Abstract
Bacterial proteases are considered virulence factors and it is presumed that by abrogating their activity, host endogenous protease inhibitors play a role in host defense against invading pathogens. Here we present data showing that Staphylococcus aureus cysteine proteases (staphopains) are efficiently inhibited by Squamous Cell Carcinoma Antigen 1 (SCCA1), an epithelial-derived serpin. The high association rate constant (kass) for inhibitory complex formation (1.9×104 m/s and 5.8×104 m/s for staphopain A and stapho-pain B interaction with SCCA1, respectively), strongly suggests that SCCA1 can regulate staphopain activity in vivo at epithelial surfaces infected/colonized by S. aureus. The mechanism of staphopain inhibition by SCCA1 is apparently the same for serpin interaction with target serine proteases whereby the formation of a covalent complex result in cleavage of the inhibitory reactive site peptide bond and associated release of the C-terminal serpin fragment. Interestingly, the SCCA1 reactive site closely resembles a motif in the reactive site loop of native S. aureus-derived inhibitors of the staphopains (staphostatins). Given that S. aureus is a major pathogen of epithelial surfaces, we suggest that SCCA1 functions to temper the virulence of this bacterium by inhibiting the staphopains.
Keywords: cathepsin-like protease, inhibition, pathogenicity, proteolysis regulation, serpin, staphylococcal infection, virulence factor
Introduction
Squamous Cell Carcinoma Antigen 1 and 2 (SCCA1 and SCCA2), members of the intracellular SERPINB clade (ovserpins) of protease inhibitors (Silverman et al., 2004), were originally purified from squamous cell carcinoma of the uterine cervix as the major component of the TA-4 antigen (Kato and Torigoe, 1977). Subsequently, the SCCA serpins were found in numerous healthy tissues of epithelial origin, most notably in some areas of the skin and the tongue epithelium (Cataltepe et al., 2000). Although they lack the signal sequence they are present in biological fluids, including respiratory system mucus and saliva of healthy individuals, probably as a result of passive secretion during desquamation (Cataltepe et al., 2000). The expression of SCCA serpins is regulated by pro-inflammatory cytokines and they participate in the regulation of the immune response to infections and restoration of homeostasis via control of proteolysis in inflamed epithelial tissues (Cataltepe et al., 2000; Suminami et al., 2001).
Despite the high degree of amino acid sequence identity (92%), SCCA serpins differ in their inhibitory specificity. SCCA1 targets papain-like cysteine proteases including papain and cathepsins S, K, and L, whereas SCCA2 interacts with cathepsin G, mast cell chymase and house dust mite allergens (Der p1 and Der f1), which are also proteases (Schick et al., 1997; Schick et al., 1998; Sakata et al., 2004). Although the archetypical suicide substrate mechanism of inhibition was initially suggested (Schick et al., 1997; Schick et al., 1998), subsequent reports implied an alternative non-covalent interaction model, unusual for the serpin family of inhibitors, as a mechanism of inhibition of the cysteine cathepsins by the SCCA serpins (Masumoto et al., 2003; Sakata et al., 2004).
The broad spectrum of inhibited proteases of two catalytic classes suggests that the SCCA serpins might play a role in the control of bacterially-derived proteases such as those produced by Staphylococcus aureus, a frequent pathogen of epithelial surfaces. S. aureus secretes two papain-like cysteine proteases of the papain-like fold (family C47 of clan CA of cysteine peptidases). These enzymes can directly or indirectly damage the epithelium and underlying connective tissue. Specifically, ScpA not only exerts strong elastinolytic activity but it also degrades fibrinogen, fibronectin and high molecular weight kininogen; furthermore, it inactivates α-1-protease inhibitor and α-1-antichymotrypsin (Potempa et al., 1986; Potempa et al., 1988; Massimi et al., 2002). By contrast, SspB has been shown to interact with cells of the host immune system. Through shedding of CD31 from the neutrophil surface, SspB might affect the clearance of apoptotic neutrophils at sites infected by S. aureus and thus disturb the re-establishment of homeostasis in the inflamed tissue (Smagur et al., 2009a).
In the context of the tissue-damaging, pro-inflammatory activity of staphopains, it is clear that their local inhibition by the extracellular SCCA serpins could have beneficial effects. Consequently, we investigated interactions between the SCCA serpins and staphopains and found that SCCA1 is a potent inhibitor of both staphopains. The kinetic parameters of the inhibitory complex formation strongly suggest that this reaction might occur in vivo.
