Abstract
The protein LC3 is indispensible for the cellular recycling process of autophagy and plays critical roles during cargo recruitment, autophagosome biogenesis, and completion. Here, we report that LC3 is phosphorylated at threonine 50 (Thr50) by the mammalian sterile-20 kinases STK3 and STK4. Loss of phosphorylation at this site blocks autophagy by impairing fusion of autophagosomes with lysosomes, and compromises the ability of cells to clear intra-cellular bacteria, an established cargo for autophagy. Strikingly, mutation of LC3 mimicking constitutive phosphorylation at Thr50 reverses the autophagy block in STK3/4-deficient cells and restores their capacity to clear bacteria. Loss of STK3/4 impairs autophagy in diverse species, indicating that these kinases are conserved autophagy regulators. We conclude that phosphorylation of LC3 by STK3/4 is an essential step in the autophagy process. Since several pathological conditions, including bacterial infections, display aberrant autophagy, we propose that pharmacological agents targeting this novel regulatory circuit hold therapeutic potential.
Keywords: Ste20 kinases, Mst1/2, Hippo kinase, autophagic flux, MAP1LC3, group A streptococcus
INTRODUCTION
Autophagy is a conserved, catabolic process that plays an essential role in cellular homeostasis by facilitating lysosomal degradation of cytosolic components such as defective macromolecules and organelles. Autophagy was first characterized in yeast as a survival mechanism induced by nutrient deprivation. Since then, elegant studies in multicellular organisms have established key roles for autophagy in a variety of processes ranging from development to aging. Consistent with its critical function in diverse processes, autophagy is often perturbed in disease states such as cancer, diabetes, neurodegeneration, and immune-related disease (Huang and Klionsky, 2007).
Autophagy comprises a highly intricate and complex series of events in which cytosolic material, or cargo, is sequestered within double-membrane vesicles known as autophagosomes, which then fuse with acidic lysosomes to form autolysosomes, where the cargo is enzymatically degraded and recycled. These steps are orchestrated by a group of proteins encoded by the autophagy genes (ATG). Chief among these is the highly conserved ATG8/LC3 ubiquitin-like protein that plays crucial roles in the formation of autophagosomes and recruitment of cargo. Upon induction of autophagy by stressors such as nutrient deprivation, LC3 is post-translationally modified via the action of ATG3/4/7 proteins, which attach phospholipid phosphatidylethanolamine (PE) to the C-terminus of LC3 to generate LC3-II. PE-conjugated LC3-II plays several crucial roles in autophagy (Nakatogawa et al., 2007). In addition to facilitating elongation and completion of the autophagosome, LC3-II facilitates delivery of cargo to these structures by interacting with adaptor proteins, including p62/SQSTM1. p62 thus acts as a bridge by binding ubiquitinated proteins and organelles marked for destruction (Komatsu and Ichimura, 2010). Following cargo recruitment and maturation, completed autophagosomes go on to fuse with lysosomes to form autolysosomes, in which the cytosolic cargo is degraded. The complex process of fusion between the two compartments is mediated by a set of multi-protein complexes, that include Rab small GTPases (Bento et al., 2013), soluble N-ethylmaleimide sensitive factor attachment protein receptors [SNARES (Moreau et al., 2012)], and homotypic fusion and protein sorting complexes [HOPS (Jiang et al., 2014)], whose individual roles in fusion are yet to be fully characterized.
While a detailed understanding of the functional roles played by LC3 during autophagy exists, the signaling events that coordinate the various functions of LC3 remain poorly understood. To bridge this gap, a genome-wide analysis of the human autophagy-interaction network identified multiple signaling and regulatory factors that interact with LC3, among which are the sterile-20 kinase STK4/MST1 and its closely related functionally redundant paralog, STK3/MST2 (Behrends et al., 2010). STK3/STK4 are the mammalian homologs of Hippo, a major inhibitor of cell proliferation in the fruit fly Drosophila (Halder and Johnson, 2011). These conserved kinases are components of a multistep signaling pathway that controls the expression of genes associated with cell growth and survival in numerous species. Consistent with this role, STK3/STK4 function as tumor suppressors in flies and mice. More recent studies have implicated STK4 in the regulation of antibacterial- and antiviral immunity in mammals. Stk4−/− mice display progressive loss of B- and T lymphocytes due to excessive apoptosis, and humans with Stk4 deficiency are prone to recurrent infections, likely due to neutropenia and lymphopenia (Abdollahpour et al., 2012; Dong et al., 2009; Nehme et al., 2011). It is unknown if the pro-immune function of STK4 is linked to its role as a tumor suppressor, or occurs via a different and novel mechanism.
In this study, we describe a novel role for STK3/STK4 in promoting autophagy via direct phosphorylation of LC3. This function is conserved, since we find that STK3/STK4 deficiency impairs autophagy in mammalian cells, the nematode Caenorhabditis elegans, and the yeast Saccharomyces cerevisiae. Mechanistically, we observe that STK3/STK4 phosphorylation of LC3 on threonine 50 (Thr50) is critical for fusion between autophagosomes and lysosomes. We also demonstrate a vital role for STK3/STK4 in antibacterial immunity. STK3/STK4-deficient mouse-embryonic fibroblasts (MEFs) are unable to efficiently clear intracellular group A streptococci, an established cargo for autophagic degradation, but reconstitution of the deficient cells with an LC3 phosphomimetic (T50E) reverses the autophagy block and bacterial killing defect in these cells. Taken together, our findings uncover a novel and conserved regulatory axis for autophagy with implications for human disease.
RESULTS
Loss of STK3/STK4 results in autophagy defects in mammalian cells
Human STK3 and STK4 have previously been reported to interact with several ATG8 isoforms, including MAP1LC3B (Behrends et al., 2010). To begin our investigation of the function of STK3/STK4 in autophagy, we asked whether a similar interaction is observed in mouse cells. For these experiments, we focused our analysis on the interaction between STK3/STK4 and the LC3B isoform (hereafter LC3) in MEFs derived from wild-type (WT) and Stk3+/−;Stk4−/− mice. MEFs from Stk3+/−;Stk4−/− mice harbor full deletion of Stk4 and a hemizygous deletion of Stk3 (Song et al., 2010). Consistent with the observations in human cells (Behrends et al., 2010), we found that endogenous STK4 co-immunoprecipitated with transfected green fluorescent protein-tagged LC3 (GFP::LC3) from WT MEFs, but not from Stk3+/−;Stk4−/− MEFs (Figure S1A). Moreover, we found that both STK3 and STK4 could phosphorylate LC3 in in vitro kinase assays (Figure S1B), thereby establishing LC3 as a substrate for both STK3 and STK4.
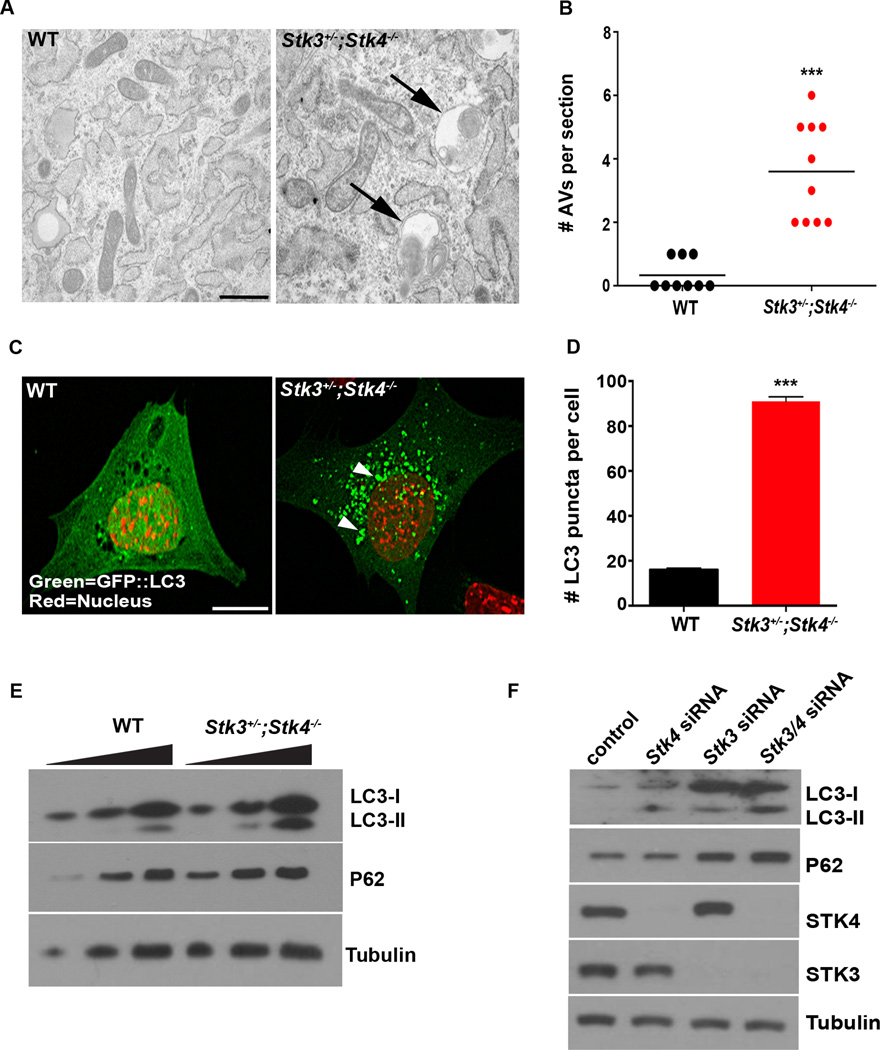
The finding that STK3/STK4 interact with and phosphorylate LC3 raised the possibility that these kinases might play a role in modulating autophagy. To examine this, we analyzed several key components of the autophagy process in WT and Stk3+/−;Stk4−/− MEFs. Transmission electron microscopic (TEM) analysis of cells under basal (non-stressed) conditions revealed that Stk3+/−;Stk4−/− MEFs contained ∼4 times more autophagic vesicles (double- or single-membrane–bound vesicles containing cytoplasmic cargo) than observed in WT MEFs (Figures 1A, 1B & S2A). We also examined the subcellular localization and biochemical properties of LC3 in these cells. During autophagy, PE-conjugated LC3-II tightly associates with autophagosomal membranes, altering its subcellular localization from diffuse distribution in the cytoplasm to distinct punctate structures that can represent either incomplete pre-autophagosomes or completed autophagosomes. These autophagic structures can be readily visualized by expressing GFP-tagged LC3 in cells (Klionsky et al., 2012). In agreement with our findings by TEM, the number of GFP::LC3 puncta present in Stk3+/−;Stk4−/− MEFs was much greater than that in WT cells (Figures 1C & D). Total levels of endogenous LC3-II and endogenous LC3 puncta were also both dramatically induced, even under fully-fed conditions in Stk3+/−;Stk4−/− MEFs (Figures 1E & S2B). Crucially, reintroduction of active STK4, but not kinase-dead STK4 (Glantschnig et al., 2002), restored the number of LC3 puncta in Stk3+/−;Stk4−/− MEFs to that seen in WT cells (Figure S2C). These data demonstrate that STK3/STK4 deficiency has marked effects on autophagy phenotypes, as shown by an increased abundance of autophagic structures and LC3 puncta.
