | 320.2 | 2.41 | 3,472 | Phosphate ABC transporter (binding protein) | 109.7 | 1.1 | |
| S1052 (pipA) | 255.3 | 2.35 | 3,011 | Unknown | –3.9 | –1.6 | |
| phoR (3′) | 252.6 | 2.49 | 3,190 | Two-component sensor kinase | NA | NA | |
| phoB* | 216.3 | 1.88 | 3,360 | σE | Alkaline phosphatase III | 162.3 | 1.2 |
| ygzD/trnSL-gly1 | 199.7 | 1.97 | 2,676 | Putative HTH-type transcriptional regulator/tRNA-Gly | –1.5 | –1.8 | |
| wapA | 194.6 | 1.78 | 2,950 | YvrGHb, DegSU, WalRK | Contact-dependent inhibition system | –19.8 | –17.4 |
| yvyD* | 186.3 | 2.1 | 2,691 | σB, σH | Dimerization of ribosomes in the stationary phase | 3.3 | 3.4 |
| yomG | 178.6 | 1.9 | 2,307 | Unknown | –1.2 | –1.0 | |
| pgsB (capB)/rbsR | 165.1 | 1.77 | 2,594 | DegU/AbrB, CcpA | Poly-gamma-glutamate synthetase/transcriptional repressor (ribose) | –1.2 | –1.1 |
| –1.2 | –1.6 | ||||||
| comQ | 152 | 1.62 | 2,381 | Prenyltransferase, regulation of quorum sensing | 4.2 | 4.3 | |
| dacA | 150.6 | 1.8 | 2,645 | d-Alanyl-d-alanine carboxypeptidase (Pbp 5*) | –1.5 | –3.2 | |
| srfAA* | 149.4 | 1.91 | 2,202 | Abh, CodY, ComA, PerR, Spx | Surfactin synthetase | 2.2 | 1.6 |
| codV | 114.3 | 1.45 | 2,214 | CodY | Site-specific integrase/recombinase | –1.6 | –2.0 |
| phoA* | 112.9 | 1.72 | 2,246 | Alkaline phosphatase A | 107.0 | –1.4 | |
| yydBCD [3] | 103.5 | 1.1 | 2,941 | WalRK | Putative phosphohydrolase (yydB) | –1.0 | –1.1 |
| ykoL* | 96.2 | 1.44 | 1,820 | TnrA | Unknown | 91.8 | 1.5 |
| phoD* | 70.7 | 1.34 | 1,448 | Phosphodiesterase/alkaline phosphatase | 98.6 | –1.1 | |
| glpQ* | 60.7 | 1.28 | 1,329 | CcpA, GlpP | Glycerol phosphate diester phosphodiesterase | 76.6 | –2.6 |
| rapK [1] | 57.5 | 0.98 | 2,144 | AbrB, σH | Response regulator aspartate phosphatase | 5.6 | 2.9 |
| yjdB* | 54.9 | 1.08 | 1,782 | AbrB | Unknown | 14.4 | 2.2 |
| yurI (bsn)* | 48.6 | 1.15 | 1,450 | AbrB | Extracellular RNase | 61.6 | 1.2 |
*, genes previously identified as members of the PhoPR regulon. A slash signifies genes that are divergently transcribed; numbers in brackets are numbers of structural genes covered by PhoP∼P. Gene functions are as given in the Subtiwiki (http://www.subtiwiki.uni-goettingen.de), GenoList (http://genodb.pasteur.fr), and PubMed databases.
Calculation of the score and the maximum level is described in Materials and Methods. The score is the sum of the signals (log ratios) of each probe within the designated peak.
Max is the highest measure of binding signal within the peak.
Size indicates the length of chromosomal DNA covered by the probes that have a signal (log ratios).
Fold change in RNA abundance between exponentially growing and phosphate-limited cells in either the WT or ΔphoPR strain (data were obtained from reference 9). NA, not applicable.
Twelve of the chromosomal loci at which PhoP∼P binds overlap the promoters of known PHO regulon genes (see genes marked with asterisks in Table 2; see also Fig. S1 in the supplemental material). Ten of the remaining 13 chromosomal PhoP∼P binding loci overlap promoter-containing intergenic regions, suggesting a regulatory role for PhoP∼P binding at these sites. However, expression of the genes at these loci has not been associated with PhoPR-dependent regulation in any previous study (7, 9, 11). Two of the newly identified PhoP∼P binding loci are located within open reading frames (yydC and yomG). The location of the remaining PhoP∼P binding locus is intriguing, being positioned at the 3′ end of the phoPR operon between the phoR open reading frame and the transcriptional terminator (Fig. 2B; see also Fig. S2 in the supplemental material). Multiple transcriptional regulators are involved in controlling the expression of five operons (wapA, pgsB, codV, yydBCD, and rapK), while five genes (ggaAB, yqgS, wapA, and dacA) encode proteins that participate in cell wall metabolism (Table 2).
The profile of PhoP∼P binding to chromosomal DNA displays novel features at several of the 13 newly identified chromosomal loci. PhoP∼P binding is enriched throughout a 28-kb region of the chromosome that encodes genes involved in cell wall polymer synthesis (tuaABCDEFGH, ggaAB, tagABC tagDEF tagGH), autolysin expression (lytRABC and lytD) and nucleotide sugar metabolism (gtaB, yvzE, and mnaA) (Fig. 2A). The peaks of enriched PhoP∼P binding are centered at the tuaA-H, ggaAB, and tagAB promoter regions. However, PhoP∼P also clearly binds throughout the ggaAB operon and to several other open reading frames (e.g., tagABDEF, lytRABCD, mnaA, and gtaB) in this region. It is interesting that ggaAB (minor teichoic acid synthesis) expression has not been reported to be PhoPR dependent in previous transcriptome studies (7, 9).
