Abstract
Background Cereal diseases cause tens of billions of dollars of losses annually and have devastating humanitarian consequences in the developing world. Increased understanding of the molecular basis of cereal host–pathogen interactions should facilitate development of novel resistance strategies. However, achieving this in most cereals can be challenging due to large and complex genomes, long generation times and large plant size, as well as quarantine and intellectual property issues that may constrain the development and use of community resources. Brachypodium distachyon (brachypodium) with its small, diploid and sequenced genome, short generation time, high transformability and rapidly expanding community resources is emerging as a tractable cereal model.
Scope Recent research reviewed here has demonstrated that brachypodium is either susceptible or partially susceptible to many of the major cereal pathogens. Thus, the study of brachypodium–pathogen interactions appears to hold great potential to improve understanding of cereal disease resistance, and to guide approaches to enhance this resistance. This paper reviews brachypodium experimental pathosystems for the study of fungal, bacterial and viral cereal pathogens; the current status of the use of brachypodium for functional analysis of cereal disease resistance; and comparative genomic approaches undertaken using brachypodium to assist characterization of cereal resistance genes. Additionally, it explores future prospects for brachypodium as a model to study cereal–pathogen interactions.
Conclusions The study of brachypodium–pathogen interactions appears to be a productive strategy for understanding mechanisms of disease resistance in cereal species. Knowledge obtained from this model interaction has strong potential to be exploited for crop improvement.
Keywords: Brachypodium distachyon, barley stripe mosaic virus, cereal–pathogen interaction, ecotypes, functional genomics, Fusarium, Magnaporthe, model species, mutants, plant defence, Puccinia, Pyrenophora, Rhizoctonia, Stagonospora, Xanthomonas
INTRODUCTION
The relevance of plant models to crop research
The study of a relatively small number of organisms chosen based on their suitability for research, frequently referred to as ‘model’ organisms, has provided the majority of our current fundamental biological knowledge (Müller and Grossniklaus, 2010). Nevertheless, what constitutes a model organism is not well defined. Ankeny and Leonelli (2011) have suggested that a model organism might be more specifically characterized as one that is representative of a target species at the ‘whole-organism’ level. They highlight that ‘modern’ model organism systems are built upon extensive infrastructure, including germplasm and in silico resources, that is openly accessible to the research community and facilitates diverse research approaches. It follows that the value of a model organism is dependent upon at least two factors: (1) the ease with which diverse research questions can be addressed, and (2) the relevance of information obtained from the model system to the target system(s).
Ideally research should be performed on the organism of primary interest. However, this is often neither practical nor achievable for many reasons. Rapid technological advances are facilitating the application of powerful ‘omics’ approaches (e.g. transcriptomics and metabolomics) to crop plants, allowing for unprecedented dissection of physiological processes in these species. Nevertheless, major challenges for conducting fundamental research on crop plants still exist. Many crops possess inherent characteristics that impede research, including large size, long generation times, and large and complex genomes. Additionally, dedicated model species have proved remarkably effective in stimulating community-driven research, as exemplified by resources developed and exploited for Arabidopsis thaliana (arabidopsis), whereas intellectual property and quarantine restrictions can deter such efforts in crop plants (Jung et al., 2008).
Brachypodium as a monocot model
Arabidopsis has proven to be a highly effective model plant providing extensive insight into numerous fundamental processes. Nevertheless, the development of a model monocot species is seen by many as necessary, in part because of the major physiological, morphological and/or molecular differences between the dicotyledonous arabidopsis and monocot crops (Brkljacic et al., 2011). Rice (Oryza sativa) possesses many characteristics of a monocot model. However, significant challenges including large plant size, demanding growth requirements, long generation time and constraints upon access to germplasm due to intellectual property and quarantine restrictions (as posited above) appear to have impeded its full use in this capacity (Jung et al., 2008).
In the last decade, Brachypodium distachyon (brachypodium) has emerged as an effective model for monocot species. As reviewed previously, brachypodium possesses characteristics required for an effective plant model, including small stature, self-fertilization (but able to be cross-fertilized), rapid generation time, a compact genome and high transformation efficiency (Garvin, 2008; Vogel and Bragg, 2009; Vain, 2011; Brkljacic et al., 2011).
Brachypodium as a model to study cereal diseases
Cereal grains, as food or animal feed, provide approx. 60 % of human calories globally (Cassman et al., 2003). Diseases are a chronic problem that can seriously limit cereal production; direct production losses in the major cereal crops rice, wheat and maize due to diseases have been estimated at approx. 10 % of total production worldwide (Oerke, 2006). Robust genetic resistance is an ideal solution for combating disease; however, both the incorporation of disease resistance into crops and the durability of this resistance over time can be challenging. Difficulties associated with the identification of effective resistance and its deployment in elite cultivars can be particularly overwhelming for pathogens to which the plant displays partial or ‘quantitative’ resistance (St. Clair, 2010). Conversely, the ability of pathogen populations to rapidly overcome incorporated resistance appears to be especially problematic with major ‘qualitative’ resistance (Dangl et al., 2013), such as seedling resistance to rust pathogens. However, improved understanding of the mechanisms employed by pathogens to cause disease and the defences employed by plants to negate them should aid attempts to improve resistance in cereal crops.
Much has been learnt already about the molecular basis of the plant immune system, largely using arabidopsis. This information is a valuable platform upon which strategies for increased resistance in all crop species, including cereals, can be built. However, although some fundamental aspects of plant immunity appear to be strongly conserved, it is also clear that many features of plant–pathogen interactions can be highly pathosystem-specific (Boyd et al., 2013). Therefore, to facilitate the translation of information obtained from a model pathosystem into crop improvement, in many instances it would be beneficial if (1) the model was infected by the same pathogen in a similar manner as the crop; and (2) the model and crop were genetically similar, such that orthologues of immune components characterized in the model could be readily identified in the crop. Brachypodium shares a close phylogenetic relationship with major cereal crops, including wheat and barley with which it diverged from a common ancestor less than 40 Mya (The Brachypodium Initiative, 2010). Additionally, as reviewed here, it is increasingly becoming apparent that brachypodium is also susceptible to many important cereal pathogens.
BRACHYPODIUM–CEREAL PATHOGEN MODEL PATHOSYSTEMS: FUNGAL PATHOGENS
As described, several experimental brachypodium pathosystems have been recently established. An overview of descriptions of cereal pathogens infecting brachypodium is given in Table 1. Symptoms of brachypodium infected with selected cereal pathogens are demonstrated in Fig. 1.
Table 1.
An overview of cereal pathogens demonstrated to infect brachypodium
| Pathogen | Cereal host | Reference |
|---|---|---|
| Bacterial | ||
| Xanthomonas translucens | Wheat, barley | T. L. Fitzgerald et al. (unpubl. res.) |
| Fungal | ||
| Colletorichum cereale | Rye, wheat, oat | Sandoya and Buanafina (2014) |
| Fusarium culmorum | Wheat, barley | Peraldi et al. (2011) |
| Fusarium graminearum | Wheat, barley | Peraldi et al. (2011) |
| Fusarium pseudograminearum | Wheat, barley | J. J. Powell et al. (unpubl. res.) |
| Gaeumannomyces graminis | Wheat | Sandoya and Buanafina (2014) |
| Bipolaris sorokiniana | Wheat, barley | Falter and Voigt (2014) |
| Magnaporthe oryzae | Rice | Routledge et al. (2004) |
| Oculimacula acuformis | Wheat | Peraldi et al. (2013) |
| Oculimacula yallundae | Wheat | Peraldi et al. (2013) |
| Puccinia graminis f. sp. tritici | Wheat | Ayliffe et al. (2013) |
| Puccinia striiformis f. sp. avenea | Oat | Ayliffe et al. (2013) |
| Puccinia striiformis f. sp. tritici | Wheat | Ayliffe et al. (2013) |
| Puccinia triticina | Wheat | Ayliffe et al. (2013) |
| Ramularia collo-cygni | Barley | Peraldi et al. (2013) |
| Rhizoctonia solani AG 8 | Wheat, barley | Schneebeli et al. (2014) |
| Pyrenophora teres | Barley | Falter and Voigt (2014) |
| Stagonospora nodorum | Wheat | Falter and Voigt (2014) |
| Oomycete | ||
| Pythium aphanidermatum | Maize | Sandoya and Buanafina (2014) |
| Viral | ||
| Barley stripe mosaic virus | Barley | Cui et al. (2012) |
| Panicum mosaic virus | Pearl millet | Mandadi and Scholthof (2012) |
| Wheat streak mosaic virus (WSMV) | Maize, wheat, barley | Mandadi et al. (2014) |
| Brome mosaic virus (BMV) | Barley, wheat, maize | Mandadi et al. (2014) |
| Sorghum yellow banding virus (SYBV) | Maize, sorghum | Mandadi et al. (2014) |
| Foxtail mosaic virus (FoMV) | foxtail, sorghum, wheat | Mandadi et al. (2014) |
Fig. 1.
