Abstract
Background
RNA-binding protein Translocated in LipoSarcoma/FUsed Sarcoma (TLS/FUS) is one of causative genes for familial amyotrophic lateral sclerosis (ALS). We previously identified that TLS was associated with protein arginine methyltransferase 1 (PRMT1), and four arginine residues within TLS (R216, R218, R242 and R394) were consistently dimethylated. Protein arginine methylation is involved in various cellular events such as signal transduction, transcriptional regulation and protein-protein interactions.
Results
To understand the biological role of arginine methylation of RNA-binding protein, we prepared and characterized a mouse monoclonal antibody against asymmetric dimethylarginine of TLS. By cloning and screening, one stable hybridoma cell clone (2B12) producing anti-asymmetric dimethylated TLS on R216 and R218 antibody was established. The monoclonal antibody 2B12 is specific for the asymmetrically dimethylated arginine peptide and does not react with the same peptide sequence containing unmodified and symmetrically dimethylated arginine residues by dot-blot analysis. 2B12 was also validated GST tagged TLS with PRMT1 by in vitro arginine methylation assays. Since methylated TLS in HeLa cells and mouse and human brain protein extracts was immunoprecipitated with 2B12, we performed RNA-binding protein immunoprecipitation assays using HeLa cell lysate and this antibody. We demonstrated that the long noncoding RNA (lncRNA) transcribed from cyclin D1 promoter binds methylated TLS.
Conclusions
A monoclonal antibody that is capable of detecting the methylarginine status of TLS will facilitate the molecular and cellular analysis of transcriptional regulation by lncRNA through methylated TLS, and can be used as a favorable tool for clinical diagnosis of ALS caused by TLS dysregulation.
Electronic supplementary material
The online version of this article (doi:10.1186/2045-3701-4-77) contains supplementary material, which is available to authorized users.
Keywords: TLS/FUS, Arginine methylation, RNA-binding protein, Long noncoding RNA, Monoclonal antibody
Background
Translocated in LipoSarcoma/FUsed in Sarcoma (TLS/FUS) was originally identified in malignant liposarcoma as a part of the chimeric fusion protein TLS-CHOP [1]. Recently, it was reported that TLS is one of causative genes for familial amyotrophic lateral sclerosis (ALS) [2, 3]. TLS is also implicated in various cellular programs such as transcription, RNA processing and DNA repair [4]. We have demonstrated that the long noncoding RNAs (lncRNAs) transcribed from the cyclin D1 (CCND1) promoter (promoter-associated noncoding RNAs: pncRNAs) bind TLS and inhibit the histone acetyltransferase activities to repress the transcription of CCND1 gene [5]. Recent studies reveal that lncRNAs regulate the transcription of target genes [6]. The precise mechanisms of transcriptional regulation by lncRNAs, however, are still unclear.
Arginine methylation is one of posttranslational modifications, and accomplished by protein arginine methyltransferases (PRMTs). Arginine residues can be monomethylated or dimethylated, and dimethylation can be both symmetric (me2s) and asymmetric (me2a). Asymmetric dimethylarginine (aDMA) is catalyzed by the type I class of PRMTs (PRMT1, 3, 4, 6 and 8), and symmetric dimethylarginine (sDMA) is catalyzed by the type II class (PRMT5 and 7). In regarding to histone arginine modification, H4R3me2a and H4R3me2s are basically linked to transcriptional activation and repression, respectively [7, 8]. We have shown that TLS is associated with PRMT1, and four arginine residues within TLS (R216, R218, R242 and R394) are constitutively dimethylated [9]. However, the functional role of arginine methylation of RNA-binding proteins still needs to be studied. RNA-binding proteins often contain glysine-arginine-rich motifs and are considered substrates for PRMTs. In fact, FMRP, EWS, which are also related with diseases, are dimethylated [10, 11]. Therefore, it is believed that methylation of RNA-binding proteins could influence RNA-protein and/or protein-protein interactions.