Figure 1. Loss of STK3/STK4 results in autophagy defects in mouse cells.
(A) Representative electron micrographs of wild-type (WT) and Stk3+/−;Stk4−/− mouse embryonic fibroblasts (MEFs). Arrows indicate autophagic structures containing cytosolic material undergoing degradation. Scale bar = 1 µm. See Figure S2A for additional images.
(B) Quantification of autophagic vesicles (double-membrane autophagosomes and single-membrane autolysosomes) in sections of MEFs represented in (A). Horizontal bar indicates the mean of 10 sections, 5 each from 2 independent experiments. ***P < 0.001, Student’s t-test.
(C) Representative confocal fluorescence micrographs of WT and Stk3+/−;Stk4−/− MEFs expressing GFP::LC3. Nuclei were stained with DAPI and false colored red. Arrows indicate individual GFP::LC3 puncta. Scale bar = 10 µm.
(D) Quantification of GFP::LC3 puncta in MEFs represented in (C). Mean ± SEM of puncta from at least 15 cells. ***P < 0.001, Student’s t-test. Data are representative of 3 independent experiments.
(E) Western blot analysis of LC3 and p62 levels in WT and Stk3+/−;Stk4−/− MEFs. Lysates containing 5, 10, or 20 µg total protein (left to right) were resolved and blotted with antibodies against the indicated proteins. Tubulin was probed as a loading control. Data are representative of 3 independent experiments.
(F) Western blot analysis of C2C12 myoblasts transfected with scrambled siRNA (control), or siRNA against Stk3, Stk4, or both. Lysates were prepared 72 h post-transfection and samples (20 µg total protein) were blotted with antibodies against the indicated proteins. Tubulin was probed as a loading control. Data are representative of 2 independent experiments.
See also Figure S1.
The observed phenotypes in STK3/STK4-deficient cells could result from either an induction of autophagy or a block in the turnover of LC3-bound autophagosomes (Klionsky et al., 2012). To distinguish between these possibilities, we examined levels of p62/SQSTM1 protein in WT and STK3/STK4-deficient MEFs. During autophagy, cargo-bound p62 is recruited by LC3 into the autophagosomes and is eventually degraded in lysosomes. Thus, when autophagy is blocked, p62 accumulates due to inefficient autophagosome turnover (Klionsky et al., 2012). We found that Stk3+/−;Stk4−/− MEFs contained significantly higher basal levels of p62 compared with WT MEFs (Figure 1E), suggesting that the increase in LC3 puncta in these cells (Figure 1C) was due to a block in autophagy rather than its induction. The effect of STK3/STK4 deficiency on autophagy is not specific for MEFs, but was also observed in the murine myoblast line, C2C12. In these cells, siRNA-mediated silencing of Stk3 and/or Stk4 dramatically increased the levels of LC3-II and p62 protein compared with control cells (Figure 1F). Collectively, these observations are consistent with depletion of STK3/4 resulting in a block in autophagy.
Autophagic flux is perturbed in MEFs lacking STK3/STK4
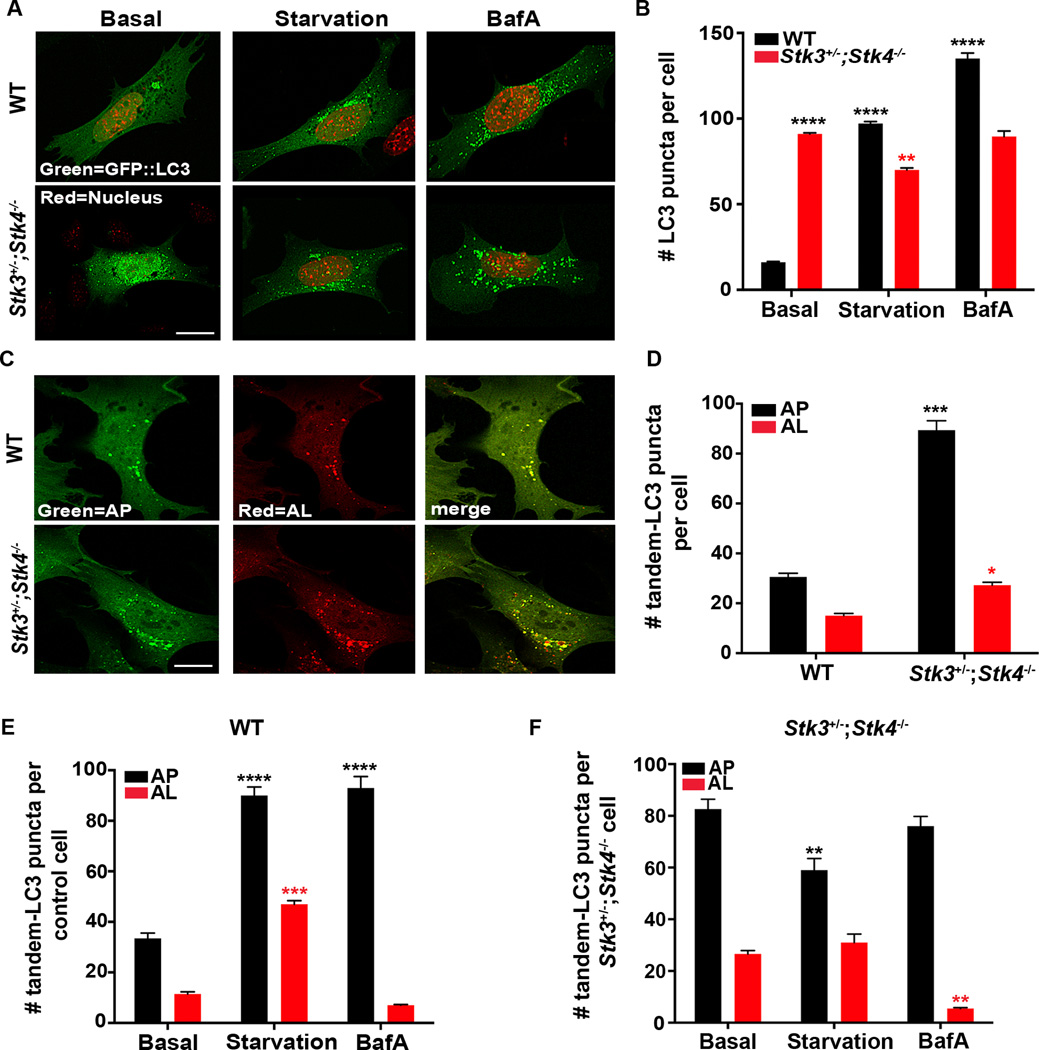
Because autophagy is induced under stress conditions such as nutrient deprivation, we next determined whether STK3/STK4 deficiency might abrogate the autophagy response of MEFs subjected to starvation, mimicked by serum withdrawal (Klionsky et al., 2012). WT and Stk3+/−;Stk4−/− MEFs transiently expressing GFP::LC3 were incubated for 2 h in serum-free medium. Notably, GFP::LC3 puncta were increased nearly 10-fold in starved WT MEFs, consistent with induction of autophagy, but Stk3+/−;Stk4−/− MEFs failed to increase LC3 puncta beyond the numbers seen under basal conditions (Figures 2A & B). Likewise, GFP::LC3 puncta increased dramatically in WT but only mildly so in Stk3+/−;Stk4−/− MEFs treated with rapamycin (Figure S2D), which induces autophagy by inactivating the conserved nutrient sensor TOR (Jung et al., 2010).