Brachypodium symptoms upon infection with selected cereal pathogens (permission obtained for figure modifications). (A) Puccinia graminis f. sp. tritici infection of spray-inoculated BdTr3b leaves at 28 dpi (modified from Ayliffe et al., 2013). (B) Magnoporthe oryzae infection of ecotype Bd21 leaves by spray inoculation (modified from Wang et al., 2012). Left to right: duplicate leaves at 3, 5 and 7 dpi. (C) Rhizoctonia solani AG 8 infection of Bd3-1 by growth in infested soil (modified from Schneebeli et al., 2014) at 18 days post planting. Left: representative root lengths of inoculated plants; right: representative root lengths of uninoculated plants. (D) Fusarium graminearum infection of spray-inoculated Bd21 florets at 7 dpi (modified from Peraldi et al., 2011). (E) Fusarium graminearum infection of coleoptile (left) and root (right) tissue of Bd21 by spray and agar plug inoculation, respectively (modified from Peraldi et al., 2011). Root tissue at 48 dpi inoculated (left) and mock-inoculated is shown (right). (F) Oculimacula yallundae stem base infection of ecotype Bd3-1 at 28 dpi with a colonized agar slurry (modified from Peraldi et al., 2014). (G) Ramularia collo-cygni infection of spray-inoculated Bd3-1 leaves at 21 dpi of whole plants (modified from Peraldi et al., 2014). (H) Barley stripe mosaic virus ND18 infection of Bd3-1 (resistant) and Bd21 (susceptible) (modified from Cui et al., 2012). Left to right: uninoculated Bd3-1, inoculated Bd3-1, inoculated Bd21. (I) Panicum mosaic virus (PMV) and Panicum mosaic satellite virus (SPMV) infection of ecotype Bd21-3 (modified from Mandadi and Scholthof, 2012) at 42 dpi. Left to right: uninfected, PMV-infected, PMV + SPMV-infected.
Biotrophic fungal pathogens
Rusts (Puccinia spp.).
Rust diseases are caused by obligate, biotrophic fungal pathogens that are members of the Basidiomycota. Rusts infect a wide range of plant species, including most cereals (e.g. wheat, barley, maize, oats, triticale, sorghum and millet) and many agricultural grasses (e.g. sugarcane, fescue and phalaris). Interestingly, rice is the only cereal for which no rust pathogen has been identified (Ayliffe et al., 2011). Rust pathogens are considered the major disease threat to wheat production, a plant crop that singularly provides 20 % of the world’s calorific intake. Wheat is a host for three rust pathogen species, Puccinia graminis f. sp. tritici, P. striiformis f. sp. tritici and P. triticina, which are responsible for wheat stem rust, stripe rust and leaf rust diseases, respectively. The emergence of a new wheat stem rust race, Ug99, that was virulent on 80 % of the world’s wheat crops is a recent reminder of the constant threat these pathogens represent to global food security (Singh et al., 2011).
Brachypodium, like many grasses, is a host to a rust pathogen: P. brachypodii. In addition to B. distachyon, this pathogen infects a number of other Brachypodium species (Barbieri et al., 2011, 2012). Phylogenetic analyses indicate that P. brachypodii is most similar to the wheat stripe rust pathogen P. striiformis, when compared with other cereal rust pathogens including P. graminis and P. triticina (Zambino and Szabo, 1993). Genetic analyses in brachypodium suggest that resistance to P. brachypodii is quantitatively inherited, with multiple quantitative trait loci (QTL) providing additive resistance in both seedlings and adult plants (Barbieri et al., 2012).
Brachypodium is considered to be a non-host to infection by cereal rust pathogens (Ayliffe et al., 2013). Non-host resistance is the ability of a plant species to be resistant to all variants, i.e. races, isolates and/or pathovars, of a particular pathogen. Currently, very little is known about the molecular mechanisms of non-host resistance in plants (Mysore and Ryu, 2004). Interestingly, the response of brachypodium to infection by cereal rusts has been reported for several rust species and formae speciales, and in some brachypodium accessions partial susceptibility to these cereal pathogens is observed (Ayliffe et al., 2013; e.g. Fig. 1A). Infection of brachypodium accessions with P. striiformis f. sp. tritici, hordei and bromi (wheat, barley and brome stripe rust, respectively) resulted in symptoms on different accessions ranging from the formation of small sporulating uredinia, to macroscopic lesion formation, to apparent immunity (Draper et al., 2001; Barbieri et al., 2011; Ayliffe et al., 2013). A similar range of macroscopic symptoms, from sporulation to immunity, was also observed upon infection of brachypodium accessions with P. graminis f. sp. tritici, lolii, phlei-pratensis, avena and phalaridi, which are stem rust pathogens of wheat, ryegrass, timothy grass, oat and phalaris, respectively (Ayliffe et al., 2013; Figueroa et al., 2013). Interestingly, the last four stem rust formae speciales, which all have Poeae hosts, sporulated on most brachypodium accessions tested, demonstrating that rusts of the Poeae appear more adapted to brachypodium than are rusts of the Triticeae. In contrast, infection of brachypodium with wheat and barley leaf rust pathogens, P. triticina and P. hordei, did not produce as obvious macroscopic symptoms, although small lesions were observed in some instances (Draper et al., 2001; Ayliffe et al., 2013).
Microscopic analysis of stem, stripe and leaf rust infection on brachypodium lines that showed no macroscopically visible lesions identified a distribution of infection sites that ranged from only a sub-stomatal vesicle to infection sites that contain hyphae within the plant apoplast and haustoria formation within plant mesophyll cells (Ayliffe et al., 2013). In contrast, a significant amount of prehaustorial resistance to wheat stem rust in brachypodium was reported by Figueroa et al. (2013), suggesting either pathogen race specificity or significant environmental effects for these interactions. Brachypodium lines with macroscopically visible lesions and/or pustule development when infected by cereal rust pathogens showed extensive underlying fungal colonization of plant mesophyll cells with frequent haustoria formation at these sites (Ayliffe et al., 2013).
In general, cell death was not common at most cereal rust infection sites on brachypodium, regardless of whether the accession allowed extensive or restricted fungal colonization (Ayliffe et al., 2013). Brachypodium callose deposition patterns showed similarity to the callose deposition observed during the wheat basal defence response against these same rust species (Ayliffe et al., 2013). In both plant species, larger rust infection sites appeared capable of suppressing callose production in some cells, suggesting a mechanistic overlap between the brachypodium response to cereal rust infection and the wheat basal defence response (Ayliffe et al., 2013). No change in salicylic acid (SA) levels in brachypodium leaf tissue was observed upon infection with P. graminis f. sp. tritici (Ayliffe et al., 2013).
Generally, genetically related brachypodium accessions (based on phylogenetic analysis of Vogel et al., 2009) showed similar infection phenotypes when challenged with cereal rust pathogens. Subsequent genetic analyses have indicated that segregation of extensive/restricted wheat stripe rust growth is simply inherited in some brachypodium mapping families and in one case controlled by a single dominant gene (Ayliffe et al., 2013). This makes the positional cloning of the underlying gene(s) responsible for these differential brachypodium stripe rust infection phenotypes a real possibility, which would lead to a better understanding of potential molecular mechanisms behind non-host resistance in plants.
Powdery mildew (Blumeria graminis).
Powdery mildew is a disease affecting a wide range of plant species, caused by various fungal species within the order Erysiphales (Glawe, 2008). Powdery mildew of cereals is caused by formae speciales of Blumeria graminis; wheat powdery mildew, caused by f. sp. tritici, is a disease of major economic significance globally (Huang and Röder, 2004).