PhoP∼P binds to the 3′ end of the phoPR operon in vivo, while no detectable binding was observed within the operon promoter, a novel and intriguing feature of the bindome (Fig. 2B and Table 2). The presence of four putative PhoP∼P DNA binding motifs located between the 3′-end of phoR and the operon terminator supports the validity of PhoP∼P binding at this location (Fig. 2B and 4A; see also Fig. S2 in the supplemental material).
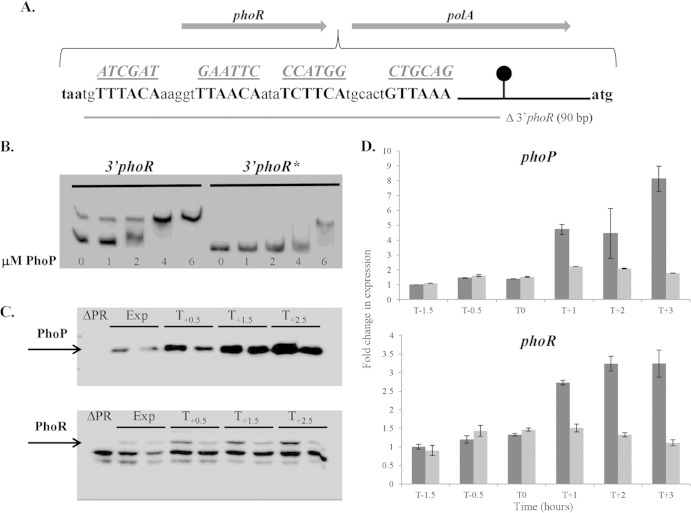
FIG 4.
Binding of PhoP∼P to the 3′ end of the phoPR operon is required for positive autoregulation. The role of PhoP∼P binding to the 3′ end of the phoPR operon was examined by gel shift (B), Western (C), and RT-qPCR (D) analyses. (A) The sequence of the PhoP boxes (boldface) positioned between the phoR stop codon (taa, in boldface) and the beginning of polA (atg, in boldface). The putative PhoP boxes are shown in boldface capital letters, while the sequences to which they are mutated are shown underlined above each motif. The region deleted in strain LSB046 is shown by a gray bar below the sequence. The phoPR transcriptional terminator is represented by a black line and circle. (B) An electrophoretic mobility shift assay was performed to verify PhoP∼P binding to DNA encoding the PhoP∼P boxes at the 3′ end of the phoPR operon. 3′phoR, a DNA fragment containing the wild-type PhoP boxes; 3′phoR*, a DNA fragment containing mutated (as shown in panel A) PhoP boxes. Each reaction mixture contained 2 ng of biotin-labeled DNA fragment, and the concentration of PhoP used in each reaction is indicated below the figure (0 to 6 μM). (C) Western blot analysis of PhoP and PhoR protein levels in strains 168 (wild-type) and LSB088 (mutated PhoP∼P boxes at the 3′ end of the phoPR operon). Both strains were grown in LPDM, and samples were harvested during exponential growth (Exp) and at 0.5 h (T0.5), 1.5 h (T1.5), and 2.5 h (T2.5) after the onset of phosphate limitation. For each time point, the band on the left is from wild-type strain 168, and the band on the right is from strain LS088. The PhoP and PhoR bands are indicated by arrows. Twenty micrograms of total protein was loaded in each lane. (D) The phoP (top panel) and phoR (bottom panel) transcript levels in cells of wild-type strain 168 (dark gray bars) and strain LSB088 (containing mutated PhoP binding boxes at the 3′ end of the phoPR operon) (light gray bars) growing in LPDM were measured by RT-qPCR. The level of each transcript was normalized to the level of transcript for the wild-type (WT) sample at time point T−1.5, which was assigned a value of 1.
A bioinformatics analysis was undertaken to identify potential PhoP∼P DNA binding motifs within the promoter regions of the newly identified bindome members. It is known that PhoP∼P binding motifs are in opposite orientations (relative to the initiation point of transcription) within promoters that are activated (e.g., tuaA) or repressed (e.g., tagA) by PhoP∼P (see Fig. S2 in the supplemental material) (11, 20). Putative PhoP∼P motifs were identified within 12 of the newly identified chromosomal binding regions (see Fig. S2 in the supplemental material). However, only the sites at the 3′ end of the phoPR operon and those within the S1052/pipA promoter region are in the orientation associated with PhoP∼P activation with the remainder being in the orientation associated with repression (see Fig. S2 in the supplemental material).
Expression of newly identified members of the PhoPR bindome.
