Abstract
Background and Aims Allelopathy may drive invasions of some exotic plants, although empirical evidence for this theory remains largely inconclusive. This could be related to the large intraspecific variability of chemically mediated plant–plant interactions, which is poorly studied. This study addressed intraspecific variability in allelopathy of Heracleum mantegazzianum (giant hogweed), an invasive species with a considerable negative impact on native communities and ecosystems.
Methods Bioassays were carried out to test the alleopathic effects of H. mantegazzianum root exudates on germination of Arabidopsis thaliana and Plantago lanceolata. Populations of H. mantegazzianum from the Czech Republic were sampled and variation in the phytotoxic effects of the exudates was partitioned between areas, populations within areas, and maternal lines. The composition of the root exudates was determined by metabolic profiling using ultra-high-performance liquid chromatography with time-of-flight mass spectrometry, and the relationships between the metabolic profiles and the effects observed in the bioassays were tested using orthogonal partial least-squares analysis.
Key Results Variance partitioning indicated that the highest variance in phytotoxic effects was within populations. The inhibition of germination observed in the bioassay for the co-occurring native species P. lanceolata could be predicted by the metabolic profiles of the root exudates of particular maternal lines. Fifteen compounds associated with this inhibition were tentatively identified.
Conclusions The results present strong evidence that intraspecific variability needs to be considered in research on allelopathy, and suggest that metabolic profiling provides an efficient tool for studying chemically mediated plant–plant interactions whenever unknown metabolites are involved.
Keywords: Allelopathy, compound identification, germination bioassay, giant hogweed, Heracleum mantegazzianum, Apiaceae, invasion ecology, invasive species, metabolic profile, novel weapons hypothesis, OPLS analysis, plant metabolomics, Plantago lanceolata, root exudates, UHPLC–TOF–MS
INTRODUCTION
According to the novel weapons hypothesis, some invasive exotic species possess novel chemical compounds that have allelopathic effects in new ranges, which facilitate their invasion (Callaway and Aschehoug, 2000; Hierro and Callaway, 2003; Callaway and Ridenour, 2004). However, many studies failed to prove allelopathy as an important driver of invasion (Blair et al., 2006; Wille et al., 2013; Del Fabbro et al., 2014), which may have resulted from methodological difficulties of investigations concerning allelopathy. First, when allelopathy is studied experimentally, the examined plant extract or leachate contains putative allelochemicals, but it also causes changes in pH, osmotic potential and nutrient concentrations and availability, making it difficult to distinguish allelopathy from resource competition (Inderjit and Callaway, 2003). Second, laboratory bioassays do not directly describe ecological complexity. For example, phytotoxicity found in vitro is not necessarily applicable under field conditions, because it can be significantly modified by the soil biota (Kaur et al., 2009; Inderjit and van der Putten, 2010; Lankau, 2010). Third, effects of bioactive compounds are often concentration-dependent, though the estimation of environmental concentrations of allelochemicals is not straightforward, as they are subject to differential adsorption and degradation (Barto and Cipollini, 2009).
Another source of inconclusive results of studies on allelopathy is likely to be intraspecific variation in the production of allelochemicals. Previous studies have mostly considered allelopathy as a species trait, and when intraspecific differences were tested they were focused on biogeographical comparisons (e.g. Thorpe et al., 2009). However, within individual plant species there are different levels of spatial and temporal variation in metabolite composition and concentration (Macel et al., 2010). This intraspecific metabolic variability often reflects community dynamics and ecosystem structure. For example, Lankau et al. (2009) demonstrated that allelopathy strength was related to the invasion history of the invader, with younger populations being more phytotoxic, likely due to a higher probability of competition with native heterospecifics.
Heracleum mantegazzianum (Apiaceae, giant hogweed) is a very well studied invasive plant that has a strong negative impact on the structure of native communities (Dostál et al., 2013) and the properties of entire ecosystems (Jandová et al., 2014a). Junttila (1976) and Baležentienė (2012) reported invasive conspecifics of H. mantegazzianum to be allelopathic; however, Wille et al. (2013) found only limited evidence of its allelopathic effect on the germination of native plants. In a previous study (Jandová et al., 2014b) we found that the allelopathic effects of H. mantegazzianum exudates in the introduced range varied across several seasons. Apart from possible seasonal variation and despite the origin of all populations sampled for seeds being the Czech Republic, the source of such variability could be considerable intraspecific variation. We suggest that this variation is likely due to differences in the presence and concentrations of allelochemical compounds.
Since the allelochemicals of H. mantegazzianum are unknown, we employed untargeted metabolomics to reveal differences in metabolic profiles between maternal lines within the species. Recently, untargeted metabolomics has been used in ecology and environmental science. Atkinson et al. (2012) used untargeted metabolomics to test an assumption of life history theory that expects slower-growing species to invest proportionally more resources in storage, and compared the metabolic profiles of storage organs of slower- and faster-growing plants. Davey et al. (2008, 2009) described the effect of a latitudinal climate gradient on the metabolic profile of different populations of Arabidopsis lyrata and later tested the effect of cold on their metabolic changes in a manipulative experiment. Moreover, untargeted metabolomics has proved to be a good tool for identifying bioactive metabolites in the field of chemical ecology (Nylund et al., 2011; Marti et al., 2013).
Here, we addressed the intraspecific variability of allelopathy in H. mantegazzianum using bioassays of its root exudates in combination with metabolic profiling based on ultra-high-performance liquid chromatography (UHPLC) with time-of-flight (TOF) mass spectrometry (MS) detection. Specifically, our aims were (1) to estimate the allelopathic effects of root exudates on germination and root development of assay species, (2) to partition the variation in allelopathic effect of H. mantegazzianum between areas, populations within areas and maternal lines within populations, and (3) to link the allelopathic effects to metabolic profiles and tentatively identify compounds responsible for allelopathy.
