Abstract
Purpose
The ability of bacteria in the intestinal microbiome to convert naturally occurring primary ginsenosides in red ginseng extract or RGE to active secondary ginsenosides was investigated.
Methods
The anti-proliferative activity of ginsenosides was tested using the mouse lung cancer LM1 cells. Their permeabilities were evaluated in Caco-2 cell monolayers. Systemic exposure of secondary ginsenosides was determined in A/J mice. 16S rRNA gene pyrosequencing was used to determine membership and abundance of bacteria in the intestinal microbiome.
Results
Secondary ginsenoside C-K exhibited higher anti-proliferative activity and permeability than primary ginsenosides, and significant amounts of secondary ginsenosides (F2 and C-K) were found in the blood of A/J mice following oral administration of the primary ginsenoside Rb1. Because mammalian cells did not hydrolyze ginsenoside, we determined the ability of bacteria to hydrolyze ginsenosides and found that the primary ginsenoside Rb1 underwent stepwise hydrolysis to Rd, F2, and then C-K. Formation of F2 from Rd was the rate-limiting step in the biotransformation of Rb1 to C-K.
Conclusion
This is the first study to characterize the A/J mouse intestinal microbiome and reveal the presence of certain bacterial families capable of efficiently converting inactive primary ginsenosides to active secondary ginsenosides in vivo.
Keywords: ginsenosides Rb1, Rd, F2 and C-K; ginseng; permeability; pharmacokinetic profile; stepwise metabolism; rate-limiting step; microbiome; 16S rRNA gene sequencing
1. Introduction
Lung cancer remains the leading cause of cancer deaths for both men and women in the United States (1). The administration of medication or alteration of diet to prevent or delay the development of cancer is coined “chemoprevention” and has recently drawn public attention due to the low cure rates of advanced lung cancer using traditional approaches, such as chemotherapy and surgery (2–13).
The efficacy of red ginseng extract (RGE) as a chemopreventive agent has been examined over the past 30 years (6, 14–18), and accumulated evidence supports the notion that RGE is a potent agent for the prevention of lung cancer. A recent study by You, et al. indicated that RGE (10 mg/ml in drinking water) prevented benzopyrene induced lung carcinogenesis with a significant reduction (70%) of tumor load (6).
The primary, or most abundant, naturally occurring ginsenosides present in RGE include Rb1 and Rd (19, 20). However, secondary, less abundant, ginsenosides such as Rh2 demonstrate excellent inhibitory activities in both lung adenocarcinoma cells (21) and in A/J mice (22). Additionally, secondary ginsenosides appear to have better permeability in Caco-2 cells and hence better bioavailability (see later). Mammalian cells do not express enzymes that hydrolyze ginsenosides (23–26) suggesting that the in vivo transformation of secondary ginsenosides occurs via the action of the intestinal microbiota.
The intestinal microbiome produces different types of glycosidases, including β-glucosidases, the predominant enzymes responsible for the hydrolysis of ginsenosides. Bacterial β-glucosidases are hydrolytic enzymes that release terminal glucose residues successively (27). Primary ginsenosides are hydrolyzed stepwise to produce secondary ginsenosides and finally the aglycone. Secondary ginsenosides exhibit the much higher anti-proliferative activity against lung cancer cells and possess higher permeability across membranes of mammalian cells. Therefore, enhancing the production of secondary ginsenosides will likely benefit the lung cancer chemoprevention efficacy of RGE.
Recent investigations showed that the primary ginsenoside Rb1 is hydrolyzed stepwise to different secondary ginsenosides, such as Rg3, Rh2, F2, and C-K, by fecal extracts and specific microorganisms (28–36). However, these studies failed to investigate the kinetics of RGE stepwise hydrolysis necessary to elucidate the rate-limiting step in the production of active ginsenosides. Identification of the rate-limiting step and the microorganism that catalyzes it could allow us to manipulate the rate and production of desired ginsenosides with anti-cancer activity. Furthermore, the microorganisms studied do not represent the whole spectrum of the intestinal microbiome. Studies using fecal specimens failed to define the gut microbiota capable of ginsenoside hydrolysis (37–39). Therefore, these studies leave open the potential to discover probiotic candidates that could be administered concurrently with RGE to improve its efficacy.
In the present study we (1) determine the rate-limiting step in the kinetics of stepwise metabolism from ginsenoside Rb1 to C-K in A/J mouse fecal matrix; and (2) use 16s rRNA gene sequencing to define the A/J mouse intestinal microbiome to aid future studies investigating bacterial metabolism of ginsenosides from this and other laboratories.
2. Materials and Methods
Chemicals and reagents
Ginsenosides Rb1, Rd and F2 (>99% pure) were purchased from LKT Laboratories (St. Paul, MN). Ginsenoside C-K (>95% pure) was kindly provided by Dr. Zhi-Hong Jiang from Hong Kong Baptist University. Red Ginseng powder and LM1 cells were provided by Dr. Ming You from Medical College of Wisconsin. Testosterone was purchased from Sigma-Aldrich (St. Louis, MO). Simulated intestinal fluid was purchased from VWR (Houston, TX). BCA protein assay kit was purchased from Thermo Scientific (Rockford, IL). Other chemicals (analytical grade or better) were used as received.
LM1 cell culture model and anti-proliferative assays
LM1 cells were grown in plastic tissue culture flasks in minimal essential medium (MEM) mixed with 2 mM glutamine, 50 units/ml penicillin, 50 pg/ml streptomycin, 1% nonessential amino acids from Cellgro (Catalog number: 10-010-CV), and supplemented with 10% fetal bovine serum from HyClone (Catalog number: SH30088-03). Cells were incubated at 37°C and 5% CO2, with media changes every two days. Cells were passaged 10 times, and one split (1:20) was used each week.
LM1 cells were seeded onto 96-well plates at a density of 5 × 103 per well and incubated in MEM without red phenol from Cellgro (Catalog number: 17-305-CV). After 24 hr, ginsenosides, RGE, 5-flurouracil (positive control) or 1% DMSO (negative control) was added to the medium for 48 hr. Cells were then incubated with MTT (0.5 mg/ml) for 4 hr. The formazan precipitate was dissolved in 100 μl DMSO, and the absorbance at 570 nm was detected with a Benchmark Microplate Reader (Bio-Rad, California). Cell survival was calculated by the following formula: % cell survival = (mean absorbency in test wells)/(mean absorbency in control wells) × 100. The effective dose to inhibit 50% growth (ED50) was calculated for the ginsenosides and the positive control, using non-linear regression analysis. Each test was performed in triplicate.
Caco-2 cell culture and transcellular transport experiments
Caco-2 cells were cultivated as described previously (40). Porous polycarbonate cell culture inserts (3 μm pore size) from Corning (Catalog No: 3414) were used to seed cells at a density of 100,000 cells/cm2. After the cell monolayers reach maturity and become ready to use in 19–21 days, they were tested for integrity (minimal transepithelial electrical resistance value of 465 Ω/cm2), and then used for the transcellular transport as described previously (40). Briefly, 2.5 ml of ginsenoside solution (2 or 10 μM) in Hanks’s balanced salt solution or HBSS was loaded on apical side of the cell monolayer and 2.5 ml of blank HBSS onto the basolateral side. Five sequential samples (0.5 ml) were taken at time points 0, 1, 2, 3 and 4 hr from both sides of the cell monolayer. Ginsenoside solution and HBSS media was added to donor or receiver side immediately to compensate for sampling volume lost. The pH of HBSS in both the apical and basolateral side was 7.4. A volume of 125 μl of an internal standard (1 μM formononetin in 100% acetonitrile) was added to the samples right after sampling. Caco-2 samples were blown dried by purified air and reconstituted with 200 μl of 100% methanol for UPLC-MS/MS analysis.