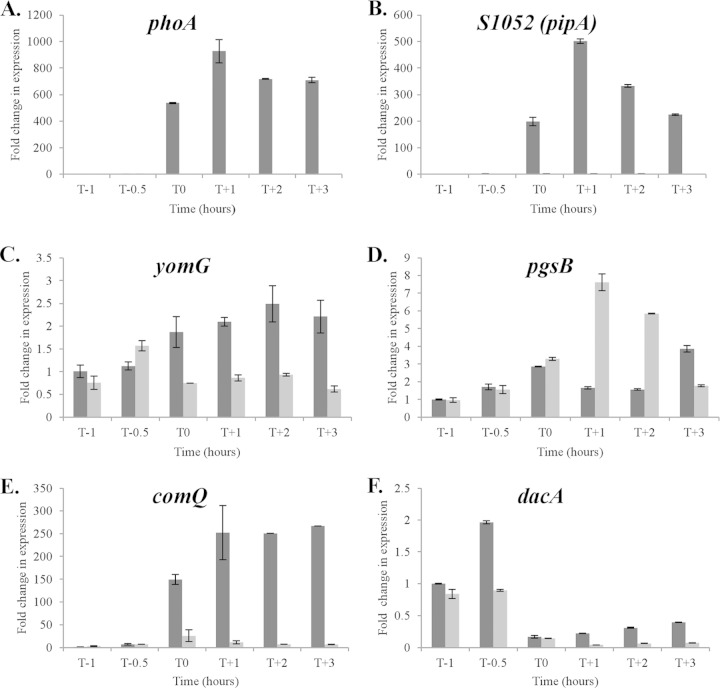
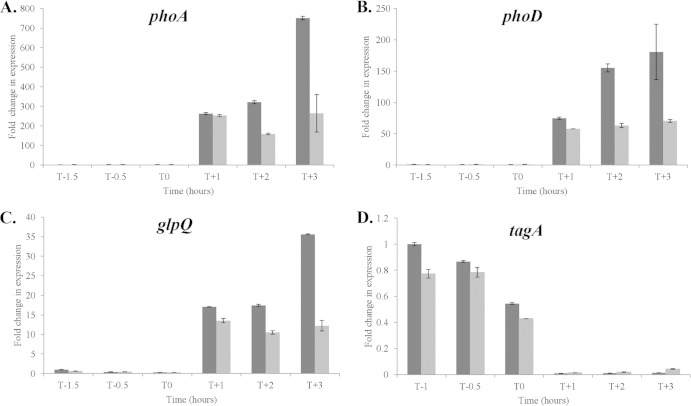
The observation that 12 of the 13 newly identified binding sites overlap promoter regions suggests that PhoP∼P plays a regulatory role in gene expression at these loci. To address the possibility that previous studies were not of sufficient sensitivity or temporal refinement to detect PhoPR regulated expression, we established the relative transcript level of genes at 10 loci by RT-qPCR in cells of strains 168 (wild-type 168) and AH024 (ΔphoPR) growing in LPDM. Our results show that five newly identified bindome members are differentially expressed in wild-type 168 and AH024 (ΔphoPR) strains (Fig. 3). The newly identified S1052 small RNA and the comQ gene encoding isoprenyl transferase (that modifies the competence pheromone ComX) are induced 200- to 500-fold and 150- to 250-fold, respectively, in a PhoPR-dependent manner upon phosphate limitation (Fig. 3). PhoPR is required to maintain yomG (∼2-fold) and dacA (∼2-fold) expression in phosphate-limited cells, while PhoP∼P represses pgsB expression ∼6 to 8-fold at T1 and T2 during phosphate limitation. However, expression of the other newly identified members of the bindome is PhoPR independent. It is especially interesting that ggaAB, yqgS, and wapA expression is PhoPR independent in view of the extensive PhoP∼P binding to their promoter regions (Table 2).
FIG 3.
The relative transcript level of five newly identified members of the PhoP bindome whose expression is PhoPR dependent. Strains 168 (WT) (dark gray) and AH024 (ΔphoPR) (light gray) were grown in LPDM and RNA prepared from cells at the designated times. The relative transcript levels were measured by RT-qPCR and normalized to the level of transcript in the T−1 sample of strain 168 (data not shown), which was assigned a value of 1. Time is shown in hours before (T minus) and after (T plus) the point at which cells enter the phosphate-limited state (T0). The level of phoA transcript (A) was determined using the same RNA samples to establish the time point at which cells became phosphate limited.
PhoP∼P binding at the 3′ end of phoPR operon is required for positive autoregulation upon phosphate limitation.
ChIP-chip analysis showed that PhoP∼P binds to the intergenic region between the phoPR (3′-end) and polA (5′-end) operons (Fig. 2B). There are four putative PhoP∼P binding motifs located between the phoR stop codon (TAA) and the phoPR transcriptional terminator but none between the terminator and the polA start codon (Fig. 4A; see Fig. S2 in the supplemental material). Thus, it is likely that PhoP∼P binding at this locus is associated with the phoPR operon rather than with polA expression. This was verified by showing (using RT-qPCR) that polA expression is PhoPR independent (data not shown).
To establish whether these motifs at the 3′ end of the phoPR operon contribute to PhoP∼P binding, we performed gel mobility shift assays on a biotinylated PCR-generated fragment with intact motifs and on a similarly generated fragment in which these motifs are mutated as shown in Fig. 4A. Our results show that the proficiency with which PhoP∼P retards the mobility of these fragments is diminished by mutation of the four motifs (Fig. 4B, compare the 3′phoR and 3′phoR* profiles). We conclude that these four motifs contribute to PhoP∼P binding at the 3′ end of the phoPR operon, thereby supporting the results obtained in the in vivo bindome study (i.e., results of the ChIP-chip analysis shown in Fig. 2B).
To determine whether PhoP∼P binding to the 3′ end of the phoPR operon affects its expression, we compared the phoP and phoR transcript levels in wild-type strain 168 to those in strains LSB088 (PhoP boxes mutated) and LSB046 (PhoP boxes deleted), in which the motifs are mutated (as shown in Fig. 4A) and deleted (not shown), respectively. Our results show that in cells growing exponentially the level of phoP and phoR transcript is unaffected by mutation (Fig. 4D, T−1.5, T−0.5, and T0) or deletion (data not shown) of the four motifs. However, the 2- to 4-fold increase in each transcript that occurs when wild-type cells become phosphate limited is abolished by mutation (Fig. 4D, T1, T2, and T3) or by deletion (data not shown) of these motifs. It is notable that in strains in which the PhoP∼P binding motifs at the 3′ end of the phoPR operon have been mutated, a similar level of phoPR transcript is observed in exponentially growing and phosphate-limited cells (Fig. 4D). Therefore, we conclude that PhoP∼P binding to sequences located between the 3′ end of phoR and the operon terminator is required for the positive autoregulation of phoPR expression that occurs at the onset of phosphate limitation.