MATERIALS AND METHODS
Plant material
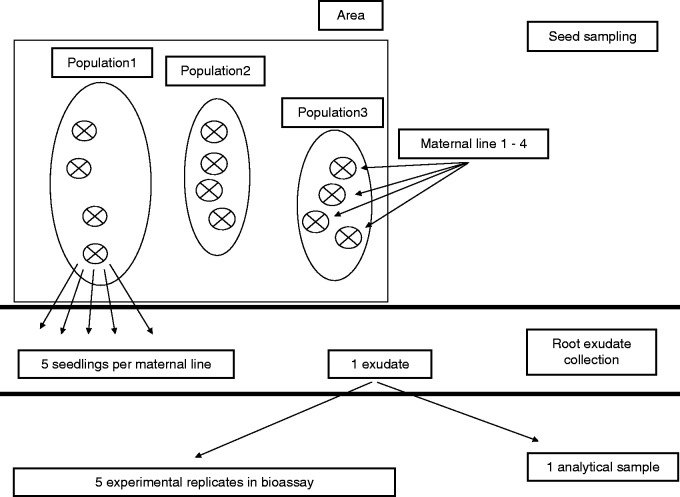
Seeds of Heracleum mantegazzianum were collected from separate maternal plants from 12 populations in six areas. Five of the six areas were in the western part of the Czech Republic, the average distance between these areas was 7 km, and the average distance between populations within these areas was 500 m. One area containing one population was in the central part of the Czech Republic. Seeds were collected at sites invaded for different periods of time ranging from 10 to 50 years and mostly with mesic grasslands (pre-invasion dominants: Poa trivialis, Dactylis glomerata, Festuca rubra and Agrostis capillaris; Dostál et al., 2013). In each population, we sampled seeds from four randomly selected maternal plants (at a minimum distance of 5 m from each other), reported hereafter as maternal lines (for details of the experimental design see Fig. 1 and Supplementary Data Fig. S1). Altogether we had 41 different maternal lines. All seeds were collected in summer 2012, air-dried and stored in paper bags (seeds from one maternal line in one paper bag) at room temperature. Later, they were cold-stratified in wet sterilized sand at 4 °C in the dark for 2½ months, and then allowed to germinate on wet sterilized sand in the greenhouse with a 20°/10 °C temperature regime. The experiment took place at the Institute of Botany, Průhonice, Czech Republic (322 m asl; 49 °99′N, 14 °57′E). After 2 weeks of growth, the seedlings were transplanted into 50-mm mesh pots and transferred in randomized order to aeroponic growing systems (Amazon 32, Nutriculture, Skelmersdale, UK) placed in the same greenhouse and grown in 0·25-strength Hoagland solution for 1 month. Finally, the plants were collected, and five individuals of one maternal line were placed in one glass bottle with 200 mL of distilled water covered with aluminium foil to keep out the light. The roots were soaked for ∼20 h. The water enriched with root exudates was vacuum-filtered on glass fibre filters (0·7 µm mesh; Vitrum, Czech Republic), and 4 mL of each sample was taken for a bioassay. The rest was freeze-dried and kept at –20 °C until metabolic profiling using UHPLC–TOF–MS. This procedure yielded on average 35 mg of dry material per maternal line.
Fig. 1.
Experimental design of the study.
Bioassays
Allelopathic effects of root exudates were assayed on commercially purchased seeds of Arabidopsis thaliana (Columbia-0 ecotype) and Plantago lanceolata (Planta Naturalis, Markvartice, Czech Republic). These species were chosen based on the facts that the former represents the standard target species and that the latter represents a species common in grassland communities documented to be invaded by H. mantegazzianum (Dostál et al., 2013). Distilled water was used as the control. Petri dishes (6 cm diameter) were fitted with qualitative filter paper (Papírna Perštejn, Czech Republic), ten seeds of the assay species per dish were placed in, 4 mL of root exudate or the control was pipetted in, and the Petri dishes were placed in a chamber with a 25/10 °C temperature and 12/12 h light regime. Bioassays started immediately after the exudates were collected. After 7 d we recorded the number of germinated seeds of A. thaliana. For P. lanceolata we recorded the number of germinated seeds and the root length of three largest seedlings, both scored 10 d after the experiment was initiated. We used five replicates per maternal line in the bioassay.
Bioassay data analyses
Within-study meta-analysis of allelopathic effects
We used a within-study meta-analytical approach to compare allelopathic effects between 41 maternal lines of H. mantegazzianum. Effect sizes were expressed as log-response ratios, calculated as the natural logarithm of the mean bioassay response to the respective exudate divided by the mean bioassay response to the control. Thus, negative values of log-response ratios indicated that the maternal line was allelopathic whereas positive values indicated that the maternal line stimulated germination or root growth of the assay species. The log-response ratios were calculated based on five replications per maternal line and for each bioassay response (A. thaliana germination, P. lanceolata germination, P. lanceolata root length) separately. The analyses were performed in the ‘metafor’ package (Viechtbauer, 2010) in R 3.0.2 (R Development Core Team, 2013). First, we calculated the effect sizes (log-response ratios) from the bioassay responses using the ‘escalc’ function. Second, the effect sizes were analysed with random models to estimate the overall allelopathic effect. Third, the mixed-effects models with the exudates’ dry weights entered as moderator (i.e. covariate) were fitted to estimate the allelopathic effect independently of the amount of root exudate produced.
Partitioning of variation in allelopathic effects
We carried out a variance component analysis to partition the overall variation in allelopathic effects of H. mantegazzianum between areas, populations within areas and maternal lines within populations using the ‘MCMCglmm’ function in the ‘MCMCglmm’ package (Hadfield, 2010) in R 3.0.2 (R Development Core Team, 2013). The areas, populations and maternal lines were entered as random terms and we used default priors for the G and R variance structures. The variance was extracted from the models together with the 95 % confidence intervals. The variance component analysis was calculated for bioassay responses of P. lanceolata only since there was not an allelopathic effect of exudates on A. thaliana. Germination and root length were analysed separately, based on five replications per maternal line and the root length was log-transformed before analyses to meet the model assumptions.
Metabolite extraction and UHPLC–TOF–MS-based metabolic profiling
The extraction procedure proposed by De Vos et al. (2007) for untargeted metabolomics studies was followed, except for different extraction ratios. Before metabolite extraction, the freeze-dried root exudates were weighed to allow later concentration correction. Then, 2·4 mL of chilled 50 % methanol, 0·1 % formic acid in water was added. After immediate vortexing, the extracts were sonicated for 15 min at room temperature and then filtered through 0·2 µm nylon filters (Costar, USA) in a centrifuge (10 min, 2347 g). The extracts were prepared freshly at the beginning of analysis.
The samples were analysed on an Acquity UPLC system with an LCT premier XE TOF mass spectrometer (Waters) using an LC Column Acquity UPLC BEH C18 (50 mm × 2·1 mm internal diameter, particle size 1·7 µm, Waters) and a two-component mobile phase. Mobile phases A and B consisted of 0·1 % formic acid in water and 0·1 % formic acid in acetonitrile, respectively. The analyses were performed under a linear gradient programme (min/%B) of 0/5, 1·5/5, 15/70, 21/95, 23/98 followed by a 1-min equilibration (5 % B). The total analysis time was 25 min. The column temperature was set at 40 °C and flow rate at 0·4 mL min–1, and the injection volume was 5 µL. The mass spectrometer operated in W mode with capillary voltage set at +2800 (positive ionization mode) or –2500 V (negative ionization mode), cone voltage +40 or –40 V, desolvation gas temperature 350 °C; ion source block temperature 120 °C, cone gas flow 50 L h–1, desolvation gas flow 800 L h–1, ion guide 1 and 2 radio frequencies (RFs) 200 and 300 V, respectively, and hexapole RF 150 V. Ions in the m/z range 100–1500 were detected using a scan time of 0·1 s and an inter-scan delay of 0·01 s. The mass accuracy was kept below 5 ppm using lock spray technology with leucine enkephalin as the reference compound (2 ng µl–1, 5 µl min–1). Fragmentation by collision-induced dissociation (CID) was triggered by setting the aperture 1 value at 30–50 V. The CID fragmentation spectra were recorded under the chromatographic conditions described earlier (system 1) and using the same chromatographic system modified by replacing acetonitrile with methanol as part B of the mobile phase (system 2). Since no significant differences among triplicates of initially analysed samples were observed, only one replicate per sample was measured. Samples were run in random order in a single batch and MS detection was performed in both negative and positive modes in independent runs. The data were processed using MassLynx V4.1 (Waters).
To confirm compound identification, we compared retention times and fragmentation patterns with commercially available standards of bergapten (Aldrich), xanthotoxin (Fluka) and tryptophan (Sigma-Aldrich), which were dissolved in methanol and analysed using the same protocol as described above.