Figure 2. Loss of STK3/STK4 kinases impairs autophagic flux.
(A) Representative confocal fluorescence micrographs of wild-type (WT) and Stk3+/−;Stk4−/− MEFs transiently expressing GFP::LC3. Cells were transfected for 36 h and incubated in growth medium (basal), starvation medium (starvation), or medium containing 50 nM bafilomycin A1 (BafA) for 2 h. Nuclei were stained with DAPI and are false colored red. Scale bar = 10 µm.
(B) Quantification of GFP::LC3 puncta in cells represented in (A). Mean ± SEM of puncta from at least 15 cells. ****P < 0.0001, **P < 0.01 by one-way ANOVA. Black and red asterisks indicate comparisons to WT and Stk3+/−;Stk4−/− MEFs under basal conditions, respectively. Data are representative of 4 independent experiments. A small but significant reduction in the number of GFP::LC3-positive puncta was observed for Stk3+/−;Stk4−/− MEFs exposed to starvation. At this time, we do not fully comprehend the cause for this effect, which could be triggered by downstream compensatory mechanisms.
(C) Representative confocal fluorescence micrographs of WT and Stk3+/−;Stk4−/− MEFs transfected with mCherry::GFP::LC3 for 36 h. Scale bar = 10 µm.
(D) Quantification of autophagosomes (AP, black bars) and autolysosomes (AL, red bars) in cells represented in (C). Mean ± SEM of puncta from at least 15 cells. ***P < 0.001, *P < 0.05, Student’s t-test. Black and red asterisks indicate comparisons to the number of autophagosomes and autolysosomes, respectively, in WT cells.
(E) Wild-type or (F) Stk3+/−;Stk4−/− MEFs were transfected with mCherry::GFP::LC3 for 36 h and incubated in basal-, starvation-, or medium containing 50 nM BafA for 2 h. Autophagosomes (AP) and autolysosomes (AL) were visualized as described for (C). Mean ± SEM of puncta from at least 15 cells. ****P < 0.0001, ***P < 0.001, **P < 0.01, one-way ANOVA. Black and red asterisks indicate comparisons to the number of APs and ALs, respectively, in cells incubated under basal conditions. Data are representative of 3 independent experiments.
See also Figure S2.
The abundance of LC3 puncta in mammalian cells is also increased by exposure to lysomotropic reagents such as Bafilomycin A1 (BafA), which inhibit lysosomal acidification and block the degradation of LC3-II. BafA treatment therefore allows quantification of autophagic flux; that is, the delivery of LC3-positive autophagosomes to lysosomes for degradation (Klionsky et al., 2012). We found that treatment of WT MEFs with BafA led to a robust accumulation of GFP::LC3 puncta, indicative of efficient delivery of LC3 to autophagosomes (Figures 2A & B). However, this was not observed with Stk3+/−;Stk4−/− MEFs, which showed no change in LC3 puncta compared with basal levels (Figures 2A & B). To ensure that endogenous LC3-II protein behaved similar to GFP-tagged LC3, we also measured the accumulation of LC3-II upon exposure to starvation and BafA. As expected, WT cells responded by robustly accumulating LC3-II in a time-dependent manner. In sharp contrast, the high levels of basal LC3-II protein observed Stk3+/−;Stk4−/− cells only slightly increased over the time course (Figure S2E). This compromised ability of Stk3+/−;Stk4−/− MEFs to induce LC3 foci/LC3-II in response to starvation or treatment with rapamycin and BafA is characteristic of a block in autophagy.
To elucidate whether the increase in LC3-positive foci observed in Stk3+/−;Stk4−/−cells reflects an increase in autophagosomes or in autolysosomes, we utilized a pH-sensitive reporter generated by tandem fusion of LC3 with GFP and the red fluorescent protein mCherry [mCherry::GFP::LC3,(Klionsky et al., 2012)]. Both sensors fluoresce in the neutral environment of the autophagosome, which renders them visible, resulting in yellow fluorescent structures; however, GFP fluorescence is rapidly quenched in the acidic lysosome, resulting in only red fluorescence. Therefore, the tandem sensor allows autophagosomes and autolysosomes to be distinguished and enumerated. We found that WT MEFs transiently expressing the mCherry::GFP::LC3 reporter emitted low yellow or red fluorescence, reflecting basal numbers of autophagosomes and autolysosomes, respectively. In contrast, Stk3+/−;Stk4−/− MEFs displayed much higher basal numbers of autophagosomes than the WT cells, whereas autolysosomes were only slightly increased (Figure 2D). Moreover, starvation of WT MEFs caused a robust increase in the number of autophagosomes and autolysosomes, indicating successful delivery of LC3 to these compartments, whereas treatment with BafA significantly increased the number of autophagosomes, but not autolysosomes, as expected (Figure 2E). In contrast, Stk3+/−;Stk4−/− cells were compromised in their ability to alter the levels of both autophagic compartments in response to starvation and BafA treatments (Figures 2E & F). Indeed, the number of GFP::LC3 positive puncta in Stk3+/−;Stk4−/− MEFs under basal conditions was comparable to that of WT MEFs treated with BafA. These data indicate a requirement for STK3/STK4 activity primarily at the step when autophagosomes fuse with lysosomes to form autolysosomes, a premise that we test in-depth below.
Role of STK4 in autophagy is conserved across taxa
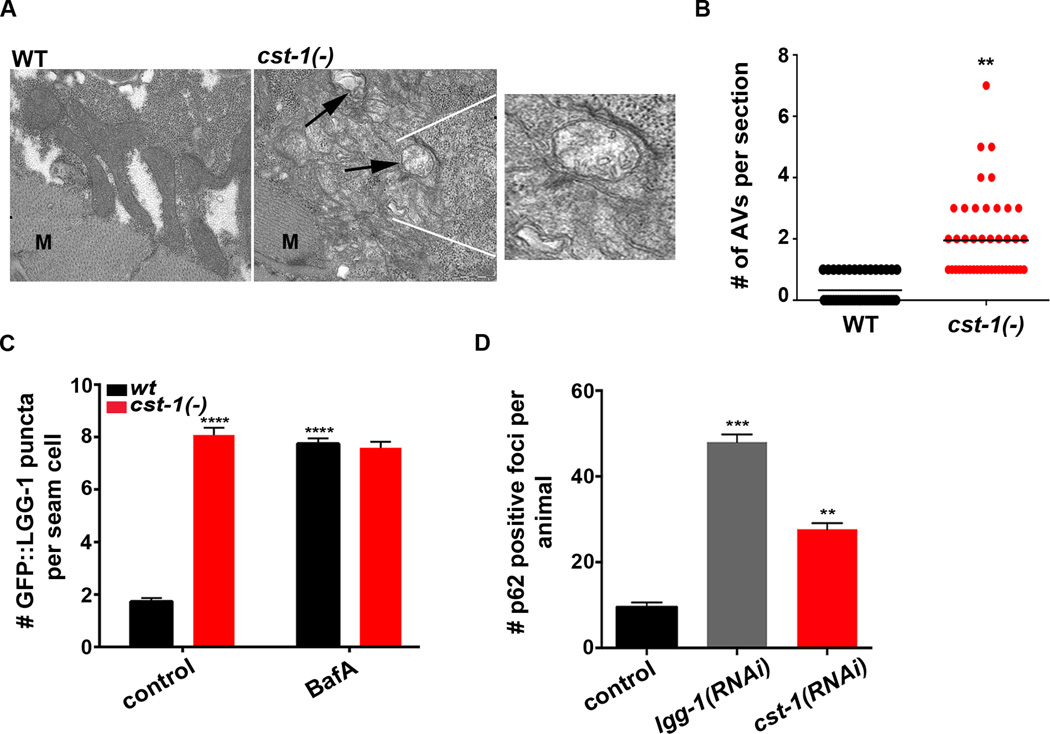
STK3 and STK4 show strong evolutionary conservation, raising the possibility that their roles in autophagy may also be conserved. To test this, we examined autophagy in the nematode C. elegans lacking the STK4 ortholog, cst-1 (Yang and Hata, 2013). Using TEM to visualize autophagic vesicles in the muscle (Figures 3A & B) and the intestine (not shown), we detected a significantly higher number of structures in cst-1(tm1900) deletion mutants than in WT animals (Figures 3A & B). We next examined transgenic C. elegans WT and cst-1(−) strains stably expressing GFP-tagged LGG-1/LC3, which allows autophagosomes to be visualized as GFP::LGG-1-positive puncta (Melendez et al., 2008). Compared with WT animals, transgenic cst-1(−) mutants contained 3-fold more GFP::LGG-1 puncta in hypodermal seam cells (Figure 3C, see control), a tissue commonly used for monitoring autophagy in C. elegans. RNAi against beclin-1, a gene important for autophagy initiation, abolished basal LGG-1 puncta in cst-1(−) animals (data not shown), suggesting that the accumulation of autophagic structures in cst-1(−) mutants is dependent on autophagy.
Figure 3. Loss of STK4 causes autophagy defects in C. elegans.
(A) Representative electron micrographs of muscle cells from 1-day-old wild-type (WT) and cst-1(tm1900)/Stk4 animals. Arrows indicate autophagic vesicles; ‘M’ indicates myofibers. Scale bar = 1 µm. Inset is an enlargement of the boxed area showing double membrane-bound autophagosomes.