Several brachypodium ecotypes have been assessed for infection by Blumeria graminis f. sp. tritici, f. sp. avena and f. sp. hordei (Draper et al., 2001) and all showed a similar, high level of resistance. Brachypodium-infecting powdery mildew has previously been reported (Braun, 1995); however, we are not aware of anyone currently working with powdery mildew strains that are fully compatible with brachypodium.
Hemibiotrophic and necrotrophic fungal pathogens
Rice blast (Magnaporthe oryzae).
Magnaporthe grisea is a species complex of ascomycete fungal plant pathogens that cause disease on many grass species (Couch and Kohn, 2002). Within this complex, the hemibiotrophic pathogen M. oryzae causes blast disease in rice and 10–30 % of global rice harvest is lost to rice blast each year (Skamnioti and Gurr, 2009). Additionally, some M. grisea isolates infect other major cereal crops including wheat and barley, and can cause substantial yield loss (Talbot, 2003). Draper et al. (2001) first demonstrated the susceptibility of brachypodium to M. oryzae and noted variation in the response of brachypodium ecotypes to the pathogen. Several additional articles dealing with the interaction of M. oryzae isolates with brachypodium (e.g. Fig. 1B) have subsequently been published, making this the most mature brachypodium–cereal pathogen model system. Parker et al. (2008) provided an optimized protocol for study of the brachypodium–Magnoporthe interaction.
Routledge et al. (2004) assessed the interaction of four diverse M. grisea isolates with 21 diverse brachypodium ecotypes. A range of responses to each of the isolates was observed with two of the assessed ecotypes universally resistant and one universally susceptible. Symptoms exhibited by susceptible ecotypes (rapidly spreading lesions) and resistant ecotypes (highly localized necrotic flecks) resembled those displayed by susceptible and resistant rice varieties, respectively. These similarities were also comparable at the microscopic level. In a susceptible brachypodium ecotype (ABR1), primary infected cells became filled with secondary hyphae, with subsequent penetration of a second cell at approx. 48 h after inoculation. In contrast, in a resistant ecotype (ABR5), after formation of infection hyphae between 24 and 48 h, lesion development appeared to result primarily from cell death and no substantial hyphal growth occurred beyond 48 h following inoculation. Segregation of resistance to M. oryzae in F2 progeny from a cross between ABR1 and ABR5 was consistent with a single dominant resistance gene.
In related work, Allwood et al. (2006) compared metabolomic responses of ABR1 and ABR5 and found that phosphatidic acid phospholipid metabolism varied substantially between the ecotypes in response to M. oryzae challenge. Different phosphatidic acid phospholipids were detected at significantly higher and lower abundance in the resistant and susceptible ecotypes, respectively. Parker et al. (2009) extended the study of the metabolomic effect of M. oryzae infection in rice, barley and brachypodium. The authors initially compared the global metabolomic profile of healthy tissue with tissue at 1–5 d after inoculation in each of the hosts and similar metabolomic changes were detected in response to infection in all three species. Further analysis showed that changes observed in key metabolites throughout infection were consistent with suppression of the defensive reactive oxygen species (ROS) and lignification responses.
Recently, Wang et al. (2012) assessed the virulence on brachypodium of several M. oryzae mutants with attenuated or abolished pathogenicity on rice. For all mutants, alteration of virulence on brachypodium was comparable to that on rice, suggesting that defence against this pathogen is conserved between brachypodium and rice, and/or that M. oryzae deploys similar pathogenicity strategies during infection of both hosts.
Rhizoctonia root rot (Rhizoctonia solani AG 8).
The basidiomycete species Rhizoctonia solani (teleomorph Thanatephorus cucumeris) is currently divided into 14 anastomosis groups and contains strains that have varying levels of pathogenicity and host specificity (Carling et al., 2002). Rhizoctonia solani is a necrotrophic pathogen of major economic importance for wheat and barley production in regions including Australia and the Pacific Northwest of the US (Cook et al., 2002; Murray and Brennan, 2009). One strain, AG 8, causes the majority of disease on cereals worldwide and was first described by Neate and Warcup (1985). This pathogen invades roots of young seedlings, producing a range of enzymes that are ostensibly used to destroy root tissue, although pathogen virulence has not yet been linked to a particular enzyme (O’Brien and Zamani, 2003). In the field, the disease is often obvious as distinct empty patches (‘bare patch’), but may also result in a more general reduction in crop growth (Paulitz et al., 2009).
Cereal varieties with substantial resistance to R. solani are not available. Therefore, brachypodium offers the possibility for discovery of new genetic variation in resistance to the pathogen. Towards this aim, Schneebeli et al. (2014) developed a brachypodium–R. solani AG 8 pathosystem, demonstrating that infection by R. solani occurs similarly in wheat and brachypodium. When grown in soil containing approx. 0·1 R. solani propagules per gram, total root length of wheat and brachypodium was reduced by an average of 39 and 49 %, respectively, compared with uninoculated controls (Fig. 1C). Preliminary evidence of quantitative differences in resistance to R. solani was identified among seven brachypodium ecotypes.
An inherent advantage of using brachypodium as a model for root diseases is that it can be grown in small pots to a later stage of development than wheat (Watt et al., 2009). There is some indication that nodal roots, which appear during the vegetative phase, may play a role in compensating for early primary root loss due to disease. Schroeder and Paulitz (2008) noted that in barley more nodal roots appeared following infection with R. solani AG 8 and that these were less affected by disease than primary roots.
Fusarium head blight (Fusarium spp.).
Fusarium head blight (FHB) is one of the most devastating wheat diseases globally, responsible for pronounced losses in wheat production throughout growing regions in the US, Canada, Asia, Europe and South America. For example, FHB caused an estimated loss of approx. US$2 billion in the Northern Great Plains and Central US region in 1993–2001 (Nganje et al., 2004). FHB is predominantly caused by Fusarium graminearum (O’Donnell et al., 2004), but other Fusarium species including F. culmorum (Kollers et al., 2013) and F. pseudograminearum (Chakraborty et al., 2010) can also cause the disease. FHB infection commences at anthesis and leads to development of two main symptoms: necrotic lesion formation on the spike (scab) and bleaching of florets (blight) (Leonard and Bushnell, 2003). FHB-affected plants have reduced yield and produce grain of relatively poor quality. Significantly, FHB-infected grain can also be contaminated by mycotoxins (Desjardins, 2006) that can make the grain unsuitable for human or animal consumption. Mycotoxins produced by pathogenic Fusarium species can also be FHB virulence factors, with the production of tricothecenes including deoxynivalenol (DON) shown to promote head blight infection in wheat (Jansen et al., 2005). DON has been shown to inhibit protein synthesis in eukaryotic cells (Rocha et al., 2005) and production of DON within wheat stimulates production of ROS and defence gene expression, and induces host cell death (Desmond et al., 2008).
Recent work has established that brachypodium is readily infected by F. graminearum and F. culmorum via the application of methods typically used to perform head blight infection assays in wheat (Peraldi et al., 2011). Spray and point inoculation of floral tissues were optimized to produce consistently high levels of infection in brachypodium, with humidity and developmental stage found to be determining factors for successful infection, consistent with previous work in wheat and barley. Macroscopic symptom development was observed as development of necrotic lesions on the lemma between 12 and 36 hours post inoculation (hpi) followed by characteristic bleaching of florets between 48 and 96 hpi. Observation of infected tissues by confocal microscopy identified the base of macro-hairs as the probable site of infection on lemma tissue. Localized symptom development was associated with the formation of globose structures by the fungus at the base of macro-hairs, with extensive hyphal growth surrounding these and associated with extensive browning and tissue collapse at later stages of infection.