Metabolomic data processing
The LC–MS data were automatically processed using MetAlign version 3 (Lommen and Kools, 2012). Baseline and noise calculations were performed from scan 10 to scan 3500, corresponding to retention times of 1·0–16·0 min. The maximum amplitude was set to 40 000, and peaks below twice the local noise were discarded. For subsequent data analyses, the data matrices (retention time × accurate m/z × peak intensity) from positive and negative ionization modes were used separately. The matrices were reduced by discarding variables with maximum signal below the intensity of 100. All data matrices were then normalized to total signal intensity by observation and then log-transformed and centred by variables. Orthogonal partial least-squares analysis (OPLS) was used to test whether allelopathy of exudates could be explained by their metabolic profile and to mine out metabolomic peaks (characterized by retention time × accurate m/z) most related to the magnitude of the allelopathic effect in the bioassay from significant models. The OPLS method is designed to separate the predictive part of the data related to the y variable from variation that is not related to the response (Trygg and Wold, 2002). Here, we used effect sizes of responses in the bioassay as y variables. We used OPLS cross-validation–analysis of variance (CV-ANOVA) (Eriksson et al., 2008) to test the significance of each model. To indicate compounds associated with allelopathy, loading column plots were produced, and peaks (characterized by retention time × accurate m/z) most related to the magnitude of the allelopathic effect were extracted. Furthermore, the models were examined to identify possible influential outliers using Hotelling’s T2, which is the sum over all components of scores squared divided by their squared standard deviation. All multivariate data analyses were performed using SIMCA-P software (Umetrics AB, Umeå, Sweden). In addition, we tentatively identified compounds corresponding to extracted metabolomic peaks by processing the LC–MS data. Elemental compositions were proposed based on accurate mass and isotopic pattern recognition using MassLynx V4.1 (Waters). The resulting molecular formulae were submitted to the Reaxys and SciFinder databases. The proposed molecular structures were then confronted with our MS fragmentation experiments.
RESULTS
Within-study meta-analysis of allelopathic effects
Germination of A. thaliana was stimulated by H. mantegazzianum root exudates (Table 1, Fig. 2A). Conversely, germination and root length of P. lanceolata were significantly inhibited by the root exudates of H. mantegazzianum (Table 1, Fig. 2B, C). The effect of root exudates was thus species-specific. The maternal lines differed substantially in allelopathic effects (Table 1, Fig. 2); the maternal lines that suppressed germination the most also suppressed root length (t = 4·1891, d.f. = 39, P < 0·001, r = 0·557). To examine qualitative attributes only, i.e. exudate composition, we analysed the effect sizes together with exudate dry weight as a moderator (i.e. covariate). When the amount of root exudate produced was incorporated in the analyses, we failed to find any effect of exudates on germination of A. thaliana (Table 1). The exudate concentration did not modify the effect on P. lanceolata germination; however, the effect on root length of P. lanceolata was significantly and negatively dependent on the dry weight of root exudates, and as a result, incorporation of the moderator inverted the final effect size to positive values (Table 1). Our results therefore suggest that allelopathic effects are caused by both qualitative (composition) and quantitative (amount) properties of the root exudates.
Table 1.
Overall effect sizes estimated with random-effects models and effect sizes with dry weight of root exudates used as a moderator estimated with mixed-effects models, both by the restricted maximum-likelihood method. Shown are degrees of freedom (d.f.) as well as model estimates of effect sizes with their standard errors (s.e.) and Z-values and levels of significance for each response measured in the bioassay
| d.f. | Estimate | s.e. | Z-value | |
|---|---|---|---|---|
| Without moderator | ||||
| Germination of A. thaliana | 40 | 0 ·0601 | 0 ·0255 | 2 ·3618* |
| Germination of P. lanceolata | 40 | −0 ·5509 | 0 ·0410 | −13 ·4402*** |
| Root length of P. lanceolata | 40 | −0 ·2415 | 0 ·0736 | −3 ·2803** |
| With dry weight of exudates used as a moderator | ||||
| Germination of A. thaliana | ||||
| Intercept | 39 | 0 ·1174 | 0 ·0658 | 1 ·7823NS |
| Exudate dry weight | 1 | −1 ·4318 | 1 ·519 | −0 ·9426NS |
| Germination of P. lanceolata | ||||
| Intercept | 39 | −0 ·4444 | 0 ·1028 | −4 ·3245*** |
| Exudate dry weight | 1 | −2 ·7711 | 2 ·4568 | −1 ·1279NS |
| Root length of P. lanceolata | ||||
| Intercept | 39 | 0 ·3715 | 0 ·1598 | 2 ·3255* |
| Exudate dry weight | 1 | −15 ·4328 | 3 ·7478 | −4 ·1178*** |
*P < 0 ·05, **P < 0 ·01, ***P < 0 ·001; NS, not significant.
Fig. 2.
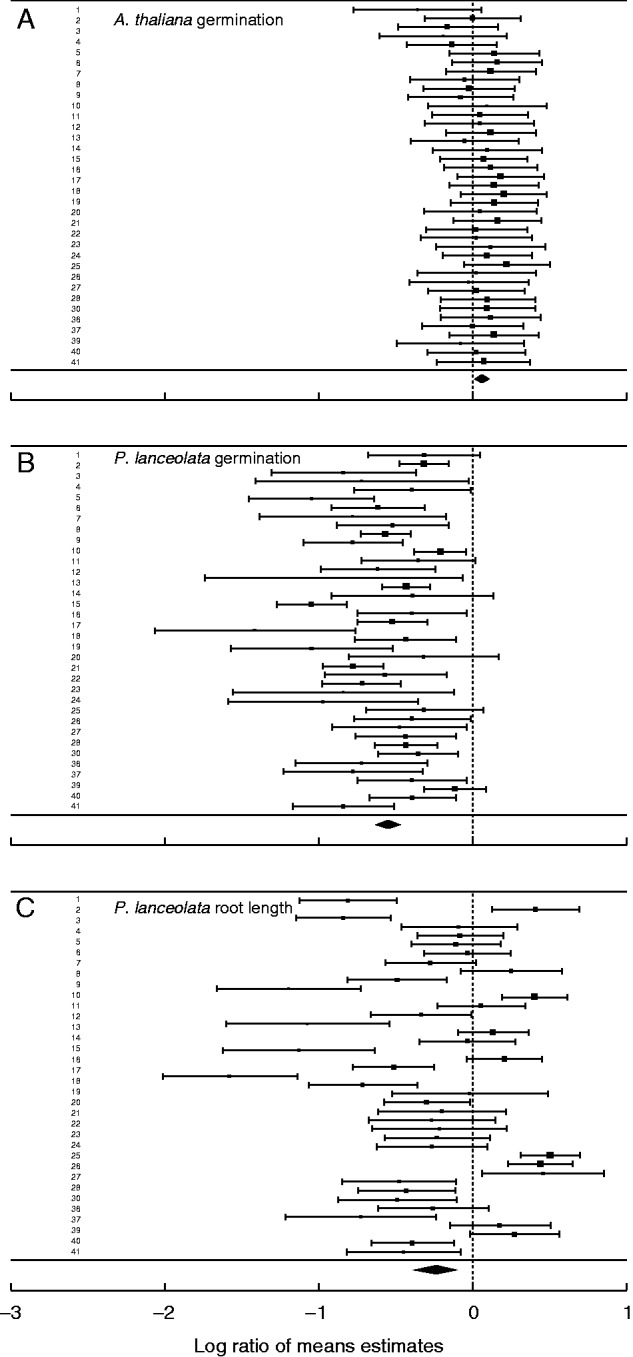
Overview of allelopathic effects. Effect sizes of root exudates of 41 maternal lines of H. mantegazzianum on germination of A. thaliana (A) and germination (B) and root length (C) of P. lanceolata. The figures show the log-response ratios with corresponding 95 % confidence intervals in the individual maternal lines and the estimates of overall effect size based on random-effects models fitted by the restricted maximum-likelihood method.