(B) Quantification of autophagic vesicles (double-membrane autophagosomes and single-membrane autolysosomes) per section in the muscle of 1-day-old WT and cst-1(tm1900) animals, as shown in (A). Horizontal line indicates the mean. Data are the average of 3 independent experiments. **P < 0.01, Student’s t-test.
(C) Quantification of GFP::LGG-1-positive puncta in seam cells of WT and cst-1(tm1900). Day 1 adults were injected with 50 µM BafA or DMSO, and 2 h later, GFP::LGG-1 puncta were quantified. Mean ± SEM of ∼200 cells. Data are representative of 3 independent experiments. DMSO on its own has negligible effect on LGG-1 punta under several conditions tested (data not shown). RNAi of autophagy gene bec-1 decreased the number of GFP::LGG-1-positive puncta in cst-1(−) animals, consistent with these structures representing autophagic events (data not shown).
(D) Quantification of p62::GFP foci in animals represented in Figure S3A. See Figure S3A for region of quantification in head region. Mean ± SEM of ∼20 animals. Data are representative of 3 biological repeats.
For C & D, ***P < 0.001, **P < 0.01 to WT control, one-way ANOVA.
See also Figure S3.
To ascertain whether the basal increase in LGG-1 puncta was a consequence of blocked autophagy, we established a new methodology to measure autophagy flux in C. elegans. WT or cst-1(−) C. elegans expressing GFP::LGG-1 were injected with BafA at day 1 of adulthood, and two hours later the number of GFP::LGG-1-positive puncta were quantified in hypodermal seam cells. Injection of WT animals with BafA lead to a ∼4-fold increase in GFP::LGG-1-positive puncta compared to control-injected animals (Figure 3C, compare black bars). In contrast, cst-1(−) animals displayed high levels of basal puncta and failed to further upregulate the number of LGG-1::GFP-positive foci in response to BafA (Figure 3C, compare black bars), consistent with a block in autophagy in these mutants. We also examined the effects of RNAi-mediated depletion of cst-1 [which also downregulates cst-2/Stk3 (Lehtinen et al., 2006)], in animals expressing GFP-tagged SQST-1/p62 (Tian et al., 2010). As in mammalian cells, accumulation of GFP::p62-positive foci in C. elegans is indicative of a block in autophagy (Lapierre et al., 2013; Tian et al., 2010). As expected, RNAi-mediated inhibition of lgg-1/Lc3 blocked autophagy and led to a marked increase in p62 foci in the anterior pharyngeal region of adult animals (Figures 3D & S3A). Consistent with a critical role for the STK4 ortholog in autophagy in C. elegans, we also found a significantly higher number of p62-positive foci in animals subjected to cst-1/Stk4 RNAi (Figures 3D & S3A), corroborating an autophagy block in cst-1/2(RNAi) mutants. To further confirm the evolutionarily conserved function of STK3/STK4, we also examined autophagy regulation in the unicellular organism S. cerevisiae; here too, genetic deletion of a putative STK4 ortholog, Sps1, blocked autophagy (Figure S3B & C). Collectively, these data highlight an evolutionarily conserved function of STK kinases in regulating autophagy from yeast to mammals.
STK4 phosphorylates LC3 at threonine 50
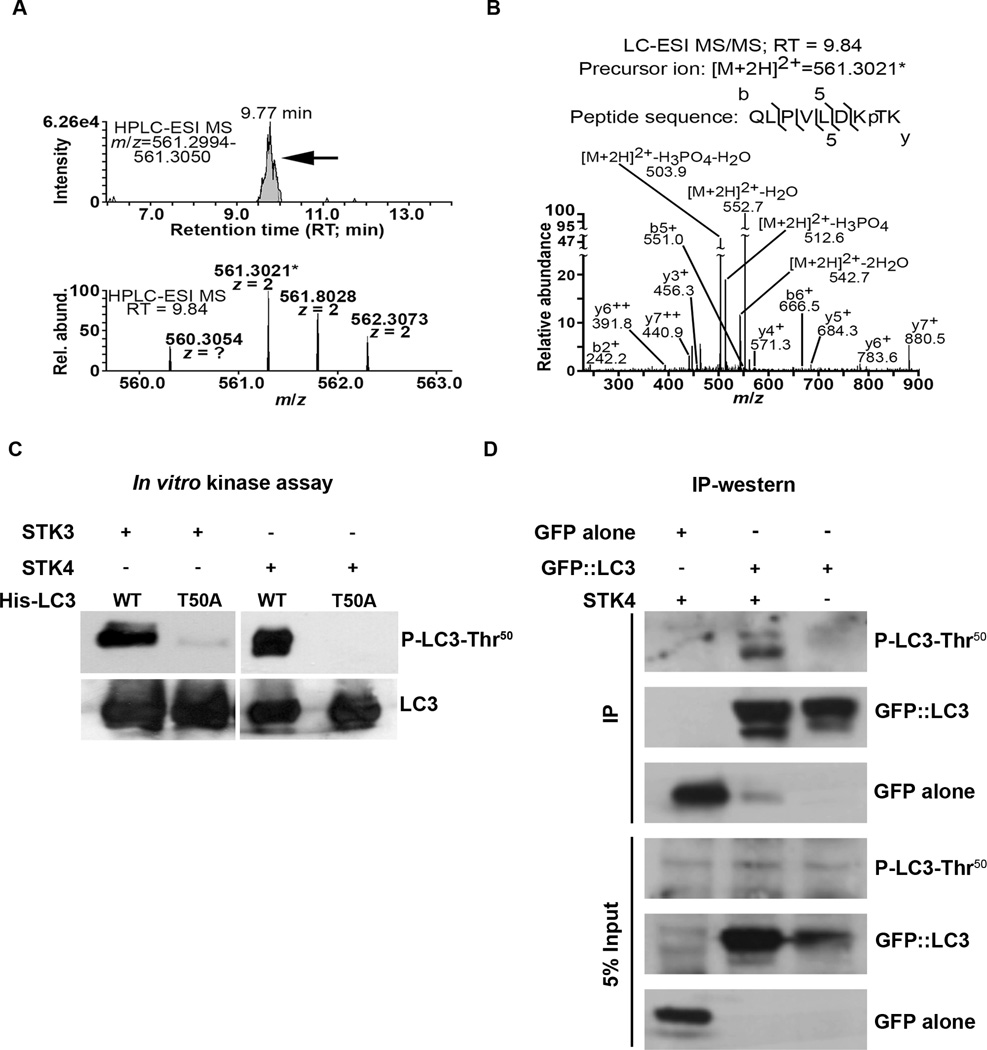
Having established a regulatory role for STK3/STK4, we next sought to identify possible STK3/STK4 phosphorylation sites on LC3. For this, we performed in vitro kinase assays with epitope-tagged, recombinant mouse LC3B in the presence or absence of recombinant STK4, and analyzed LC3 phosphopeptides by TiO2-HPLC-MS/MS. In two independent experiments, we successfully detected LC3 peptides containing threonine 50 (Thr50) that were phosphorylated only when LC3 was incubated with STK4 (Figure 4A and data not shown); the spectrum for one of these, QLPVLDKpTK, is shown (Figure 4B). To determine whether this site on LC3 is phosphorylated in vitro and in vivo, we generated a rabbit polyclonal antibody against the phosphorylated peptide. Following in vitro kinase assays, the antibody recognized STK4- and STK3-phosphorylated WT LC3, but not LC3 carrying a threonine to alanine ‘phospho-null’ mutation (LC3-Thr50A) (Figure 4C). Similarly, GFP::LC3 immunoprecipitated from transfected WT MEFs was an excellent substrate for STK4 in vitro, whereas GFP:: LC3-Thr50A was not detectably phosphorylated (Figure S4). Finally, we detected Thr50 phosphorylation of GFP::LC3 immunoprecipitated from WT MEFs overexpressing STK4 (Figure 4D). Collectively, these analyses demonstrate that LC3B is phosphorylated on at least one specific site, Thr50, by STK3/STK4 kinases, both in vitro and in vivo.
Figure 4. STK3/STK4 phosphorylate LC3 at threonine 50.
(A) HPLC-ESI-MS extracted-ion chromatograms (8 min retention time (RT) and 5 ppm m/z windows) of LC3 (B isoform) incubated with STK4 in vitro. A distinct peak with RT 9.77 min (upper panel, arrow) is suggestive of the precursor ion of an LC3 phosphopeptide (predicted m/z = 561.3022) containing phosphorylated threonine50 (p-Thr50). A doubly-charged (2+) precursor ion with a measured m/z of 561.3021 (*; mass differs from the predicted m/z of 561.3022 by only 178 ppb) was detected in the HPLC-ESI-MS peak (lower panel shows precursor ions from a representative peak with RT 9.84 min). Note the peaks at m/z of 561.8028 and 562.3073 contain one and two naturally occurring heavy atoms respectively, most likely C13 (these peaks enable the mass spectrometer to determine the 2+ charge state of the precursor ion).
(B) Annotated MS/MS scan of the fragmented precursor ion (m/z of 561.3021*) isolated from the scan shown in panel A, demonstrating that the ion was from LC3 and contained p-Thr50. The b, y, and neutral loss product ions are marked. As expected, no comparable MS/MS scans were derived from samples containing LC3 only (data not shown).