Results
Stoichiometry of inhibition of staphopains by the SCCA serpins
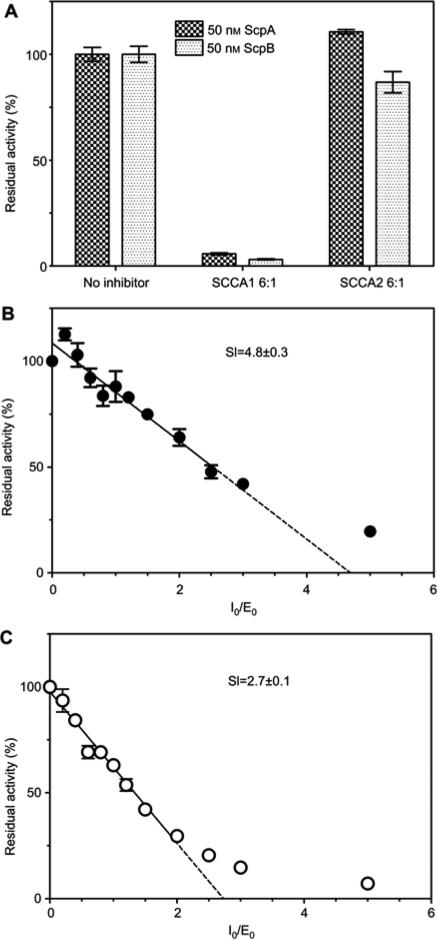
At a 6-fold molar excess, SCCA2 did not affect ScpA activity and only weakly inhibited SspB, whereas SCCA1 effectively inhibited both staphopains. The significant difference in residual activity of the two enzymes in the presence of SCCA1 indicated that SCCA1 is a more efficient inhibitor of SspB than ScpA (Figure 1A). To further investigate the interaction between staphopains and SCCA1, we determined the stoichiometry of inhibitory complex formation, that is, the number of SCCA1 molecules necessary for formation of a stable staphopain-SCCA1 inhibitory complex (Figure 1). The analysis revealed a stoichiometry index (SI) of 4.8±0.3 for ScpA, compared to only 2.7±0.1 for SspB. This difference explains the more efficient inhibition of SspB by SCCA1 when compared to ScpA.
Figure 1.
Determination of SCCA1-staphopains interaction stoichiometry. (A) Inhibition of staphopains by GST-SCCA1. Staphopains were preincubated with six molar excess of GST-SCCA1 or GST-SCCA2 for 30 min at 37°C before substrate (suspension of Azocoll) was added. The residual staphopain activity was determined as an increase of absorbance at λ=520 nm after 30 min of incubation with the substrate. Results were normalized as a percentage of the uninhibited control. Bars represent mean±SEM from three experiments. (B, C) Stoichiometry of GST-SCCA1 – staphopain A (B) and staphopain B (C) inhibition. Increasing amounts of GST-SCCA1 were preincubated for 30 min with a constant concentration of staphopain, resulting in molar ratios of the inhibitor to enzyme in the range of 0–5. The residual enzyme activity was measured using fluorescent substrates specific for each staphopain. The data were plotted as the residual activity (Vi/V0) vs. the inhibitor:enzyme molar ratio. The stoichiometry of inhibitory complex formation (SI) was determined by linear regression to the initial points of inhibitory curves. The results presented are mean±SEM from three experiments.
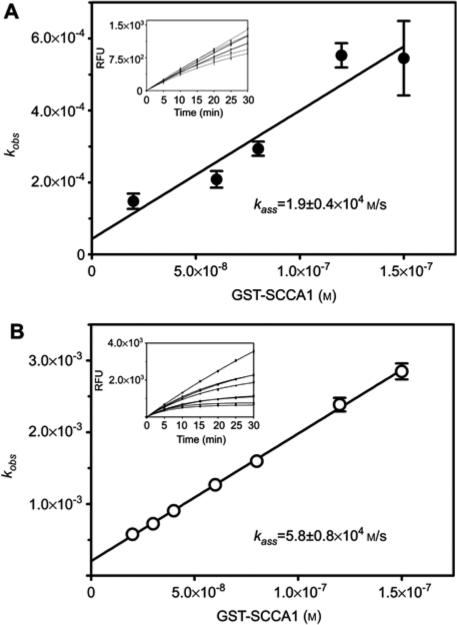
Apart from the stoichiometry, physiological efficacy of serpin as an inhibitor of a targeted protease is dependent on the rate of stable complex formation. Therefore, to determine the potential biological relevance of staphopain inhibition by SCCA1, the kinetics of interaction of this serpin with staphopains was investigated. Using progress curve analysis of the inhibitory reaction under pseudo-first order conditions (Morrison and Walsh, 1988), the kass value was determined as 1.9±0.4×104 m/s and 5.8±0.8×104 m/s for ScpA and SspB inhibition by SCCA1, respectively (Figure 2).
Figure 2.
Determination of the secondary rate constant (kass) of staphopains inhibition by GST-SCCA1.
Constant amounts of staphopain A (panel A) and staphopain B (panel B) were added to samples containing increasing concentrations of GST-SCCA-1 and a fixed amount of substrate. The samples were mixed immediately and the time dependent increase in fluorescence due to substrate hydrolysis was recorded (insets). The data were analyzed by the progress curve method and obtained values of kobs were plotted against inhibitor concentration in the sample (panels A and B). The secondary rate constant was determined by linear regression to the data points and corrected for substrate competition and the stoichiometry factor. The data represent results from triplicate experiments±SD.