The apparent unidirectional permeability, from apical to basolateral side (Pa–b), was obtained according to the following equation (Eq.1):
| (1) |
where is the rate of concentration change in the receiver chamber (equals to the slope of the regression line derived for the amount transported vs. time profile), V is the chamber volume (2.5 ml), S is the surface area of the monolayer (4.65 cm2), and C0 is the initial concentration in the donor side. Permeability from apical to basolateral side (Pa–b) was calculated according to the above equation.
Oral pharmacokinetic dosing studies using Rb1 in A/J mice
The study was approved by the Institutional Animal Care and Use Committee at the University of Houston. Male A/J mice (20–25 g) were purchased from Harlan Laboratory (Indianapolis, IN) at 6–8 weeks of age. They were housed individually in an environmentally controlled room (temperature: 25 ± 2°C, humidity: 50 ± 5%, 12-hr light-dark cycle) for one week before the experiments. Drinking water and diet were supplied ad libitum. The body weight of mice was measured every other day for the duration of the study.
Ginsenoside Rb1 dispersed in oral suspending vehicle was given by oral gavage to 4 A/J mice at a dose of 20 mg/kg. Blood samples (20–25 μl per sample) for each mouse were collected in heparinized tubes at 0.25, 0.5, 1, 2, 3, 4, 6, 8, 12 and 24 hr by snipping the tail and stored at −20°C until analysis. The internal standard solution (200 μl of 1 μM formononetin in methanol) was added to each 20 μl aliquot of blood. The samples were vortexed for 30 sec and centrifuged at 15, 000 rpm for 15 min. The supernatant was blown dried by purified air and reconstituted in 100 μl of 100% methanol, and 10 μl of the sample was injected into the UPLC-MS/MS for analysis.
Hydrolysis of ginsenosides in fecal lysate
Fecal lysate preparation
Feces of nine A/J mice were collected and stored at −80°C until use. Feces (500 mg) were mixed with 5 ml ice-cold 0.1 mM phosphate buffer solution or PBS (pH 7.4) and vortexed for 5 sec followed by centrifugation at 1,000 rpm and 4°C for 15 min. The pellet was further washed twice using 0.1 mM PBS, followed by resuspension in 10 ml ice-cold PBS, sonicated in ice water bath for 45 min and centrifuged at 15,000 rpm and 4°C for 30 min. The supernatant was aliqoted and stored at −80°C. Protein concentrations were determined by the BCA protein assay kit using bovine serum albumin as the standard.
Stepwise metabolism of ginsenoside Rb1
Thawed fecal lysate (200 μl) was transferred to disposable glass vials from VWR (Houston, TX) and diluted with ice-cold 0.1 mM PBS to 2 ml. Ginsenoside Rb1 was added to the samples to a final concentration of 45 μM. The mixture was incubated at 37°C and 120 rpm in an orbital shaker from Thermo Scientific (Asheville, NC) for 24 hr. Samples (100 μl) were collected with low adhesion surface tips from VWR (Houston, TX) to minimize binding to the micro-centrifuge tubes from Corning (Pittston, PA) at 0, 0.5, 1, 2, 4, 8, 12 and 24 hr. The reaction was stopped with the addition of 500 μl of 2.5 μM testosterone (internal standard) in 100% acetonitrile. The samples were vortexed for 15 sec and centrifuged at 15,000 rpm for 15 min. A 540 μl portion of supernatant was air dried and reconstituted with 100 μl of 30% acetonitrile, and 10 μl of the sample was injected into the UPLC for analysis. The experiments were performed in triplicate.
Determination of the rate-limiting step in stepwise metabolism of ginsenosides
Fecal lysate protein concentration, incubation conditions, and sampling time were optimized to ensure less than 30% of the metabolite would appear such that the calculated metabolic rate closely approximates the initial rate. Ginsenosides Rb1, Rd and F2 and C-K were added to the diluted fecal lysate described in previous section, making the final substrate concentration of 20 or 5 μM. The sample processing procedures were performed as described above, in triplicates. Amounts of metabolite formed were determined by UPLC and protein concentrations were determined using BCA protein assay. The appearance rates of Rb1, Rd, F2, and C-K were normalized by protein concentration and reaction time. Recovery of total ginsenosides was determined to ascertain that the mass balance fell in the range of 80% to 120% (i.e., no other metabolites were present in significant quantities)
Quantitation of ginsenosides in biological matrices
For Caco-2 and mouse blood samples, a UPLC/MS-MS method was used to quantify ginsenosides. The LC conditions for the analysis were the same as those for the quantitation of ginsenoside Rh2 (41). A triple quadruple mass spectrometer (API 3200 Qtrap, Applied Biosystems, Foster City, CA) was used to perform the analysis of the eluent from the UPLC. The ion spray voltage and ion source temperature were set to 5500 kV and 600°C, respectively. Nebulizer gas, turbo gas, and curtain gas were optimized to 40, 40, and 20 psi, respectively. Multiple reactions monitoring (MRM) mode was used to monitor ginsenoside Rb1, Rd, F2, C-K and formononetin (internal standard). The compound dependent parameters were listed in Table 1.
Table 1.
Compound dependent parameters of ginsenoside Rb1, Rd, F2, C-K and internal standard testosterone in MRM mode LC/MS analysis
| Compound | Q1 | Q3 | Time(msec) | DP | CEP | CE | CXP |
|---|---|---|---|---|---|---|---|
| Rb1 | 1110.0 | 325.3 | 100 | 36 | 62 | 38 | 5 |
| Rd | 946.7 | 161.0 | 100 | −54 | −42 | −64 | −1 |
| F2 | 807.6 | 627.5 | 100 | 125 | 50 | 53 | 5 |
| C-K | 645.4 | 202.9 | 100 | 96 | 34 | 45 | 3 |
| IS | 269.2 | 197.1 | 100 | 10 | 14 | 49 | 3 |
Processed samples were injected into an Acquity UPLC BEH C18 column (50 mm×2.1 mm, 1.7 μm, Waters, Milford, MA) and run for 6.5 min with a flow rate of 0.5 ml/min. The elution gradient was as follows: initial, 80% A (100% water) and 20% B (100% acetonitrile), 0–0.5 min, 20% B, 0.5–1 min, 20–35% B, 1–2 min, 35% B, 2–3 min, 35–50% B, 3–4 min, 50% B, 4–4.5 min, 50–75%, 4.5–5 min, 75–90% B, 5–5.2 min, 90–100% B; 5.2–6 min 100% B, 6–6.5 min 100–20% B. UPLC analysis was performed on a Acquity UPLC (Waters, Milford, MA) with photodiode-arrayed (PDA) detector. Quantitation was performed at 200 nm wavelength. The column temperature and sample temperature were set to 45°C and 20°C, respectively. The injection volume was 10 μl. The chromatograph and UV spectrum of Rb1, Rd, F2, C-K, and testosterone are shown in Supplemental Figure 1.