Peraldi et al. (2011) also assessed whether brachypodium exhibits type II FHB susceptibility (i.e. the bidirectional spread of disease directly between florets within the grain head; Schroeder and Christensen, 1963). Similar to wheat, brachypodium was shown to be susceptible to spread of infection within the head after point inoculation, and the authors also noted variation in type II FHB resistance between two brachypodium ecotypes, with Bd3-1 exhibiting significantly greater disease progression than Bd21. Additionally, brachypodium was assessed for sensitivity to DON. The application of DON after wounding in a detached leaf assay resulted in a zone of necrosis spreading from the point of inoculation, and exogenous application of DON to infected tissues yielded greater disease symptoms during brachypodium infection by both F. graminearum and F. culmorum. Furthermore, DON was found to accumulate to high levels in brachypodium spikes after spray inoculation with F. graminearum (Peraldi et al., 2011). Furthermore, a recent study has found that brachypodium plants infected with a DON-minus F. graminearum mutant triggered attenuated levels of defence gene expression and tryptophan-derived metabolites, and that this mutant had a reduced ability to colonize the plant (Pasquet et al., 2014). This study, together with previously discussed findings in wheat (Desmond et al., 2008), suggests that DON is an elicitor of defence responses and also acts as a virulence factor in both wheat and brachypodium.
Eyespot disease (Oculimacula spp.) and Ramularia leaf spot (Ramularia collo-cygni).
Eyespot, caused by the necrotrophic pathogens O. acuformis and O. yallundae, is a disease of major economic significance for wheat (Wei et al., 2011). Eyespot presents as elliptical, necrotic lesions on the stem base. The disease inhibits nutrient transport and can induce lodging and premature grain ripening (Lucas et al., 2000), with typical yield losses of 10–15 % and up to 50 % losses reported (Murray, 2010).
Ramularia leaf spot (RLS), caused by the necrotrophic pathogen R. collo-cygni, has recently been identified as an important pathogen of barley in northern Europe (Walters et al., 2008). RLS initially presents as brown/black leaf spots, with the surrounding leaf area subsequently becoming rapidly chlorotic then necrotic (Walters et al., 2008). The reduction in photosynthetic capacity due to rapid leaf senescence induced by RLS can result in considerable yield loss and loss of grain quality (Oxley and Havis, 2004). Although currently RLS is only economically significant for barley cultivation, it has also been identified on wheat and oats (Walters et al., 2008).
Peraldi et al. (2014) have demonstrated that brachypodium can be infected with eyespot and RLS, and exhibits similar symptoms (Fig. 1F, G). Lesions were present at 28 d following inoculation of the stem base of ecotypes Bd21 and Bd3-1 with O. acuformis or O. yallundae. On Bd3-1, lesions strongly resembled the characteristic ‘eye-shaped’ lesions occurring on wheat (Lucas et al., 2000), while symptoms exhibited by Bd21 were a more non-specific browning. Spray inoculation of Bd21 and Bd3-1 with R. collo-cygni produced brown necrotic spots strongly resembling RLS of barley. In brachypodium, Oculimacula spp. were found to form hyphal aggregates (infection plaques) under which penetration holes are formed, which is strikingly similar to the infection process described in wheat (Daniels et al., 1991). Infection of brachypodium with R. collo-cygni resulted in hyphal emergence from stomata on the abaxial surface, as occurs in barley (Walters et al., 2008). Peraldi et al. (2013) reported variation in resistance of the two ecotypes assessed, with Bd3-1 more susceptible than Bd21 to eyespot caused be either O. acuformis or O. yallundae.
Anthracnose (Colletotrichum cereale), stalk rot (Pythium aphanidermatum) and take-all (Gaeumannomyces graminis).
Sandoya and Buanafina (2014) have assessed the response of multiple brachypodium ecotypes to several insects as well as fungal and oomycete pathogens. In addition to reporting infection of brachypodium by Rhizoctonia solani and Magnoporthe oryzae/grisea, as previously described, the authors demonstrated the infection of brachypodium with known cereal pathogens Colletotrichum cereale, Pythium aphanidermatum and Gaeumannomyces graminis.
Colletotrichum cereale is of most significance as a pathogen of turf grass. However, it can also cause anthracnose disease on cereals such as sorghum, rye, wheat and oats (Crouch and Beirn, 2009). Spray inoculation of brachypodium foliage with a C. cereale spore suspension resulted in chlorotic/necrotic foliar lesions similar to those exhibited by other cereal and grass species (Crouch and Beirn, 2009). Spreading necrosis occurred in response to inoculation of the brachypodium stem base with oomycete pathogen P. aphanidermatum. Gaeumannomyces graminis is the causal agent of take-all, which is the most important root disease in wheat globally (Freeman and Ward, 2004). Inoculation of brachypodium roots with autoclaved, G. graminis-colonized oat seeds resulted in chlorosis and/or die-back of the above-ground tissue, similar to take-all symptoms in wheat (Cook et al., 2002).
BRACHYPODIUM–CEREAL PATHOGEN MODEL PATHOSYSTEMS: VIRAL PATHOGENS
Barley stripe mosaic virus
Barley stripe mosaic virus (BSMV) is a single-stranded, tripartite RNA virus, which can cause severe losses in its primary host, barley (Sastry, 2013). BSMV infects several other cereals naturally, and a range of plant species (including some dicots) under experimental conditions (Jackson et al., 2009). While the virus is highly mechanically transmissible facilitating rapid spread in the field, seed transmission is required for survival across seasons. Therefore, BSMV has been effectively controlled in developed countries by the use of diagnostic screening to detect and eliminate infected seed stocks (Jackson et al., 2009). However, it remains a significant problem in developing nations. Demircan and Akkaya (2010) initially demonstrated the ability of BSMV to infect and induce gene silencing in brachypodium. More recently, Cui et al. (2012) have assessed the resistance of diverse brachypodium inbred lines to the North Dakota 18 (ND18) strain of BSMV (e.g. Fig. 1H). Substantial variation was identified, and analysis of populations developed from a highly resistant (Bd3-1) and a highly susceptible (Bd21) ecotype led to the identification of a single dominant source of BSMV resistance, designated Bsr1. The authors observed a high recombination rate in a recombinant inbred line (RIL) population developed from Bd21 and Bd3-1 and performed fine-mapping of Bsr1 to a 23-kb interval using 165 RILs. Among the candidate genes identified was a nucleotide-binding site leucine-rich repeat (NBS-LRR) encoding gene (Bradi3g00757). Given that NBS-LRR proteins constitute a major class of R-proteins (Eitas and Dangl, 2010), Bradi3g00757 appears to be a good candidate for Bsr1. In related work, Lee et al. (2012) demonstrated that while Bd3-1 is highly resistant to ND18 and several additional BSMV strains, the Norwich strain of the virus is virulent on Bd3-1. Amino acid residues within the triple gene block 1 protein encoded by Norwich BSMV were shown to be responsible for its virulence on Bd3-1.
Panicum mosaic virus and Panicum mosaic satellite virus
Panicum mosaic virus (PMV) is a positive sense single-stranded RNA virus that infects some Poaceae species. Panicum mosaic satellite virus (SPMV), also a positive sense single-stranded RNA virus, commonly co-infects hosts infected with PMV. The PMV–SPMV co-infection (PMV + SPMV) is unusual for virus–satellite virus interactions in that it is synergistic; SPMV infection enhances accumulation of PMV and exacerbates disease symptoms (Scholthof, 1999). PMV and PMV + SPMV can cause crop loss in pearl millet (Thottappilly, 1992) and in turf grass causing a major disease called St. Augustine Decline (Cabrera and Scholthof, 1999). These two viruses can also be transmitted to maize and wheat, although this does not result in economically significant disease. Mandadi and Scholthof (2012) have recently demonstrated that PMV infection causes chlorosis and necrosis of leaves and stunting in brachypodium. Furthermore, PMV + SPMV infection of brachypodium resulted in exacerbated symptoms relative to infections by the individual viruses (Fig. 1I). Disease development in brachypodium is strongly similar to that of PMV and PMV + SPMV previously studied in pearl millet (Pennisetum glaucum) and foxtail millet (Setaria italica), and the systemic movement of both viruses within leaf and root tissue in brachypodium was demonstrated by immunoblot analysis (Scholthof, 1999). Mandadi and Scholthof (2012) also studied the transcriptomic host response to infection using microarray analysis. Upregulation of putative SA signalling components and downregulation of jasmonic acid and ethylene signalling components were observed in response to infection by both PMV and PMV + SPMV. Numerous genes demonstrated additive alteration of expression in PMV + SPMV compared with PMV infection alone. Interestingly, however, several pathogenesis-related (PR) gene homologues that were strongly induced by PMV showed attenuated upregulation upon infection with PMV + SPMV, suggesting that the host defence response may be partially repressed by co-infection.