ALS is a fatal neurodegenerative disease caused by degeneration of motor neurons. Identification of several mutations in the TLS gene from ALS patients suggested that disruption of RNA metabolism might be one of key events in ALS pathogenesis. Interestingly, natural arginine mutation (R216C), one of methylated arginine we identified, of TLS from ALS patients was reported [12]. Moreover, it was an interesting report that the RNA-binding ability of TLS is essential for the neurodegenerative phenotype in vivo of mutant TLS although it was unclear whether direct contact with RNA or through interactions with other RNA-binding proteins [13]. Taken together, these findings suggest that arginine methylation of TLS might play an important role in the lncRNA-dependent transcriptional regulation and the disruption of RNA binding could be implicated in the pathogenesis of ALS.
In this study, we attempt to establish hybridoma cell lines that can stably produce anti-methylated TLS monoclonal antibodies. Here we show one monoclonal antibody (2B12) can specifically recognize arginine-methylation of TLS. Our generated antibody could detect selectively the asymmetrically dimethylated TLS by western blotting. Moreover, 2B12 was suitable for RNA-binding protein immunoprecipitation (RIP) assays to show the interplay between lncRNA and methylated TLS.
Results
Generation of asymmetric dimethylarginine-specific antibody and antibody specificity
We have recently demonstrated that PRMT1 asymmetrically methylates TLS/FUS on arginine (R) residues [9]. Using mass spectrometry, we identified which residues of TLS are methylated in vivo[9]. To investigate the biological role of methylated TLS, we attempted to develop mouse monoclonal antibodies that specifically recognized TLS symmetrically or asymmetrically dimethylated on R216 and R218. We prepared TLS peptides that were contained unmodified, symmetrically modified (me2s), or asymmetrically modified arginines (me2a) at R216 and R218 (Figure 1A). Unmodified peptide was used for producing polyclonal antibody in rabbits, and the antiserum was obtained (hereafter referred as A1). Modified peptides were used for immunization of mice, and hybridoma clones were screened by enzyme-linked immunosorbent assay (ELISA). We obtained a few positive clones. The purified antibody (hereafter referred as 2B12) was selected for further analysis. To access antibody specificity, we tested 2B12 using synthetic peptides by dot-blot analysis. As shown in Figure 1B, A1 reacts with all of synthesized peptides equally. In contrast, the monoclonal antibody 2B12 specifically recognizes the asymmetrically methylated peptide and does not react with the same peptide sequence containing unmodified and symmetrically dimethylated arginine residues by dot-blot analysis (Figure 1C), confirming the specificity of 2B12 for asymmetric arginine methylation of TLS. Unfortunately we were not able to obtain a monoclonal antibody for detecting R216/R218me2s in this study.
Figure 1.
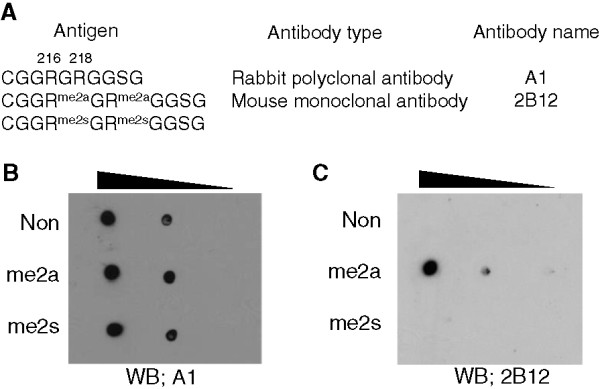
The monoclonal antibody specificity tested by dot-blot analysis. (A) Summary of peptide sequences. Three TLS peptides containing either no modification (Non) or R216/R218me2a (me2a) or R216/R218me2s (me2s) were synthesized. TLS peptide containing no modification was used for producing polyclonal antibody in rabbit, and TLS peptides containing R216/R218me2a or R216/R218me2s were used for the immunization of mice and hydridoma development. (B and C) Antibody specificity was tested by dot-blot analysis. Diluted peptides (B; 0.2, 1, 5 ng, C; 20, 100, 500 ng) were blotted onto the nitrocellulose membrane and the dot-blotted membranes were incubated with a rabbit polyclonal antibody A1 (B) or a mouse monoclonal antibody 2B12 (C). Note that A1 reacted equally with TLS peptides either no modification or symmetrical or asymmetrical dimethylation, and 2B12 recognized only TLS peptide containing asymmetrical dimethylated arginines.
