ABSTRACT
Mutations that cause the constitutive expression of the PHO regulon of Escherichia coli occur either in the pst operon or in the phoR gene, which encode, respectively, a high-affinity Pi transport system and a histidine kinase sensor protein. These mutations are normally selected on glycerol-2-phosphate (G2P) as the carbon source in the presence of excess Pi. The emergence of early PHO-constitutive mutants, which appear after growth for up to 48 h on selective medium, depends on the presence of phoA, which codes for a periplasmic alkaline phosphatase, while late mutants, which appear after 48 h, depend both on phoA and on the ugp operon, which encodes a glycerophosphodiester transport system. The emergence of the late mutants hints at an adaptive mutation process. PHO-constitutive phoR mutants appear only in a host that is mutated in pitA, which encodes an alternative Pi transport system that does not belong to the PHO regulon. The conserved Thr217 residue in the PhoR protein is essential for PHO repression.
IMPORTANCE One of the principal ways in which bacteria adapt to new nutrient sources is by acquiring mutations in key regulatory genes. The inability of E. coli to grow on G2P as a carbon source is used to select mutations that derepress the PHO regulon, a system of genes involved in the uptake of phosphorus-containing molecules. Mutations in the pst operon or in phoR result in the constitutive expression of the entire PHO regulon, including alkaline phosphatase, which hydrolyzes G2P. Here we demonstrate that the ugp operon, another member of the PHO regulon, is important for the selection of PHO-constitutive mutants under prolonged nutritional stress and that phoR mutations can be selected only in bacteria lacking pitA, which encodes a secondary Pi transport system.
INTRODUCTION
The PHO regulon of Escherichia coli consists of more than 50 genes and operons that respond to orthophosphate (Pi) limitation (1, 2). The best-documented ones are the phoA and phoE genes, the pstSCAB-phoU (or pst) and ugpBAECQ operons, and the regulatory phoBR operon. phoA encodes a periplasmic alkaline phosphatase (AP), phoE encodes an anion-specific porin, and the first four genes of the pstSCAB-phoU operon code for an ABC-type high-affinity Pi transport system which, together with phoU, also plays a role in the regulation of the PHO genes. Similar to the pst operon, the ugpBAEC genes encode an ABC transport system for sn-glycerol-3-phosphate and glycerophosphodiesters, while the fifth gene of the operon, ugpQ, encodes a phosphodiesterase that hydrolyzes glycerophosphoryl diesters (3, 4). The phoBR operon codes for the two-component system that controls the PHO regulon. Null mutations in phoB show a PHO-negative phenotype, while phoR mutations cause the constitutive expression of the PHO regulon (5).
Instead of the −35 sequences, members of the PHO regulon carry in their promoters a sequence known as the PHO box (6). Under conditions of Pi limitation, the histidine kinase PhoR autophosphorylates and transfers the Pi moiety to the regulatory protein PhoB. The phosphorylated PhoB binds to the PHO boxes, where it interacts with Eσ70 (σ70 associated with the core RNA polymerase) (7). Under conditions of Pi excess, PhoR dephosphorylates PhoB, thereby ending the induced expression of the PHO regulon (8). Null mutations, polar and nonpolar alike, in any of the five pst genes result in the constitutive expression of the PHO regulon. The mechanism by which the Pst transport system and PhoU repress PHO is still unclear. It has recently been shown that PhoR interacts with PstB and PhoU, suggesting that the external signal of Pi availability is transduced through direct contact between these proteins (9).
In addition to Pst, E. coli possesses a constitutive, high-velocity, low-affinity Pi transport system, PitA, that does not belong to the PHO regulon. A third cryptic Pi transporter, PitB, is not functional under Pi starvation or in a PHO-constitutive background (6, 10, 11).
Wild-type E. coli is unable to grow on glycerol-2-phosphate (G2P) as the sole carbon source in the presence of excess Pi. Mutants that grow under these conditions are constitutive for the entire PHO regulon, including AP (12, 13). Most of the mutations appear in pstSCAB or in phoU, and some are phoR mutants, whose constitutive level of AP is usually lower (5). The postulated mechanism for the selection of these PHO-constitutive mutants is associated with the constitutive expression of AP (13), whose high periplasmic concentration hydrolyzes G2P into glycerol and Pi. Glycerol enters the cell with the help of the glycerol facilitator GlpF (14), and Pi is taken up either via PitA, in the case of a pst mutation, or via PitA and Pst, if the mutation occurred in phoR. It was later reported that besides transporting glycerol-3-phosphate (G3P), the Ugp transport system is also able to take up G2P (4). This observation raises an alternative possibility, namely, that the uptake of G2P by Ugp (which is also constitutively expressed in a PHO-constitutive mutant) is a contributing factor for the selection of the PHO-constitutive mutants. In the present study, the role of Ugp in this process was examined. In addition, in the course of this study it was noticed that phoR mutants are considerably less frequent than expected. Investigation of this phenomenon revealed that phoR mutants are preferentially isolated in pitA-negative strains.