Other viral pathogens.
More recently, Mandadi et al. (2014) demonstrated that brachypodium can be infected with a number of other cereal viruses (Table 2). Comparative analyses of defence responses triggered by virus infection in brachypodium and Seteria viridis, a C4 cereal species, have revealed both conserved and unique responses to these viruses. One of the conserved responses observed was the suppression of PHYTOALEXIN DEFICIENT4 encoding a regulator of SA signalling by virus infection in both species (Mandadi et al., 2014).
Table 2.
An overview of recent efforts to map wheat and barley resistance genes assisted by comparative genomic analysis with brachypodium
| Host | Disease | Gene | Brief description | Reference |
|---|---|---|---|---|
| Wheat | Rust/powdery mildew | Lr34/Yr18 | Multi-pathogen resistance gene Lr34/Yr18 (Krattinger et al., 2009) was mapped to ∼0·5 cM region on wheat 7DS by incorporating brachypodium-derived markers. Genes flanking Lr34/Yr18 region in wheat are separated by only 5 kb in brachypodium. | Spielmeyer et al. (2008) |
| Wheat | Powdery mildew | Ml3D233 | Eight markers co-segregating with powdery mildew resistance gene Ml3D233 were identified on wheat 5BL by incorporating brachypodium-derived markers. Markers correspond to 314 kb region in brachypodium containing 29 annotated genes. One brachypodium gene (Bd4g36980) is a NBS-LRR RGA, and a homologous wheat EST co-segregates with resistance. | Zhang et al. (2010) |
| Barley | Stem rust; spot blotch | Rpg1; rpg4; Rpg5; Rcs5 | Regions in brachypodium syntenic to those harbouring known barley resistance genes were inspected for putative brachypodium orthologues. Potential orthologues were identified in syntenic regions for Rpg1, rpg4 and Rpg5. Additionally, a 2·8 cM region on wheat harbouring the Rcs5 locus (yet to be cloned) was found to be highly collinear with an ∼300 kb region in brachypodium. | Drader and Kleinhofs (2010) |
| Wheat | Powdery mildew | Pm6 | Powdery mildew resistance gene Pm6 was fine-mapped on wheat 2BL by incorporating brachypodium-derived markers, and two markers spanning the locus were identified; syntenic region in brachypodium corresponds to ∼190 kb, containing two LRR-receptor-like protein kinase RGAs. | Qin et al. (2011) |
| Wheat | Powdery mildew | PmAS846 | Brachypodium-derived markers were used to assist fine-mapping of powdery mildew resistance gene PmAS846 to a 0·8 cM region in wheat collinear to a 197 kb region in brachypodium; 28 annotated brachypodium genes were found in this region including multiple RGAs. | Xue et al. (2012) |
| Wheat | Powdery mildew | MlIW170 | Powdery mildew resistance gene MlIW170 was mapped to a 2·69 cM region on wheat 2BS by incorporating brachypodium-derived markers; region collinear with 131 kb region in brachypodium; four RGA homologues were identified in wheat and brachypodium in this collinear region. | Liu et al. (2012) |
| Barley | Powdery mildew | QTL on 7HL and 7HS | Incorporation of markers derived from brachypodium allowed fine-mapping of two barley powdery mildew QTL on 7HL and 7HS, respectively, to regions of ∼0·6 and ∼0·7 cM. Brachypodium RGAs were not identified in syntenic regions. | Silvar et al. (2012) |
| Wheat | Tan spot | Tsc2 | Wheat tan spot susceptibility gene Tsc2 mapped to a 3·3 cM region on wheat 2BS by incorporating brachypodium-derived markers. Region syntenic to 390 kb in brachypodium containing 43 annotated genes was found but no RGAs were identified. | Abeysekara et al. (2010) |
| Wheat | Stripe rust | Yr26 | Yr26 was fine-mapped to 0·25 cM on wheat 1BL by incorporating brachypodium-derived markers. 1·92 Mb syntenic region in brachypodium harbours two RGAs. | Zhang et al. (2013) |
BRACHYPODIUM–CEREAL PATHOGEN MODEL PATHOSYSTEMS: ADDITIONAL INTERACTIONS
Stagonospora glume blotch (Stagonospora nodorum) and tan spot (Pyrenophora tritici-repentis)
Pyrenophora tritici-repentis and Stagonospora nodorum cause tan spot (also known as yellow spot) and Stagonospora glume blotch, respectively. Both diseases are of major economic importance for wheat production (Oliver et al., 2012; Kollers et al., 2014). Resistance to these diseases is largely governed by the interaction of specific ‘necrotrophic effectors’ (NEs) from the pathogen and host ‘susceptibility genes’ (Oliver and Solomon, 2010). These specific P. tritici-repentis and S. nodorum NEs have previously been referred to as host-specific or host-selective toxins (HSTs) and the interaction of HSTs and host susceptibility genes can be described as an ‘inverse’ gene-for-gene relationship (Faris et al., 2013), with reference to the classical plant/pathogen R/Avr interactions that have been extensively studied (Hammond-Kosack and Jones, 1997).
To study a ‘compatible’ interaction of wheat-infecting P. tritici-repentis or S. nodorum strains with brachypodium, brachypodium accessions or species harbouring corresponding HST susceptibility gene(s) are required. Stagonospora blotch has been reported to affect Brachypodium sylvaticum (Halbritter et al., 2012). Additionally, Falter and Voigt (2014) have reported colonization of brachypodium by S. nodorum and the economically important barley-infecting Pyrenophora teres (Liu et al. 2011) using a detached leaf assay. Thus, study of the interaction of brachypodium with cereal-infecting Stagonospora and Pyrenophora species may be possible under certain laboratory conditions.
Fusarium crown rot (Fusarium pseudograminearum)
In addition to FHB described above, Fusarium crown rot (FCR) is a disease of substantial economic importance. In Australia, FCR is a more substantial problem for wheat growers than FHB, causing an average annual loss estimated at $79 million (Murray and Brennan, 2009). Although F. graminearum and F. culmorum can cause FCR, the most common FCR pathogen is F. pseudograminearum (Chakraborty et al., 2010). We have observed that brachypodium infected with F. pseudograminearum in glasshouse and laboratory-based assays exhibits symptoms that are highly similar to those of wheat (e.g. Fig. 2A).
Fig. 2.
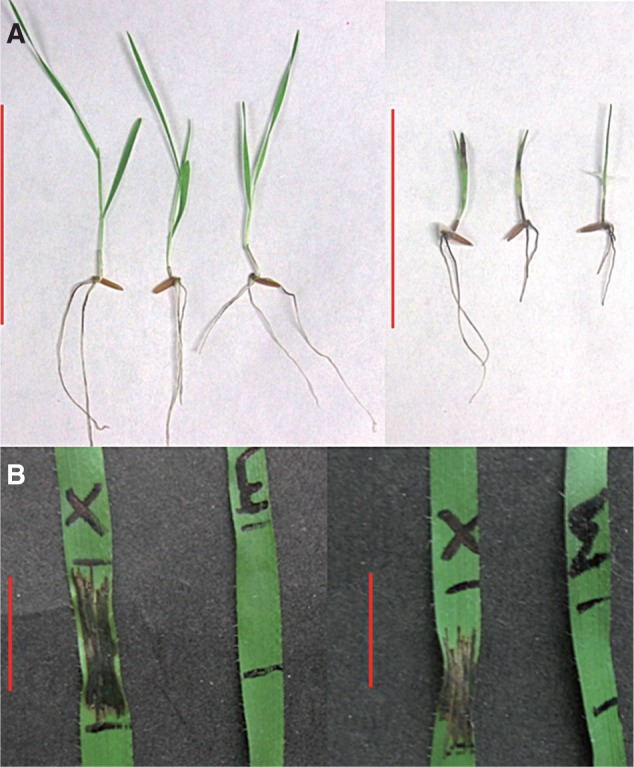
Brachypodium infected with Xanthomonas translucens and Fusarium pseudograminearum. (A) Symptoms of ecotype Bd21-3 inoculated with Fusarium pseudograminearum using the method of Yang et al. (2010) with minor modifications. Representative symptoms at 14 days post-F. pseudograminearum inoculation are shown (right) in comparison with mock inoculated seedlings (left). Scale bar = 10 cm. (B) Symptoms of ecotypes Bd21 (left) and Bd21-3 (right) inoculated with Xanthomonas translucens isolate DAR61454 using the method outlined by Gardiner et al. (2014) with minor modifications. Representative symptom development in a leaves inoculated with DAR61454 (X) and mock inoculated (M) at 6 days post-inoculation is shown. Scale bar = 2 cm.