The standard curves for Rb1 and Rd in fecal lysate were linear in the concentration range of 0.39 – 50 μg/ml with correlation coefficient values >0.999. The lower limit of quantification (LLOQ) was 0.39 μg/ml for both Rb1 and Rd. The standard curves for F2 were linear in low concentration range of 0.306 – 4.9 μg/ml and in high concentration range of 4.9 – 39.2 μg/ml. As for C-K, standard curves were linear in low concentration range of 0.243 – 3.89 μg/ml and in high concentration range of 3.89 – 31.1 μg/ml. The correlation coefficient values were 0.9994, 0.9937 for F2 and 0.9999, 0.9916 for C-K, respectively. The LLOQ was 0.306 μg/ml and 0.243 μg/ml for F2 and C-K, respectively. Intra-day and Inter-day precision and accuracy were well within the 15% acceptance range for all quality control (QC) samples at three concentrations levels in fecal lysate (Supplemental Table 1). The mean extraction recoveries determined using three replicates of QC samples at three concentration levels in fecal lysate fell in the range of 51.4% to 98.1% (Supplemental Table 1). The stability of 20 μM Rb1, Rd, F2, and C-K in PBS (37°C, 4 hr), A/J mouse liver, small intestine, and colon S9 (37°C, 24 hr), the long term stability of glycosidases in fecal lysate (−80°C, 1 month), and three cycles of freeze-thaw effect were evaluated in triplicates. All the samples displayed 90–120% recoveries in the stability tests (Supplemental Fig. 2).
Pyrosequencing of gut microbiome using 16s rRNA
Fecal samples (500 μl) were homogenized in 1.5 ml of Fecal Bead Solution (MoBio, Carlsbad, CA), centrifuged for 5 min at 2000× g, and 500 μl supernatant was transferred to PowerBead Tubes (MoBio, Carlsbad, CA). Samples were heated for 10 min at 65°C and 95°C to aid bacterial lysis. Genomic DNA was isolated using PowerSoil DNA Isolation Kit purchased from MoBio Laboratories (Carlsbad, CA) starting at step 2 of the manufacturer’s protocol. DNA concentration and purity was determined by spectrometry on the NanoDrop ND-2000 (Thermo Scientific). Variable regions 3 to 5 (V3–V5) of the 16S rRNA gene were amplified using barcoded primers 357F (CTGCTGCCTCCCGTAGG) and 926R (CCGTCAATTCMTTTRAGT). Polymerase chain reaction (PCR) (total volume 20 μl) contained AccuPrime PCR Buffer II (Invitrogen, Carlsbad, CA), AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA), 4 nM barcoded primers, and 2 μl of template DNA. PCR was performed on Mastercycler ep gradient S (Eppendorf, Hamburg, Germany) with the following cycling conditions: 2 min at 95°C and 30 cycles of 20 sec at 95°C, 30 sec at 50°C, and 5 min at 72°C. DNA concentration of PCR products was determine by PicoGreen (Invitrogen, Carlsbad, CA) and on an Agilent Bioanalyzer 2100 DNA 1000 chip (Agilent Technologies, Santa Clara, CA). Pyrosequencing was performed on Roche 454 FLX Titanium platform (Branford, CT) according to the manufacturer’s instructions.
Sequences were pre-processed using Mothur (42). Reads were removed from subsequent analysis if they had a quality score of <35 over a 50 bp window, contained ambiguous bases or homopolymer repeats of >8 bp, had >1 bp mismatch from the barcode sequence, or had >2 bp mismatch from the primer sequence. Sequences were binned based on barcode followed by trimming of the barcode and primer sequences. Sequences were analyzed using the CloVR 16S pipeline (43).
Statistical analysis
The data in this paper are presented as mean ± SD, if not specified otherwise. For enzyme function studies, significance is assessed by one way ANOVA and Student’s-test. A p-value of < 0.05 was considered statistically significant. Relative abundance and mean relative abundance were determined for each taxa found in the microbial communities.
3. Results
Anti-proliferation activities of ginsenosides
We determined the anti-proliferative activities of related primary and secondary ginsenosides in LM1 cells, a metastatic lung cancer cell line derived from the A/J mouse (44). While primary ginsenosides are well represented in RGE, secondary ginsenosides are derived from the primary ginsenosides by the metabolic action of glycosidases (Fig.1). The MTT assay showed that secondary ginsenosides, but not primary ginsenosides, significantly inhibit proliferation of LM1 cells (Fig. 2). No anti-proliferation activity was observed for RGE, where the major species are primary ginsenosides. The IC50 value of ginsenoside F2 was more than 100 μg/ml in the LM1 cell line, while the activity of C-K was the highest among the tested ginsenosides, with a IC50 value of approximately 13 μg/ml. These results show that the anti-proliferation activities of ginsenosides may be closely correlated to the number of sugars attached.
Figure 1.
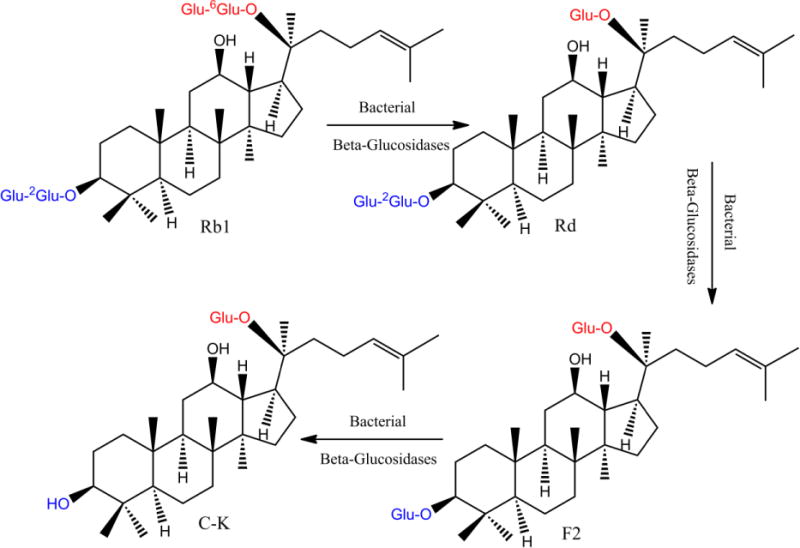
The proposed metabolic pathway for production of active ginsenoside C-K in A/J mouse fecal lysate. The superscript denotes the position of the hydroxyl group attached to the adjacent glucose.
Figure 2.