Initiation of phoPR transcription is directed by up to five promoters recognized by several different sigma factors (40). To investigate whether PhoP∼P binding to the 3′ end of the phoPR operon affects the points of initiation or termination of phoPR transcription, we performed 5′ and 3′ RACE on RNA isolated from wild-type cells and from cells in which the PhoP∼P binding motifs at the 3′ end of phoPR were mutated as shown (Fig. 4A). There was no detectable difference in the profile of the initiation points of transcription between wild-type cells and cells in which the motifs are mutated (data not shown). Moreover, the phoPR transcript terminates at the same base in both wild-type cells and in cells in which the PhoP∼P cognate boxes are mutated (data not shown).
To ascertain whether cellular PhoP and PhoR protein levels are affected by mutation or deletion of the PhoP∼P boxes at the 3′ end of the phoPR operon, we performed Western blot analysis on lysates prepared from exponentially growing and phosphate-limited wild-type (strain 168) and mutant (strain LSB088) cells. Our results show that the cellular level of both proteins increases when wild-type cells become phosphate limited as previously reported (compare the Exp lanes with the T0.5, T1.5, and T2.5 lanes for strain 168 in Fig. 4C). However, the level of PhoR does not increase in strain LSB088 upon phosphate limitation (lower panel Fig. 4C), while the level of PhoP increases but to a significantly lower level than that observed in wild-type cells (upper panel Fig. 4C). Thus, the PhoP∼P binding sites at the 3′ end of the phoPR operon are required to increase the level of PhoPR protein when cells become phosphate limited. In summary, these data show that PhoP∼P binding to cognate motifs positioned between the 3′ end of phoR cistron and the operon terminator are required for positive autoregulation of phoPR transcription and increased PhoPR protein levels upon phosphate limitation.
PhoP∼P binding at the 3′ end of phoPR operon is required for full induction of the PHO response.
To establish the contribution of PhoP∼P binding at the 3′ end of the phoPR operon to induction of PHO regulon genes upon phosphate limitation, we determined the phoD, phoA, glpQ, and tagA transcript levels by RT-qPCR in cells of wild-type strain 168 and in cells of strain LSB088 (the PhoP∼P binding sites are mutated). Our results show that the timing of phoD, phoA, and glpQ induction and tagA repression is unaffected by mutation of these PhoP∼P binding motifs (see T1 to T3 in Fig. 5). However, the extent to which expression of these genes is induced upon phosphate limitation differs in the two strains. Mutation of these PhoP∼P boxes results in significantly lowered levels of phoA, phoD, and glpQ transcript at 2 (T2) and 3 (T3) hours after the onset of phosphate limitation (Fig. 5A to C). The difference in induction of these genes at T1 is lower than is observed at T2 and T3 (Fig. 5A to C), consistent with the finding of Botella et al. (2, 9) that full induction of the PHO response is not achieved until ∼90 min after the onset of phosphate limitation. However, only a small difference in PhoP∼P-mediated repression of tagAB was discernible between the two strains (Fig. 5D). We conclude that PhoP∼P binding to the 3′ end of the phoPR operon does not affect the timing of PhoPR activation upon phosphate limitation but is required for full induction of PHO regulon gene expression.
FIG 5.
Mutation of PhoP∼P boxes at the 3′ end of the phoPR operon reduces the transcript levels of PhoPR regulon genes upon induction. Strains 168 (wild-type) (dark gray bars) and LSB088 (mutated PhoP∼P boxes at the 3′ end of the phoPR operon) (light gray bars) were grown in LPDM, and cells were harvested at the designated times. The level of each transcript was measured by RT-qPCR and normalized to the level of transcript in wild-type strain 168 at time point T−1.5 (A, B, and C) or T−1 (D), which was assigned a value of 1. Time is indicated in hours before (T minus) or after (T plus) the point at which cells enter the phosphate-limited state (T0).
The S1052 small RNA encodes a 40-amino-acid protein named PipA.