Common root rot/leaf spot (Bipolaris sorokiniana)
Common root rot and leaf spot caused by B. sorokiniana is responsible for major losses in wheat and barley globally (Kumar et al., 2002). In addition to their study of brachypodium interactions with S. nodorum and P. teres described above, Falter and Voigt (2014) observed colonization of detached brachypodium leaves with B. sorokiniana. However, it remains unknown whether this pathogen can infect intact brachypodium plants.
Bacterial leaf streak (Xanthomonas translucens)
Three Xanthomonas species, including rice-infecting Xanthomonas oryzae pv. Oryzae, are considered to belong to a ‘Top 10’ of bacterial plant pathogens in terms of their economic impact and research significance (Mansfield et al., 2012). Xanthomonas translucens is a pathogen of wheat and/or barley (Adhikari et al., 2011) causing bacterial leaf streak (BLS), or ‘black chaff’, which is a significant wheat disease globally with up to 40 % yield losses (Forster and Schaad, 1988). Current understanding of cereal–bacterial pathogen interactions is limited (Gardiner et al. 2014) and a brachypodium–X. translucens model system may help to improve such understanding.
Recently, the genome of a wheat and barley-infecting X. translucens isolate ‘DAR61454’ has been sequenced (Gardiner et al., 2014). We have observed that DAR61454 is pathogenic on a range of brachypodium ecotypes, including major community ecotypes Bd21-3 and Bd21 (Fig. 2B). DAR61454 possesses a Type III secretion system and is capable of delivering heterologous proteins into wheat cells (Gardiner et al., 2014). This delivery system may prove to be useful for the characterization of cereal pathogen effectors. Given that X. translucens causes massive cell death upon infiltration into brachypodium leaves, DAR61454-mediated protein delivery may be particularly well suited to the study of heterologous proteins with putative roles in repressing host cell death.
In addition to the brachypodium–cereal pathogen experimental systems described above, the interaction of brachypodium with other grass pathogens that are not known to cause significant disease on cereals has been described. Falter and Voigt (2014) observed colonization of detached brachypodium leaves with Fusarium sacchari [native host (NH): sugarcane; Saccharum interspecific hybrid), Phaeosphaeria caricis (NH: Typha spp.), Pithomyces chartarum (NH: ryegrass; Lolium perenne), Stagonospora macropycnidia (NH: common reed; Phragmites spp.), Stagonospora paspali (NH: bahiagrass; Paspalum notatum) and Stagonospora tainanensis (NH: sugarcane). Furthermore, Sandoya and Buanafina (2014) observed infection of brachypodium with Ophiosphaerella agrostis (NH: creeping bentgrass; Agrostis stolonifera), Ophiosphaerella korrae (NH: bermudagrass; Cynodon dactylon) and Sclerotinia homoeocarpa (NH: several turf grass species). Additionally, Mandadi and Scholthof (2013) have recently presented a summary of reported pathogen interactions with brachypodium and/or its perennial relative Brachypodium sylvaticum that is currently being developed as a model for perennial grasses (Steinwand et al., 2013).
FUNCTIONAL ANALYSIS OF DEFENCE-ASSOCIATED BRACHYPODIUM GENES
Compared with arabidopsis and rice, functional analysis of brachypodium genes is still very much in its infancy. Preliminary work performed in this area is described here.
Mur et al. (2004) identified putatively wound-induced transcripts in brachypodium. Among the transcripts detected was a serine proteinase inhibitor homologue designated as Bdpin1 which was induced both locally and systemically in response to wounding as well as challenge by M. grisea in both a resistant (ABR3) and a susceptible (ABR1) ecotype. Bdpin1 was also shown to be induced by the application of methyl jasmonate but not the SA analogue benzothiadiazole. Although the exact function of Bdpin1 in pathogen defence is unknown, proteinase inhibitors have been shown to increase plant resistance to fungal and bacterial pathogens (Charity et al., 2005).
Uridine diphosphate glycosyltransferases (UGTs) are encoded by a large gene family in plants and glycosylate a vast range of low-molecular-weight compounds, including plant hormones and secondary metabolites (Bowles et al., 2005). UGTs that detoxify DON have been identified (AtUGT73C5, Poppenberger et al., 2003; HvUGT13248, Schweiger et al., 2010). By analysis of the Bd21 genome sequence, Schweiger et al. (2013) identified 159 brachypodium genes encoding UGTs that included two putative homologues of the gene encoding the DON-inactivating arabidopsis enzyme UGT73C5. These genes were found to be highly responsive to both inoculation with F. graminearum and treatment with DON. However, heterologous expression of the enzymes in yeast failed to induce DON resistance, suggesting that the enzymes are not able to detoxify DON. Additionally, six brachypodium UGT-encoding genes with high homology to a barley UGT (UGT13248) that detoxifies DON (Schweiger et al., 2010) were highly responsive to F. graminearum infection and DON treatment. Two of these genes, Bradi5g03300.1 and Bradi5g02780.1, induced resistance to DON when expressed in yeast. Further biochemical experiments confirmed that yeast strains expressing the encoded enzymes detoxified DON to DON-3-O-glucoside. These genes would appear to be high-priority candidates for additional experiments to assess in planta roles of these enzymes during FHB infection of brachypodium and also for heterologous expression in wheat to reduce FHB.
The plant cell wall is a mechanical barrier that presents as a first line of defence against pathogens (Hématy et al., 2009). Pogorelko et al. (2013) found that transgenic brachypodium lines (ecotype Bd21) expressing a xylan-specific acetylesterase from Aspergillus nidulans (designated AnAXE) showed significantly decreased cell-wall acetyl content, indicating that cell-wall xylans were deacetylated in the transgenic lines. Cell-wall morphology and thickness was not altered in the AnAXE-expressing transgenics compared with the wild-type Bd21, as assessed by microscopic examination. However, in response to infection by Bipolaris sorokiniana, symptom development was significantly delayed and in planta accumulation of the pathogen was significantly reduced.
Small RNAs (sRNAs) are 18- to 24-bp ribonucleotides that represent a core regulatory component of the plant transcriptome (Axtell, 2013). Lucas et al. (2014) have recently assessed the brachypodium genome for micro-RNAs (miRNAs) that may be involved in the regulation of host R genes. Brachypodium genes with NBS-LRR domains were identified via a search of the Conserved Domain Database (Marchler-Bauer et al., 2011). Of the 121 putative R genes identified, possible miRNA target sites were identified for 33 genes. For five of these genes, the expression profiles of both the gene and its putative regulatory miRNA were assessed in response to infection by the cereal pathogen Fusarium culmorum, and SA treatment; however, no correlations could be found between the expression of gene and miRNA. Nevertheless, further exploration of the regulation of components of host resistance in brachypodium by endogenous sRNAs seems warranted (Balmer and Mauch-Mani, 2013).
EXPLOITING NATURAL VARIATION IN BRACHYPODIUM TO CHARACTERIZE GENES AFFECTING RESISTANCE
A large collection of diverse brachypodium ecotypes and accessions has been established (Filiz et al., 2009; Vogel et al., 2009; Mur et al., 2011). As outlined above, variation in the resistance of ecotypes has been observed for several cereal pathogens. This provides the opportunity to identify loci contributing to resistance within this material by the development and analysis of populations from ecotypes with contrasting resistance; the previously discussed BSMV work of Cui et al. (2012) exemplifies this approach.