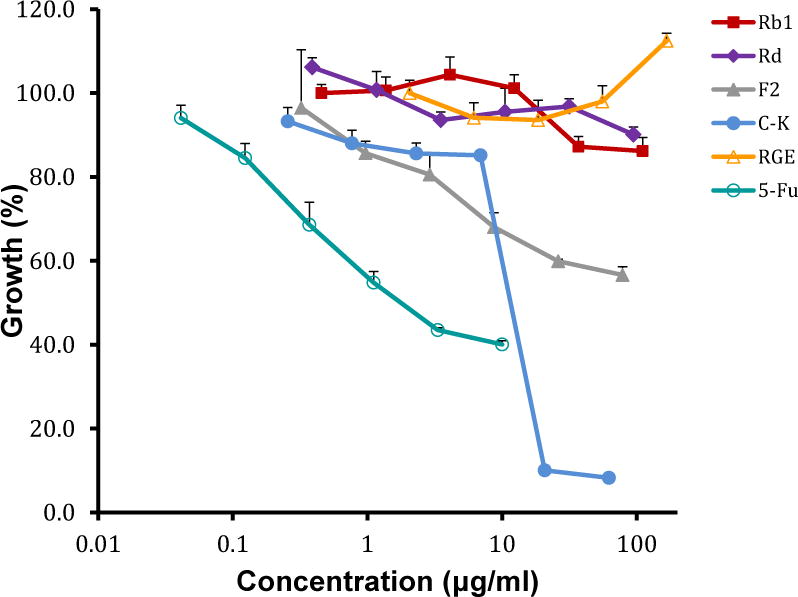
Anti-proliferation activity of ginsenosides Rb1, Rd, F2, C-K, and RGE in the lung cancer LM1 cell line. 5-fluorouracil was used as the positive control and 1% DMSO was used as the negative control. The concentration range of tested agents was 0.46–110.80 μg/ml for Rb1, 0.39–94.6 μg/ml for Rd, 0.32–78.4 μg/ml for F2, 0.26–62.2 μg/ml for C-K, 2.06–500 μg/ml for RGE and 0.041–10 μg/ml for 5-fluorouracil, respectively.
Transcellular transport of ginsenosides across Caco-2 cell monolayers
We determined the permeability of ginsenosides Rb1, Rd, F2, and C-K in Caco-2 cell monolayers, a model employed to mimic human intestinal absorption characteristics (45, 46). Transport of 10 μM Rb1 and Rd, 2 μM F2 and C-K from apical side to basolateral side (A–B) in Caco-2 cells were studied at pH 7.4. The results from Figure 3 show that the permeability of ginsenosides Rb1, Rd and F2 were less than 1 × 10−6 cm/sec (corresponding to incomplete absorption in humans(46), indicating that ginsenosides with more than one glucose were poorly permeable. Whereas ginsenoside C-K, with only one glucose attached, exhibited moderate permeability (Papp between 1 ×10−6 cm/s and 10 ×10−6 cm/s, corresponding to good absorption in humans(46). This pattern of distinctive permeabilities between primary and second ginsenosides was similar to the correlation of ginsenoside anti-proliferation activity and the number of sugar moieties.
Figure 3.

Transcellular transport of ginsenosides Rb1, Rd, F2, and C-K from apical side to basolateral side in Caco-2 cell monolayers. Data are presented as mean ± S.D.; n=3.
Oral pharmacokinetics of Rb1 in A/J mice
The plasma concentrations of ginsenoside Rb1 and its metabolites in A/J mice were determined following oral administration of Rb1 at 20 mg/kg. The results from Figure 4 show that significant amounts of secondary ginsenosides F2 and C-K were observed in blood, reaching peak concentrations higher than 1 μM. A/J mouse liver, small intestine, and colon S9 fraction (which contains epithelium-derived enzymes) did not hydrolyze Rb1 (Supplemental Fig. 2). This is consistent with the fact that β-glucosidases are responsible for hydrolysis of ginsenosides Rb1 and none of the β-glucosidases identified so far are from mammalian cells (23–26). Therefore, the presence of secondary ginsenosides F2 and C-K were attributed to the action of bacterial glycosidases in the intestinal microbiome.
Figure 4.
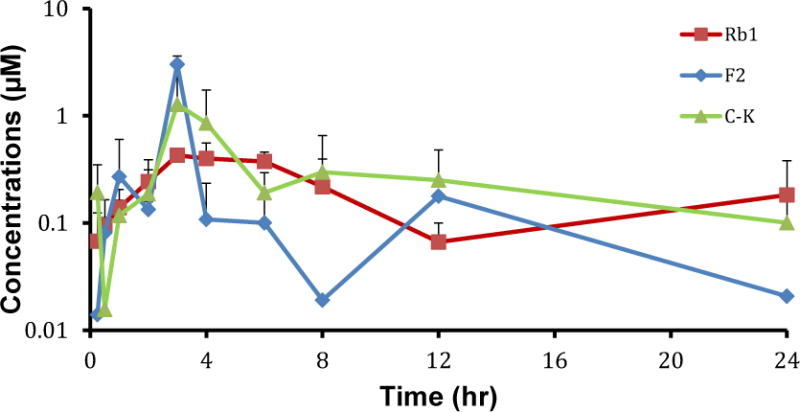
Pharmacokinetic profiles of Rb1 and its metabolites F2 and C-K following Rb1 oral administration (20mg/kg) in male A/J mice. Data are presented as mean ± S.D.; n=4.
Stepwise metabolism of ginsenoside Rb1
An in vitro hydrolysis study of ginsenoside Rb1 by A/J mouse fecal lysate was performed to correlate with the pharmacokinetic profiles seen in Figure 4. Primary ginsenoside Rb1 was hydrolyzed to ginsenoside Rd, as indicated by the rapid disappearance of Rb1 and appearance of Rd within 1 hr, while F2 and C-K were rarely found at this time point (Fig.5). Concentration of Rd plateaued around 2 hr and then began to drop. However, Rd’s metabolite F2 accumulated only slightly during the course of incubation, suggesting that the formation of F2 from Rd was the slowest step. C-K’s concentration paralleled F2 concentration initially but increased significantly after 8 hr, compensating for the dramatic loss of Rd. Also, this pattern of hydrolysis indicated that the formation of C-K from F2 was rapid. Taken together, primary ginsenoside Rb1 was hydrolyzed stepwise to Rd, F2 and finally C-K as shown in Figure 1.
Figure 5.
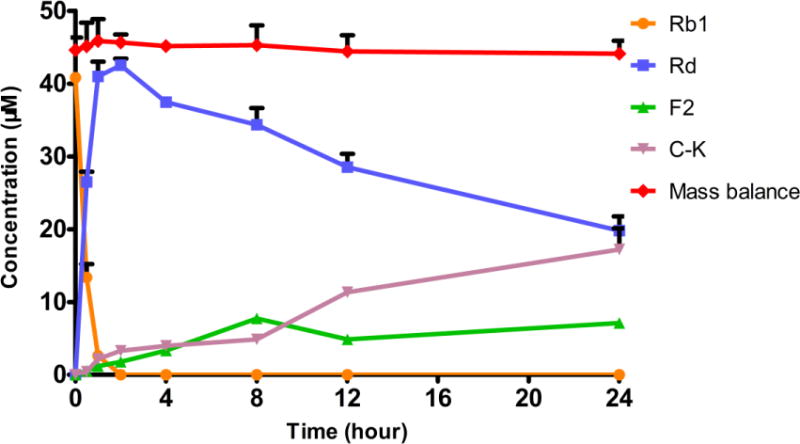
Metabolic profile of ginsenoside Rb1 (45 μM) in A/J mouse fecal lysate. Mass balance stands for the total amount of ginsenosides recovered. Data are presented as mean ± S.D.; n=3.