In vitromethylation of TLS
To validate whether 2B12 can detect methylated TLS, we performed in vitro methylation assays by incubating GST tagged TLS (GST-TLS) with protein arginine methyltransferase 1 (PRMT1) as we reported previously [9]. Western blotting using 2B12 was performed, and the signal was detected in GST-TLS methylated by PRMT1 in the presence of S-adenosyl methionine (SAM) (Figure 2). No signal was observed in the absence of methylation (i.e. without SAM) (Figure 2). Interestingly, the interaction between TLS and PRMT1 was enhanced by the methylation of TLS (Figure 2). These results suggest that 2B12 specifically reacts with TLS methylated by PRMT1 (i.e. asymmetrical dimethylation), and methylation of TLS may effect protein-protein interactions.
Figure 2.
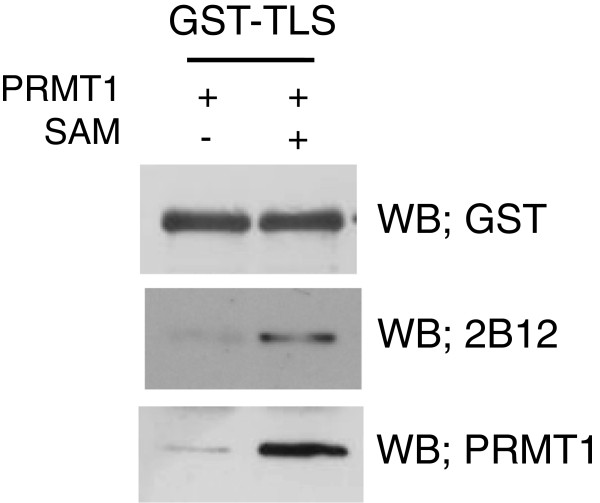
In vitro methylation of the recombinant GST-TLS. GST-TLS was in vitro methylated using PRMT1 in the presence or absence of SAM (20 μM). Reaction products were analyzed by SDS-PAGE followed by western blotting with the indicated antibodies: anti-GST (top), 2B12 (middle), and anti-PRMT1 (bottom). Note that 2B12 specifically reacts with TLS methylated by PRMT1 only in the presence of SAM, and methylated TLS strongly associates with PRMT1.
TLS is arginine methylated in HeLa cells
To examine whether 2B12 can detect methylated TLS in vivo, we carried out immunoprecipitation (IP) experiments on HeLa cells. We should note that TLS was not immunoprecipitated with a rabbit polyclonal antibody A1 (data not shown). Thus, we used a rabbit polyclonal anti-TLS antibody commercially available. To verify the specificity of 2B12, HeLa cells were treated with a methyltransferase inhibitor adenosine-2’,3’-dialdehyde (AdOx) (3 μM) for 24 hours. Reduced recognition of TLS by 2B12 was observed for the AdOx-treated cell extracts, indicating that the treatment significantly reduced TLS methylation and 2B12 specifically recognized methylated TLS (Figure 3A). Somehow unmethylated TLS was immunoprecipitated with TLS polyclonal antibody efficiently although the expression levels of TLS were almost same between control and AdOx-treated cells (Figure 3A). We also assessed if 2B12 could immunoprecipitate methylated TLS in vivo. To test cross reactivity of 2B12, peptide inhibition assays were done. Cell extracts were immunoprecipitated with 2B12 in the presence of competing peptides used for immunization as shown in Figure 1A, and the presence of TLS was revealed using an anti-TLS polyclonal antibody. The immunoprecipitation of 2B12 was clearly inhibited by the excess of R216/R218me2a peptide in a dose-dependent manner, not by other peptides (Figure 3B and Additional file 1), indicating that a monoclonal antibody 2B12 specifically immunoprecipitated asymmetrically dimethylated TLS. These results suggest that 2B12 can be valuable to identify and investigate methylated TLS in vivo.
Figure 3.