Partitioning of variation in allelopathic effects of H. mantegazzianum between areas, populations within areas and maternal lines within populations
Variance partitioning showed that the highest biological variation in allelopathic effects lay in variation within populations. Of the total variance in germination and root length of P. lanceolata, 11·62 and 39·62 %, respectively, was attributed to allelopathic differences between maternal lines. Differences in allelopathy between areas or between populations made negligible contributions to variation in the response variables (Table 2). The uncertainty was expressed as the 95 % confidence intervals of the estimates of variance proportions. Since our design for variance partitioning was not fully balanced and some of our areas were represented only by one population, we performed an additional analysis that omitted the level ‘area’, i.e. it partitioned the variance only between and within populations. Importantly, this did not change the main finding in any substantive way and the most biological variance was found again within populations (Supplementary Data Table S1).
Table 2.
Proportions of total variance (%) in allelopathic effects of H. mantegazzianum attributed to variation between areas, populations within areas, maternal lines within populations and residuals, together with the variance measures and their 95 % confidence intervals (CI)
| Between areas | Between populations | Within populations | Residuals | |
|---|---|---|---|---|
| Germination of P. lanceolata | ||||
| Proportion of variance (%) | 0.48 | 0.22 | 11.62 | 87.69 |
| Variance | 0.0172 | 0.00772 | 0.417 | 3.15 |
| Lower bound of 95 % CI | 1.38 × 10–16 | 9.45 × 10–17 | 2.10 × 10–10 | 2.40 |
| Upper bound of 95 % CI | 0.0892 | 0.0322 | 1.03 | 3.91 |
| Root length of P. lanceolata | ||||
| Proportion of variance (%) | 1.12 | 0.21 | 39.62 | 59.05 |
| Variance | 0.00538 | 0.001 | 0.190 | 0.283 |
| Lower bound of 95 % CI | 3.55 × 10–17 | 7.08 × 10–17 | 0.0915 | 0.225 |
| Upper bound of 95 % CI | 0.0317 | 0.00365 | 0.324 | 0.344 |
Linkage of allelopathic effects to metabolic profiles
Principal components analysis was performed as an exploratory step of data analysis to provide an unsupervised overview of the UHPLC–TOF–MS metabolic profiles (Supplementary Data Fig. S2). We used OPLS to model the respective bioassay response by the metabolic profile and to indicate peaks associated with allelopathy. We modelled both ionization modes and each bioassay response separately, resulting in the six particular models described in Table 3A. No statistically significant models were obtained to explain the effect of the metabolic profiles on A. thaliana germination. By contrast, the germination and root length of P. lanceolata were always significantly explained by the metabolic profile in OPLS (P < 0·05). Metabolomic peaks most related to the magnitude of the allelopathic effect in the bioassay were extracted from significant models (Fig. 3). We found one maternal line with significantly greater influence (P < 0·01) in our models according to Hotelling’s T2 (Supplementary Data Fig. S3). When this maternal line was excluded from the OPLS analyses, only the model explaining root length of P. lanceolata by the metabolic profile measured in negative ionization mode remained significant (Table 3B). Nonetheless, the peaks most related to the allelopathic effect in the latter model led to the same compounds as the peaks in models based on all observations (Supplementary Data Fig. S4).
Table 3.
Orthogonal partial least-squares (OPLS) models used to model the respective bioassay response (y variable) by the metabolic profile. Each electrospray ionization mode (positive, ESI+; negative, ESI–) and each bioassay response were modelled separately. Log-response ratios were used as effect sizes. (A) Models with all observations. (B) Model that remained significant after exclusion of outlying maternal line
| ESI | y variable | N | K | R2X(cum) | R2X(pred) | R2X(ortho) | R2Y | Q2Y | F | P |
|---|---|---|---|---|---|---|---|---|---|---|
| (A) All observations | ||||||||||
| ESI+ | Effect sizes of A. thaliana germination | 41 | 2248 | 0·319 | 0·035 | 0·884 | 0·195 | 0·008 | 0·048 | 0·999 |
| ESI+ | Effect sizes of P. lanceolata germination | 41 | 2248 | 0·291 | 0·091 | 0·200 | 0·736 | 0·297 | 3·795 | 0·011 |
| ESI+ | Effect sizes of P. lanceolata root length | 41 | 2248 | 0·298 | 0·108 | 0·189 | 0·769 | 0·494 | 8·794 | <0·001 |
| ESI– | Effect sizes of A. thaliana germination | 41 | 3649 | 0·296 | 0·0413 | 0·255 | 0·903 | 0·187 | 1·304 | 0·282 |
| ESI– | Effect sizes of P. lanceolata germination | 41 | 3649 | 0·265 | 0·094 | 0·171 | 0·751 | 0·298 | 3·825 | 0·011 |
| ESI– | Effect sizes of P. lanceolata root length | 41 | 3649 | 0·269 | 0·121 | 0·148 | 0·807 | 0·491 | 8·686 | <0·001 |
| (B) After exclusion of outlying maternal line | ||||||||||
| ESI– | Effect sizes of P. lanceolata root length | 40 | 3649 | 0·332 | 0·078 | 0·254 | 0·989 | 0·526 | 4·297 | 0·002 |
N, number of observations; K, number of variables; R2X(cum), fraction of variation of metabolomic data explained by the model; R2X(pred), fraction of systematic variation in the metabolomic data related to the y variable; R2X(ortho), fraction of systematic variation in the metabolomic data non-related to the y variable; R2Y, fraction of variation of the y variables explained by the model; Q2Y, total variation of the y variable predicted by the model.
F values and levels of significance (P) were computed by CV-ANOVA.
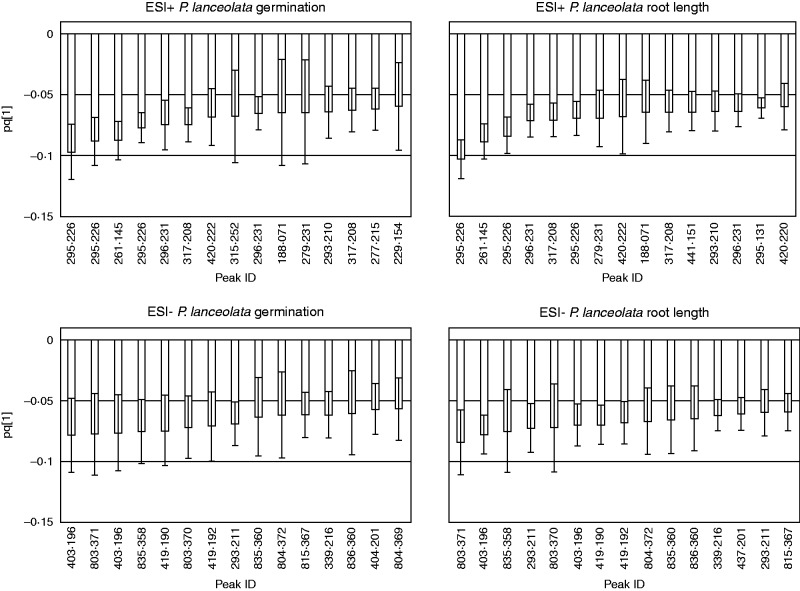
Fig. 3.