Rate-limiting step in Rb1 stepwise hydrolysis
Metabolite formation rates of ginsenosides normalized for protein concentration are presented in Fig. 6. The hydrolysis rates of Rb1 to Rd, Rd to F2, and F2 to C-K were 1.13 ± 0.04, 0.09 ± 0.003, 1.36 ± 0.27 nmol/min/mg at 20 μM concentration, and 0.60 ± 0.01, 0.09 ± 0.01, 0.52 ± 0.01nmol/min/mg at 5 μM concentration, respectively. The hydrolysis rate of Rd to F2 was approximately 11 fold and 14 fold lower than the formation of Rd and C-K (from their corresponding substrate) at 20 μM, respectively. At 5 μM concentration, a similar hydrolysis pattern was observed. The formation rate of F2 was around 5 fold less than the formation of Rd or C-K. Formation of aglycone protopanaxadiol (PPD) from C-K was not observed during incubation, which could be attributed to the low solubility of the aglycone (data not shown). However, the amount of aglycone formed must be small, since the recovery was within 80–110% at two concentrations.
Figure 6.
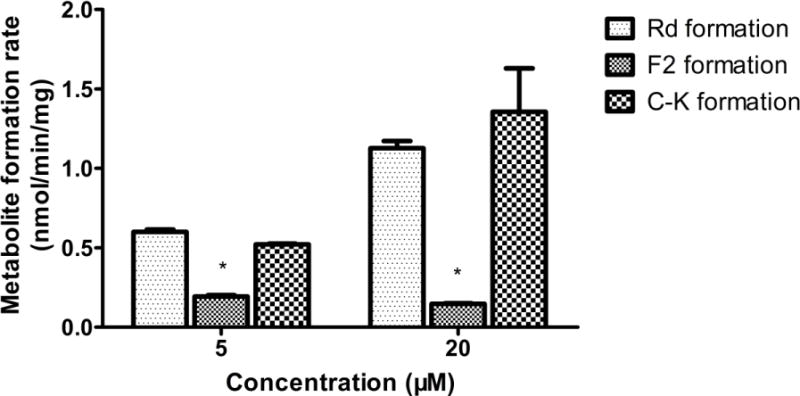
Stepwise metabolite formation rates of ginsenosides Rb1, Rd, and F2 in A/J mouse fecal matrix. Data are presented as mean ± S.D.; n=3. With 20 or 5μM ginsenosides as substrates, fecal lysates were incubated for 30 min. Metabolite formation rates were calculated as the metabolite concentration divided by protein concentration of the fecal lysate and reaction time. The “*” symbol indicated p<0.05, analyzed by one-way ANOVA and Student’s t-test.
Membership and relative abundance of the A/J mouse intestinal microbiota
Presence of bacteria capable of ginsenoside hydrolysis, we performed 16S rRNA gene pyrosequencing to determine membership and relative abundance of the A/J mouse intestinal microbiota and identify bacterial species capable of hydrolyzing ginsenosides. Relative abundance at the family level of 9 A/J mice demonstrates the low inter-animal variability of the intestinal microbiome (Figure 7a). The mean relative abundance in Figure 7b demonstrates that 5 major families (“Lachnopiraceae”, “Ruminococcaceae”, Bacteroidaceae, Porphyromonadaceae, and Prevotellaceae) comprise approximately 80 percent of the A/J mouse intestinal microbiota. An additional 15 minor families account for 10% of the intestinal microbiome.
Figure 7.
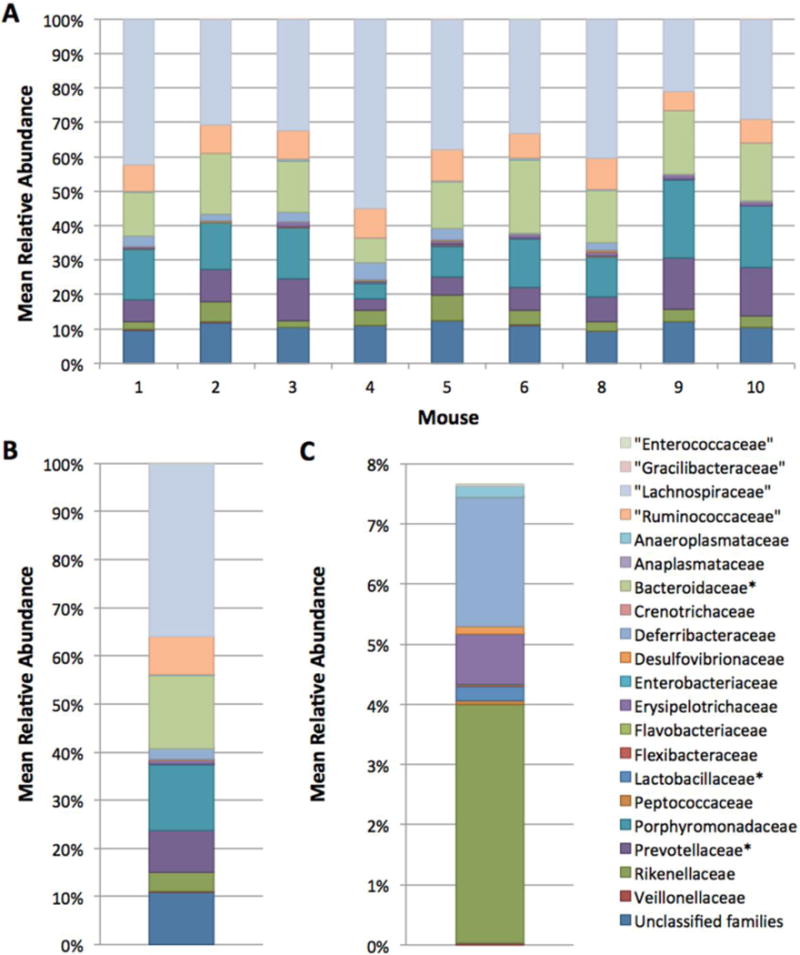
Relative abundance of families in the A/J mouse intestinal microbiome. A: Relative abundance for individual mice at the family level. B: Mean relative abundance of families in the intestinal microbiome of 9 mice. C: Mean relative abundance after the 5 most abundant families are removed. The “*” symbol denotes families containing species capable of hydrolyzing ginsenosides.
4. Discussion
RGE shows promise in preventing lung cancer (6). However, the ginsenosides responsible for the activity of RGE are poorly defined as primary ginsenosides are inactive (Fig.2). In this study, we systematically investigate the role of intestinal microbiome in activating primary ginsenosides in RGE using A/J mouse fecal lysate. To our knowledge, this is the first comprehensive study demonstrating the formation of ginsenoside F2 from Rd is the rate limiting step in ginsenoside Rb1 stepwise hydrolysis in a mouse intestinal microbiota. Furthermore, we have begun identifying the intestinal bacteria potentially responsible for this pattern of stepwise hydrolysis, which should facilitate future studies of this type.