Mechanism of staphopains inhibition by SCCA1
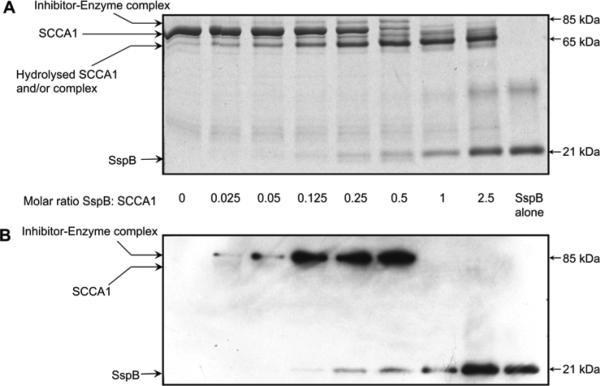
The reaction of serpin with a target serine protease results in the formation of a covalent complex in which the enzyme and inhibitor are linked by an ester bond. Such complexes are resistant to separation during SDS-PAGE and stable during boiling in reducing conditions. Cysteine proteases significantly differ in that regard, as the thioester bond formed between catalytic cysteine and an inhibitor are labile and highly sensitive to reducing conditions. Therefore, to prevent the breakdown of the complex, the reaction mixtures containing a constant amount of SCCA1 and an increasing concentration of staphopains preincubated for 30 min were treated with non-reducing SDS-PAGE sample buffer at 37°C in the presence of 0.1 mm E-64 and then resolved by SDS-PAGE. The analysis revealed a characteristic banding pattern (Figure 3). The intensity of the band corresponding to intact
Figure 3.
Detection of the SDS-stable GST-SCCA1-SspB inhibitory complex.
To identify the SDS-stable complex of GST-SCCA1-SspB, increasing concentrations of SspB were incubated together with a constant amount of GST-SCCA1 resulting in molar ratios of the inhibitor to the enzyme in the range of 1:0.025–1:2.5. (A) Coomasie G-250-stained gels. (B) Western blot analysis using the mouse monoclonal anti-SspB antibody.
GST-SCCA1 (~70 kDa) gradually decreased proportionally to the SspB concentration and practically disappeared at a 1:1 molar ratio. This change correlated with a proportional increase in intensity of a 65 kDa band apparently corresponding to GST-SCCA1 cleaved within the RLS. Most notably, however, the band with molecular weight of approximately 85 kDa appeared in a manner proportionate to the SspB concentration. The intensity of this band was highest at an optimal SI of SspB-SSCA1 interaction (between 1:2 and 1:4), as determined above. At higher enzyme concentrations, the band disappeared. The molecular mass of the band (85 kDa) suggested that it represents the SDS-stable complex composed of SspB (21 kDa) and RLS-truncated GSTSCCA1 (65 kDa) (Figure 3A).
To confirm the presence of staphopain in the 85 kDa complex, we performed Western blot analysis using a specific mouse monoclonal anti-SspB antibody. The 85 kDa band strongly reacted with the mAb indicating the presence of SspB in the complex (Figure 3B). The intensity of the reaction correlated with the Coomassie Blue-stained band (Figure 3A) and was highest in the sample which had a 1:2 molar mixture of SspB and SCCA1. This result unambiguously confirmed the presence of SspB in the higher molecular weight product resulting from the SspB-SCCA1 interaction, thus demonstrating the formation of a SDS-stable serpin-enzyme complex.
As can be expected for the mechanism of protease inhibition by serpins, the complex formation between SspB and SCCA1 was accompanied with the release of a 4.5 kDa peptide (Figure 4) due to the reactive site loop (RSL) cleavage of the inhibitor by the protease. Edman degradation analysis of the peptide yielded the same N-terminal sequence of Ser-Ser-Pro-Thr-Ser-Thr-Asn for SCCA1 interacting with both SspB and ScpA, indicating that both enzymes cleaved the peptide bond between Gly354-Ser355. This bond was identified as the P1-P1’ peptide bound of the inhibitor reactive site in the same manner as for the inhibition of cathepsins (Schick et al., 1998).
Figure 4.
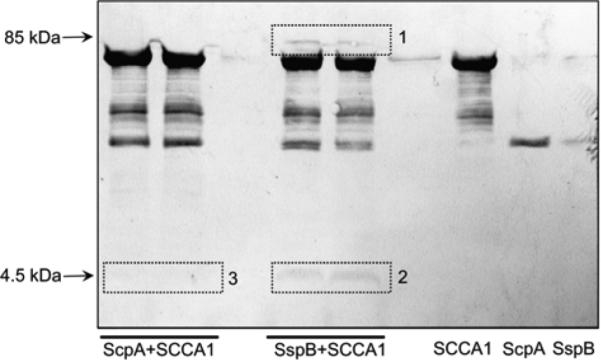
Determination of the cleavage site at the RSL during SCCA1 interaction with staphopains.