(C) Non-radiolabelled in vitro kinase assays of purified His-tagged WT LC3 or LC3-T50A mutant incubated with recombinant STK4 or STK3. Samples were resolved by SDS-PAGE and subjected to western blotting with anti-P-LC3-Thr50 antibody (upper panel) and anti-LC3 antibody (lower panel). Data are representative of 4 independent experiments.
(D) In vivo phosphorylation of LC3-Thr50 detected by western blot analysis. Wild-type MEFs were co-transfected with STK4 and either GFP or GFP::LC3. GFP was immunoprecipitated from lysates 48 h post-transfection, and the complexes were resolved by SDS-PAGE and subjected to western blotting with anti-P-LC3-Thr50, anti-LC3, or, anti-GFP antibodies, as indicated. Lysates equivalent to 5% of IP input were probed as controls. Data are representative of 3 independent experiments.
See also Figure S4.
LC3 Thr50 phosphorylation is essential for autophagy flux in vivo
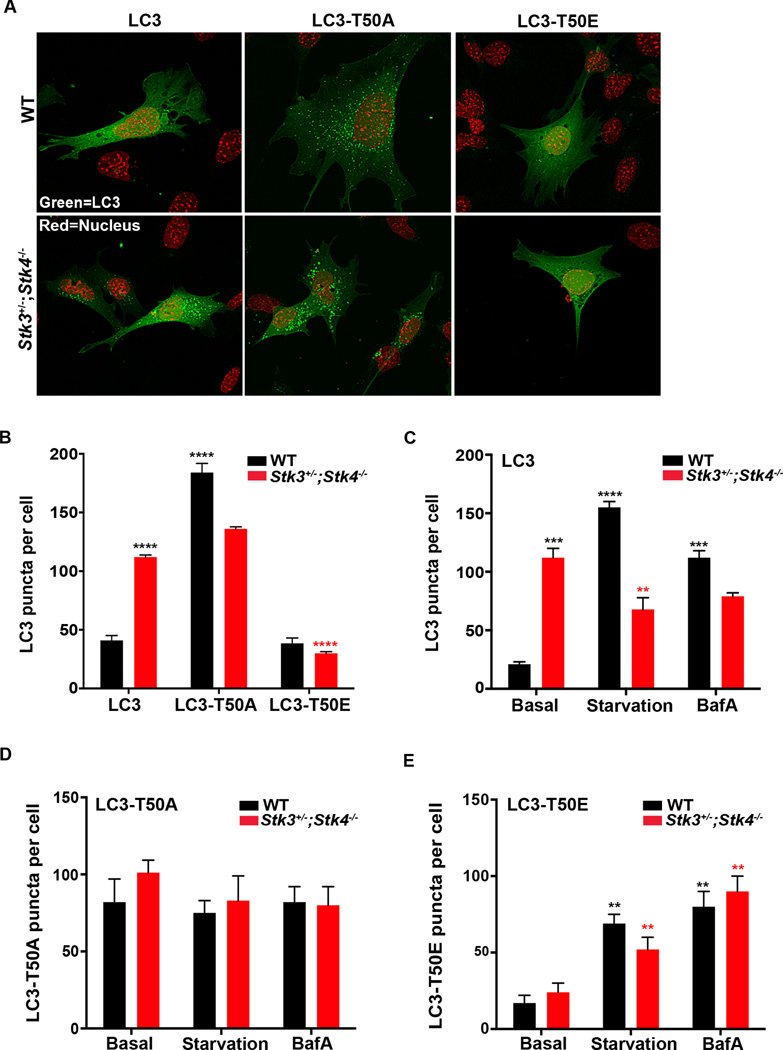
We next sought to determine the relevance of STK3/STK4-mediated LC3-Thr50 phosphorylation for the function of LC3 in vivo. We transiently expressed GFP-tagged constructs of wild-type (LC3), the phospho-mimetic mutant (LC3-Thr50E), or the phospho-null mutant (LC3-Thr50A) in both WT and Stk3+/−;Stk4−/− MEFs. All of these constructs were expressed to similar levels in WT MEFs, whereas the expression levels of LC3-Thr50A and LC3-Thr50E was slightly lower in Stk3+/−;Stk4−/− MEFs (Figure S5A). To monitor autophagy in WT cells, we counted the number of GFP::LC3 foci and found that introduction of the phospho-null mutant LC3-Thr50A caused a dramatic accumulation of GFP::LC3 puncta in WT cells (Figure 5A & B, compare black bars), whereas no further increase in LC3 puncta was observed in Stk3+/−;Stk4−/− MEFs (Figure 5A & B, compare red bars). In contrast, cells expressing the phosphomimetic, LC3-Thr50E, had normal numbers of LC3 puncta that were diffuse in both WT and Stk3+/−;Stk4−/− MEFs, mirroring the cellular distribution pattern of WT LC3 in WT MEFs (Figure 5A & B). These data strongly suggest a key role for phosphorylation of LC3-Thr50 in STK3/4-mediated autophagic flux. Consistent with this possibility, we observed that WT and Stk3+/−;Stk4−/−MEFs expressing LC3-Thr50A remained refractory to both starvation and BafA treatment (Figure 5D), whereas both insults upregulated LC3 puncta in LC3-Thr50E-expressing MEFs (Figure 5E). Of note, LC3-Thr50E puncta were consistently less abundant than puncta formed with WT LC3 (Figure 5C & E, note difference in scale bars), suggesting that the phosphomimetic LC3 only partially recapitulated the functions of WT LC3. We corroborated our LC3 puncta analysis by exposing MEFs stably expressing the LC3 point mutants to a combined starvation and BafA treatment (as in Figure S2E), and measuring endogenous LC3-II accumulation. First, we noted that stable introduction of LC3-Thr50E in Stk3+/−;Stk4−/− MEFs diminished basal LC3-II levels compared to introduction of WT-LC3 (Figure S5D). Furthermore, exposure of Stk3+/−;Stk4−/− MEFs expressing LC3-Thr50E to starvation and BafA led to accumulation of LC3-II over time in a manner comparable to the kinetics observed for WT cells expressing LC3-Thr50E (Figure S5D), whereas stable introduction of LC3-Thr50A failed to rescue the autophagy flux defect observed in Stk3+/−;Stk4−/− cells (Figure S5C). Interestingly, introduction of LC3-Thr50A in WT cells appeared to modestly diminish the kinetics of LC3-II accumulation when compared to WT-LC3 (Figure S5C), potentially due to dominant-negative effects of the phosphomutant. Taken together, these data demonstrate that STK3/STK4 promote autophagic flux in vivo by phosphorylating Thr50 of LC3.
Figure 5. STK3/STK4 phosphorylation of LC3 at threonine 50 is essential for autophagic flux.
(A) Representative confocal fluorescence micrographs of wild-type (WT) and Stk3+/−;Stk4−/− MEFs transiently expressing GFP-tagged WT LC3, LC3-T50A (phospho null), or LC3-T50E (phosphomimetic). Nuclei were stained with DAPI and are false colored red. Scale bar = 10 µm.
(B) Quantification of GFP::LC3-positive puncta in cells represented in (A). Data are representative of 4 independent experiments.
(C-E) Quantification of GFP::LC3–positive puncta in WT and Stk3+/−;Stk4−/− MEFs expressing WT LC3 (C), LC3-T50A (D), or LC3-T50E (E) incubated in basal-, starvation-, or basal medium containing 50 nM BafA. Note that the y-axis scale for C, D, and E differs. Data are representative of 3 independent experiments.
For B-E, ****P < 0.0001, ***P < 0.001, **P < 0.01, one-way ANOVA. Black and red asterisks indicate comparisons to WT and Stk3+/−;Stk4−/− cells expressing WT LC3 under basal conditions, respectively. Mean ± SEM from at least 15 cells per condition.
See also Figure S5.
LC3 Thr50 phosphorylation is essential for fusion of autophagosomes with lysosomes
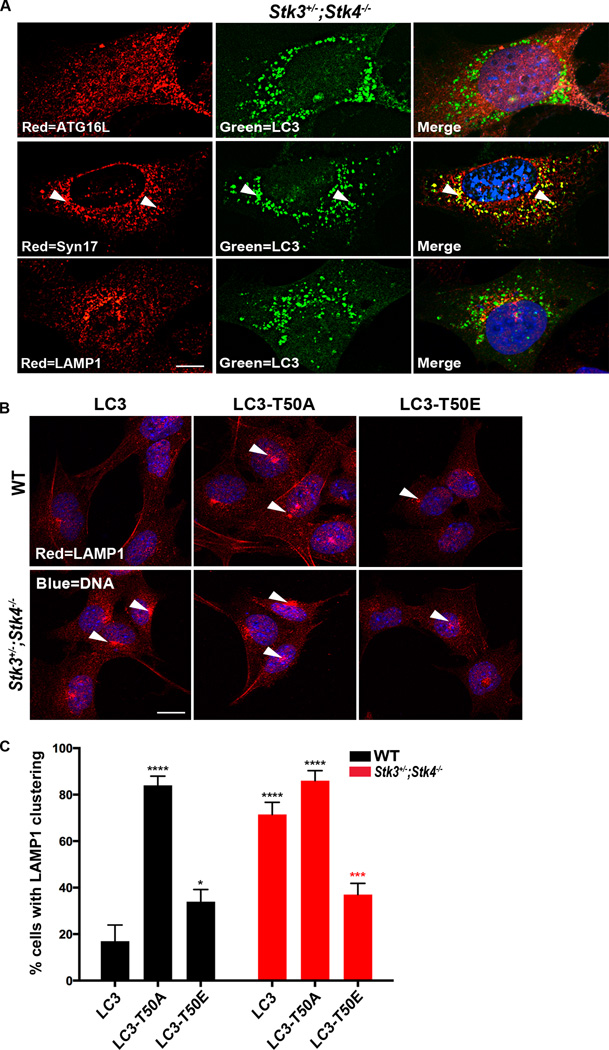
Having established that LC3-Thr50 phosphorylation is critical for autophagic flux, we next sought to clarify if a particular step of the autophagic process was targeted by this modification. Since LC3 family members have known roles in autophagosome completion (Weidberg et al., 2010), we first tested whether the autophagy block in Stk3+/−;Stk4−/− cells could result from incomplete autophagosome formation. To this end, we stained Stk3+/−;Stk4−/− cells expressing GFP::LC3 with antibodies against ATG16L [solation membrane marker (Klionsky et al., 2012)] and against STX17 [late autophagosome marker (Itakura et al., 2012)]. This immunohistochemical analysis showed that GFP::LC3 puncta in Stk3+/−;Stk4−/− cells exhibited weak colocalization with ATG16L and strong co-localization with STX17 (Figure 6A), supporting the premise that Stk3+/−;Stk4−/− cells accumulate fully formed autophagosomes.