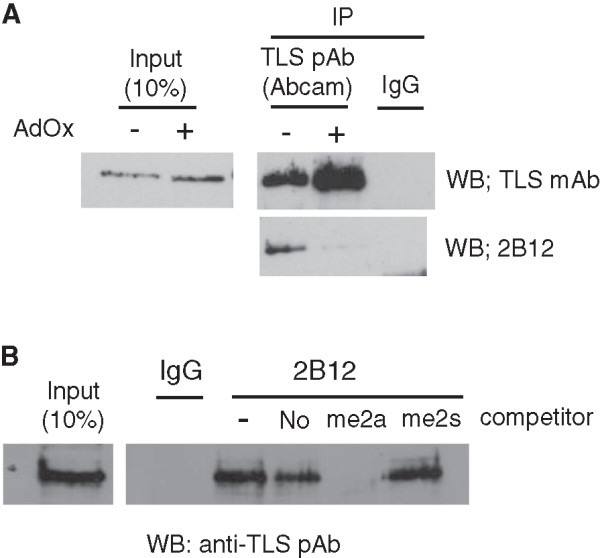
Detection of in vivo methylation of TLS. (A) Endogenous TLS is methylated in HeLa cells. Cell extracts from HeLa treated or not with 3 μM of the general methylation inhibitor AdOx for 24 h were used for immunoprecipitations. The extracts were immunoprecipitated with rabbit normal IgG or rabbit polyclonal anti-TLS antibody. The immunoprecipitated TLS were analyzed by western blotting with 2B12 or mouse monoclonal anti-TLS antibody. The input lane shows 10% of the protein used in each immunoprecipitation. Note that TLS methylation was inhibited by AdOx, and 2B12 specifically recognized methylated TLS. (B) Immunoprecipitation of endogenous methylated TLS from HeLa cell extracts was performed with 2B12 in the presence or absence of competing peptides used for immunization. Bound methylated TLS was eluted with SDS sample buffer resolved by SDS-PAGE, and analyzed by western blotting with rabbit polyclonal anti-TLS antibody.
Assessment of antibody suitability for immunoprecipitation and RIP assays
The antibody for detecting methylated TLS may be a valuable tool for analyzing the ALS pathogenesis caused by TLS dysregulation using IP and the function of TLS methylation in vivo using RNA-binding protein immunoprecipitation (RIP) assays. We have shown that TLS binds the lncRNAs transcribed from CCND1 promoter (CCND1 pncRNAs) [5]. The importance of arginine methylation of TLS for RNA-protein interactions needs to be studied. RIP assay is a powerful technique for studying RNA-binding proteins and their RNA partners. We demonstrated the specificity of 2B12 in Figures 1, 2 and 3. Thus, we carried out IP assays using mouse and human brain samples. 2B12 was able to specifically precipitate methylated TLS from mouse and human brain extracts (Figure 4). We further examined RIP assays using 2B12 for detecting the interplay between methylated TLS and lncRNA. RIP was conducted using HeLa cell lysate and either 2B12 or normal mouse IgG. Purified RNA was then analyzed by RT-PCR using the specific primers for the D region of CCND1 pncRNA (CCND1-pncRNA-D). As shown in Figure 5, PCR product was observed in the input and not in the normal mouse IgG RIP. CCND1-pncRNA-D could be detected in 2B12 RIP by RT-PCR, suggesting that CCND1-pncRNA-D binds methylated TLS in vivo.
Figure 4.
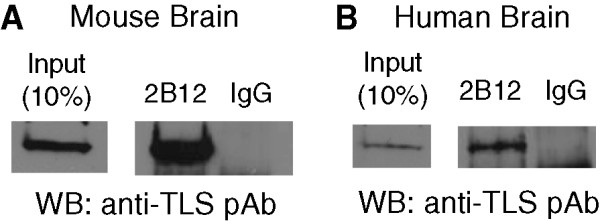
2B12 is suitable for immunoprecipitation analysis. 2B12 was used to immunoprecipitate methylated TLS in the total cell lysate from mouse brain (A) and human brain (B). Bound methylated TLS was eluted with SDS sample buffer resolved by SDS-PAGE, and analyzed by western blotting with rabbit polyclonal anti-TLS antibody. The input lane shows 10% of the protein used in each immunoprecipitation.