MATERIALS AND METHODS
Bacteria strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. LB/L agar was the standard rich medium (15). TGP is a minimal medium composed of T salts (12) supplemented with 0.2% glucose and 1 mM KH2PO4. TG2P medium is composed of T salts supplemented with 0.2% glycerol-2-phosphate and 1 mM KH2PO4. TG2P plates were also supplemented with 40 μg/ml of the AP chromogenic substrate 5-bromo-4-chloro-3-indolyl-phosphate p-toluidine (X-P). Medium A is a semirich Pi-limited medium (Pi concentration, ∼0.2 mM) (16). When required, 100 μg/ml ampicillin, 20 μg/ml chloramphenicol, 50 μg/ml kanamycin, or 100 μg/ml spectinomycin was added.
TABLE 1.
Bacterial strains, plasmids, and DNA oligomers used in this study
| Strain, plasmid, or oligomer | Description or sequence | Reference or source |
|---|---|---|
| Strains | ||
| MG1655 strains | ||
| MG1655 | Wild-type E. coli | 39 |
| BS7 | ΔpstSCAB-phoU::Km | 40 |
| BS8 | proC::Tn10 | 41 |
| EG2 | pitA::Km | This study |
| RI05 | ΔphoA::Km | This study |
| RI06 | ΔugpBAEC::Cm | This study |
| RI07 | ΔugpBAEC::Cm ΔpstSCAB-phoU::Km | This study |
| RI08 | ΔphoA::Cm ΔpstSCAB-phoU::Km | This study |
| RI21 | ΔpitA::Km ΔugpBAEC::Cm | This study |
| RI65 | ΔpitA::Km phoR217, high-AP phoR mutant | This study |
| RI81 | ΔpitA::Km pst mutant | This study |
| TP01 | pitA+ phoR129, transduction from strain JV1 to MG1655 proC::Tn10 | This study |
| Other strains | ||
| K10 | Hfr garB10 fhuA22 ompF627 fadL701 relA1 pitA10 spoT1 rrnB2 mcrB1 rob-1 creC510 | CGSC |
| C3 | K10 phoR69 high-AP phoR mutant | 12 |
| JV1 | K10 phoR129 low-AP phoR mutant | This study |
| JV2 | K10 phoR130 low-AP phoR mutant | This study |
| JV6 | K10 pst mutant | This study |
| E2348/69 | Enteropathogenic E. coli (EPEC) Nalr | 42 |
| KM32 | ΔrecBCD::Ptac-gam-bet-exo cat | 18 |
| KM44 | ΔrecBCD::Ptac-gam-bet-exo kan | 18 |
| Plasmids | ||
| pBS6 | pstSCAB-phoU+ cloned in the low-copy-no. plasmid pGB2 | 43 |
| pGEM-T Easy | Cloning vector | Promega |
| pKD3 | Carries the cat gene | 17 |
| pKD4 | Carries the Kmr cassette | 17 |
| pUC4K | Carries the Kmr cassette | 44 |
| Oligomers | ||
| phoA71 | AAGAAGTTATTGAAGCATCCTCGTCAGTAAAAAGTTAATCGTGTAGGCTGGAGCTGCTTC | |
| phoA1701 | TTTCATAGCACCATCCCTCTTCATGTTTTAACCATGAGCGCATATGAATATCCTCCTTAG | |
| phoA-721 | CTTTGGAGATTATCGTCACTG | |
| phoA-2707 | CAGGCAATCACTCATGTAGG | |
| ugp350 | AACGATGAAACCGTTACATTATACAGCTTCAGCACTGGCGGTGTAGGCTGGAGCTGCTTC | |
| phoR_S_FE | TTTAACGCCTTGCTCATCGG | |
| phoR_S_RE | CAGCATCGACTGGCTTATGG | |
| phoR_S_FI | TGAGTTACGTACGCCATTGAC | |
| phoR_S_RI | CGGCTGCTCATTCATCATCTC | |
| ugp4480 | CCAGATGCAGCCACAGCGTGCTGCCTGCCGTCGGGCGCTCCATATGAATATCCTCCTTAG | |
| ugp-858 | ACCGCCTTGTCATCTTTCTG | |
| ugp-5210 | CTCGTTGTCCTGTTTCACC |
Construction of mutants.
The phoA and ugpBAEC deletions (ΔphoA and ΔugpBAEC, respectively) were constructed using the bacteriophage λ-red recombinase system essentially as described previously (17, 18). Primers phoA71/phoA1701 and ugp350/ugp4480 were used to construct the deletion of phoA and ugpBAEC, respectively. PCR was performed using plasmid pKD3 (carrying a chloramphenicol resistance [Cmr] cassette) or pKD4 (carrying a kanamycin resistance [Kmr] cassette) as the template. The Cmr and Kmr cassettes flanked by 40-bp phoA or ugp DNA sequences were transformed into strains KM32 and KM44, respectively, which carry the bacteriophage λ-red genes in the chromosome, and the recombinants were selected on the appropriate antibiotic plate. The mutations were then transferred into strain MG1655 by P1 transduction. The deletions were confirmed by PCR with primers phoA-721/phoA-2707 for ΔphoA and ugp-858/ugp-5210 for ΔugpBAEC. In the case of phoA, the deletion was also confirmed by an AP activity assay, as described previously (16). Double mutants were constructed by transferring antibiotic resistance markers between strains using P1 transduction. For the construction of the pitA::Km mutant, the pitA gene was amplified by PCR with primers pitAF and pitAR and cloned into plasmid pGEM-T Easy. The resulting plasmid was digested with HincII and ligated to a Kmr cassette obtained from plasmid pUC4K digested with HincII. The pitA::Km fragment was amplified by PCR using the same primers, and the PCR product was introduced into strain KM32 by recombineering, as described above. The insertion of Kmr into pitA was confirmed by PCR.