Loading column plots of orthogonal partial least-squares (OPLS) models. The models are described in Table 3. The plots show metabolomic peaks (characterized by retention time × accurate m/z) most associated with allelopathic effects that were extracted from the analyses of full metabolomic data. On the x axes the peaks are represented by their m/z values. The y axes represent the predictive components of the OPLS models (pq[1]). Negative values of predictive components are associated with negative values of effect sizes in the bioassay, i.e. allelopathy. The error bars were calculated by jack-knifing (Efron and Gong, 1983) in SIMCA-P software.
Characterization of compounds associated with allelopathy
We subsequently examined the peaks most related to the magnitude of allelopathic effect and the associated UHPLC–TOF–MS data, which led us to the 15 individual compounds summarized in Table 4. Several compounds were identified in the OPLS models by more than one peak. Interestingly, 14 out of 15 compounds can be categorized into three groups according to their chemical structure: (1) two dipeptides and one amino acid; (2) six C18 oxylipins; and (3) five malonyl monoglycosides. It should also be noted that several compounds, each of them identified by at least one peak, are chromatographically separable isomers of identical elemental composition; e.g. compounds IV, V and VI listed in a common row in Table 4. Moreover, peaks extracted from OPLS modelling of either bioassay response mostly led to the same compounds, suggesting that both germination and root length of P. lanceolata are affected by the same compounds.
Table 4.
Compounds associated with allelopathy of H. mantegazzianum revealed in the bioassay with P. lanceolata. Peaks extracted from OPLS models based on all observations are presented in bold. Bioactivity shows whether the peaks were identified based on inhibition of P. lanceolata germination (g) or root length (r). In addition, compounds indicated based only on the model explaining root length of P. lanceolata by the metabolic profile measured in negative ionization mode that remained significant after exclusion of the outlier are presented in bold.
| Compound number* | Retention time (min) in system 1: acetonitrile mobile phase | Molecular formula | Adduct/fragment identified in MS spectrum |
Bioactivity | Tentative identification** |
||
|---|---|---|---|---|---|---|---|
| ESI+ | ESI– | DBE, double bond equivalent; O, number of oxygens besides carboxyl group | Compound category | ||||
| I | 1·15 | C11H20N2O5 | 261·1450 [M+H]+ | 259·1302 [M-H]– | g, r |
Amino acids/dipeptides |
|
| II | 2·04 | C11H20N2O3 | 229·1544 [M+H]+ | 227·1375 [M-H]– | g | ||
| III | 1·46 | C11H12N2O2 | 188·0807 [M-NH3+H]+ | 249·076 [M+HCOOH-H]– | g | tryptophan | |
| 205·0969 [M+H]+ | |||||||
| IV, V, VI | 12·63; 12·90; 13·13 | C18H30O3 | 277·2147 [M–H2O+H]+ | 293·2117 [M-H]– | g, r | DBE 4 |
C18 oxylipins |
| 295·2282 [M+H]+ | 339·2154 [M+HCOOH-H]– | O 1 | |||||
| 296·2315 A+1 isotope of [M+H]+ | |||||||
| 317·2075 [M+Na]+ | |||||||
| 333·1820 [M+K]+ | |||||||
| 358·2338 [M+Na+CH3CN]+ | |||||||
| VII | 10·5 | C18H34O4 | 279·2305 [M–2H2O+H]+ | 313·2353 [M-H]– | g, r | DBE 2 | |
| 297·2412 [M–H2O+H]+ | O 2 | ||||||
| 315·2519 [M+H]+ | |||||||
| 337·2339 [M+Na]+ | |||||||
| 353·2079 [M+K]+ | |||||||
| VIII | 12·04 | C18H28O3 | 293·2099 [M+H]+ | 291·1954 [M-H]– | g, r | DBE 5 | |
| 315·1921 [M+Na]+ | 337·2010 [M+HCOOH-H]– | O 1 | |||||
| IX | 12·38 | C18H32O3 | 279·2333 [M–H2O+H]+ | 295·2268 [M-H]- | g, r | DBE 3; O 1 | |
| X, XI, XII | 7·23; 7·41; 7·01 | C19H30O9 | 420·2209 [M+NH4]+ | 403·1967 [M+H2-H]– | g, r | R-O-malonyl-O-monoglycoside |
Malonyl monoglycosides |
| 425·1761 [M+Na]+ | 404·2014 A+1 isotope of [M+H2-H]– | R = C10H17 | |||||
| 441·1495 [M+K]+ | 803·3701 [2M-H]-– | ||||||
| 804·3745 A+1 isotope of [2M-H]– | |||||||
| 357·1907[M-CO2-H]– | |||||||
| XIII, XIV | 4·64; 4·75 | C19H30O10 | 401·1816 [M+H]+ | 419·1939 [M+H2-H]– | g, r | R-O-malonyl-O-monoglycoside | |
| 441·1690 [M+Na]+ | 420·1950 A+1 isotope of [M+H2-H]– | R = C10H17O | |||||
| 457·1441 [M+K]+ | 835·3622 [2M-H]– | ||||||
| 836·3633 A+1 isotope of [2M-H]– | |||||||
| XV | 4·78 | (C39H60O18)*** | ND | 815·3700 [M-H]– | g | Unknown | |
ESI, electrospray ionization mode: positive (ESI+), negative (ESI–); ND, not detected.
*Isomers of the same molecular formula but differing in their retention times are listed in one common row.
**Based on LC–MS data, including CID fragmentation.
***The identity of the pseudomolecular ion was not confirmed by the presence of an adduct or fragment; the prediction of elemental composition cannot therefore be considered reliable.
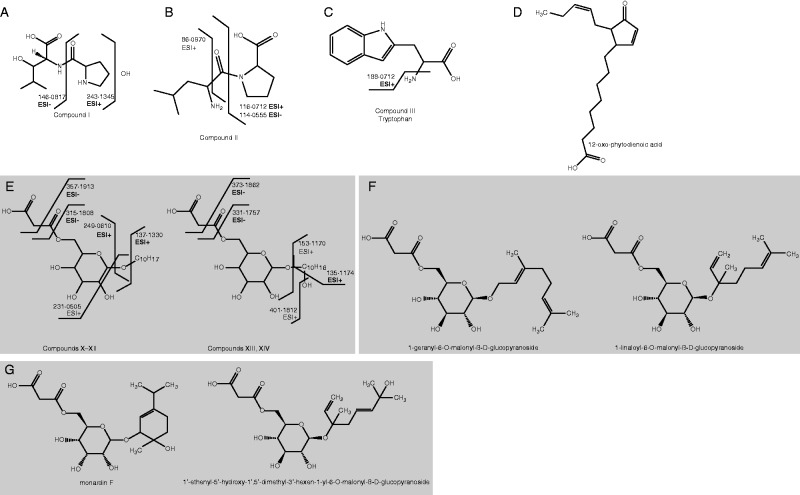
We carried out CID MS fragmentation experiments to obtain a more detailed insight into the compound structures. The MS data for compounds I and II correspond to dipeptides. Compound I is tentatively composed of common yet non-proteinogenic amino acids (hydroxyleucine or hydroxyisoleucine and hydroxyproline) as depicted in Fig. 4A. We have not found such a dipeptide, i.e. a dipeptide that is consistent with both the elemental composition and fragmentation data in the databases. Compound II was tentatively identified as leucyl-proline or isoleucyl-proline (Fig. 4B). Compound III was identified as tryptophan based on its elemental composition and comparison of retention times and fragmentation patterns in the CID MS spectra with those of the authentic standard (Fig. 4C).