PhoP∼P binds at a high level to the chromosomal region between the yrzF (encodes a putative Ser/Thr kinase) and yrzH (a gene of unknown function) genes (Fig. 1 and Table 1; see Fig. S2 in the supplemental material). This region encodes a 197-nucleotide RNA (S1052) first identified by transcriptome analysis (41). To establish the activity and induction profile of the S1052 promoter, expression of a PS1052 gfp transcriptional fusion was determined in cells of strain LSB434 growing in LPDM. The promoter is highly induced (>1,000 activity units) in a PhoPR-dependent manner at the onset of phosphate limitation and continues at this level of activity (∼800 to 1,000 activity units) while the stimulus persists (Fig. 6B). This profile is consistent with the presence of several potential PhoP∼P DNA recognition sequences within the S1052 promoter (see Fig. S2 in the supplemental material). Further analysis suggested that the S1052 RNA may encode a 40-amino-acid open reading frame (beginning at nucleotide 39) that is preceded by a strong ribosome binding site. The putative protein lacks a signal peptide and is highly conserved in several Bacillus species (subtilis, mojavensis, vallismortis, siamensis, amyloliquefaciens, and atrophaeus) (Fig. 6A). This 40-amino-acid open reading frame also constitutes the C-terminal end of a 79-amino-acid open reading frame found in some B. amyloliquefaciens (e.g., FZB42 and CAU B946) strains (Fig. 6A). To establish whether this open reading frame is translated, strain LSB448 was generated containing a PS1052 orf′-gfp translational fusion (the 5′ half of the open reading frame was fused in-frame with gfp). Our results show that expression of this translational fusion is induced at the onset of phosphate limitation and continues while the stimulus is maintained (data not shown). We have renamed this locus pipA (PhoPR-induced protein A). The PipA′-′Gfp fusion protein was uniformly distributed throughout the cytoplasm (data not shown). To confirm that this open reading frame is translated, strain LSB450 was generated in which the putative open reading frame was extended at the 3′ end to encode a 3×FLAG tag. Cells of strain LSB450 were harvested during growth in LPDM, and lysates were prepared for Western blot analysis with an anti-FLAG antibody to detect the protein. Our results (Fig. 6C) show a protein of appropriate size (∼14 kDa) expressed only in phosphate-limited cells (T0.5 to T2.5) of strain LS450. The specificity of detection is shown by the failure to detect a band at this position when the protein is not tagged with 3×FLAG (Fig. 6C, top panel, sample 168), while the onset of phosphate limitation is confirmed by the increase in PhoP levels in samples T0.5 to T2.5. Thus, expression of the 40-amino-acid protein intracellular PipA protein is induced as part of the PHO response in B. subtilis.
FIG 6.
PipA is a small protein whose expression is induced by activation of the PHO response. An analysis of the newly identified S1052/pipA locus was performed. (A) Alignment of PipA from various strains: B. subtilis (Bsub) strains 168, W23, and TU-B-10; B. mojavensis (Bmo); B. vallismortis (Bval); B. siamensis (Bsiam); B. amyloliquefaciens (Bam) strains FZB42, DSM7, TA208l, plantarum NAU-B3, and plantarum CAU; and B. atrophaeus (Batro). Dark gray shading, amino acids conserved in all proteins; light gray shading, conserved in at least 10 of the 12 proteins. Alignments were performed with CLUSTAL W and shaded with the Genedoc program (http://genedoc.software.informer.com). (B) Determination of S1052/PpipA promoter activity. The DNA region 400 bp upstream of the pipA RBS was fused to a promoterless gfpmut3 reporter gene, and expression was determined by measuring the GFP activity in cells grown in LPDM. Filled squares, growth monitored by OD600; unfilled squares, promoter activity. (C) PipA is a small peptide expressed in phosphate-limited cells. Western blot analysis was performed on cell extracts prepared from cells of strain LSB450 (pipA-3×FLAG) grown in LPDM. The top panel shows that FLAG-tagged PipA was detected with a M2 mouse anti FLAG antibody. The bottom panel shows that, as a positive control, PhoP protein was detected in the same extracts with a rabbit polyclonal anti-PhoP antibody. The times at which cells were harvested are indicated above the sample; time is shown in hours after (T plus) the point at which cells enter phosphate limitation (T0). The 168 sample is a negative-control extract, harvested at T+1.5, from cells that do not contain a FLAG-tagged PipA.
Physiological role of PipA.
To determine the physiological role of PipA, we examined the growth, morphology, and PHO response (phoA, phoB, tuaA, and glpQ expression) in a pipA-null mutant strain (LSB396) and in cells of strain LSB435 [thrC::Pxyl S1052(pipA)] that express PipA heterologously from a xylose-inducible promoter. However, these characteristics did not differ significantly between wild-type and mutant strains (data not shown).
To identify the protein(s) with which PipA interacts in the cell, phosphate-limited cells of wild-type strain 168 and strain LSB450 (pipA′-′3×FLAG) were treated with formaldehyde to cross-link proteins interacting with PipA-3×FLAG. Immunoprecipitation was carried out with an anti-FLAG antibody, and coprecipitated proteins were identified by LTQ-Orbitrap Velos mass spectrometry as described in Materials and Methods. The identities of the proteins enriched in the immunoprecipitate from two separate experiments are presented in Table 3. PipA peptide fragments were identified by mass spectrometry, further confirming that the S1052 transcript is translated, yielding the PipA protein. Other proteins identified in the immunoprecipitate had a spectrum of functions, with catalase 2 (KatE) and RsbT (member of the SigB stressosome) being the most abundant. A second protein involved in SigB activation (RsbV) and three proteins involved in motility/chemotaxis (McpA, McpC, HemAT, and FliD) were also enriched in the immunoprecipitate (Table 3). Two-hybrid analysis was used in an attempt to verify an interaction between PipA and all 18 proteins listed in Table 3. However, none of the 18 proteins yielded a positive interaction using this technique (data not shown). To further explore a possible role for PipA in the SigB general stress response, we examined expression of a Pctc gfp transcriptional fusion in wild-type strain 168 and in a S1052-null mutant. However, mutation of pipA did not affect induction of the SigB response.
TABLE 3.