Selection of PHO-constitutive mutants.
PHO-constitutive mutants were selected on TG2P plates essentially as described previously (13). Bacteria were grown overnight in TGP, washed, and suspended in 0.9% NaCl. Approximately 109 cells were plated on TG2P medium supplemented with the AP substrate X-P. In parallel, bacterial dilutions were plated on L agar to record the exact cell concentration. The number of blue colonies on the G2P plates was recorded daily. The mutation frequency was calculated by dividing the number of colonies on the G2P plates by the number of CFU on L agar. For each strain, 8 to 20 independent replicates were performed.
Determination of constitutive genotype.
PHO-constitutive pst mutations were determined by electroporation of the mutant with a plasmid that expresses the entire pst operon (pBS6), followed by plating on L agar containing spectinomycin. The loss of AP constitutivity pointed to the presence of a mutation in one of the pst operon genes. phoR mutations were assessed by transducing the mutant with a P1 lysate of proC::Tn10 cells (strain BS8). Selected tetracycline-resistant colonies on X-P plates that showed approximately similar numbers of blue and white colonies (cotransduction frequency of 60%) indicated a phoR mutant genotype.
AP assays.
Colony plate assays were performed either by including the chromogenic AP substrate X-P in the plates or by flooding the colonies with a mixture of α-naphthyl phosphate and tetrazotized o-dianisidine chloride (Fast Blue) as described previously (19). For quantitative assays, cells were grown overnight in medium A without Pi or medium A supplemented with 1 mM KH2PO4 (medium A with Pi). p-Nitrophenyl-phosphate (pNPP) was used as a substrate. The reaction was stopped by the addition of 0.25 M Na2HPO4, and the AP specific activity was calculated according to the equation A410 × time (min)−1 × cell density (optical density at 600 nm)−1.
P1 transduction.
Transductions of chromosomal markers were performed as described previously (15).
DNA sequencing.
The phoR open reading frame was amplified by PCR using primers phoR_S_FE and phoR_S_RE. PCR products were purified from agarose gels with a Wizard DNA purification system (Promega), and sequencing reactions were performed using the BigDye Terminator (v.3.1; Applied Biosystems) kit, according to the manufacturer's instructions. For the sequencing reactions, oligonucleotides phoR_S_FE, phoR_S_RE, phoR_S_FI, and phoR_S_RI were used. The sequencing products were resolved and analyzed in an automatic sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems/Hitachi, Warrington, United Kingdom).
RESULTS
To test the roles of phoA and of the ugpBAEC operon (henceforth referred to as ugp) in the selection of PHO-constitutive mutants, we constructed deletion mutants of phoA and ugp each associated with an antibiotic resistance gene, namely, ΔphoA::Cm and Δugp::Cm (see Materials and Methods). Each construct was introduced into the wild-type strain MG1655. A test of the growth of these mutants along with that of the proper controls was performed by streaking them on TG2P (in which G2P is the sole carbon source and excess Pi is supplemented with the AP chromogenic substrate X-P), which is the same medium used for the selection of PHO-constitutive mutants (13). Figure 1 shows that, as expected, the wild-type strain MG1655 (Fig. 1A and E) and its phoA and ugp single mutant derivatives (Fig. 1B and F, respectively) failed to grow, while the PHO-constitutive Δpst mutant derivative grew well on the plate (Fig. 1C and G). The central role of phoA in the selection of PHO-constitutive strains became evident from the inability of a Δpst ΔphoA double mutant to grow on TG2P (Fig. 1D). On the other hand, the normal growth of the Δpst Δugp double mutant (Fig. 1H) hinted that the ugp operon is apparently not required for the growth of PHO-constitutive mutants on TG2P.
FIG 1.
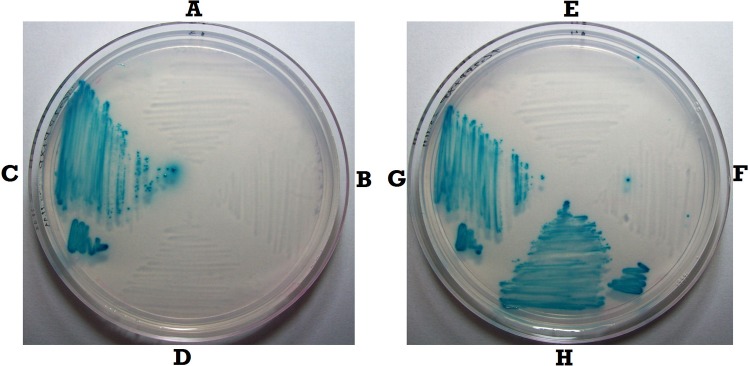
Growth of ΔphoA and Δugp mutants on TG2P plates. Bacteria were streaked on TG2P (minimal medium with G2P as the sole carbon source supplemented with the AP substrate X-P) and grown for 48 h at 37°C. (A) MG1655; (B) ΔphoA::Cm; (C) Δpst::Km; (D) ΔphoA::Cm Δpst::Km; (E) MG1655; (F) Δugp::Cm; (G) Δpst::Km; (H) Δugp::Cm Δpst::Km.