Fig. 4.
Proposed chemical structures and fragmentation patterns of some compounds identified by our metabolic profiling to be associated with allelopathy (A, B, C, E) and compounds found in databases that correspond to putatively allelopathic compounds (D, F, G). ESI+/– (in bold), fragment identified in both chromatographic systems (acetonitrile as well as methanol mobile phase systems); ESI+/–, fragment identified only in one chromatographic system.
Compounds IV–IX are C18 fatty acids according to their elemental composition and corresponding hits in the databases. This assumption was supported by the presence of [M-H-CO2]– fragments in the negative ionization CID MS spectra, confirming the presence of a carboxyl group in the structures. Furthermore, all the structures contain at least one additional oxygen besides those of the carboxyl group, which specifies these compounds as C18 oxylipins. Unfortunately, the MS data did not allow us to localize double bonds and/or oxygen-containing functionalities in the structures. Nevertheless, several compounds that correspond to the MS data were found in the interrogated databases. One of them, corresponding to compound VIII, is 12-oxo-phytodienoic acid (Fig. 4D), an important precursor of a plant hormone, which we discuss below. The other oxylipins we found in root exudates of H. mantegazzianum may represent derivatives of 12-oxo-phytodienoic acid or a similar compound.
Compounds X–XIV are malonyl monoglycosides, all of them composed of a malonyl moiety attached to a saccharide unit, which is further ornamented with an aliphatic or alicyclic side chain, as shown in Fig. 4E. The MS data do not enable us to determine the type of saccharide or the exact structure of the side-chain. However, for isomers X–XII and XIII–XIV the interrogated databases revealed specific compounds of the respective elemental compositions, which contain a malonyl monosaccharide moiety. These comprised 1-geranyl-6-O-malonyl-β-d-glucopyranoside (GMGP) and 1-linaloyl-6-O-malonyl-β-d-glucopyranoside (LMGP) (Fig. 4F), corresponding to isomers X–XII and monardin F, and 1′-ethenyl-5′-hydroxy-1′,5′-dimethyl-3′-hexen-1-yl-6-O-malonyl-β- d-glucopyranoside (Fig. 4G), corresponding to isomers XIII–XIV. Except for monardin F, the compounds contain a glucose unit bearing an aliphatic side-chain. Interestingly, the monardin F molecule shows different properties because it is composed of an allose unit and an alicyclic side-chain. We propose that compounds X–XIV contain the same saccharide moiety; however, it remains unclear whether it is glucose, which we consider more likely, or allose or any other saccharide moiety. It is even more difficult to predict the exact structure of the side-chain, which may be either aliphatic or alicyclic. In addition, we assume that the isomeric compounds differ in the position of a double bond and/or hydroxyl group.
We were unable to reliably identify even its pseudomolecular ion of compound XV; therefore, more detailed information on this substance is not available.
The insufficient amounts of root exudate extracts did not enable purification of the compounds for nuclear magnetic resonance analysis in order to further elucidate the compound structures.
In addition, we searched for fouranocoumarins, which are typical secondary metabolites in H. mantegazzianum (Tiley et al., 1996) and were previously reported to be responsible for allelopathic effects (Macías et al., 1993; García et al., 2002). We found two furanocoumarins, xanthotoxin and bergapten, present in the exudates according to elemental composition and comparison of retention times and fragmentation patterns in the CID MS spectra with those of authentic standards. However, these were not related to the magnitude of the allelopathic effect.
DISCUSSION
Allelopathy may contribute to the invasive success of some exotic species (Callaway and Aschehoug, 2000; Hierro and Callaway, 2003; Callaway and Ridenour, 2004). Most research carried out so far has focused on species-level patterns in allelopathy, and little is known about intraspecific variation of allelopathy (but see Lankau et al., 2009). Here we provide strong evidence that one of the worst invaders in Europe (DAISIE European Invasive Alien Species Gateway, 2008), H. mantegazzianum, is phytotoxic towards the native plant P. lanceolata, although the effect is subject to considerable variation between maternal lines. Generally, the phytotoxicity of root exudates against P. lanceolata was dependent on both the qualitative (composition) and quantitative (amount) properties of the exudate produced.
Biological variation in allelopathic effects was greatest at the maternal line level and very low between different areas or populations in which we sampled seeds. It has to be born in mind that our estimates of variation proportions had quite wide 95 % confidence intervals. Nonetheless, the lack of spatial structure in allelopathic effects of plants from different populations and even areas has already been observed and may be due to growing plants under common conditions (Kunin et al., 2009). Had the differences in allelopathic effects resulted from environmental conditions such as the temperature or water regime, we would have observed much greater total variability and a stronger spatial pattern in plants sampled directly in the field. Here, we can neither assess the sources of intraspecific variation nor give the reason why this variation is sustained in our system. We are also lacking information on the costs of allelopathy in H. mantegazzianum, as shown by the example of Brassica nigra (Lankau and Strauss, 2007; Lankau, 2008), or whether the species itself is negatively affected by its own root exudates, as shown for other species (Canals et al., 2005; Alías et al., 2006), these being the mechanisms that could select for a decrease in allelopathic potential. Trade-offs related to the production of allelochemicals in H. mantegazzianum need to be estimated in order to explain why the variability is sustained in our system.
The variation in allelopathic effects on P. lanceolata germination and root length within different populations turned out to be determined by the metabolic profiles of the respective root exudates. However, one maternal line had a strong influence on our models, and its exclusion led to loss of significance in all but the model explaining root length of P. lanceolata by the metabolic profile measured in negative ionization mode. The weak predictive power could have resulted from the fact that the predictive component represented only about one-third of the systematic variation. We assume that there was another important source of metabolic variation in our maternal lines apart from allelopathic effect. Such metabolic variation could be related, for example, to chemical defence against herbivores or pathogens, since it has been shown not to relate to allelochemicals in other species (Uesugi and Kessler, 2013). Nonetheless, we tentatively identified 15 compounds associated with allelopathic effects based on models containing all observations and further confirmed three out of six C18 oxylipins (compounds IV–VI) and all the malonyl glycosides (compounds X–XIV) when we based our conclusions only on the model that remained significant after the exclusion of the outlying maternal line. Compounds of particular interest are discussed below.
Plant oxylipins were shown to be involved in signalling and defence mechanisms against various herbivores or pathogens (Mosblech et al., 2009). For instance, 12-oxo-phytodienoic acid is a precursor of the plant hormone jasmonic acid and it has also been reported to be a regulator of defence responses (Blée, 2002). To the best of our knowledge, there is only one study of the phytotoxicity of plant-derived oxylipins; Fiorentino et al. (2008) found several hydroxylated unsaturated C18 oxylipins in extracts from the Mediterranean shrub Cestrum parqui that inhibited the germination and growth of Lactuca sativa.