The staphopains were preincubated with SCCA1, the reaction was stopped by the addition of non-reducing sample buffer and proteins were separated by SDS-PAGE and subsequently electrotransferred onto PVDF membrane. Bands stained with CBB G250 corresponding to the SCCA1-SspB complex (box 1) and the released C-terminal peptide (box 2 and 3) were excised and subjected to N-terminally sequence analysis.
Discussion
For years serpins have been assigned a potential role in controlling the activity of proteases from microbial pathogens, however, no data have yet been published to support this. On the contrary, numerous reports show that serpins are readily inactivated by microbial proteases (Potempa and Pike, 2009). Accordingly, we present here the interaction of the S. aureus cysteine protease, staphopains, with epithelial-origin SCCA1, which is to our knowledge the first ever described example of efficient inhibition of pathogen-derived proteases by human serpin. With kass in the range of 104 m/s, the inhibition of the S. aureus cysteine protease, SCCA1 might be fast enough to efficiently abrogate staphopain activity in vivo.
Although SCCA1 lacks a signal sequence and is present in the cytoplasm of cells of epithelial origin, substantial amounts of this inhibitor are found in extracellular locations, resulting from the keratinisation and desquamation of epithelial cells (Kato, 1996; Izuhara et al., 2008). Therefore, this inhibitor might play an essential role in restricting staphopain activity at three different levels of host colonization by S. aureus. On the epithelial cell surface, SCCA1 can protect essential connective tissue components, including elastin and collagen, from degradation by the staphopains (Potempa et al., 1988; Ohbayashi et al., 2011). If the epithelium is compromised and S. aureus invades subepithelial tissues, SCCA1 can prevent homeostasis disruption in this extracellular environment by inhibiting staphopain-dependent kinin generation (Imamura et al., 2005) and protect immune cells from the damaging activity of staphopains (Smagur et al., 2009a; Smagur et al., 2009b). Finally, SCCA1 could play an important role in S. aureus growth inside macrophages (Kubica et al., 2008; Koziel et al., 2009), epithelial cells (Balwit et al., 1994) and keratinocytes (Mempel et al., 2002). For example, it has been shown that overexpression of SCCA1 in epithelial cells protects against damage-induced apoptosis, possibly by inhibition of cathepsins leaking into the cytoplasm (Kato et al., 1987; Kato, 1996; Suminami et al., 2001; Pontisso et al., 2004). Therefore, it is tempting to speculate that the consumption of intracellular SCCA1 by staphopain secreted within the cell could allow S. aureus to hijack the apoptosis regulation system, allowing initial proliferation of S. aureus within epithelial cells. This would then be followed by the induction of apoptosis, leading to subsequent dissemination of invading bacteria (Kahl et al., 2000).
SCCA1 was previously reported to inhibit target proteases, cysteine cathepsins, via formation of a non-covalent enzyme-inhibitor complex (Masumoto et al., 2003; Sakata et al., 2004). Our results challenge this contention strongly, arguing for the typical serpins suicide substrate mechanism for the SCCA1-SspB interaction followed by formation of an 85 kDa covalent inhibitory complex which is stable in SDS-PAGE. Significantly, complex formation was accompanied by the release of a C-terminal, approximately 4.5 kDa, peptide generated by the cleavage of the RSL at the Gly354-Ser355 peptide bond, identified previously as the P1-P19 residues for SCCA-1 interaction with human cathepsins (Schick et al., 1998). The covalent mode of staphopain inhibition by the SCCA serpins is further supported by the finding that the cysteine protease inhibiting serpin, MENT (Irving et al., 2002a) and an antitrypsin/SCCA-1 chimera (Irving et al., 2002b) also form classic ‘serpin-like’ covalent complexes with cysteine proteases.
Notably, the sequence (Phe-Gly-Ser-Ser) at the SCCA-1 RSL is strikingly similar to those in the reactive site of the staphostatins (Leu-Gly-Thr-Ser and Ile-Gly-Thr-Ser for staphostatin A and B, respectively) (Figure 5), which are natural inhibitors of the staphopain enzymes (Rzychon et al., 2003b). This observation suggests the adaptation of the SCCA-1 serpin for inhibition of staphylococcal cysteine proteases. In this context, it is important to emphasize that staphopain genes are strictly conserved in all clinical isolates of S. aureus characterized to date (Karlsson and Arvidson, 2002). Taking into account that SCCA1 is predominantly expressed in epithelial tissues including respiratory pathways, hair follicles and skin (Kato, 1996); all of which are regularly colonized by S. aureus, the physiological relevance of SCCA1-staphopain B interaction as a defense mechanism seems to be very well substantiated.
Figure 5.
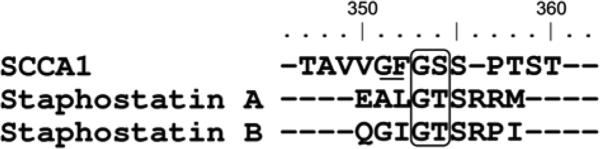
Alignment of the reactive site loop sequences of SCCA1 and staphostatins with the positions of the primary P1 and P1’ residues at cleavage sites by target proteases.