As described above, Sandoya and Buanafina (2014) recently assessed the resistance of several brachypodium accessions and ecotypes to a range of cereal and grass pathogens. Of eight lines that were included in their study, the authors found one accession (PI 227011) to be comparatively susceptible, and another (PI 245730) to be comparatively resistant to most fungal pathogens and both insect pests assessed. The authors also assessed changes in defence-associated gene expression in response to S. homoeocarpa infection, in the genome-sequenced inbred line Bd21 (The International Brachypodium Initiative, 2010), which was highly susceptible to this pathogen, and the resistant PI 245730 accession. A significantly higher induction of a LOX3 homologue putatively involved in jasmonate (JA) biosynthesis was detected in PI 245730 than Bd21. In contrast, induction of an NPR1 homologue putatively involved in activation of SA-dependent defences was significantly higher in Bd21 than PI 245730. This provides preliminary evidence that alterations in JA/SA defence signalling known to play a core role in modulating host defence in arabidopsis (Bari and Jones, 2009) may contribute to altered levels of resistance in brachypodium ecotypes.
MUTANT RESOURCES FOR THE STUDY OF PLANT–PATHOGEN INTERACTIONS IN BRACHYPODIUM
Large-scale pathogen resistance screening of arabidopsis mutant accessions, followed by genetic characterization of mutants with altered resistance (‘forward genetics’) has contributed substantially to our understanding of plant–pathogen interactions (Glazebrook et al., 1997; Glazebrook, 2001).
A substantial collection of brachypodium T-DNA mutants has been created (Thole et al., 2010; Bragg et al., 2012), and efforts are underway to develop homozygous lines for insertions in or near genes. Since publication of the initial description of the WRRC brachypodium T-DNA collection (Bragg et al., 2012), the resource has expanded to contain approx. 22 000 mutants from which >25 000 insertion sites have been identified (http://brachypodium.pw.usda.gov/TDNA/). Additionally, a brachypodium TILLING population has been developed (Dalmais et al., 2013).
COMPARATIVE GENOMICS IN BRACHYPODIUM FOR THE CHARACTERIZATION OF CEREAL RESISTANCE GENES
Comparative genomics can be broadly defined as the comparison of the structure and function of genomes. Brachypodium belongs to the subfamily Pooideae along with the major cereal crops wheat and barley. Thus, the synteny and sequence conservation between brachypodium, wheat and barley is particularly high (The International Brachypodium Initiative, 2010). Both barley and wheat feature large, repeat-rich genomes that present significant challenges for traditional map-based cloning approaches. The genome sequence of brachypodium can assist this process by allowing development of new markers for fine-mapping in wheat and barley, based on genes within the syntenic region of interest. In addition, once a locus has been finely mapped in wheat and barley, candidate genes based upon those present in the syntenic region in brachypodium can be identified. However, caution must be used when undertaking such a strategy as rearrangements and/or gene losses or expansions can occur at this fine scale despite generally strong ‘macrosynteny’ between genomic regions (Luo et al., 2012).
In recent years, a substantial number of studies have exploited the brachypodium genome sequence to aid the mapping of resistance genes in wheat and barley (Table 1). Markers derived from brachypodium have proven extremely useful for fine-mapping of the targeted resistance genes. Frequently, resistance gene analogues (RGAs) have been identified within relatively small genomic regions in brachypodium corresponding to regions harbouring resistance QTL in wheat and barley. In contrast, comparative genomic approaches to identify wheat disease resistance genes using the rice genome sequence have often been limited by low collinearity in regions harbouring such genes (Keller et al., 2005).
Several comparative approaches have used brachypodium to assist with fine-mapping of powdery mildew (Blumeria graminis) resistance (PMR) genes in cereals (Table 1). For example, Zhang et al. (2010) reported the use of wheat–brachypodium synteny for fine-mapping of a broad-spectrum PMR gene (Ml3D232), introgressed into bread wheat from wild emmer wheat (Triticum turgidum var. dicoccoides). Ml3D232 was initially mapped to the chromosome 5BL bin 0·59–0·76 by identification of polymorphic microsatellite markers between susceptible/resistant F2 bulks and subsequent bin mapping of these polymorphic microsatellites using Group 5 Chinese Spring deletion accessions (Endo and Gill, 1996). Wheat expressed sequence tags (ESTs) mapped to this location were then used to identify the brachypodium genomic region homologous to the region harbouring Ml3D232 in wheat. Subsequently, primers targeting additional wheat ESTs homologous to brachypodium genes within this region were used to map Ml3D232 in an F2 population. Eight markers co-segregated with Ml3D232 and these markers correspond to a 314 kb region in brachypodium harbouring 29 annotated genes. Among these is an NBS-LRR resistance gene homologue Bd4g36980; a wheat EST (CJ683537) homologous to Bd4g36980 was found to co-segregate with Ml3D232. Clearly, where crop homologues of brachypodium RGAs co-segregate with resistance, as was demonstrated in this study, both the crop and the brachypodium gene present as strong candidates for further analysis.
Despite the effectiveness of comparative genomics in brachypodium for improving the efficiency of fine-mapping in cereals, with the emergence of techniques that can allow rapid characterization of QTL without the need for map-based cloning (e.g. Takagi et al., 2013; Liu et al., 2012; as discussed below) the degree to which this traditional approach is employed may decrease in coming years. In this case, comparative genomic approaches using brachypodium may more frequently involve functional characterization of RGAs and homologues of other classes of genes known to modulate resistance, as exemplified by the work of Schweiger et al. (2013) on brachypodium UGTs outlined above. Tan and Wu (2012) performed an in silico genome-wide assessment of NBS RGAs in brachypodium, identifying 239 NBS-encoding genes. Similarly, Tripathi et al. (2012) performed in silico analysis of brachypodium WRKY transcription factors, known to possess critical roles in the modulation of numerous plant functions, including the response to biotic and abiotic stresses; the authors have developed a publicly accessible database for comparative analysis of these transcription factors. Such resources provide a useful platform for the selection of brachypodium genes to assess for roles in resistance to cereal diseases.
FUTURE PROSPECTS FOR BRACHYPODIUM AS A MODEL FOR CEREAL–PATHOGEN INTERACTIONS
Genome-wide association studies (GWAS), which seek to identify markers associated with a trait by large-scale genotyping of individuals differing for that trait, have recently been applied to cereal crop species with impressive results (Tian et al., 2011; Huang et al., 2012; Jia et al., 2013). GWAS may also be highly effective for the identification of brachypodium genes with roles in resistance to cereal disease. Currently an initiative to re-sequence 54 brachypodium accessions is underway (http://Brachypodium.pw.usda.gov/; Gordon et al., 2014), and this will provide a platform for GWAS efforts in brachypodium. With substantial variation detected in the resistance of brachypodium ecotypes to a range of cereal diseases as described above, GWAS to identify loci contributing to resistance appears to be a powerful future strategy. Additionally, the potential of different brachypodium species, including polyploid species [e.g. allotetraploid B. hybridum (Catalán et al., 2012)] and perennial diploid B. sylvaticum (Steinwand et al. 2013) to complement the use of B. distachyon as a model, has recently been recognized. A number of polyploid brachypodium species that may differ from diploid species for various adaptive traits such as drought tolerance have been described (Manzaneda et al., 2012). Future characterization of diploid and polyploid brachypodium species may similarly reveal new traits associated with biotic stress tolerance.
Over the last two decades, map-based cloning approaches have made a substantial contribution to our understanding of plant genetics (Peters et al., 2003). Nevertheless, traditional map-based cloning is a notoriously labour-intensive and time-consuming process (Hall et al., 2010). Bulked segregant analysis (BSA; Michelmore et al., 1991), wherein DNA markers specific to individuals possessing a characteristic of interest are identified by comparison of pooled DNA from progeny segregating for the characteristic, has seen widespread use in plant genetic research. Recently, BSA approaches exploiting high-throughput sequencing technologies have been shown to be highly effective for the identification of causal genes (Schneeberger and Weigel, 2011), circumventing the need for traditional map-based cloning. Both DNA (e.g. Takagi et al., 2013) and RNA (e.g. Liu et al., 2012) sequencing-based BSA approaches have been developed and applied to the identification of natural allelic variants (e.g. Takagi et al., 2013) and induced mutations (Abe et al., 2012; Nordstrom et al., 2013). Brachypodium’s small, diploid genome is particularly amenable to such approaches, and they should therefore facilitate rapid identification of genes involved in brachypodium resistance to cereal pathogens.