PipA interaction targets determined by immunoprecipitation and mass spectrometry analysis
| Target | Fold enrichment (LSB450 vs. 168)a | Function(s) | Transcriptional regulation |
|---|---|---|---|
| KatE | 7.75 | Catalase 2, detoxification of hydrogen peroxide | σB |
| RbsT | 6.67 | PP2C activator, protein serine kinase, control of σB activity | |
| YdaP | 4 | Putative pyruvate oxidase | σB |
| McpA | 3.25 | Methyl-accepting chemotaxis protein, control of chemotaxis | σD |
| YtpP | 3 | Unknown, similar to thioredoxin H1 | Spx |
| RbfA | 2.47 | Ribosome-binding factor A | Stringent response |
| YorD | 2.33 | Unknown, SPβ-derived stress response protein | AbrB |
| LytB | 2.31 | Modifier protein of major autolysin LytC | σD, YvrGHb, SlrR |
| McpB | 2.27 | Methyl-accepting chemotaxis protein, control of chemotaxis | σD |
| YolA | 2.2 | Unknown, SPβ-derived protein | AbrB |
| RsbV | 2.1 | Anti-anti-σB: control of σB activity | |
| RplL | 1.98 | Ribosomal protein L12 (BL9) | Stringent response |
| YabO | 1.93 | Unknown, similar to heat shock protein Hsp15 | |
| YhdF | 1.9 | Unknown, similar to glucose 1-dehydrogenase | σB |
| HemAT | 1.89 | Soluble chemotaxis receptor, heme-containing O2 sensor protein | σD |
| Spo0F | 1.89 | Phosphotransferase of the sporulation initiation phosphorelay | Spo0A, σH |
| FliD | 1.88 | Filament cap, serves as an extracytoplasmic chaperone required for polymerization of flagellin into the helical filament | σD |
| YabJ | 1.78 | 2-Iminobutanoate/2-iminopropanoate deaminase, required for regulation of purine biosynthesis | PurR |
Values are the averages from two experiments. Protein functions and transcriptional regulation are those ascribed in the Subtiwiki (http://www.subtiwiki.uni-goettingen.de) database.
We conclude that PipA is induced in B. subtilis cells upon phosphate limitation and is retained within, and uniformly distributed throughout, the cytoplasm. Cross-linking experiments suggest it may interact with several stress-related proteins in the cell.
DISCUSSION
We report here three novel features of the PHO response in B. subtilis that have emerged from the establishment of the PhoP∼P bindome: (i) an enlarged regulon, (ii) an increased involvement in cell wall metabolism, and (iii) a new regulatory feature of the mechanism by which the response is induced upon phosphate limitation. Together, these observations show that the regulation and physiology of the PHO response in B. subtilis is significantly more complex than previously described.
PhoP∼P binds to 25 regions of the chromosome.
A genome-wide analysis shows that PhoP∼P binds to 25 regions of the B. subtilis chromosome. Thirteen of these loci are newly identified PhoP∼P binding sites, with ten being positioned within promoters. However, expression of the genes at only 4 of these 10 promoter loci is PhoPR dependent. The comQ and pipA (S1052) genes are highly induced (200- to 500-fold) upon phosphate limitation, while the expression of yomG and pgsB changes ≤2-fold in a PhoPR-dependent manner. The failure to identify pipA (S1052) in previous studies can be attributed to its recent annotation (41). It is unclear why comQ was not identified in previous studies: perhaps the cellular level of comQ transcript is below that required for robust detection by transcriptomic analysis. However, expression of the genes under the control of the remaining six newly identified promoters is not PhoP∼P dependent under the conditions of the present study. A similar observation was made in studies of WalRK, a TCS closely related to PhoPR (18, 20, 21). Only seven genes were ascribed to the WalRK regulon by transcriptome analysis, whereas ChIP-chip analysis showed that WalR∼P binds to 15 additional chromosomal sites (18, 20, 21). The majority of newly identified binding sites for both TCS are positioned within promoter regions, suggesting that WalR∼P and PhoP∼P perform a regulatory function at these loci (21; the present study). However, such regulatory roles must be manifest only in combination with other regulators or only under particular environmental or nutritional conditions.
The newly identified PipA protein (40 amino acids) is highly induced upon phosphate limitation and distributed uniformly throughout the cytoplasm. Nicolas et al. (41) first annotated this locus as encoding a new short RNA (S1052). However, we show here, using translational fusions, Western blotting of FLAG-tagged PipA, and mass spectrometry, that S1052 is translated to produce a short protein, which we have named PipA. PipA is encoded only in some Bacillus species (subtilis, amyloliquefaciens, atrophaeus, vallismortis, siamensis, and mojavensis) and may form the C-terminal region of a larger (79 amino acid) peptide in some strains of B. amyloliquefaciens. We sought to identify proteins with which PipA interacts on the premise that most cytoplasmically located PHO regulon proteins are involved in cell wall metabolism. PipA can be reproducibly cross-linked in vivo with catalase (KatE), with proteins involved with the control of SigB activity (RsbT and RsbV), and with proteins involved with chemotaxis and motility (McpA, McpB, HemAT, and FliD). However, since these interactions could not be independently verified by bacterial two-hybrid analysis, they must be viewed with some caution. Moreover, deletion of pipA has no effect on the PHO response, the SigB response, or cellular morphology.
Positive autoregulation requires PhoP∼P binding at the 3′ end of the phoPR operon.
Positive autoregulation of phoPR expression is a well-established feature of the PHO response in B. subtilis with transcript and PhoPR protein levels increasing 3- to 4-fold upon induction (20, 40, 42). However, this regulation displays some unusual features. Here, we show that PhoP∼P binds to the 3′ end of the phoPR operon in vivo (i.e., between the phoR stop codon and the transcriptional terminator), while binding is not detected within the promoter region (Fig. 2B). PhoP∼P binding is supported by the presence of four cognate binding motifs within the 3′ region of the phoPR operon and is attenuated when they are mutated (Fig. 2B and 4B; see Fig. S2 in the supplemental material). The physiological importance of PhoP∼P binding to the 3′ end of the phoPR operon is shown by the fact that when prevented by mutation, phoPR transcript and PhoPR protein levels do not increase upon phosphate limitation but remain at the level found in exponentially growing, phosphate-replete cells (Fig. 4C and D and 5). Furthermore, mutation of these binding sites prevents full induction of PHO regulon genes, although the timing of induction is unaffected (Fig. 4C and D and 5).