Staphopains (all sequences) and cathepsin S (SCCA1) are boxed (Schick et al., 1998; Filipek et al., 2003; Rzychon et al., 2003a). The secondary, inactivating cleavage site identified for papain in SCCA1 RSL is underlined (Masumoto et al., 2003). Residues are numbered according to the SCCA1 amino acid sequence.
Taken together, our results reveal a novel, potentially physiologically important, interaction of an epithelial serpin and two S. aureus cysteine proteases. It presents controversial findings with regard to the mechanism of cysteine pro-tease inhibition by SCCA1 and also lays the groundwork for further investigations of S. aureus survival in the human host.
Materials and methods
Staphopains
Staphylococcal enzymes were purified according to procedures described previously (Drapeau, 1978; Potempa et al., 1988; Massimi et al., 2002). ScpA was active site titrated using human α2-macro-globulin, and staphostatin was used to calibrate the concentration of active SspB (Filipek et al., 2003; Rzychon et al., 2003a; Rzychon et al., 2003b). Therefore, the concentrations of staphopains used in this work refer not to the protein concentration but to the concentration of the active enzymes.
Purification of GST-SCCA1 and GST-SCCA2 fusion proteins
The recombinant GST-SCCA1 and GST-SCCA2 fusion proteins were synthesized in a bacterial expression system and purified using glutathione-Sepharose 4B beads (Amersham Biosciences, Piscataway, NY, USA) using modifications of previously described methods (Schick et al., 1997; Pak et al., 2004). Briefly, 50 ng of the pGEX-SCCA1 (or pGEX-SCCA2) plasmid was transformed into competent Escherichia coli BL21 cells. After overnight growth at 37°C on LB agar plates supplemented with 100 μg/ml ampicillin, transformants were washed off using 5 ml of LB broth. The culture was then used to inoculate 1 1 of LB broth supplemented with 100 μg/ml ampicillin and allowed to grow at 37°C until the culture reached an OD of 0.5 at 600 nm. Following a 5 min incubation period in an ice bath, protein expression was induced by the addition of isopropyl-1-thio-β-D-galactopyranoside to a final concentration of 0.5 mM and incubated at 25°C for 4 h. Cultures were harvested by centrifugation at 5000 g for 10 min and lysed using 60 ml of lysis buffer (100 mM NaCl, 100 mM Tris-HCl, pH 8.0, 50 mm EDTA, 2% Triton X-100, 1.5 mg/ml lysozyme and protease inhibitor cocktail). The bacterial lysate was clarified by centrifugation at 12 000 g for 20 min. Forty-eight milliliters of supernatant was transferred to a fresh tube containing 2 ml of 50% glutathione-Sepharose 4B beads and incubated for 30 min at 4°C to facilitate binding. The beads were collected by centrifugation at 500 g and washed twice in lysis buffer (minus lysozyme) and twice in PBST (10 mm phosphate buffer, 27 mM KCl, 137 mM NaCl, 0.1% Tween 20, pH 7.4). The GST-SCCA1 and GST-SCCA2 proteins were eluted from the beads using three 1 ml washes of glutathione elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl, pH 8.0). GST-SCCA concentrations in the eluates were determined by Bradford analysis and protein purity was checked by SDS-PAGE analysis.
GST-SCCA-1/GST-SCCA-2: staphopain A and B inhibition assays
In preliminary experiments, the staphopains A and B were activated for 15 min in freshly prepared 0.1 M Tris-HCl, 5 mm EDTA, pH 7.6, 2 mM DTT at 37°C. The activated enzymes were mixed with SCCA serpin solutions at final concentrations of 50 nm enzyme and 300 nM serpin (1:6 molar ratio) in a total volume of 200 μl. After 30 min at 37°C 100 νl of azocoll (Calbiochem, San Diego, CA, USA) suspension (15 mg/ml in 0.5 m sucrose, 0.05% Tween-20) was added to each sample. Mixtures were incubated at 37°C with shaking for 30 min, undigested azocall was removed by centrifugation and the absorbance of the supernatant at 520 nm was measured using a SpectraMAX microplate reader (Molecular Devices, Sunnyvale, CA, USA). The activity of the enzyme incubated in parallel without the inhibitor was taken as 100% and the inhibitory effect of the SCCA serpins on staphopains was shown as the mean of residual activity (±SD) calculated from experiments performed in triplicate.