Isomers X–XII represent 6-O-malonyl-glycosides of certain monoterpenols. These compounds have not been previously reported in plants from the Apiaceae family, although they have been found in different fruits and vine leaves (Withopf et al., 1997), whole plants of Monarda punctata (Yamada et al., 2010), flower buds of Jasminum sambac (Moon et al., 1994) and flowers of Boronia megastigma (Cooper et al., 2011). Isomers XIII and XIV represent 6-O-malonyl-glycosides of monoterpendiols that are likely hydroxyderivatives of isomers X–XII and that were previously found in Boronia megastigma (Cooper et al., 2011). In general, terpenoid glycosides represent transport and storage forms of terpenoids in plant tissues, i.e. they serve as precursors of aglycone terpenoids (Winterhalter and Skouroumounis, 1997). Volatile terpenoids play many ecological roles in plant chemical defences and communication (Gershenzon and Dudareva, 2007). Importantly, Yang et al. (2013) found compounds putatively identified as linalool malonyl glucoses in leaves of Chrysanthemum morifolium, where they functioned as deterrents of herbivorous insects. Their study therefore suggested for the first time that linalool glycosides are bioactive. We can hypothesize that the monoterpenoid 6-O-malonyl-glycosides we found in root exudates would function also as allelochemicals; however, direct proof of this effect will require purification of these compounds and direct bioassays.
We also found two furanocoumarins, xanthotoxin and bergapten, in the exudates; however, these were not connected to the magnitude of the allelopathic effect. This contradicts previous studies on Pilocarpus goudotianus leaves (Macías et al., 1993) or on seeds of Ammi majus (García et al., 2002), where furanocoumarines were responsible for the allelopathy of these species. In general, phenolics are considered to be responsible for allelopathy in many plants (Inderjit, 1996; Kim and Lee, 2010; Li et al., 2010), however, here we indicate that compounds belonging to different structural groups are associated with allelopathy.
This study presents the results of a comprehensive investigation of the metabolic profile of root exudates of the invasive species H. mantegazzianum together with bioassays testing the effects of root exudates on germination and root development of other native species. We suggest that intraspecific variation in the production of allelochemicals should be included in studies of the novel weapons hypothesis. In addition, we employed state-of-the-art techniques in allelopathy research and suggest that environmental metabolomics is an efficient tool for studying plant–plant interactions whenever unknown metabolites are involved or already described metabolites fail to explain the full effect observed in nature.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: alternative analysis of proportions of total variance in allelopathic effects of H. mantegazzianum attributed to variation between and within populations. Fig. S1: scheme of areas (with GPS co-ordinates), populations within areas and maternal lines within populations sampled for seeds. Fig. S2: principal components analysis of the UHPLC–TOF–MS metabolic profiles of the maternal lines analysed in positive and negative ionization mode. Fig. S3: scores plot of an orthogonal partial least-squares model explaining effect sizes of P. lanceolata root length in the bioassay by metabolomic data measured in positive ionization mode. Fig. S4: loading column plot of an orthogonal partial least-squares model that remained significant after exclusion of the outlying maternal line.
ACKNOWLEDGEMENTS
Frederick Rooks kindly improved our English. We are grateful for the assistance of Tereza Klinerová, Dana Parysová, Daniel Samek, Alexandra Špaldoňová and Tereza Tylová. We thank Hans Albert Pedersen for insightful comments on the manuscript. This work was supported by grants from the Czech Science Foundation (P504/10/0132) to P.D., the Charles University Grant Agency (GAUK512712) to K.J. and T.C. and the Ministry of Education, Youth and Sports of the Czech Republic (CZ.1.07/2.3.00/30.0003) to Z.K. This study was also supported by long-term research development project no. RVO 67985939 of the Academy of Sciences of the Czech Republic.
LITERATURE CITED
- Alías JC, Sosa T, Escudero JC, Chaves N. 2006. Autotoxicity against germination and seedling emergence in Cistus ladanifer L. Plant and Soil 282: 327–332. [Google Scholar]
- Atkinson RRL, Burrell MM, Osborne CP, Rose KE, Rees M. 2012. A non-targeted metabolomics approach to quantifying differences in root storage between fast- and slow-growing plants. New Phytologist 196: 200–211. [DOI] [PubMed] [Google Scholar]
- Baležentienė L. 2012. Inhibitory effects of invasive Heracleum sosnowskyi on rapeseed and ryegrass germination. Allelopathy Journal 30: 197–208. [Google Scholar]
- Barto EK, Cipollini D. 2009. Half-lives and field soil concentrations of Alliaria petiolata secondary metabolites. Chemosphere 76: 71–75. [DOI] [PubMed] [Google Scholar]
- Blair AC, Nissen SJ, Brunk GR, Hufbauer RA. 2006. A lack of evidence for an ecological role of the putative allelochemical (+/–)-catechin in spotted knapweed invasion success. Journal of Chemical Ecology 32: 2327–2331. [DOI] [PubMed] [Google Scholar]
- Blée E. 2002. Impact of phyto-oxylipins in plant defense. Trends in Plant Science 7: 315–322. [DOI] [PubMed] [Google Scholar]
- Callaway RM, Aschehoug ET. 2000. Invasive plants versus their new and old neighbors: a mechanism for exotic invasion. Science 290: 521–523. [DOI] [PubMed] [Google Scholar]
- Callaway RM, Ridenour WM. 2004. Novel weapons: invasive success and the evolution of increased competitive ability. Frontiers in Ecology and the Environment 2: 436. [Google Scholar]
- Canals RM, Emeterio LS, Peralta J. 2005. Autotoxicity in Lolium rigidum: analyzing the role of chemically mediated interactions in annual plant populations. Journal of Theoretical Biology 235: 402–407. [DOI] [PubMed] [Google Scholar]
- Cooper CM, Davies NW, Motti CA, Menary RC. 2011. Glycosidic conjugates of C13 norisoprenoids, monoterpenoids, and cucurbates in Boronia megastigma (Nees). Journal of Agricultural and Food Chemistry 59: 2610–2617. [DOI] [PubMed] [Google Scholar]
- DAISIE European Invasive Alien Species Gateway. 2008. Available from: http://www.europe-aliens.org/ (24 September 2014). [Google Scholar]
- Davey MP, Burrell MM, Woodward FI, Quick WP. 2008. Population-specific metabolic phenotypes of Arabidopsis lyrata ssp. petraea. New Phytologist 177: 380–388. [DOI] [PubMed] [Google Scholar]
- Davey MP, Ian Woodward F, Paul Quick W. 2009. Intraspecfic variation in cold-temperature metabolic phenotypes of Arabidopsis lyrata ssp. petraea. Metabolomics 5: 138–149. [Google Scholar]
- Dostál P, Müllerová J, Pyšek P, Pergl J, Klinerová T. 2013. The impact of an invasive plant changes over time. Ecology Letters 16: 1277–1284. [DOI] [PubMed] [Google Scholar]
- Efron B, Gong G. 1983. A leisurely look at the bootstrap, the jackknife, and cross-validation. American Statistician 37: 36–48. [Google Scholar]
- Eriksson L, Trygg J, Wold S. 2008. CV-ANOVA for significance testing of PLS and OPLS® models. Journal of Chemometrics 22: 594–600. [Google Scholar]