However, despite PhoP∼P binding to the phoPR promoter not being observed in vivo in the present study, it has been shown in vitro by footprinting analysis (17, 40). The footprints reported are not completely concordant but two regions in the vicinity of the activated promoters are footprinted by PhoP∼P in both studies (17, 40). Unusually, the sequences designated as PhoP∼P boxes within the footprinted regions conform very poorly to the established consensus, suggesting that PhoP∼P binding within the phoPR promoter is weak (11, 17, 40). Together, these results suggest that PhoP∼P binding at the 3′ end of the phoPR operon is significantly stronger than it is within the phoPR promoter (Fig. 2B).
Instances are rare in bacteria of transcription being activated by the binding of a transcription factor to the 3′ end of an open reading frame/operon (43, 44). The cases documented involve σ54 (SigL in B. subtilis)-dependent promoters. For example, RocR activates the expression of both rocG and rocABC operons by binding to a common enhancer-type element (also called the upstream activation sequence [UAS]) (45). Although this UAS is located upstream of the rocABC promoter, it is positioned ∼1.5 kb downstream of the rocG promoter (45). Moreover, the UAS can still mediate transcriptional activation when placed a further 15 kb downstream of the rocG promoter (45). Such activation involves binding of an AAA+-type transcription factor to the distant UAS followed by interaction with the poised RNA polymerase/σ54 complex to stimulate open complex formation (44). However, all phoPR promoters are of the SigA type, making its activation by PhoP∼P binding to the 3′ end of the operon a unique case (40).
It is interesting to speculate therefore on how phoPR expression is increased by such a PhoP∼P binding profile and to what physiological advantage. The basal cellular level of PhoP and PhoR protein is governed by the combined transcriptional activity of the five promoters of the phoPR operon that have been characterized in vitro (40). We propose that PhoP∼P binding within the phoPR promoter region alone is too weak to effect increased transcription. However, binding within the promoter region may be potentiated by the presence of PhoP∼P bound at the 3′ end of the operon. Potentiation may be effected by a looping-type mechanism, similar to that by which expression of the araBAD operon in regulated by AraC Escherichia coli (46). These observations suggest that positive autoregulation of phoPR expression occurs only when PhoP∼P is bound to the 3′ potentiator site and free activated PhoP∼P is at a level sufficient to bind to the relatively weak binding sites within the promoter region. Presumably, this will happen only when all binding sites with higher affinity for PhoP∼P binding are filled. The effects of ablating positive autoregulation are manifest most clearly between 2 and 3 h after the onset of phosphate limitation (Fig. 5), suggesting that positive autoregulation of phoPR transcription contributes to amplification of the PHO response only when phosphate limitation is persistent and/or extreme.
Model for amplification of the PHO response.
The high phosphate content of WTA in B. subtilis dictates that cell wall metabolism is an integral feature of the PHO response. However, because it entails significant changes in cell wall composition, the extent to which the PHO response is activated must be proportionate to the level of phosphate deficiency. Failure to achieve this balance is clearly illustrated by the aberrant cellular growth and morphology that results from inappropriate activation of the PHO response under phosphate-replete conditions (2). We propose that proportionality is achieved through amplification of a basal response by two mechanisms: (i) the switch from WTA to teichuronic acid synthesis and (ii) positive autoregulation of phoPR expression (2, 9, 11). The mechanism by which the PHO response is triggered upon phosphate limitation is not yet known. However, a low-level basal response can be achieved in the absence of amplification that persists while the stimulus is maintained (2). The onset of teichuronic acid synthesis is the principal amplification mechanism and is followed by the repression of enzymes (TagAB) involved in WTA synthesis. The consequences of ablating either process are manifest within ∼30 min of the onset of phosphate limitation, showing that these contribute to the initial amplification that occurs immediately after the PHO response is triggered (2). Our data suggest that positive autoregulation of phoPR expression contributes to the amplification process at a later stage: binding of PhoP∼P to the phoPR promoter appears to be very weak, while the effects of ablating this binding are manifest most clearly between 1 and 2 h after the onset of phosphate limitation (Fig. 5). Thus, it appears that (free) Pho∼P levels must first rise above a certain threshold before phoPR autoregulation makes a significant contribution to further amplify the PHO response by 2- to 3-fold (2; the present study). Together, these mechanisms have the potential to enable a graded PHO response that is appropriate to the prevailing level of phosphate deficiency.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Science Foundation Ireland principal investigator awards 08/1N.1/B1859 and 12/IP/1532 to K.M.D.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02570-14.