Figure 6. Phosphorylation of threonine50 is important for fusion of autophagosomes to lysosomes.
(A) Representative confocal fluorescence micrographs of Stk3+/−;Stk4−/−MEFs transiently expressing GFP::LC3 (green) and stained with the indicated antibodies (red: top, ATG16; middle, Syntaxin17 (Syn17); bottom, LAMP1). Nuclei were stained with DAPI. Arrows indicate colocalization events between LC3 and Syn17. At least 30 cells were imaged for each repeat. Scale bar = 10 µm. Data are representative of 3 biological repeats.
(B) Representative confocal fluorescence micrographs of Stk3+/−;Stk4−/−MEFs stained with LAMP1 antibody (red). Nuclei were stained with DAPI. ∼ 50 cells were imaged for each repeat. Scale bar = 10 µm. Data are representative of 3–5 biological repeats.
(C) Quantification of LAMP1 staining in Stk3+/−;Stk4−/−MEFs represented in (B), and in wild-type (WT) MEFs. Mean ± SEM from at least 50 cells. ****P < 0.001, ***P < 0.001, *P < 0.05, one-way ANOVA. Black and red asterisks indicate comparisons to WT and Stk3+/−;Stk4−/− cells expressing WT LC3, respectively. Data are representative of 3 independent experiments.
See also Figure S6.
Since Thr50 is localized within a pocket that participates in cargo recruitment to the autophagic membrane (Shvets et al., 2008), we next tested whether loss of Thr50 phosphorylation hampered recruitment of the cargo-binding protein p62. In co-immunoprecipitation assays, p62 appeared equally precipitated from WT versus Stk3+/−;Stk4−/− MEFs expressing GFP::LC3 (Figure S6A), suggesting that cargo recruitment, at least of p62, was not significantly compromised in Stk3+/−;Stk4−/− cells (consistent with this notion, equal recruitment of bacterial cargo is reported below, Figure S7E). Collectively, these data indicate that the accumulated autophagosomes in Stk3+/−;Stk4−/− cells are fully formed and capable of sequestering cargo.
Our analysis of autophagic flux using the tandem-tagged LC3 reporter revealed that the majority of LC3 in Stk3+/−;Stk4−/− cells exists as foci bound to autophagosomes (Figure 2D). Furthermore, no change in LC3::GFP-positive foci occurred when Stk3+/−;Stk4−/− cells were challenged in a manner to either activate or block autophagy (Figure 2F). These results coupled with our findings that autophagosome formation and cargo recruitment were largely unaffected in Stk3+/−;Stk4−/− cells (Figures 6A & S6A) led us to test whether LC3-Thr50 phosphorylation promotes fusion of autophagosomes to lysosomes. We addressed this question by analyzing colocalization between GFP::LC3 foci and LAMP1 protein [lysosomal marker (Klionsky et al., 2012)] in Stk3+/−;Stk4−/− cells. This analysis revealed that autophagosomes (marked by GFP::LC3) were minimally colocalized with lysosomes (Figure 6A). Moreover, induction of autophagy by starvation induced robust colocalization of GFP::LC3 foci and LAMP1 protein in WT cells, whereas Stk3+/−;Stk4−/− cells only displayed partial colocalization (Figure S6B & C), further supporting the notion of a block in fusion with autophagosomes.
Strikingly, LAMP1 staining of Stk3+/−;Stk4−/− cells revealed a dramatic clustering of lysosomes around the perinuclear region in the cell even under basal conditions, a staining pattern not evident in WT MEFs (Figure 6B). Of note, we never observed such a clustering of LC3-positive autophagosomes around the perinuclear region in Stk3+/−;Stk4−/− MEFs, using several different assays (Figures 2, 5 & 6A). This observation may imply a defect in the transport of autophagosomes to the lysosomes for fusion (see Discussion). Although their cellular distribution was altered, lysosomes in Stk3+/−;Stk4−/−cells appeared to be functional as we found no measurable difference in activity of cathepsin B, a lysosome-residing enzyme, between WT and Stk3+/−;Stk4−/− MEFs (Figure S6D). Importantly, stable introduction of LC3-Thr50E into MEFs restored the normal distribution pattern of lysosomes in Stk3+/−;Stk4−/− cells, whereas cells expressing the Thr50A mutation phenocopied the strong perinuclear clustering of lysosomes under basal conditions (Figures 6B & C). Together, these data support the notion that loss of LC3-T50 phosphorylation triggers an autophagy block due to a potential defect in the recruitment of factors essential for fusion of autophagosomes with lysosomes, although we cannot rule out other mechanisms participating in the autophagy block.
STK3/STK4 phosphorylation of LC3 is important for autophagy-mediated elimination of intracellular bacteria
Finally, we investigated whether LC3-Thr50 phosphorylation is important for the physiological function of autophagy in pathogen defense. STK4 deficiency in humans is associated with increased susceptibility to bacterial infections (Abdollahpour et al., 2012; Dong et al., 2009; Nehme et al., 2011). Given that autophagy is known to play a role in clearance of intracellular pathogens (Sumpter and Levine, 2010), our discovery that STK3/STK4 deficiency severely compromises autophagy raises the possibility that STK4 plays a role in anti-microbial immunity through its essential role in autophagy.
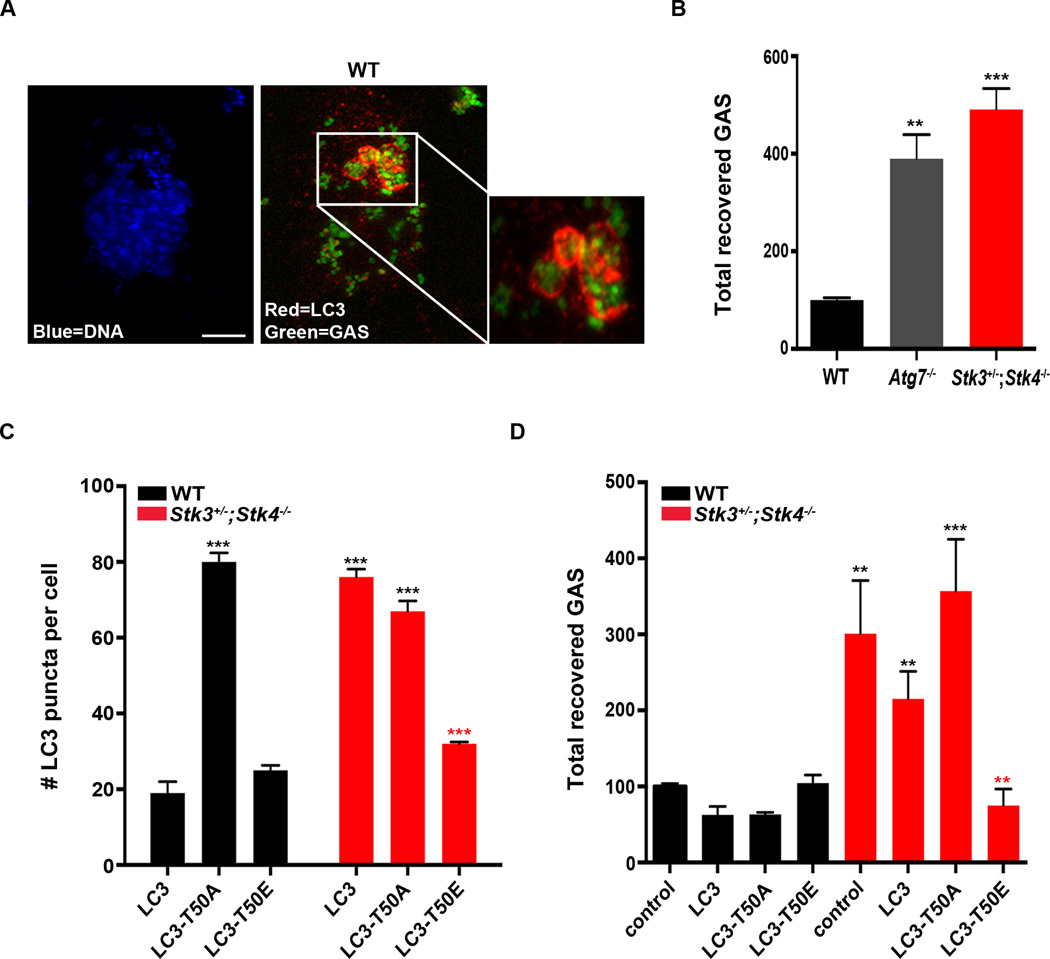
To address this, we examined the ability of Stk3+/−;Stk4−/− MEFs to clear infection with the intracellular bacterial pathogen group A streptococcus (GAS). GAS are Gram-positive bacteria that are known to be cleared by autophagy and can be detected sequestered within LC3-coated autophagosomes in infected mammalian cell lines, including MEFs (Barnett et al., 2013; Nakagawa et al., 2004). Therefore, we examined LC3 puncta formation and bacterial clearance In WT and Stk3+/−;Stk4−/− MEFs infected with FITC-labeled GAS for 2 h. Indeed, fluorescent bacteria were readily observed encapsulated within LC3-coated autophagosomes (Figure 7A & S7E), indicative of successful capture by autophagy. GAS clearance was assessed by enumerating colony-forming units (CFU) recovered from WT, autophagy-deficient Atg7−/−, and Stk3+/−;Stk4−/− MEFs 2 h post-infection. GAS survival was significantly higher in Atg7−/− MEFs than in WT MEFs, underscoring the important role of autophagy in bacterial clearance. Notably, GAS survival was even higher in Stk3+/−;Stk4−/− MEFs (Figure 7B), supporting the critical role of LC3 phosphorylation by STK3/STK4 in autophagy-mediated clearance of this pathogen.