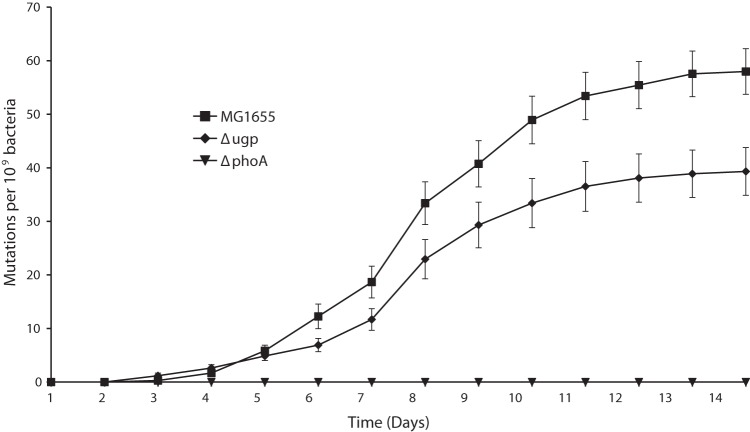
However, these observations do not rule out a function for Ugp during the selection of PHO-constitutive mutations. Since the Ugp system can take up G2P (4), the constitutive expression of Ugp could benefit the bacteria by providing extra glycerol or Pi for the nascent mutant. To test whether ugp plays a role in the mechanism of selection of PHO-constitutive mutants, 20 separate cultures each containing 109 cells of wild-type strain MG1655, ΔphoA mutant RI05, and Δugp mutant RI06 were plated on TG2P. The number of emerging blue colonies was recorded for the following 14 days (Fig. 2). Both the wild-type strain and the Δugp mutant showed a similar pattern of accumulation of constitutive mutants on TG2P, but with one important difference; as of day 5, the wild-type strain started to accumulate more PHO-constitutive colonies than did the Δugp mutant. At the end of 2 weeks, there were, on average, 25% fewer PHO-constitutive colonies derived from the Δugp mutant than from its wild-type parental strain. No colonies were observed on the plates seeded with ΔphoA cells, confirming that the emergence of PHO-constitutive mutants on TG2P is dependent on phoA. This agrees with the suggested AP-dependent mechanism for the selection of PHO-constitutive mutants (13). However, the authors of the previous study monitored the appearance of AP-constitutive colonies for only 2 to 3 days, while the present results suggest that the bulk of the PHO-constitutive mutants, which emerged later, are partially dependent on Ugp. It seems that the PHO-constitutive mutants that appeared at later times did not derive from preexistent mutations but emerged in response to the prolonged incubation in the presence of G2P. Their accumulation pattern resembles an inflection curve common in other cases of stress-induced (adaptive) mutations (see Discussion).
FIG 2.
Selection of PHO-constitutive mutants on TG2P. Bacteria were seeded on TG2P plates and incubated for 14 days at 37°C. The appearance of colonies was monitored daily. The data points were calculated by dividing the number of colonies observed on each day by the effective number of CFU seeded on each plate (about 109). The data represent the mean ± SEM from 20 independent experiments.
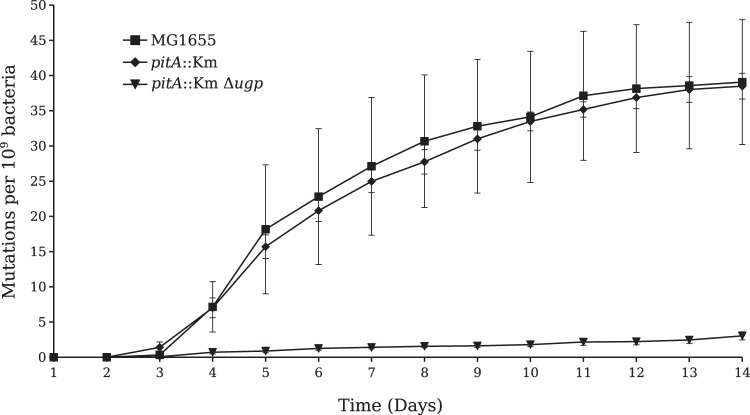
When pst mutants are selected on TG2P, the supply of Pi is expected to depend only on the low-affinity PitA transporter. However, in the absence of PitA, Ugp, as a G2P transporter, might confer on the emerging PHO-constitutive mutants an advantage by providing an alternative source of Pi. To test this hypothesis, a pitA::Km mutation was introduced into the wild-type strain and into the Δugp mutant. Each organism, along with the wild-type control, was plated on TG2P, and the formation of AP-constitutive colonies was scored as described above. Figure 3 shows that, compared to the findings for the wild type, inactivation of pitA alone only slightly reduced the frequency of PHO-constitutive mutants, whereas the mutation frequency of the pitA::Km Δugp double mutant was drastically reduced. Since the majority of the mutants derived from the pitA::Km strain had mutations in the pst operon (see below), these results strongly suggest that in the absence of a functional Pi transport system, the constitutively expressed Ugp system provides most of the Pi required for growth. In fact, a pitA::Km pst double mutant was unable to grow on TGP (minimal medium containing 0.2% glucose and 1 mM Pi) (see Fig. S2 in the supplemental material), as both Pi transport systems were eliminated, but it could grow on TG2P (minimal medium containing 0.2% G2P and 1 mM Pi) because G2P serves as a carbon as well as a Pi source (see Fig. S1 in the supplemental material). If this is the case, a ΔpitA Δpst Δugp triple mutant would be unable to grow on G2P. To test this possibility, we pursued the construction of the triple mutant. However, despite many attempts using different combinations of P1 transduction, a triple mutant could not be obtained. The inability to get a ΔpitA Δpst Δugp triple mutant was also reported elsewhere (20).