- Del Fabbro C, Güsewell S, Prati D. 2014. Allelopathic effects of three plant invaders on germination of native species: a field study. Biological Invasions 16: 1035–1042. [Google Scholar]
- Fiorentino A, D’Abrosca B, Dellagreca M, et al. 2008. Chemical characterization of new oxylipins from Cestrum parqui, and their effects on seed germination and early seedling growth. Chemistry & Biodiversity 5: 1780–91. [DOI] [PubMed] [Google Scholar]
- García C, Moyna P, Fernández G, Heinzen H. 2002. Allelopathic activity of Ammi majus L. fruit waxes. Chemoecology 12: 107–111. [Google Scholar]
- Gershenzon J, Dudareva N. 2007. The function of terpene natural products in the natural world. Nature chemical biology 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Hadfield JD. (2010). MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. Journal of Statistical Software 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Hierro JL, Callaway RM. 2003. Allelopathy and exotic plant invasion. Plant and Soil 256: 29–39. [Google Scholar]
- Inderjit 1996. Plant phenolics in allelopathy. Botanical Review 62: 186–202. [Google Scholar]
- Inderjit, Callaway RM. 2003. Experimental designs for the study of allelopathy. Plant and Soil 256: 1–11. [Google Scholar]
- Inderjit, van der Putten WH. 2010. Impacts of soil microbial communities on exotic plant invasions. Trends in Ecology & Evolution 25: 512–519. [DOI] [PubMed] [Google Scholar]
- Inderjit, Wardle DA, Karban R, Callaway RM. 2011. The ecosystem and evolutionary contexts of allelopathy. Trends in Ecology & Evolution 26: 655–662. [DOI] [PubMed] [Google Scholar]
- Jandová K, Klinerová T, Müllerová J, et al. 2014a. Long-term impact of Heracleum mantegazzianum invasion on soil chemical and biological characteristics. Soil Biology and Biochemistry 68: 270–278. [Google Scholar]
- Jandová K, Dostál P, Cajthaml T. 2014b. Searching for Heracleum mantegazzianum allelopathy in vitro and in a garden experiment. Biological Invasions. doi 10.1007/s10530-014-0771-5. [Google Scholar]
- Junttila O. 1976. Allelopathic inhibitors in seeds of Heracleum laciniatum. Physiologia Plantarum 36: 374–378. [Google Scholar]
- Kaur H, Kaur R, Kaur S, Baldwin IT, Inderjit 2009. Taking ecological function seriously: soil microbial communities can obviate allelopathic effects of released metabolites. PloS One 4: e4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YO, Lee EJ. 2010. Comparison of phenolic compounds and the effects of invasive and native species in East Asia: support for the novel weapons hypothesis. Ecological Research 26: 87–94. [Google Scholar]
- Kunin WE, Vergeer P, Kenta T, et al. 2009. Variation at range margins across multiple spatial scales: environmental temperature, population genetics and metabolomic phenotype. Proceedings of the Royal Society B: Biological Sciences 276: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankau R. 2008. A chemical trait creates a genetic trade-off between intra- and interspecific competitive ability. Ecology 89: 1181–1187. [DOI] [PubMed] [Google Scholar]
- Lankau R. 2010. Soil microbial communities alter allelopathic competition between Alliaria petiolata and a native species. Biological Invasions 12: 2059–2068. [Google Scholar]
- Lankau RA, Strauss SY. 2007. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science 317: 1561–1563. [DOI] [PubMed] [Google Scholar]
- Lankau RA, Nuzzo V, Spyreas G, Davis AS. 2009. Evolutionary limits ameliorate the negative impact of an invasive plant. Proceedings of the National Academy of Sciences of the USA 106: 15362–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-H, Wang Q, Ruan X, Pan C-D, Jiang D-A. 2010. Phenolics and plant allelopathy. Molecules 15: 8933–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A, Kools HJ. 2012. MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics 8: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macel M, Van Dam NM, Keurentjes JJB. 2010. Metabolomics: the chemistry between ecology and genetics. Molecular Ecology Resources 10: 583–593. [DOI] [PubMed] [Google Scholar]
- Macías FA, Galindo JC, Massanet GM, Rodriguez-Luis F, Zubia E. 1993. Allelochemicals from Pilocarpus goudotianus leaves. Journal of Chemical Ecology 19: 1371–1379. [DOI] [PubMed] [Google Scholar]
- Marti G, Erb M, Boccard J, et al. 2013. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant, Cell & Environment 36: 621–639. [DOI] [PubMed] [Google Scholar]
- Moon J-H, Watanabe N, Sakata K, et al. 1994. Linalyl β-d-glucopyranoside and its 6′-O-malonate as aroma precursors from Jasminum sambac. Phytochemistry 36: 1435–1437. [Google Scholar]
- Mosblech A, Feussner I, Heilmann I. 2009. Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiology and Biochemistry 47: 511–517. [DOI] [PubMed] [Google Scholar]
- Nylund GM, Weinberger F, Rempt M, Pohnert G. 2011. Metabolomic assessment of induced and activated chemical defence in the invasive red alga Gracilaria vermiculophylla. PloS One 6: e29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2014. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. www.r-proj ect.org. [Google Scholar]
- Reaxys. Application version 2.18008.4; Elsevier, 2014; RN 969209 (30 May 2014). www.reaxys.com/.
- SciFinder. Application version 26.050714; Chemical Abstracts Service, Columbus, OH, 2014; RN 58-08-2 (13 June 2014). scifinder.cas.org.
- Thorpe AS, Thelen GC, Diaconu A, Callaway RM. 2009. Root exudate is allelopathic in invaded community but not in native community: field evidence for the novel weapons hypothesis. Journal of Ecology 97: 641–645. [Google Scholar]
- Tiley G, Dodd F, Wade P. 1996. Heracleum mantegazzianum Sommier & Levier. Journal of Ecology 84: 297–319. [Google Scholar]
- Trygg J, Wold S. 2002. Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics 16: 119–128. [Google Scholar]
- Uesugi A, Kessler A. 2013. Herbivore exclusion drives the evolution of plant competitiveness via increased allelopathy. New Phytologist 198: 916–924. [DOI] [PubMed] [Google Scholar]
- Viechtbauer W. 2010. Conducting meta-analyses in R with the metafor package. Journal of Statistical Software 36: 1–48. [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD. 2007. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nature protocols 2: 778–791. [DOI] [PubMed] [Google Scholar]
- Wille W, Thiele J, Walker EA, Kollmann J. 2013. Limited evidence for allelopathic effects of giant hogweed on germination of native herbs. Seed Science Research 23: 157–162. [Google Scholar]
- Winterhalter P, Skouroumounis GK. 1997. Glycoconjugated aroma compounds: occurrence, role and biotechnological transformation. Advances in Biochemical Engineering/Biotechnology 55: 73–105. [DOI] [PubMed] [Google Scholar]
- Withopf B, Richling E, Roscher R, Schwab W, Schreier P. 1997. Sensitive and selective screening for 6‘-O-malonylated glucoconjugates in plants. Journal of Agricultural and Food Chemistry 45: 907–911. [Google Scholar]
- Yamada K, Murata T, Kobayashi K, Miyase T, Yoshizaki F. 2010. A lipase inhibitor monoterpene and monoterpene glycosides from Monarda punctata. Phytochemistry 71: 1884–1891. [DOI] [PubMed] [Google Scholar]
- Yang T, Stoopen G, Thoen M, Wiegers G, Jongsma MA. 2013. Chrysanthemum expressing a linalool synthase gene ‘smells good’, but ‘tastes bad’ to western flower thrips. Plant Biotechnology Journal 11: 875–882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.