Our results showed that secondary ginsenosides exhibited high anti-proliferation potency in LM1 cells, while primary ginsenosides and RGE did not possess this activity (Fig.2). In addition, secondary ginsenosides exhibited better intestinal permeability in the Caco-2 cell monolayers (Fig.3). In contrast to their higher activity and better permeability, secondary ginsenosides are normally present in very low abundance in RGE (less than 1%). To explain this apparent discrepancy, we performed a hydrolysis study of primary ginsenoside Rb1 using A/J mouse fecal lysate, since A/J mice were used to demonstrate the efficacy of RGE in vivo (6). Ginsenoside hydrolyzing enzymes are β-glucosidase, β-xylosidase, α-L-arabinofurano-sidase, and α-L-rhamnosidase since the sugars attached to ginsenosides are glucose, L-arabinopyranoside, L-arabinofuranoside, D-xylose, and/or L-rhamnose (23). Mammalian cells do not express these enzymes (24–26), which is consistent with our result that ginsenoside Rb1 was stable in freshly prepared A/J mouse liver, small intestine, and colon S9 fraction(Supplemental Fig. 2). The results of in vivo pharmacokinetic study of ginsenoside Rb1 validated the concept that microbial hydrolysis of primary ginsenosides to bioactive secondary ginsenoside were occurring in vivo, since oral administration of Rb1 resulted in clear blood level of C-K (Fig. 4). Therefore, the efficacy of RGE demonstrated previously in A/J mice is largely explained by enzymatic activity of the intestinal microbiota.
The peak concentration of C-K in vivo (1.5 μM in Fig. 4) following Rb1 oral administration at 20 mg/kg is around 7% of its in vitro IC50 value (13 μg/ml in Fig. 2, equivalent to 20.9 μM, MW is 622 g/mol) in LM1 cells. Yang et. al. also reported that the peak concentration of C-K could reach as high as 1 μM following oral administration of C-K at 10 mg/kg in FVB mice (47). C-K’s concentration in vivo is expected to increase more than proportion at higher doses since it is a substrate of P-gp and it serves as a P-gp inhibitor itself (47). Therefore, it is possible to enhance the efficacy of RGE by increasing the bioavailability of C-K.
To enhance the therapeutic efficacy of RGE, recent investigations focused on maximizing the production of active ginsenosides by microorganisms (29, 30, 32, 33). However, these microorganisms may not be present in the intestinal microbiome, rendering these results less physiologically relevant. Furthermore, two key questions need to be addressed before trying to enhance the efficacy of RGE: (1) what is the rate limiting step of RGE hydrolysis and (2) which bacterial β-glucosidases catalyze this reaction. In a sequential reaction system, the rate-limiting step is defined as the slowest step. Here, we determined the metabolite formation rates in an in vitro hydrolysis assay by optimizing protein concentration, incubation time, and sampling time such that only one metabolite would be detected. Formation of ginsenoside F2 from Rd was found to be the rate-limiting step. It has been reported that Rb1 can be metabolized to Rh2, Rg3, C-K and gypenoside LXXV by different microorganisms (48), and the results of our study indicated that we should pay attention to the conversion to C-K and especially the formation of F2 in mice The intestinal microbiome structure is provided here because this activity may change in different labs and animal models, due to differences in species, food, and etc. Hence, the publication of our microbiome structure allows others to compare their results with our results. Taken together, the formation rate of ginsenoside F2 would likely determine the rate and extent of active ginsenoside production and hence greatly impacts the efficacy of RGE. Future studies are ongoing to identify specific bacterial β-glucosidases hydrolyzing ginsenoside Rd to F2.
A viable approach to enhance therapeutic efficacy of RGE is the manipulation of bacterial β-glucosidases responsible for ginsenoside F2 formation. Recent studies have shown that the abundance of Lactobacillus and Bifidobacteria as well as activities of β-glucosidase(s) can be significantly increased by probiotic intervention (49–51). Since members of these genera have been frequently used as probiotics and are known to produce β-glucosidases (49), an appropriately designed regimen combining probiotic intervention and RGE administration may serve as a novel approach for the chemoprevention of lung cancer.
In this study, we demonstrated the metabolic pathway of ginsenoside Rb1 and the rate limiting reaction of the stepwise hydrolysis of RGE in A/J mouse fecal lysates. However, different metabolic pathways have been reported using various individual microorganisms (30, 31, 52–55) with unknown in vivo relevance. Therefore, it is imperative to characterize the intestinal microbiome of relevant mouse models for pre-clinical studies involving RGE and other compounds. Here, we characterized the A/J mouse intestinal microbiome (Fig. 7) to aid future studies comparing changes in the microbiome and its effect on conversion of ginsenosides.
The interest in this mouse model stems from its application as a pre-clinical model to determine the efficacy of RGE as a chemopreventive agent against lung cancer. Moreover, the mouse intestinal microbiome resembles that of the human microbiome in terms of taxa present (56). Of the 5 families that comprise over 80% of the mean relative abundance of the A/J mouse intestinal microbiome (Figure 7b), Bacteroidaceae and Prevotellaceae contain species capable of hydrolyzing ginsenosides present in RGE (35, 57). Lactobacillaceae, less than 1% of the A/J mouse intestinal microbiome, also contains Lactobacillus species with ginsenoside hydrolysis activity (57). Additionally, several Lactobacillus species have been identified as generally-recognized-as-safe (GRAS) food microorganisms by the US FDA (ref, http://www.fda.gov/Food/FoodIngredientsPackaging/ucm078956.htm). Identification of GRAS microorganism would be highly desirable for use as probiotics to be administered to humans in conjunction with RGE to enhance the production of active secondary ginsenosides, thereby improving efficacy.
5. Conclusion
In summary, we have demonstrated that in vivo conversion of primary ginsenosides in RGE to the secondary and bioactive ginsenoside C-K was only mediated by microbial glycosidases. The formation of F2 from Rd was found, for the first time, to be the rate-limiting step in the biotransformation of Rb1 to C-K. The intestinal microbiome of the A/J mouse capable of producing C-K was characterized preliminary, which formed the basis for future studies of how changes in intestinal microbiome will impact bioactivities of RGE in vivo.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health [AT-005522] to Ming Hu at University of Houston, Ming You at Medical College of Wisconsin and Zhi-hong Jiang at Hong Kong Baptist University.
Abbreviations
- C-K
ginsenoside compound K
- UPLC
ultra-performance liquid chromatography
- Pa–b
permeability from apical to basolateral side
- MTT
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- 16s rRNA
16s ribosomal RNA
Footnotes
Authorship Contribution
Participated in research design: Niu, Smith, Yang, Petrosino and Hu
Conducted experiments: Niu, Smith, Yang and Yin
Performed data analysis: Niu, Smith, Yang, Petrosino and Hu
Wrote or contributed to the writing of the manuscript: Niu, Smith, Petrosino and Hu.