Stoichiometry of staphopain A and B inhibition by SCCA1
Preactivated staphopains were mixed with SCCA1 on a black micro-plate (Nunc, Roskilde, Denmark) to yield a final concentration of the inhibitor in the range of 0–200 nm, and 40 nM enzyme in a total volume of 100 μl of 0.1 m Tris, pH 7.6, 5 mM EDTA, 2 mM DTT. The plate was incubated for 30 min at 37°C and the residual enzyme activity was assayed with Abz-Glu-Ala-Leu-Gly-Thr-Ser-Pro-Arg-Lys(Dnp)-Asp-OH (for SspB) and Abz-Glu-Gly-Ile-Gly-Thr-Ser-Arg-Pro-Lys(Dnp)-Asp-OH (for ScpA) at 5 μm and 10 μm final concentrations, respectively in a total volume of 200 μl. The enzymatic hydrolysis of the substrates was monitored for 30 min at 37°C using a SpectraMAX Gmini XS (Molecular Devices) microplate fluorimeter with the excitation and emission wavelengths set at 320 and 420 nm, respectively. The initial velocity of substrate hydrolysis (Vmax) was used to calculate the residual enzyme activity (mean±SD) as a percentage of the activity (Vmax) of uninhibited staphopain and plotted vs. the serpin/staphopain molar ratio. The stoichiometry of inhibition was determined by the linear regression of the experimental data points from three independent experiments.
Determination of kinetic parameters of inhibition by the progress curve method
Mixtures of constant (20 μm) substrate concentration (Abz-Glu-Ala-Leu-Gly-Thr-Ser-Pro-Arg-Lys(Dnp)-Asp-OH for ScpA for ScpA and 20 μm Abz-Glu-Gly-Ile-Gly-Thr-Ser-Arg-Pro-Lys(Dnp)-Asp-OH for SspB) with increasing concentrations of SCCA1 in the range of 0-300 nM were prepared in 96-well black microplates (Nunc) in a total volume of 100 μl. Then, 100 μl of the staphopains preactivated in 0.1 m Tris, 5 mm EDTA, 2 mm DTT, pH 7.6 for 15 min at 37°C, were added (0.5 nM final enzyme concentration), and the rate of substrate hydrolysis was recorded as the increase of fluorescence (λexs320 nm, λems420 nm) for 60 min using a micro-plate fluorescence reader SpectraMAX Gmini XS. Data were analyzed by non-linear fitting to the progress curve for the irreversible inhibition model, described by equation 1 (Morrison and Walsh, 1988):
| (1) |
Where vz denotes the initial velocity of the reaction, kobs is the apparent association constant and t is time. kobs values determined for each inhibitor concentration were plotted against the inhibitor concentration and the apparent association constant k’ was determined from the slope of a line fitted by linear regression. As an inhibitor in the solution competes with a substrate for a binding site and the reaction velocity is affected by the enzyme-inhibitor SI, the final kass value was calculated from k’ using equation 2 (Morrison and Walsh, 1988):
| (2) |
Where SI stands for the stoichiometry index and Km is a Michaelis constant for a given substrate. The Km for Abz-Glu-Ala-Leu-Gly-Thr-Ser-Pro-Arg-Lys(Dnp)-Asp-OH and for Abz-Glu-Gly-Ile-Gly-Thr-Ser-Arg-Pro-Lys(Dnp)-Asp-OH hydrolysis by ScpA and SspB was determined to be 79.4 μm and 53 μm, respectively. The results presented show the averaged data from three independent experiments.
SDS-PAGE and Western blot analysis of the interaction between SspB and SCCA1
For the detection of the SspB-SCCA1 complex, increasing concentrations of preactivated staphopain B (0–5 μm) were incubated with 2 μm SCCA1 in separate eppendorf tubes for 30 min at 37°C. The reaction was stopped by the addition of SDS-PAGE sample buffer (1:1 v/v) supplemented with 100 μm E-64 (Sigma-Aldrich, St. Louis, MO, USA) but without reducing agent. Mixtures were incubated for a further 30 min at 37°C and were then resolved on a 10% SDS-PAGE gel (T:C ratio, 29:1). After electrophoresis the gel was stained with Coomassie brilliant blue G-250 (Serva, Heidelberg, Germany). For Western blot analysis, SDS-PAGE resolved proteins were electrotransferred in 25 mM Tris, 192 mM glycine and 20% methanol in a semi-dry Western blot chamber (Bio-Rad Life Science Research, 2000 Alfred Nobel Drive, Hercules, CA 94547, USA) onto PVDF membrane (Millipore, Billerica, MA, USA). Non-specific binding sites were blocked overnight in a solution containing 2% bovine serum albumin (Lab Empire, Rzeszow, Poland) and 10% goat serum and the membrane was then incubated with the primary mouse monoclonal anti-SspB antibodies (clone 5E12.G9, UGA core facility for MAb development, University of Georgia, Athens, GA, USA) for 3 h at room temperature. Following a washing step with TBS-Tween, secondary goat anti-mouse IgG Fc fragment antibodies (Sigma-Aldrich) were applied for 1 h. The membrane was developed using ECL+ substrate (GE Healthcare, Little Chalfont, UK) and Kodak Biofilm plate (Eastman Kodak, Rochester, NY, USA).