FIG 3.
Selection of PHO mutants in a pitA background. Bacteria were seeded on TG2P plates as described in the legend to Fig. 2. The data represent the mean ± SEM from 8 independent experiments.
PHO-constitutive mutations may occur in the pst operon or in phoR (5). Given the length of the pst operon (5 kb) and phoR gene (1.3 kb), 25% of the PHO-constitutive mutations isolated on G2P would be expected to be in phoR. However, the vast majority of mutations isolated in this study were in the pst operon. A total of 148 spontaneous PHO-constitutive mutants (95 from strain MG1655, 36 from the enteropathogenic E. coli strain E2348/69, and 17 from strain K10) were selected and mapped. All MG1655 and E2348/69 derivatives carried mutations in pst, and only 2 out of the 17 K10 mutants proved to bear mutations in phoR. Sequencing of these two mutants (JV1 and JV2) revealed in one of them a G → T transversion at position 703 and in the other a T → A transversion at position 546, resulting in both cases in premature stop codons and an AP-constitutive genotype.
Strain K10 carries a pitA nonsense mutation (11). To test whether the lack of PitA is associated with the emergence of spontaneous phoR mutants, 14 PHO-constitutive colonies isolated from MG1655 pitA::Km (Fig. 3) were mapped. Nine isolates had mutations in the pst operon, while the other five were phoR mutants. All five phoR mutants carried the phoR217 allele, a C → A transversion at position 650 resulting in a T217K substitution in the PhoR protein. Even though they carried the same mutation, these five mutants arose independently of each other; i.e., they were not siblings of a bacterial population with a preexisting mutation plated on TG2P. This conclusion derives from the fact that they all appeared at different times on the selective plate, three on day 6, one on day 7, and another one on day 8. The fact that not a single phoR mutation was found in almost 100 PHO-constitutive isolates from wild-type strain MG1655 but phoR mutants were readily selected from this strain when pitA was inactive suggests that the presence of PitA suppresses the occurrence of spontaneous phoR mutations (see Discussion).
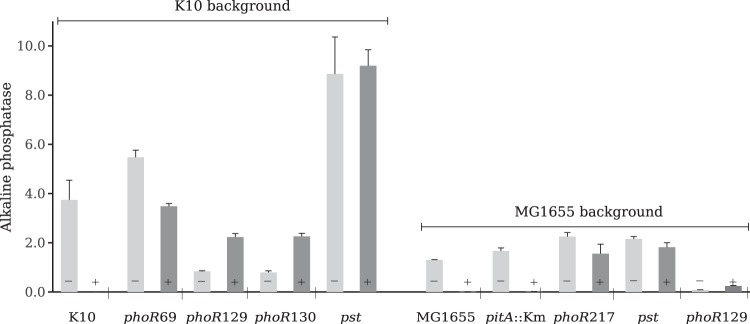
Another reason for the inability to detect phoR mutations in strain MG1655 is that, compared to the results for strain K10, the level of AP as a result of Pi starvation or in PHO-constitutive strains was much lower than that in K10 (Fig. 4). This difference became particularly striking when the phoR129 mutation was transferred to strain MG1655. The AP activity of K10 phoR129 (strain JV1) was 11 times higher than that of MG1655 carrying the phoR129 allele. Apparently, when exposed to the G2P selective medium, the low level of AP and Ugp produced by the constitutive phoR nonsense mutants in the MG1655 background was probably insufficient to provide all necessary glycerol and Pi required for their growth.
FIG 4.