REFERENCES
- 1.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botella E, Devine SK, Hubner S, Salzberg LI, Gale RT, Brown ED, Link H, Sauer U, Codée JD, Noone D, Devine KM. 2014. PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol Microbiol 94:1242–1259. doi: 10.1111/mmi.12833. [DOI] [PubMed] [Google Scholar]
- 3.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 4.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 5.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol 178:4344–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res 11:1484–1502. doi: 10.1101/gr.182801. [DOI] [PubMed] [Google Scholar]
- 7.Allenby NE, O'Connor N, Prágai Z, Ward AC, Wipat A, Harwood CR. 2005. Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J Bacteriol 187:8063–8080. doi: 10.1128/JB.187.23.8063-8080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baek JH, Lee SY. 2007. Transcriptome analysis of phosphate starvation response in Escherichia coli. J Microbiol Biotechnol 17:244–252. [PubMed] [Google Scholar]
- 9.Botella E, Hübner S, Hokamp K, Hansen A, Bisicchia P, Noone D, Powell L, Salzberg LI, Devine KM. 2011. Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology 157:2470–2484. doi: 10.1099/mic.0.049205-0. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, Huang TW, Wen SY, Chang CY, Tsai SF, Wu WF, Chang CH. 2012. Genome-wide PhoB binding and gene expression profiles reveal the hierarchical gene regulatory network of phosphate starvation in Escherichia coli. PLoS One 7:e47314. doi: 10.1371/journal.pone.0047314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulett FM. 2002. The Pho regulon, p 193–201. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and its closest relatives. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- 12.Liu W, Hulett FM. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443–1450. doi: 10.1099/00221287-144-5-1443. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Eder S, Hulett FM. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P J Bacteriol 180:753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soldo B, Lazarevic V, Pagni M, Karamata D. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol Microbiol 31:795–805. [DOI] [PubMed] [Google Scholar]
- 15.Lahooti M, Harwood CR. 1999. Transcriptional analysis of the Bacillus subtilis teichuronic acid operon. Microbiology 145:3409–3417. [DOI] [PubMed] [Google Scholar]
- 16.Bhavsar AP, Erdman LK, Schertzer JW, Brown ED. 2004. Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J Bacteriol 186:7865–7873. doi: 10.1128/JB.186.23.7865-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prágai Z, Allenby NE, O'Connor N, Dubrac S, Rapoport G, Msadek T, Harwood CR. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J Bacteriol 186:1182–1190. doi: 10.1128/JB.186.4.1182-1190.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, Devine KM. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol 49:1639–1655. doi: 10.1046/j.1365-2958.2003.03661.x. [DOI] [PubMed] [Google Scholar]
- 19.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 20.Bisicchia P, Lioliou E, Noone D, Salzberg LI, Botella E, Hübner S, Devine KM. 2010. Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate-limited Bacillus subtilis cells. Mol Microbiol 75:972–989. doi: 10.1111/j.1365-2958.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 21.Salzberg LI, Powell L, Hokamp K, Botella E, Noone D, Devine KM. 2013. The WalRK (YycFG) and σ(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol Microbiol 87:180–195. doi: 10.1111/mmi.12092. [DOI] [PubMed] [Google Scholar]
- 22.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Müller JP, An Z, Merad T, Hancock IC, Harwood CR. 1997. Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143:947–956. doi: 10.1099/00221287-143-3-947. [DOI] [PubMed] [Google Scholar]
- 25.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Bloor AE, Cranenburgh RM. 2006. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl Environ Microbiol 72:2520–2525. doi: 10.1128/AEM.72.4.2520-2525.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 28.Bisicchia P, Botella E, Devine KM. 2010. Suite of novel vectors for ectopic insertion of GFP, CFP, and IYFP transcriptional fusions in single copy at the amyE and bglS loci in Bacillus subtilis. Plasmid 64:143–149. doi: 10.1016/j.plasmid.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Botella E, Fogg M, Jules M, Piersma S, Doherty G, Hansen A, Denham EL, Le Chat L, Veiga P, Bailey K, Lewis PJ, van Dijl JM, Aymerich S, Wilkinson AJ, Devine KM. 2010. pBaSysBioII: an integrative plasmid generating gfp transcriptional fusions for high-throughput analysis of gene expression in Bacillus subtilis. Microbiology 156:1600–1608. doi: 10.1099/mic.0.035758-0. [DOI] [PubMed] [Google Scholar]
- 30.Bhavsar AP, Zhao X, Brown ED. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol 67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto H, Kurosawa S, Sekiguchi J. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol 185:6666–6677. doi: 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen S, Nielsen HB, Jarmer H. 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol 73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. 2007. Ringo: an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noone D, Howell A, Devine KM. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J Bacteriol 182:1592–1599. doi: 10.1128/JB.182.6.1592-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Howell A, Dubrac S, Noone D, Varughese KI, Devine KM. 2006. Interactions between the YycFG and PhoPR two-component systems in Bacillus subtilis: the PhoR kinase phosphorylates the non-cognate YycF response regulator upon phosphate limitation. Mol Microbiol 59:1199–1215. doi: 10.1111/j.1365-2958.2005.05017.x. [DOI] [PubMed] [Google Scholar]
- 37.Chi BK, Gronau K, Mäder U, Hessling B, Becher D, Antelmann H. 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10:M111.009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Médigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uniprot Consortium. 2007. The Universal Protein Resource (UniProt). Nucleic Acids Res 35(database issue):D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul S, Birkey S, Liu W, Hulett FM. 2004. Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EσA- and EσE-responsive promoters by phosphorylated PhoP. J Bacteriol 186:4262–4275. doi: 10.1128/JB.186.13.4262-4275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 42.Hulett FM, Lee J, Shi L, Sun G, Chesnut R, Sharkova E, Duggan MF, Kapp N. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J Bacteriol 176:1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shigler V. 2011. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol Rev 35:425–440. doi: 10.1111/j.1574-6976.2010.00255.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee DJ, Minchin SD, Busby SJW. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 45.Belitsky BR, Sonenshein AL. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci U S A 96:10290–10295. doi: 10.1073/pnas.96.18.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schleif R. 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 47.Maguin E, Duwat P, Hege T, Ehrlich SD, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 174:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.