Figure 5.
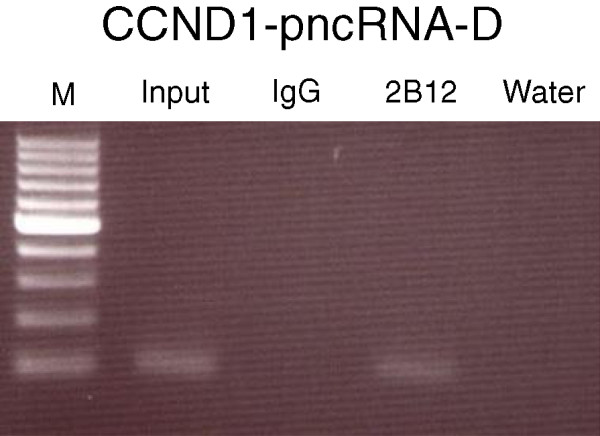
Methylated TLS binds CCND1-pncRNA-D. RIP lysate from HeLa cells were immunoprecipitated using either 2B12 or a normal mouse IgG as a negative control. RNA associated with methylated TLS was purified, and validated by RT-PCR using the specific primers for CCND1-pncRNA-D. The PCR prodcuts were ran on an agarose gel to detect the presence of CCND1-pncRNA-D. The “input” omits the immunoprecipitation step, “IgG” used an IgG antibody for the immunoprecipitation, “2B12” used a 2B12 antibody to pull down methylated TLS, and “water” lane served as a negative control for the PCR reaction. The expected size of PCR product for CCND1-pncRNA-D could be detected in 2B12 RIP. PCR product was observed in the 10% input and not in the normal mouse IgG RIP.
Discussion/conclusions
We previously demonstrated that CCND1 pncRNAs bind to TLS and inhibit the histone acetyltransferase activities to repress the transcription of CCND1 gene [5]. We also identified that four arginine residues within TLS (R216, R218, R242 and R394) were consistently dimethylated by a mass spectrometry [9]. These results suggest that arginine methylation of TLS could have an important role for the transcriptional regulation by lncRNA.
In this study, we attempted to establish hybridoma cell lines that can stably produce anti-methylated TLS monoclonal antibodies by hybridoma technique. By cloning and screening, one mouse monoclonal antibody specific to dimethylated TLS on R216 and R218 was obtained and the hybridoma cell line was named as 2B12. The characteristic of 2B12 was confirmed by dot-blot and western blot analyses (Figures 1 and 2). Methylated TLS is more associated with PRMT1 by in vitro methylation assays (Figure 2), suggesting that arginine-methylation of TLS might affect protein-protein interactions. Recently, many proteins have been reported to contain both sDMA and aDMA [14, 15]. It will be possible that TLS is also modified by the symmetric and asymmetric methylations on the same arginine residues. Since we did not obtain monoclonal antibodies against symmetrically dimethylated TLS, further studies will be required to solve this point.
TLS was originally identified as a fusion protein TLS-CHOP in myxoid liposarcoma [1]. More recently, TLS attracts attention because it was found to be a causative gene for the familial ALS [2, 3]. More than a dozen mutations were reported in amino acids sequence of TLS [16, 17]. It is interesting to note that R216, one of dimethylated arginine we identified, is the site of a naturally occurring mutation associated with ALS [12]. Thus, the posttranslational modification of TLS might be implicated in the pathogenesis of ALS. Since 2B12 was suitable to precipitate methylated TLS in mouse and human brain samples (Figure 4), 2B12 can be a favorable tool for clinical diagnosis of ALS and will gain insight into the pathogenesis of ALS caused by arginine mutations of TLS.
We also verified whether our antibody could be used for RIP assays. CCND1-pncRNA-D binds methylated TLS in vivo by RIP using 2B12 (Figure 5), suggesting that arginine-methylation of TLS can affect RNA-protein interactions. The antibody for detecting asymmetrical arginine-specific methylation of TLS can be a valuable tool for analyzing the function of TLS methylation in vivo. Further study using 2B12 will uncover the mechanism of transcriptional regulation by lncRNA via RNA-binding protein TLS.