N-terminal sequencing of staphopain-GST-SCCA1-derived fragments
Preactivated staphopains (1.41 μm) were mixed with 7.04 μm GSTSCCA1 (1:5 molar ratio) in 20 μl of 0.1 m Tris, 5 mM EDTA, 2 mm DTT, pH 7.6. The control samples, containing the same concentration of each staphopain and GST-SCCA1, were prepared separately in the same manner. All mixtures were incubated for 30 min at 37°C and the reaction was stopped by the addition of non-reducing SDS-PAGE sample buffer supplemented with 100 μm E-64 and samples were resolved on a peptide gel using Schagger and von Jagow tricine electrophoretic system (Schagger and Von Jagow, 1987) and electrotransferred onto polyvinylidene difluoride membranes in 10 mM CAPS, 10% methanol, pH 11 buffer using semi-dry blotting chamber. Protein bands were visualized by Coomassie brilliant blue staining, excised and analyzed by automated Edman degradation using a Procise 494HT amino acid sequencer (Applied Biosystems, Carlsbad, CA, USA).
Acknowledgments
This study was supported in part by the Jagiellonian University statutory funds (DS/9/WBBiB) and grant DK081422 from the NIH to G.A.S. J.P and T.K. We acknowledge support from the Foundation for Polish Science (TEAM project DPS/424-329/10). Finally, we are indebted to Drs. Les N. Shaw and Ky-Anh Nguyen (University of South Florida, Tampa, FL, USA and Westmead Centre for Oral Health, Sydney, Australia, respectively) for critical reading of the manuscript. The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficent of the structural funds from the European Union (grant No: POIG.02.01.00-12-064/08 – ‘Molecular biotechnology for health’).
References
- Balwit JM, van Langevelde P, Vann JM, Proctor RA. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 1994;170:1033–1037. doi: 10.1093/infdis/170.4.1033. [DOI] [PubMed] [Google Scholar]
- Cataltepe S, Gornstein ER, Schick C, Kamachi Y, Chatson K, Fries J, Silverman GA, Upton MP. Co-expression of the squamous cell carcinoma antigens 1 and 2 in normal adult human tissues and squamous cell carcinomas. J. Histochem. Cytochem. 2000;48:113–122. doi: 10.1177/002215540004800112. [DOI] [PubMed] [Google Scholar]
- Drapeau GR. Role of metalloprotease in activation of the precursor of staphylococcal protease. J. Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek R, Rzychon M, Oleksy A, Gruca M, Dubin A, Potempa J, Bochtler M. The staphostatin-staphopain complex: a forward binding inhibitor in complex with its target cysteine protease. J. Biol. Chem. 2003;278:40959–40966. doi: 10.1074/jbc.M302926200. [DOI] [PubMed] [Google Scholar]
- Imamura T, Tanase S, Szmyd G, Kozik A, Travis J, Potempa J. Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J. Exp. Med. 2005;201:1669–1676. doi: 10.1084/jem.20042041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Shushanov SS, Pike RN, Popova EY, Brömme D, Coetzer TH, Bottomley SP, Boulynko IA, Grigoryev SA, Whisstock JC. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J. Biol. Chem. 2002a;277:13192–131201. doi: 10.1074/jbc.M108460200. [DOI] [PubMed] [Google Scholar]
- Irving JA, Pike RN, Dai W, Brömme D, Worrall DM, Silverman GA, Coetzer TH, Dennison C, Bottomley SP, Whisstock JC. Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering a1-antitrypsin to inhibit cathepsin proteases. Biochemistry. 2002b;41:4998–5004. doi: 10.1021/bi0159985. [DOI] [PubMed] [Google Scholar]
- Izuhara K, Ohta S, Kanaji S, Shiraishi H, Arima K. Recent progress in understanding the diversity of the human ov-serpin/clade B serpin family. Cell Mol. Life Sci. 2008;65:2541–2553. doi: 10.1007/s00018-008-8049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 2000;68:5385–5392. doi: 10.1128/iai.68.9.5385-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A, Arvidson S. Variation in extracellular pro-tease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 2002;70:4239–4246. doi: 10.1128/IAI.70.8.4239-4246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. 1996;16:2149–2153. [PubMed] [Google Scholar]
- Kato H, Torigoe T. Radioimmunoassay for tumor antigen of human cervical squamous cell carcinoma. Cancer. 1977;40:1621–1628. doi: 10.1002/1097-0142(197710)40:4<1621::aid-cncr2820400435>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Kato H, Suehiro Y, Morioka H, Torigoe T, Myoga A, Sekiguchi K, Ikeda I. Heterogeneous distribution of acidic TA-4 in cervical squamous cell carcinoma: immunohistochemical demonstration with monoclonal antibodies. Jpn. J. Cancer Res. 1987;78:1246–1250. [PubMed] [Google Scholar]