Other: Hu, Petrosino and You acquired funding for the research
References
- 1.Ahmedin Jemal RS, Xu Jiaquan. Elizabeth Ward. Cancer statistics. CA CANCER J CLIN. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Clarkand J, You M. Chemoprevention of lung cancer by tea. Mol Nutr Food Res. 2006;50:144–151. doi: 10.1002/mnfr.200500135. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- 4.Tanand XL, Spivack SD. Dietary chemoprevention strategies for induction of phase II xenobiotic-metabolizing enzymes in lung carcinogenesis: A review. Lung Cancer. 2009;65:129–137. doi: 10.1016/j.lungcan.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson TE, Hermanson D, Wang L, Kassie F, Upadhyaya P, O’Sullivan MG, Hecht SS, Lu J, Xing C. Lung tumorigenesis suppressing effects of a commercial kava extract and its selected compounds in a/j mice. Am J Chin Med. 39:727–742. doi: 10.1142/S0192415X11009202. [DOI] [PubMed] [Google Scholar]
- 6.Yan Y, Wang Y, Tan Q, Hara Y, Yun TK, Lubet RA, You M. Efficacy of polyphenon E, red ginseng, and rapamycin on benzo(a)pyrene-induced lung tumorigenesis in A/J mice. Neoplasia. 2006;8:52–58. doi: 10.1593/neo.05652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhtar H, Khan N, Afaq F. Cancer chemoprevention through dietary antioxidants: Progress and promise. Antioxidants & Redox Signaling. 2008;10:475–510. doi: 10.1089/ars.2007.1740. [DOI] [PubMed] [Google Scholar]
- 8.Miller YE, Keith RL, Karoor V, Mozer AB, Hudish TM, Le M. Chemoprevention of murine lung cancer by gefitinib in combination with prostacyclin synthase overexpression. Lung Cancer. 2010;70:37–42. doi: 10.1016/j.lungcan.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohashi K, Takigawa N, Osawa M, Ichihara E, Takeda H, Kubo T, Hirano S, Yoshino T, Takata M, Tanimoto M, Kiura K. Chemopreventive effects of gefitinib on nonsmoking-related lung tumorigenesis in activating epidermal growth factor receptor transgenic mice. Cancer Res. 2009;69:7088–7095. doi: 10.1158/0008-5472.CAN-08-4205. [DOI] [PubMed] [Google Scholar]
- 10.Miller YE, Kelly K, Kittelson J, Franklin WA, Kennedy TC, Klein CE, Keith RL, Dempsey EC, Lewis M, Jackson MK, Hirsch FR, Bunn PA. A Randomized Phase II Chemoprevention Trial of 13-CIS Retinoic Acid with Or without alpha Tocopherol or Observation in Subjects at High Risk for Lung Cancer. Cancer Prevention Research. 2009;2:440–449. doi: 10.1158/1940-6207.CAPR-08-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott EN, Gescher AJ, Steward WP, Brown K. Development of dietary phytochemical chemopreventive agents: biomarkers and choice of dose for early clinical trials. Cancer Prev Res (Phila) 2009;2:525–530. doi: 10.1158/1940-6207.CAPR-08-0223. [DOI] [PubMed] [Google Scholar]
- 12.Qi LW, Wang CZ, Yuan CS. American ginseng: potential structure-function relationship in cancer chemoprevention. Biochem Pharmacol. 2010;80:947–954. doi: 10.1016/j.bcp.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Sun S, Qi LW, Du GJ, Mehendale SR, Wang CZ, Yuan CS. Red notoginseng Higher ginsenoside content and stronger anticancer potential than Asian and American ginseng. Food Chemistry. 2011;125:1299–1305. doi: 10.1016/j.foodchem.2010.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y Y, Yun TK, Han IW. Anticarcinogenic effect of long-term oral administration of red ginseng on newborn mice exposed to various chemical carcinogens. Cancer Detect Prev. 1983;6:515–525. [PubMed] [Google Scholar]
- 15.K S, Yun TK, Oh YR. Medium-term (9 weeks) method for assay of preventive agents against tumor. J Korean Cancer Assoc. 1987;19:1–7. [Google Scholar]
- 16.SH YTaK. Inhibition of development of benzo(a)-pyrene-induced mouse pulmonary adenoma by natural products in medium-term bioassay system. J Korean Cancer Assoc. 1988;20:133–142. [Google Scholar]
- 17.Yun TK. Usefulness of medium-term bioassay determining formations of pulmonary adenoma in NIH(GP) mice for finding anticarcinogenic agents from natural products. J Toxicol Sci. 1991;16(Suppl 1):53–62. doi: 10.2131/jts.16.supplementi_53. [DOI] [PubMed] [Google Scholar]
- 18.K S, Yun TK, Lee YS. Trial of a new medium-term model using benzo(a)pyrene induced lung tumor in newborn mice. Anticancer Res. 1995;15:839–845. [PubMed] [Google Scholar]
- 19.Kong H, Wang M, Venema K, Maathuis A, van der Heijden R, van der Greef J, Xu G, Hankemeier T. Bioconversion of red ginseng saponins in the gastro-intestinal tract in vitro model studied by high-performance liquid chromatography-high resolution Fourier transform ion cyclotron resonance mass spectrometry. J Chromatogr A. 2009;1216:2195–2203. doi: 10.1016/j.chroma.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Lee JH, Ha JM. Effective purification of ginsenosides from cultured wild ginseng roots, red ginseng, and white ginseng with macroporous resins. J Microbiol Biotechnol. 2008;18:1789–1791. doi: 10.4014/jmb.0800.192. [DOI] [PubMed] [Google Scholar]
- 21.Cheng CC, Yang SM, Huang CY, Chen JC, Chang WM, Hsu SL. Molecular mechanisms of ginsenoside Rh2-mediated G1 growth arrest and apoptosis in human lung adenocarcinoma A549 cells. Cancer Chemother Pharmacol. 2005;55:531–540. doi: 10.1007/s00280-004-0919-6. [DOI] [PubMed] [Google Scholar]
- 22.Yun TK, Lee YS, Lee YH, Kim SI, Yun HY. Anticarcinogenic effect of Panax ginseng C.A. Meyer and identification of active compounds. J Korean Med Sci. 2001;16(Suppl):S6–18. doi: 10.3346/jkms.2001.16.S.S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park CS, Yoo MH, Noh KH, Oh DK. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Applied Microbiology and Biotechnology. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 24.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrissatand B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293(Pt 3):781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrissatand B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316(Pt 2):695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatia Y, Mishra S, Bisaria VS. Microbial beta-glucosidases: cloning, properties, and applications. Crit Rev Biotechnol. 2002;22:375–407. doi: 10.1080/07388550290789568. [DOI] [PubMed] [Google Scholar]
- 28.K H, Akao T, Kanaoka M, Hattori M, Kobashi K. Intestinal bacterial hydrolysis is required for the appearance of compound K in rat plasma after oral administration of ginsenoside Rb1 from Panax ginseng. J Pharm Pharmacol. 1998;50:1155–1160. doi: 10.1111/j.2042-7158.1998.tb03327.x. [DOI] [PubMed] [Google Scholar]
- 29.Chi H, Kim DH, Ji GE. Transformation of ginsenosides Rb2 and Rc from Panax ginseng by food microorganisms. Biol Pharm Bull. 2005;28:2102–2105. doi: 10.1248/bpb.28.2102. [DOI] [PubMed] [Google Scholar]