Methods
Antibodies and reagents
Rabbit anti-TLS/FUS antibody (ab70381) was purchased from Abcam. Mouse monoclonal anti-TLS antibody (611384) was purchased from BD Biosciences. Rabbit anti-GST antibody (Z5; sc-459) was purchased from Santa Cruz Biotechnology. Rabbit anti-PRMT1 antibody (A33) was purchased from Cell Signaling technology. Adenosine dialdehyde (Adox, Sigma) was dissolved in DMSO. Total protein lysate from mouse brain (8–10 weeks) and human brain (66 years old) were obtained from BioChain Institute Inc (Newark, CA, USA).
Peptide synthesis and antibody preparation
Unmethylated and methylated forms of peptides derived from TLS/FUS were obtained from Scrum Inc, (Tokyo, Japan). The sequences of the peptides were identical except for the presence of symmetric or asymmetric dimethylated arginine in peptide (See Figure 1A).
Rabbit polyclonal antibody against TLS peptide containing no modification (named as A1) was produced in Scrum Inc. The mouse monoclonal antibodies against TLS peptides containing either R216/R218me2s or R216/R218me2a were produced in ITM Co. Ltd. (Nagano, Japan). After the immunization and hydridoma development, cells were screened by enzyme-linked immunosorbent assay. One specific antibody against R216/R218me2a (hybridoma clone; 2B12) was obtained and characterized.
In vitro methylation assay
In vitro methylation reactions were performed as described previously [9]. Briefly, GST tagged TLS were incubated with bacterially expressed Strep-tagged PRMT1 lysate in the presence or absence of SAM (Sigma) for 1 h at 30°C. Methylation reactions were quenched by the addition of SDS sample buffer, heated at 100°C for 2 min, and separated on SDS-PAGE followed by western blotting analysis.
Dot-blot and western blot analyses
For the dot-blot analysis, one μl of diluted peptide in sterile water was blotted onto the nitrocellulose membrane (Bio-Rad) and dried. The membrane was then blocked with freshly prepared PBS containing 5% non-fat milk for 1 h at room temperature with constant agitation. The membrane was incubated with the primary antibodies diluted in 1% freshly prepared PBS-milk solution for 1 h at room temperature. After incubating the membrane with the secondary antibody (anti-mouse HRP-conjugated IgG, Dako or anti-rabbit HRP-conjugated IgG, Cell Signaling technology). Chemiluminescent detection was performed using SuperSignal West Pico substrate (Thermo Scientific).
For western blotting analysis, samples were separated by SDS-PAGE and the proteins were transferred to a nitrocellulose membrane. The membrane was then blocked similar to that used in the dot-blot analysis as mentioned above.
Cell culture
HeLa cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, Nacalai tesque, Tokyo, Japan) supplemented with 10% fetal bovine serum (Nichirei Biosciences Inc). HeLa cells were treated with AdOx (Sigma) for 24 hours. Cells were lysed in RIPA buffer, and cell lysates were used for immunoprecipitation experiments.
Immunoprecipitation
Cell extracts from HeLa, mouse brain and human brain were incubated with appropriate antibodies as indicated. Antibodies against methylated TLS or normal IgG were incubated with Protein G magnetic Dynabeads (Life technologies) for 10 min at RT with gentle rotation. The cell extract was added to the mix and incubated for 10 min at RT with gentle rotation. Beads were collected and washed three times with WCE buffer, eluted by adding SDS-sample buffer. For peptide competition assays, cell extracts were incubated in the presence or absence of competing peptide with magnetic Dynabeads Protein G. The eluted samples were analyzed by SDS-PAGE and western blotting.
RNA-binding protein immunoprecipitation assay
To determine whether methylated TLS interacts with lncRNA, 2B12 was used to pull down methylated TLS, and then bound RNA was purified and detected the expression of lncRNA from CCND1 promoter by RT-PCR using specific primers as published [5]. Magna RIP™ RNA-binding protein Immunoprecipitation kit (Millipore) was used for RIP procedures according to the manufacture’s protocol. The precipitated RNA was subject to cDNA synthesis. The presence of CCND1-pncRNA-D in the cDNA samples was detected using PCR primers previously used [5].