AP activity of the various PHO-constitutive mutants. Bacteria grown overnight in medium A without Pi (−) and medium A with Pi (+) were assayed for AP activity. K10, wild-type K10; phoR69 (strain C3), K10 high-AP phoR69 spontaneous mutant; phoR129 (strain JV1) and phoR130 (strain JV2), K10 low-AP phoR nonsense mutants; pst (strain JV6), K10 pst spontaneous mutant; MG1655, wild-type MG1655; pitA::Km, MG1655 pitA::Km (strain EG2); phoR217 (strain RI65), MG1655 high-AP phoR spontaneous mutant; pst (strain RI81), MG1655 pitA::Km pst spontaneous mutant; phoR129 (strain TP01), MG1655 into which the phoR129 mutation was transduced. Each bar represents the mean ± SEM from 3 independent experiments.
Hence, phoR nonsense mutants derived from MG1655 are not detectable on TG2P. To test this assumption, the MG1655 phoR129 transductant strain, along with other strains, was streaked on a TG2P plate and in liquid TG2P medium (see Fig. S1 in the supplemental material). As expected, MG1655 and the K10 pst mutants as well as K10 phoR129 grew well, while MG1655 phoR129 grew very poorly in both liquid and solid media. This clearly shows that phoR nonsense mutations, such as phoR129, do not cause the production of sufficient AP to allow the growth of strain MG1655 on G2P, but the same mutation promotes the growth of K10 because this strain expresses an intrinsically higher level of AP (Fig. 4). It is also interesting to note that the pitA::Km mutant expressed 30% more AP than the wild-type strain, which may contribute to the growth of the nascent phoR mutants.
Finally, there still exists the possibility that the pst pitA::Km double mutants were able to grow on G2P due to the activation of the cryptic PitB transporter (11). To test this possibility, the 14 PHO-constitutive pitA::Km mutants described above (9 pst and 5 phoR mutants) were grown in liquid TGP (minimal medium containing 0.2% glucose and 1 mM Pi) for 24 h along with some controls. As expected, the parent (MG1655 pitA::Km) and its phoR derivatives grew well, while the nine isolates that carried mutations in the pst operon did not grow in this medium (Fig. S2 in the supplemental material shows one representative result for each isolate), demonstrating that these strains are unable to use Pi as a P source and are thus unlikely to have activated pitB. When streaked on a TGP plate, the pitA::Km pst double mutant grew very poorly, while two different types of pitA::Km phoR mutants as well as the pitA::Km parent grew perfectly well on the plate (see Fig. S2 in the supplemental material). The same growth pattern was observed for all nine pitA::Km pst mutants (not shown). Hence, none of the PHO-constitutive pitA::Km mutants activated pitB.
DISCUSSION
In this study, the rationale behind the selection of PHO-constitutive mutants on G2P was revisited. As expected, the fundamental role of AP as the major contributor in the selection of the constitutive mutants was confirmed (13). However, we showed that the Ugp system also contributes to the emergence of these mutants, apparently by providing the additional phosphorus required for bacterial growth. This conclusion is supported by two findings: the drastic drop in the frequency of PHO-constitutive mutations in a ugp pitA background (Fig. 3) and by our and others' inability to construct a Δpst ΔpitA Δugp triple mutant (20). Ugp could enhance the appearance of PHO-constitutive mutants either by taking up G2P in its intact form or by taking up Pi from the medium. It has been shown that two other organophosphate transport systems, GlpT and UhpT, are able to take up Pi as a secondary substrate (5, 21). Though Pi is not a known Ugp substrate (22), the fact that we and others could not obtain a Δpst ΔpitA Δugp triple mutant suggests that the Ugp system may be involved in the transport of Pi. Moreover, the observation that the pitA::Km Δugp double mutant gives rise to very few PHO-constitutive mutants points to a function of Ugp as a Pi transporter. Another possibility would be that G2P is converted into G3P in the periplasm and only then is transported by Ugp. This may be in agreement with a recent report that challenged the notion that Ugp is able to take up G2P in vitro (23). However, a periplasmic enzyme that catalyzes the conversion of G2P to G3P is still unknown.
PitA inhibits the formation of phoR mutants through a mechanism that is not entirely clear. A pitA pst double mutant would leave the bacterium without a proper Pi transport system. Hence, under the conditions used for the selection of PHO-constitutive mutants (G2P as the only carbon source and excess Pi), the selective pressure is tilted toward phoR, whose mutation/inactivation would have a lower cost than the inactivation of pst.
To the best of our knowledge, phoR mutants have thus far been spontaneously isolated only in strain K10 (12, 24). Apparently, K10 possesses two critical characteristics that enable the selection of phoR mutants, namely, a pitA mutation (11) as well as an intrinsic capacity to synthesize a higher level of AP than most K-12 strains tested in our laboratory (Fig. 4 and data not shown). The reason why K10 synthesizes high levels of AP may be related to the presence of the creC510 allele, which causes the constitutive expression of CreC (25). CreC is a histidine kinase that can phosphorylate PhoB (via cross talk) when phoR is absent (26, 27). Thus, the constitutive expression of CreC might boost phoA transcription in nonsense phoR mutants, enabling their growth on G2P. Another K10 characteristic is the presence of an rpoS(Am) mutation, which produces a less efficient truncated form of RpoS. Bacterial strains that carry this rpoS allele express higher levels of σ70-dependent genes, such as phoA (28, 29). MG1655 pitA::Km displayed a small but statistically significant increase in AP activity (P = 0.03, Student's t test), possibly because in the absence of PitA there is a less of an influx of Pi that would slightly alleviate the repression of the PHO regulon genes (30). An intrinsic higher AP activity in a strain lacking pitA may also contribute to the growth of the nascent phoR mutant.