- Koziel J, Maciag-Gudowska A, Mikolajczyk T, Bzowska M, Sturdevant DE, Whitney AR, Shaw LN, DeLeo FR, Potempa J. Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One. 2009;4:e5210. doi: 10.1371/journal.pone.0005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, et al. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 2008;3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel M, Schnopp C, Hojka M, Fesq H, Weidinger S, Schaller M, Korting HC, Ring J, Abeck D. Invasion of human keratinocytes by Staphylococcus aureus and intracellular bacterial persistence represent haemolysin-independent virulence mechanisms that are followed by features of necrotic and apoptotic keratinocyte cell death. Br. J. Dermatol. 2002;146:943–951. doi: 10.1046/j.1365-2133.2002.04752.x. [DOI] [PubMed] [Google Scholar]
- Massimi I, Park E, Rice K, Muller-Esterl W, Sauder D, McGavin MJ. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J. Biol. Chem. 2002;277:41770–41777. doi: 10.1074/jbc.M207162200. [DOI] [PubMed] [Google Scholar]
- Masumoto K, Sakata Y, Arima K, Nakao I, Izuhara K. Inhibitory mechanism of a cross-class serpin, the squamous cell carcinoma antigen 1. J. Biol. Chem. 2003;278:45296–45304. doi: 10.1074/jbc.M307741200. [DOI] [PubMed] [Google Scholar]
- Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- Ohbayashi T, Irie A, Murakami Y, Nowak M, Potempa J, Nishimura Y, Shinohara M, Imamura T. Degradation of fibrinogen and collagen by staphopains, cysteine pro-teases released from Staphylococcus aureus. Microbiology. 2011;157:786–792. doi: 10.1099/mic.0.044503-0. [DOI] [PubMed] [Google Scholar]
- Pak SC, Kumar V, Tsu C, Luke CJ, Askew YS, Askew DJ, Mills DR, Brömme D, Silverman GA. SRP-2 is a cross-class inhibitor that participates in postembryonic development of the nematode Caenorhabditis elegans: initial characterization of the clade L serpins. J. Biol. Chem. 2004;279:15448–15459. doi: 10.1074/jbc.M400261200. [DOI] [PubMed] [Google Scholar]
- Pontisso P, Calabrese F, Benvegnu L, Lise M, Belluco C, Ruvoletto MG, Marino M, Valente M, Nitti D, Gatta A, Fassina G. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br. J. Cancer. 2004;90:833–837. doi: 10.1038/sj.bjc.6601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Watorek W, Travis J. The inactivation of human plasma a1-proteinase inhibitor by proteinases from Staphylococcus aureus. J. Biol. Chem. 1986;261:14330–14334. [PubMed] [Google Scholar]
- Potempa J, Dubin A, Korzus G, Travis J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J. Biol. Chem. 1988;263:2664–2667. [PubMed] [Google Scholar]
- Rzychon M, Filipek R, Sabat A, Kosowska K, Dubin A, Potempa J, Bochtler M. Staphostatins resemble lipocalins, not cystatins in fold. Protein Sci. 2003a;12:2252–2256. doi: 10.1110/ps.03247703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzychon M, Sabat A, Kosowska K, Potempa J, Dubin A. Staphostatins: an expanding new group of proteinase inhibitors with a unique specificity for the regulation of stapho-pains, Staphylococcus spp. cysteine proteinases. Mol. Microbiol. 2003b;49:1051–1066. doi: 10.1046/j.1365-2958.2003.03613.x. [DOI] [PubMed] [Google Scholar]
- Sakata Y, Arima K, Takai T, Sakurai W, Masumoto K, Yuyama N, Suminami Y, Kishi F, Yamashita T, Kato T, et al. The squamous cell carcinoma antigen 2 inhibits the cysteine proteinase activity of a major mite allergen, Der p1. J. Biol. Chem. 2004;279:5081–5087. doi: 10.1074/jbc.M311585200. [DOI] [PubMed] [Google Scholar]
- Schagger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J. Biol. Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, Gornstein ER, Bromme D, Chapman HA, Silverman GA. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, Irving JA, Bird PI. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell. Mol. Life Sci. 2004;61:301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagur J, Guzik K, Bzowska M, Kuzak M, Zarebski M, Kantyka T, Walski M, Gajkowska B, Potempa J. Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol. Chem. 2009a;390:361–371. doi: 10.1515/BC.2009.042. [DOI] [PubMed] [Google Scholar]
- Smagur J, Guzik K, Magiera L, Bzowska M, Gruca M, Thøgersen IB, Enghild JJ, Potempa J. A new pathway of staphylococcal pathogenesis: apoptosis-like death induced by Staphopain B in human neutrophils and monocytes. J. Innate Immun. 2009b;1:98–108. doi: 10.1159/000181014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suminami Y, Nagashima S, Murakami A, Nawata S, Gondo T, Hirakawa H, Numa F, Silverman GA, Kato H. Suppression of a squamous cell carcinoma (SCC)-related serpin, SCC antigen, inhibits tumor growth with increased intra-tumor infiltration of natural killer cells. Cancer Res. 2001;61:1776–1780. [PubMed] [Google Scholar]