Figure 7. Phosphorylation of LC3 threonine 50 is critical for clearance of intracellular bacteria by mammalian cells.
(A) Confocal fluorescence micrographs of wild-type (WT) MEFs incubated with FITC-labeled group A streptococci (GAS, green) for 2 h and then stained for endogenous LC3 (red) and nuclei (DAPI, blue). Note that DAPI stains several GAS cells in addition to the single MEF nucleus. Scale bar = 10 µm. Inset is an enlargement of the boxed area showing FITC-labeled GAS surrounded by LC3-bound autophagosomes.
(B) Quantification of intracellular GAS recovered 2 h after infection of WT, Atg7−/−, and Stk3+/−;Stk4−/− MEFs. Mean ± SEM, **P < 0.01, ***P < 0.005 to WT, one-way ANOVA. Data are representative of 4 biological repeats.
(C) Quantification of GFP::LC3 puncta in WT and Stk3+/−;Stk4−/− MEFs stably expressing GFP-tagged WT LC3, LC3-T50A, or LC3-T50E. Mean ± SEM from at least 20 cells. ***P < 0.001, one-way ANOVA. Black and red asterisks indicate comparisons to WT and Stk3+/−;Stk4−/− cells expressing WT LC3, respectively. Data are representative of 3 biological repeats.
(D) Quantification of intracellular GAS recovered 2 h after infection of WT and Stk3+/−;Stk4−/− MEFs stably expressing WT LC3, LC3-T50A, or LC3-T50E. Mean ± SEM. ***P < 0.005, **P < 0.01, *P < 0.05, one-way ANOVA. Black and red asterisks indicate comparisons to control WT and control Stk3+/−;Stk4−/− cells, respectively. In 3 independent experiments, GAS clearance by WT cells was slightly improved by stable expression of LC3 or LC3-T50A, whereas clearance by Stk3+/−;Stk4−/− cells was slightly impaired by LC3-T50A expression.
See also Figure S7.
To confirm this notion, we examined GAS clearance in MEFs stably expressing LC3-Thr50A or LC3-Thr50E; in particular, we wished to determine whether the phosphomimetic LC3-Thr50E could reverse the autophagy block observed in Stk3+/−;Stk4−/− MEFs. Towards this goal, we generated and characterized WT and Stk3+/−;Stk4−/−MEFs stably expressing full length, phosphomimetic and phospho-null versions of GFP::LC3. These stable lines expressed similar levels of LC3 protein (Figure S7A) and number of GFP::LC3 positive foci compared to WT and Stk3+/−;Stk4−/− MEFs transiently expressing the same constructs (compare Figures 7C & S7B–S7D with Figure 5). Additionally, the stable cell lines displayed no significant difference in their ability to engulf GAS into LC3-positive autophagosomes (Figure S7E), further supporting our conclusion that cargo recruitment is largely unimpaired in Stk3+/−;Stk4−/− MEFs (Figure S6A). We found that, whereas expression of WT LC3 slightly improved the capacity of Stk3+/−;Stk4−/− MEFs to clear GAS, cells expressing LC3-Thr50E reduced the GAS load as efficiently as did WT MEFs (Figure 7D). In contrast, bacterial clearance of Stk3+/−;Stk4−/−MEFs expressing the LC3-Thr50A mutant construct was not improved, and in fact appeared slightly impaired (Figure 7D).
Collectively, the results of this study strongly support a critical and evolutionarily conserved role for STK3/STK4 in the autophagic response. STK3/4 mediates these effects, at least in part, via phosphorylation of LC3, and this mechanism contributes to the response of mammalian cells to infection with intracellular pathogens.
DISCUSSION
In this study, we have uncovered a conserved mode of autophagy regulation by identifying the kinases STK3 and STK4 as requisite factors for autophagy in organisms spanning the phylogenetic scale. In mammals, these kinases phosphorylate at least one amino acid, Thr50, of the autophagosome component LC3B, and this post-translational modification is important for mediating fusion of autophagosomes with lysosomes. This discovery expands our fundamental understanding of how kinases affect events downstream of autophagy initiation by modulating the post-translational modifications of LC3. Furthermore, we provide evidence that at least one aspect of STK4 function in immunity, namely clearance of intracellular bacteria, can be tied to its regulation of autophagy; an observation with potential clinical importance in humans.
Our data suggest that STK3/4-mediated LC3-Thr50 phosphorylation is likely not required for autophagosome completion or for cargo recruitment, but instead plays a critical role in mediating fusion of autophagosomes with lysosomes. Intriguingly, the distribution of lysosomes itself is dramatically altered in Stk3+/−;Stk4−/− cells, whereas the distribution of autophagosomes appears to be unchanged. We suggest that the lysosomal mislocalization phenotype we observe occurs as a consequence of the block in fusion. When cells perceive an autophagy stimulus, lysosomes at the periphery accumulate densely around the perinuclear region to fuse with completed autophagosomes that are delivered by microtubule-dependent transport. Hybrid organelles that are generated during fusion play an essential role in reformation of lysosomes and membrane-retrieval (Luzio et al., 2007). We propose that LC3-Thr50 phosphorylation could be essential for recruiting transport- (e.g., Rab7) and/or fusion machinery (e.g., RABs, SNAREs and HOPS complex) to the completed autophagosomes to enable the fusion process with lysosomes. Absence of fusion might result in increased accumulation of lysosomes at the perinuclear region. Indeed similar lysosomal-clustering phenotypes have been observed in cells expressing mutant versions of RIPL (Rab7-interacting lysosomal protein), and in cells overexpressing Vps18p, a member of the HOPS complex (Jordens et al., 2001; Poupon et al., 2003). To our knowledge, LC3 family members have mainly been implicated in autophagosome biogenesis and completion (Weidberg et al., 2010); our study is therefore the first to suggest that LC3, via post-translational modification, can play a role in fusion of autophagosomes with lysosomes. Characterizing the molecular circuitry of how LC3-Thr50 phosphorylation may regulate fusion of autophagosomes with lysosomes will be a key future research area.
STK3/STK4 are the central kinases of the conserved Hippo signaling cascade, which plays a critical role in organ development by controlling cell proliferation and death. Activation of STK3/STK4 and other pathway components, results in phosphorylation and retention of the transcriptional coactivator complex YAP/TAZ within the cytoplasm. However, when STK3/4 and their signaling components are inactivated, YAP/TAZ complex translocates to the nucleus and activates the expression of genes that promote proliferation and survival. Thus, the ability of STK3/STK4 to antagonize YAP/TAZ activity is consistent with the role of these kinases as tumor suppressors (Halder and Johnson, 2011). Whether activation of this canonical pathway might also contribute to the function of STK3/STK4 in autophagy remains to be determined. In support of this possibility, autophagy is also blocked by loss-of-function mutations in other members of the Drosophila Hippo (STK3/STK4) pathway; namely Fat and Warts, although the underlying mechanism by which this occurs is not known (Dutta and Baehrecke, 2008; Napoletano et al., 2011). It seems likely that the nature of the STK3/STK4-activating signal and its cellular context will determine how STK3/STK4 interact with components of the autophagy pathway. These components could include members of the Atg8 family of proteins, e.g., LC3 (LC3A/B/C) and GABARAP/GATE16 subgroups, which have been shown to bind STK3/4 kinases in vitro (Behrends et al., 2010). While our studies specifically focused on describing LC3B as an STK3/STK4 substrate, it is possible that other members of the LC3- and GABARAP/GATE16 subgroups also function as substrates; future experiments are required to address this question. While we found that STK3/STK4 kinases can act downstream of autophagy activation by targeting LC3, a recent study of a mouse model of myocardial infarction showed that STK4 can instead block autophagy by directly phosphorylating Beclin1 which is integral for activation of the autophagy pathway (Maejima et al., 2013). Taken together with the findings of our study, this raises the possibility of multifaceted roles for STK3/STK4 kinases in autophagy, possibly mediated through multiple substrates.