There are two different types of phoR mutations: those that confer a high level of AP activity (high-AP mutations) and those that confer a low level of AP activity (low-AP mutations). Strains with nonsense phoR mutations exhibit a low level of noninducible AP-constitutive activity, while strains with the other type of phoR mutations present a high level of AP activity under excess Pi, which is further induced by Pi starvation (5, 12, 31). The T217K mutant displayed the high-AP phenotype (Fig. 4), which enabled growth on the selective plate by providing high levels of AP and Ugp. Incidentally, the well-characterized phoR69 mutation is a T220N substitution (31), which is located in the same region in the HisKA domain of PhoR. The phoR69 mutation confers on K10 high AP activity compared to the level of activity conferred by the phoR nonsense mutation in the same strain (Fig. 4). Furthermore, the Thr217 residue is highly conserved among sensor histidine kinases similar to EnvZ, such as PhoR (see Fig. S3 in the supplemental material), and it was shown to be critical for the phosphatase activity of EnvZ (32). Since the only high-AP phoR mutants hitherto isolated carry mutations in each of two conserved threonine residues in the HisKA domain (the five phoR mutants isolated in the present study and the phoR69 mutant), there are apparently few targets in PhoR that result in high-AP constitutivity. This narrows considerably the spectrum of possible phoR mutations in MG1655, once phoR nonsense mutations in this strain result in low levels of AP, and explains why only two high-AP phoR mutants were isolated until now.
The bulk of PHO-constitutive mutants appeared on the selective plate between days 5 and 11. Upon restreaking on TG2P, they formed colonies within 48 h, indicating that they are not slow growers and, hence, do not contain preexistent mutations. Furthermore, the pattern of accumulation of PHO mutants in the wild-type, Δugp, and pitA::Km strains resembles the inflection curve common in other models of stress-induced (adaptive) mutations (33–36). Adaptive mutations are not formed under nonselective growth conditions (preexistent mutations), but they appear later, when the bacteria are exposed to the selective conditions. The mechanism through which these mutants arise is still under intense dispute (35, 37, 38).
Supplementary Material
ACKNOWLEDGMENTS
We thank FAPESP (grant no. 2013/19307-9) for funding this research. H.I.N. and T.F.P. were supported, respectively, by FAPESP and CAPES scholarships. E.Y. was supported by the Israel Science Foundation (grant no. 702/11) and by GIF, the German-Israeli Foundation for Scientific Research and Development (grant 1062/2008).
This article is dedicated to the memory of the late Annamaria Torriani-Gorini, who passed away in May 2013 and whose pioneer work on the regulation of phosphate metabolism inspired many scientists and served as the grounds for the present work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02566-14.
REFERENCES
- 1.Baek JH, Lee SY. 2006. Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiol Lett 264:104–109. doi: 10.1111/j.1574-6968.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida Y, Sugiyama S, Oyamada T, Yokoyama K, Makino K. 2012. Novel members of the phosphate regulon in Escherichia coli O157:H7 identified using a whole-genome shotgun approach. Gene 502:27–35. doi: 10.1016/j.gene.2012.03.064. [DOI] [PubMed] [Google Scholar]
- 3.Brzoska P, Boos W. 1988. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the Ugp transport system of Escherichia coli. J Bacteriol 170:4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K, Wang M, Metcalf WW. 2009. Uptake of glycerol-2-phosphate via the ugp-encoded transporter in Escherichia coli K-12. J Bacteriol 191:4667–4670. doi: 10.1128/JB.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 6.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component P(i) signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol 259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 8.Carmany DO, Hollingsworth K, McCleary WR. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J Bacteriol 185:1112–1115. doi: 10.1128/JB.185.3.1112-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner SG, Johns KD, Tanner R, McCleary WR. 2014. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol 196:1741–1752. doi: 10.1128/JB.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffer SM, Schoondermark P, van Veen HW, Tommassen J. 2001. Activation by gene amplification of pitB, encoding a third phosphate transporter of Escherichia coli K-12. J Bacteriol 183:4659–4663. doi: 10.1128/JB.183.15.4659-4663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris RM, Webb DC, Howitt SM, Cox GB. 2001. Characterization of PitA and PitB from Escherichia coli. J Bacteriol 183:5008–5014. doi: 10.1128/JB.183.17.5008-5014.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echols H, Garen A, Garen S, Torriani A. 1961. Genetic control of repression of alkaline phosphatase in E. coli. J Mol Biol 3:425–438. doi: 10.1016/S0022-2836(61)80055-7. [DOI] [PubMed] [Google Scholar]