- 30.Cheng LQ, Na JR, Kim MK, Bang MH, Yang DC. Microbial conversion of ginsenoside Rb1 to minor ginsenoside F2 and gypenoside XVII by Intrasporangium sp. GS603 isolated from soil. J Microbiol Biotechnol. 2007;17:1937–1943. [PubMed] [Google Scholar]
- 31.Chen G, Yang M, Song Y, Lu Z, Zhang J, Huang H, Guan S, Wu L, Guo DA. Comparative analysis on microbial and rat metabolism of ginsenoside Rb1 by high-performance liquid chromatography coupled with tandem mass spectrometry. Biomed Chromatogr. 2008;22:779–785. doi: 10.1002/bmc.1001. [DOI] [PubMed] [Google Scholar]
- 32.Zhou P, Zhou W, Yan Q, Li JY, Zhang XC. Biotransformation of Panax notoginseng saponins into ginsenoside compound K production by Paecilomyces bainier sp 229. Journal of Applied Microbiology. 2008;104:699–706. doi: 10.1111/j.1365-2672.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 33.Son JW, Kim HJ, Oh DK. Ginsenoside Rd production from the major ginsenoside Rb(1) by beta-glucosidase from Thermus caldophilus. Biotechnol Lett. 2008;30:713–716. doi: 10.1007/s10529-007-9590-4. [DOI] [PubMed] [Google Scholar]
- 34.Kim YS, Kim JJ, Cho KH, Jung WS, Moon SK, Park EK, Kim DH. Biotransformation of ginsenoside Rb1, crocin, amygdalin, geniposide, puerarin, ginsenoside Re, hesperidin, poncirin, glycyrrhizin, and baicalin by human fecal microflora and its relation to cytotoxicity against tumor cells. J Microbiol Biotechnol. 2008;18:1109–1114. [PubMed] [Google Scholar]
- 35.Hasegawa H, Sung JH, Benno Y. Role of human intestinal Prevotella oris in hydrolyzing ginseng saponins. Planta Med. 1997;63:436–440. doi: 10.1055/s-2006-957729. [DOI] [PubMed] [Google Scholar]
- 36.Bae EA, Choo MK, Park EK, Park SY, Shin HY, Kim DH. Metabolism of ginsenoside R(c) by human intestinal bacteria and its related antiallergic activity. Biol Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 37.Relman DA, Dethlefsen L, Huse S, Sogin ML. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. Plos Biology. 2008;6:2383–2400. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castagnini C, Luceri C, Toti S, Bigagli E, Caderni G, Femia AP, Giovannelli L, Lodovici M, Pitozzi V, Salvadori M, Messerini L, Martin R, Zoetendal EG, Gaj S, Eijssen L, Evelo CT, Renard CM, Baron A, Dolara P. Reduction of colonic inflammation in HLA-B27 transgenic rats by feeding Marie Menard apples, rich in polyphenols. Br J Nutr. 2009;102:1620–1628. doi: 10.1017/S0007114509990936. [DOI] [PubMed] [Google Scholar]
- 39.Bolam DN, Sonnenburg ED, Zheng HJ, Joglekar P, Higginbottom SK, Firbank SJ, Sonnenburg JL. Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations. Cell. 2010;141:1241–U1256. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Gao S, Yin TJ, Kulkarni KH, Teng Y, You M, Hu M. Biopharmaceutical and pharmacokinetic characterization of matrine as determined by a sensitive and robust UPLC-MS/MS method. Journal of Pharmaceutical and Biomedical Analysis. 2010;51:1120–1127. doi: 10.1016/j.jpba.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G S, Yang Z, Wang J, Yin T, Teng Y, Wu B, You M, Jiang Z, Hu M. Enhancement of Oral Bioavailability of Ginsenoside 20(s)-Rh2 through Improved Understanding of its Absorption and Efflux Mechanisms. Drug Metab Dispos. 2011 doi: 10.1124/dmd.111.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDoniels-Silvers AL, Herzog CR, Tyson FL, Malkinson AM, You M. Inactivation of both Rb and p53 pathways in mouse lung epithelial cell lines. Experimental Lung Research. 2001;27:297–318. doi: 10.1080/019021401300054064. [DOI] [PubMed] [Google Scholar]
- 45.Hu M, Chen J, Zhu Y, Dantzig AH, Stratford RE, Jr, Kuhfeld MT. Mechanism and kinetics of transcellular transport of a new beta-lactam antibiotic loracarbef across an intestinal epithelial membrane model system (Caco-2) Pharm Res. 1994;11:1405–1413. doi: 10.1023/a:1018935704693. [DOI] [PubMed] [Google Scholar]
- 46.Artursson P, Karlsson J. Correlation between Oral-Drug Absorption in Humans and Apparent Drug Permeability Coefficients in Human Intestinal Epithelial (Caco-2) Cells. Biochemical and Biophysical Research Communications. 1991;175:880–885. doi: 10.1016/0006-291x(91)91647-u. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, Wang JR, Niu T, Gao S, Yin T, You M, Jiang ZH, Hu M. Inhibition of p-glycoprotein leads to improved oral bioavailability of compound k, an anticancer metabolite of red ginseng extract produced by gut microflora. Drug Metab Dispos. 40:1538–1544. doi: 10.1124/dmd.111.044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou JG, Xue JJ, Sun MQ, Wang CY, Liu L, Zhang DL, Lee MR, Gu LJ, Wang CL, Wang YB, Zheng YA, Li W, Sung CK. Highly selective microbial transformation of major ginsenoside Rb(1) to gypenoside LXXV by Esteya vermicola CNU120806. J Appl Microbiol. doi: 10.1111/j.1365-2672.2012.05400.x. [DOI] [PubMed] [Google Scholar]
- 49.McBain AJ, Macfarlane GT. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J Med Microbiol. 1998;47:407–416. doi: 10.1099/00222615-47-5-407. [DOI] [PubMed] [Google Scholar]
- 50.Goossens D, Jonkers D, Russel M, Stobberingh E, Van Den Bogaard A, StockbrUgger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther. 2003;18:495–505. doi: 10.1046/j.1365-2036.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 51.Marteau P, Pochart P, Flourie B, Pellier P, Santos L, Desjeux JF, Rambaud JC. Effect of chronic ingestion of a fermented dairy product containing Lactobacillus acidophilus and Bifidobacterium bifidum on metabolic activities of the colonic flora in humans. Am J Clin Nutr. 1990;52:685–688. doi: 10.1093/ajcn/52.4.685. [DOI] [PubMed] [Google Scholar]
- 52.Chen G, Yang M, Lu Z, Zhang J, Huang H, Liang Y, Guan S, Song Y, Wu L, Guo DA. Microbial transformation of 20(S)-protopanaxatriol-type saponins by Absidia coerulea. J Nat Prod. 2007;70:1203–1206. doi: 10.1021/np070053v. [DOI] [PubMed] [Google Scholar]
- 53.Chen GT, Yang M, Song Y, Lu ZQ, Zhang JQ, Huang HL, Wu LJ, Guo DA. Microbial transformation of ginsenoside Rb(1) by Acremonium strictum. Appl Microbiol Biotechnol. 2008;77:1345–1350. doi: 10.1007/s00253-007-1258-4. [DOI] [PubMed] [Google Scholar]
- 54.Cheng LQ, Kim MK, Lee JW, Lee YJ, Yang DC. Conversion of major ginsenoside Rb1 to ginsenoside F2 by Caulobacter leidyia. Biotechnol Lett. 2006;28:1121–1127. doi: 10.1007/s10529-006-9059-x. [DOI] [PubMed] [Google Scholar]
- 55.Cheng LQ, Na JR, Bang MH, Kim MK, Yang DC. Conversion of major ginsenoside Rb1 to 20(S)-ginsenoside Rg3 by Microbacterium sp. GS514. Phytochemistry. 2008;69:218–224. doi: 10.1016/j.phytochem.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 56.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 57.Chi H, Ji GE. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol Lett. 2005;27:765–771. doi: 10.1007/s10529-005-5632-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