An important aspect of our finding is that STK3/STK4 kinases are also required for autophagy in S. cerevisiae and C. elegans. However, Thr50 is conserved only in vertebrate LC3, suggesting that in these organisms, phosphorylation of LC3 orthologs by STK3/STK4 kinases might occur at alternative site(s) or through an independent mechanism. Consistent with an autophagy-promoting function for STK4, forced expression of cst-1/Stk4 extends lifespan in C. elegans (Lehtinen et al., 2006), whereas C. elegans lacking cst-1/Stk4 are short-lived (data not shown). Considering the strong evidence that an intact autophagic machinery is integral to ensuring optimal lifespan in diverse species (Gelino and Hansen, 2012), these observations suggest that cst-1/Stk4 might modulate C. elegans lifespan via its autophagy function.
Lastly, our novel findings have exciting clinical implications, particularly in the link between STK3/STK4, autophagy, and immunity. STK4-deficient humans and mice have impaired immune function and increased susceptibility to bacterial and viral infections (Abdollahpour et al., 2012). We found that reconstitution of Stk3+/−;Stk4−/− MEFs with phosphomimetic LC3-Thr50E reversed the defect in bacterial clearance, raising the possibility that the immunodeficiency associated with STK4 loss in mammals could be related to its autophagy function. Moreover, increasing evidence supports a strong role for autophagy in the development and function of the adaptive immune system (Sumpter and Levine, 2010). Impairment in the autophagy process has been linked to many disorders in humans, including cancer, diabetes, Alzheimer’s disease, and cardiac dysfunction. The novel STK3/STK4–LC3 regulatory axis identified in this study may therefore reveal new targets for the development of drugs to treat pathologies associated with aberrant autophagy.
EXPERIMENTAL PROCEDURES
Transfections
Transient transfections were performed using 0.5–3.0 µg of target DNA and Lipofectamine 2000 in Opti-MEM (Life Technologies), according to the manufacturer's instructions. For RNAi experiments, MEFs were transfected with 20 nM SMARTpool siRNA against mouse STK3 and STK4 (Dharmacon L-040155-00-0005 and L-040619-00-0005, respectively) using RNAiMAX (Life Technologies), according to the manufacturer’s instructions.
Western blotting, Co-immunoprecipitation and In vitro kinase assays
MEFs were either left untreated or incubated in starvation medium (Earle's Balanced Salt Solution; Life Technologies) and 50 nM BafA (BafilomycinA, Sigma). At the indicated time points, cells were lysed and protein extracts were prepared and subjected to standard western analysis followed by incubation with individual primary antibodies. For Co-immunoprecipitation analysis, MEFs were transfected with vector encoding GFP::LC3 for 36 h and then lysed in cold IP-lysis buffer for 30min. 5% aliquot of the lysate was reserved as the input and the remainder was incubated with equilibrated GFP-Trap beads (Allele Biotechnology) and processed according to manufacturers instructions. For kinase assays, recombinant STK3 or STK4 (5–10 ng per reaction; Sigma) was incubated for 30 min at 30°C with 1–5 µg of recombinant His-LC3B or GFP::LC3 immunoprecipitated from cells in the presence of 1mM cold ATP or 10 µCi 32P-ATP (Perkin Elmer). The reactions were stopped by boiling in 2X SDS sample buffer and the proteins were resolved by SDS-PAGE.
Autophagy assays in mammalian cells
Mouse embryonic fibroblasts (MEFs) were plated onto glass coverslips and were subsequently transfected with the indicated plasmids for 36 h. The cells were then left untreated, incubated in starvation medium for 2 h, or in medium containing 50 nM bafilomycin A1 (BafA, Sigma) or 200 nM rapamycin (Sigma) for 2 h. Longer incubations were found to compromise the survival of Stk3+/−;Stk4−/− MEFs (data not shown). Cells were fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized in 0.2% Triton X-100 in PBS for 10 min, and then processed according to Cell Signaling immunofluorescence protocol (www.cellsignal.com). Images were acquired by confocal microscopy and LC3 puncta were quantified using IMARIS spot-counting software.
Infection with group A streptococcus
MEFs were exposed to Group A streptococcus (GAS) strain NZ131 (serotype M49) for 1 h at 37°C in a 5% CO2 incubator. For enumerating bacterial colony forming units (CFU), the treated cells were detached and lysed. The lysates were serially diluted and plated on Todd-Hewitt agar for enumeration of bacterial CFU.
Statistical analysis
Group means were compared by two-tailed Student’s t-test or one-way ANOVA using Prism 5.0 software (GraphPad). A P value of ≤0.05 was considered significant.
Additional methods can be found in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. A. O’Rourke, D. Rubinsztein, K. Thiedick, M. Wilkinson, D. Wolf, as well as members of the Hansen laboratory for helpful discussions and/or critical reading of the manuscript. We thank Drs. A.-M. Cuervo, R. Shaw, J. Chernoff and N. Mizushima for the mCherry::GFP::LC3, GFP::LC3, HA-STK4 and STX17 constructs, respectively. We thank Drs Y. Zhang and R. Johnson for Stk3+/−;Stk4−/− MEFs. We acknowledge the National Bioresource Project for the Nematode, for the cst-1(tm1900) deletion allele. We thank Dr. M. Wood (The Scripps Research Institute) for help with TEM analyses and Dr. L. Brill (SBMRI-Proteomics Facility), for mass spectrometry experiments. We thank Drs. Y. Sigal, J. Kasuboski, and J. Fitzpatrick from the Salk Institute Waitt Advanced Biophotonics Center {supported by the Waitt Foundation, the W.M. Keck Foundation, NCI (CA014195-40), and NINDS (NS072031-03A1)} for assistance with confocal microscopy. This work was funded by Glenn Foundation for Aging Research fellowship to DSW, an UNCF/Merck postdoctoral fellowship to ELA, NIH grants P41 GM103504 and P50 GM085764 to KM and TI, NIH/NCI grants CA80100 and CA82683 to TH and JM, NIH/NIAID grants AI096837 and AI077780 to VN, and NIH/NIA grants AG038664 and AG039756 to MH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
DSW and MH designed the study and interpreted the data. ELA performed and analyzed the GAS experiments in VN’s lab, KM performed and analyzed the yeast experiments in TI’s lab, and JM performed the radiolabeled in vitro kinase assays in TH’s lab. JTC performed BafA injections and flux analysis in C. elegans in MAH’s lab. DSW performed and analyzed all other experiments in AD and MH’s labs with assistance from JS. DSW and MH wrote the manuscript with feedback from co-authors.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- Abdollahpour H, Appaswamy G, Kotlarz D, Diestelhorst J, Beier R, Schaffer AA, Gertz EM, Schambach A, Kreipe HH, Pfeifer D, et al. The phenotype of human STK4 deficiency. Blood. 2012;119:3450–3457. doi: 10.1182/blood-2011-09-378158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, Davies MR, Schulz BL, Nizet V, Teasdale RD, et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe. 2013;14:675–682. doi: 10.1016/j.chom.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Puri C, Moreau K, Rubinsztein DC. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci. 2013;126:1059–1069. doi: 10.1242/jcs.123075. [DOI] [PubMed] [Google Scholar]
- Dong Y, Du X, Ye J, Han M, Xu T, Zhuang Y, Tao W. A cell-intrinsic role for Mst1 in regulating thymocyte egress. J Immunol. 2009;183:3865–3872. doi: 10.4049/jimmunol.0900678. [DOI] [PubMed] [Google Scholar]
- Dutta S, Baehrecke EH. Warts is required for PI3K–regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr Biol. 2008;18:1466–1475. doi: 10.1016/j.cub.2008.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelino S, Hansen M. Autophagy - An Emerging Anti-Aging Mechanism. J Clin Exp Pathol. 2012;(Suppl 4) doi: 10.4172/2161-0681.s4-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Hall DH, Hansen M. Monitoring the role of autophagy in C. elegans aging. Methods Enzymol. 2008;451:493–520. doi: 10.1016/S0076-6879(08)03229-1. [DOI] [PubMed] [Google Scholar]
- Moreau K, Renna M, Rubinsztein DC. Connections between SNAREs and autophagy. Trends Biochem Sci. 2012;38:57–63. doi: 10.1016/j.tibs.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Napoletano F, Occhi S, Calamita P, Volpi V, Blanc E, Charroux B, Royet J, Fanto M. Polyglutamine Atrophin provokes neurodegeneration in Drosophila by repressing fat. EMBO J. 2011;30:945–958. doi: 10.1038/emboj.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme NT, Pachlopnik Schmid J, Debeurme F, Andre-Schmutz I, Lim A, Nitschke P, Rieux-Laucat F, Lutz P, Picard C, Mahlaoui N, et al. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T-cell survival. Blood. 2011;119:3458–3468. doi: 10.1182/blood-2011-09-378364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupon V, Stewart A, Gray SR, Piper RC, Luzio JP. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol Biol Cell. 2003;14:4015–4027. doi: 10.1091/mbc.E03-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Levine B. Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol. 2010;21:699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hata Y. What is the Hippo pathway? Is the Hippo pathway conserved in Caenorhabditis elegans? J Biochem. 2013;154:207–209. doi: 10.1093/jb/mvt060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.