- 13.Torriani A, Rothman F. 1961. Mutants of Escherichia coli constitutive for alkaline phosphatase. J Bacteriol 81:835–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller KB, Lin EC, Wilson TH. 1980. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J Bacteriol 144:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p 876 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 16.Spira B, Aguena M, de Castro Oliveira JV, Yagil E. 2010. Alternative promoters in the pst operon of Escherichia coli. Mol Genet Genomics 284:489–498. doi: 10.1007/s00438-010-0584-x. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy KC, Campellone KG, Poteete AR. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321–330. doi: 10.1016/S0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 19.Bracha M, Yagil E. 1969. Genetic mapping of the phoR regulator gene of alkaline phosphatase in Escherichia coli. J Gen Microbiol 59:77–81. doi: 10.1099/00221287-59-1-77. [DOI] [PubMed] [Google Scholar]
- 20.Shao J. 2007. Finding new functions for the Ugp and PitA transport systems of Escherichia coli K-12. Ph.D. thesis Purdue University, West Lafayette, IN. [Google Scholar]
- 21.Maloney PC, Ambudkar SV, Anatharam V, Sonna LA, Varadhachary A. 1990. Anion-exchange mechanisms in bacteria. Microbiol Rev 54:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer H, Argast M, Boos W. 1982. Characteristics of a binding protein-dependent transport system for sn-glycerol-3-phosphate in Escherichia coli that is part of the PHO regulon. J Bacteriol 150:1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuttge S, Bommer M, Jäger F, Martins BM, Jacob S, Licht A, Scheffel F, Dobbek H, Schneider E. 2012. Determinants of substrate specificity and biochemical properties of the sn-glycerol-3-phosphate ATP binding cassette transporter (UgpBAEC(2)) of Escherichia coli. Mol Microbiol 86:908–920. doi: 10.1111/mmi.12025. [DOI] [PubMed] [Google Scholar]
- 24.Garen A, Garen S. 1963. Genetic evidence on the nature of the repressor for alkaline phosphatase in E. coli. J Mol Biol 6:433–438. doi: 10.1016/S0022-2836(63)80054-6. [DOI] [PubMed] [Google Scholar]
- 25.Nikel PI, de Almeida A, Pettinari MJ, Méndez BS. 2008. The legacy of HfrH: mutations in the two-component system CreBC are responsible for the unusual phenotype of an Escherichia coli arcA mutant. J Bacteriol 190:3404–3407. doi: 10.1128/JB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanner BL, Latterell P. 1980. Mutants affected in alkaline phosphatase, expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics 96:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanner BL. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol 174:2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galbiati HF, Taschner NP, Spira B. 2014. The effect of the rpoSam allele on gene expression and stress resistance in Escherichia coli. Arch Microbiol 196:589–600. doi: 10.1007/s00203-014-0994-y. [DOI] [PubMed] [Google Scholar]
- 29.Taschner NP, Yagil E, Spira B. 2004. A differential effect of sigmaS on the expression of the PHO regulon genes of Escherichia coli. Microbiology 150:2985–2992. doi: 10.1099/mic.0.27124-0. [DOI] [PubMed] [Google Scholar]
- 30.Hoffer SM, Tommassen J. 2001. The phosphate-binding protein of Escherichia coli is not essential for P(i)-regulated expression of the pho regulon. J Bacteriol 183:5768–5771. doi: 10.1128/JB.183.19.5768-5771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada M, Makino K, Amemura M, Shinagawa H, Nakata A. 1989. Regulation of the phosphate regulon of Escherichia coli: analysis of mutant phoB and phoR genes causing different phenotypes. J Bacteriol 171:5601–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta R, Yoshida T, Inouye M. 2000. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine kinase/phosphatase, in Escherichia coli. J Biol Chem 275:38645–38653. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- 33.Bridges BA. 1994. Starvation-associated mutation in Escherichia coli: a spontaneous lesion hypothesis for “directed” mutation. Mutat Res 307:149–156. doi: 10.1016/0027-5107(94)90287-9. [DOI] [PubMed] [Google Scholar]
- 34.Cohen SE, Walker GC. 2010. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr Biol 20:80–85. doi: 10.1016/j.cub.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol 60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 36.Timms AR, Bridges BA. 1998. Reversion of the tyrosine ochre strain Escherichia coli WU3610 under starvation conditions depends on a new gene tas. Genetics 148:1627–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster PL. 2007. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol 42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galhardo RS, Hastings PJ, Rosenberg SM. 2007. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol 42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 40.Spira B, Silberstein N, Yagil E. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol 177:4053–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira GM, Spira B. 2008. The pst operon of enteropathogenic Escherichia coli enhances bacterial adherence to epithelial cells. Microbiology 154:2025–2036. doi: 10.1099/mic.0.2008/016634-0. [DOI] [PubMed] [Google Scholar]
- 42.Nisa S, Hazen TH, Assatourian L, Nougayrède JP, Rasko DA, Donnenberg MS. 2013. In vitro evolution of an archetypal enteropathogenic Escherichia coli strain. J Bacteriol 195:4476–4483. doi: 10.1128/JB.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spira B, Yagil E. 1999. The integration host factor (IHF) affects the expression of the phosphate-binding protein and of alkaline phosphatase in Escherichia coli. Curr Microbiol 38:80–85. doi: 10.1007/s002849900407. [DOI] [PubMed] [Google Scholar]
- 44.Vieira J, Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.