Electronic supplementary material
Additional file 1: Immunoprecipitation of endogenous methylated TLS from HeLa cell extracts was performed with 2B12 in the presence or absence of competing peptides (No; 100 ng, me2a; 25, 50, 100 ng, me2s; 100 ng). Bound methylated TLS was eluted with SDS sample buffer resolved by SDS-PAGE, and analyzed by western blotting with rabbit polyclonal anti-TLS antibody. Note that the immunoprecipitation of 2B12 was inhibited by the excess of R216/R218me2a peptide in a dose-dependent manner, not by other peptides. (PPT 72 KB)
Acknowledgements
This study was supported by Grant-in-Aid for Scientific Research (B: nos22390057; nos25293073), Grant-in-Aid for Research Activity Start-up (24810023). This work was also supported in part by a grant-in-aid for “Support Project of Strategic Research Center in Private Universities” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to Saitama Medical University Research Center for Genomic Medicine.
Abbreviations
- TLS
Translocated in LipoSarcoma
- FUS
FUsed Sarcoma
- CCND1
Cyclin D1
- lncRNA
Long noncoding RNA
- pncRNA
Promoter-associated ncRNA
- RIP
RNA-binding protein immunoprecipitation
- PRMT
Protein arginine methyltransferase
- ALS
Amyotrophic lateral sclerosis
- AdOx
Adenosine-2’,3’-dialdehyde
- SAM
S-adenosyl methionine.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RK conceived the concept. KF designed and performed experiments. KF and RK interpreted the findings. KF and RK wrote the manuscript, and approved the final manuscript.
Contributor Information
Kenta Fujimoto, Email: fujimotk@saitama-med.ac.jp.
Riki Kurokawa, Email: rkurokaw@saitama-med.ac.jp.
References
- 1.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363(6430):640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 2.Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Nigel Leigh P, Blair IP, Nicholson G, de Belleroche J, Gallo J-M, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 4.Dormann D, Haass C. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol Cell Neurosci. 2013;56:475–486. doi: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30(8):348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. 2010;115(10):2028–2037. doi: 10.1182/blood-2009-07-236059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du K, Arai S, Kawamura T, Matsushita A, Kurokawa R. TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem Biophys Res Commun. 2011;404(4):991–996. doi: 10.1016/j.bbrc.2010.12.097. [DOI] [PubMed] [Google Scholar]
- 10.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet. 2010;19(7):1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araya N, Hiraga H, Kako K, Arao Y, Kato S, Fukamizu A. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem Biophys Res Commun. 2005;329(2):653–660. doi: 10.1016/j.bbrc.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Sorarù G, Carlomagno Y, Ghezzi S, Pensato V, Colombrita C, Gagliardi S, Cozzi L, Orsetti V, Mancuso M, Siciliano G, Mazzini L, Comi GP, Gellera C, Ceroni M, D'Alfonso S, Silani V. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet. 2010;47(3):190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 13.Daigle JG, Lanson NA, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22(6):1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirino Y, Vourekas A, Kim N, de Lima AF, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285(11):8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng S, Moehlenbrink J, Lu YC, Zalmas LP, Sagum CA, Carr S, McGouran JF, Alexander L, Fedorov O, Munro S, Kessler B, Bedford MT, Yu Q, La Thangue NB. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol Cell. 2013;52(1):37–51. doi: 10.1016/j.molcel.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34(6):812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- 17.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21(6):904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Immunoprecipitation of endogenous methylated TLS from HeLa cell extracts was performed with 2B12 in the presence or absence of competing peptides (No; 100 ng, me2a; 25, 50, 100 ng, me2s; 100 ng). Bound methylated TLS was eluted with SDS sample buffer resolved by SDS-PAGE, and analyzed by western blotting with rabbit polyclonal anti-TLS antibody. Note that the immunoprecipitation of 2B12 was inhibited by the excess of R216/R218me2a peptide in a dose-dependent manner, not by other peptides. (PPT 72 KB)