Compared with arabidopsis and rice, functional analysis of brachypodium genes is still very much in its infancy. However, genomic brachypodium resources are rapidly accumulating (Brkljacic et al., 2011); key resources include a sizable collection of T-DNA mutants (Bragg et al., 2012) and a publicly available TILLING resource (Dalmais et al., 2013), as noted above. Increasingly powerful sequencing technologies are facilitating high-throughput characterization of mutant collections (Polko et al., 2012) and rapid identification of causal mutations within TILLING populations (Nordstrom et al., 2013). This has the potential to enable extensive functional characterization of disease resistance in brachypodium in the near future.
Information regarding roles of plant hormones in modulating disease resistance or susceptibility of brachypodium is currently limited. A recent study investigating potential roles of brassinosteroids in disease resistance in monocots found that the brassinosteroid-insensitive 1 (BRI1) mutation, affecting the receptor of this hormone in brachypodium, increased resistance against necrotrophic and hemibiotrophic pathogens. Importantly, disruption of the BRI1 gene produced a similar effect on disease resistance in barley, suggesting that the effect of this mutation is mechanistically conserved between these two species (Goddard et al., 2014). Additional studies are certainly needed to reveal other components of plant hormone signalling pathways in brachypodium, which may be conserved in cereal species.
Antimicrobial proteins and metabolites are a basic component of innate immunity in plants and such compounds contribute to defence against a broad range of pathogens. There is substantial heterogeneity in plant defence compounds, with considerable variation even in closely related cereal crops. Wild cereal relatives such as brachypodium are thus likely to be a rich source of novel defence compounds that could be harnessed to enhance cereal disease resistance (Großkinsky et al., 2012). In addition, pathogens appear to evolve mechanisms to neutralize host chemical defences (Hammerschmidt, 1999) and therefore the transgenic production of brachypodium antimicrobial compounds may be a more effective approach for engineering resistance in cereal crops than boosting endogenous chemical defences (Großkinsky et al., 2012). The application of metabolomic and proteomic technologies to the discovery of compounds accumulating in brachypodium upon pathogen attack would appear to be a useful strategy to develop new insight into plant defence, which may also facilitate strategies for engineering resistance in cereal crops.
Additionally, although the native plant species would appear the best candidate for the direct study of gene function via overexpression, in some instances heterologous expression may be of utility for this purpose; an obvious example is where the native species is recalcitrant towards transformation. Even in the presence of recently developed, high-efficiency transformation protocols for specific genotypes of some cereal crop species (e.g. wheat and barley; Harwood, 2012; Richardson et al., 2014), brachypodium may be an attractive alternative for the study of the role of crop genes in disease resistance.
Based on the studies reviewed here it is clear that brachypodium and cereals share similarities in symptom development and host defence responses. Furthermore, recent studies are proving that new insights into plant–microbe interactions can be revealed using brachypodium; the mapping of non-host resistance in brachypodium to wheat stripe rust (Ayliffe et al. 2013), as outlined above, is a key example. Another important example is the very recent study of Blümke et al. (2014) on the brachypodium–FHB interaction. Although the effect of DON on eliciting defence gene expression has been demonstrated in both wheat (Desmond et al., 2008) and brachypodium (Pasquet et al., 2014), Blümke et al. (2014) have identified a new role for this mycotoxin as an inducer of resistance to FHB in brachypodium, seemingly via a ‘priming’ effect. The pretreatment of brachypodium with low concentrations of DON was been found to reduce the susceptibility to FHB by eliciting defence gene expression and altering host cell-wall composition.
CONCLUSIONS
In recent years it has become evident that brachypodium is a broadly useful model to study cereal–pathogen interactions. In many instances, brachypodium is infected by cereal crop pathogens and develops very similar symptoms to the crop host. Continued expansion of publically available resources including T-DNA lines, TILLING populations, expression data, comparative genomics tools, re-sequenced ecotypes and diverse germplasm collections (Brkljacic et al., 2011) will help brachypodium fulfil its potential as a model to study cereal diseases in the future. Furthermore, together with a mature brachypodium model platform, increasingly powerful omics and systems biology approaches (Ballereau et al., 2013) have the capacity to yield sophisticated, holistic understanding of cereal–pathogen interactions. This has great potential to guide strategies for improved cereal crop disease resistance.
ACKNOWLEDGEMENTS
CSIRO-affiliated authors gratefully acknowledge support provided by the Grains Research and Development Corporation, Australia.
LITERATURE CITED
- Abe A, Kosugi S, Yoshida K, et al. 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Abeysekara NS, Friesen TL, Liu Z, McClean PE, Faris JD. 2010. Marker development and saturation mapping of the tan spot Ptr ToxB sensitivity locus Tsc2 in hexaploid wheat. The Plant Genome 3: 179–189. [Google Scholar]
- Adhikari TB, Gurung S, Hansen JM, Bonman JM. 2011. Pathogenic and genetic diversity of Xanthomonas translucens pv. undulosa in North Dakota. Phytopathology 102: 390–402. [DOI] [PubMed] [Google Scholar]
- Allwood WJ, Ellis DI, Heald JK, Goodacre R, Mur LAJ. 2006. Metabolomic approaches reveal that phosphatidic and phosphatidyl glycerol phospholipids are major discriminatory non-polar metabolites in responses by Brachypodium distachyon to challenge by Magnaporthe grisea. The Plant Journal 46: 351–368. [DOI] [PubMed] [Google Scholar]
- Ankeny RA, Leonelli S. 2011. What’s so special about model organisms? Studies in History and Philosophy of Science Part A 42: 313–323. [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annual Review of Plant Biology 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Ayliffe M, Devilla R, Mago R, et al. 2011. Nonhost resistance of rice to rust pathogens. Molecular Plant-Microbe Interactions 24: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Ayliffe M, Singh D, Park R, Moscou M, Pryor T. 2013. Infection of Brachypodium distachyon with selected grass rust pathogens. Molecular Plant-Microbe Interactions 26: 946–957. [DOI] [PubMed] [Google Scholar]
- Ballereau S, Glaab E, Kolodkin A, et al. 2013. Functional genomics, proteomics, metabolomics and bioinformatics for systems biology. In: Prokop A, Csukás B, eds. Systems biology. Dordrecht: Springer, 3–42. [Google Scholar]
- Balmer D, Mauch-Mani B. 2013. Small yet mighty — microRNAs in plant-microbe interactions. MicroRNA 2: 73–80. [PubMed] [Google Scholar]
- Barbieri M, Marcel TC, Niks RE. 2011. Host status of false brome grass to the leaf rust fungus Puccinia brachypodii and the stripe rust fungus P. striiformis. Plant Disease 95: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Barbieri M, Marcel TC, Niks RE, et al. 2012. QTLs for resistance to the false brome rust Puccinia brachypodii in the model grass Brachypodium distachyon L. Genome 55: 152–163. [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JG. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69: 473–488. [DOI] [PubMed] [Google Scholar]
- Blümke A, Sode B, Ellinger D, Voigt CA. 2014. Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Molecular Plant Pathology, doi: 10.1111/mpp.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim E-K, Poppenberger B. 2005. Glycosyltransferases: managers of small molecules. Current Opinion in Plant Biology 8: 254–263. [DOI] [PubMed] [Google Scholar]
- Boyd LA, Ridout C, O’Sullivan DM, Leach JE, Leung H. 2013. Plant–pathogen interactions: disease resistance in modern agriculture. Trends in Genetics 29: 233–240. [DOI] [PubMed] [Google Scholar]
- Bragg JN, Wu J, Gordon SP, et al. 2012. Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS ONE 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U. 1995. The powdery mildews (Erysiphales) of Europe. Jena: VEB Gustav Fischer Verlag. [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, et al. 2011. Brachypodium as a model for the grasses: today and the future. Plant Physiology 157: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera O, Scholthof K-BG. 1999. The complex viral etiology of St. Augustine decline. Plant Disease 83: 902–904. [DOI] [PubMed] [Google Scholar]
- Carling DE, Baird RE, Gitaitis RD, Brainard KA, Kuninaga S. 2002. Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani. Phytopathology 92: 893–899. [DOI] [PubMed] [Google Scholar]
- Cassman KG, Dobermann A, Walters DT, Yang H. 2003. Meeting cereal demand while protecting natural resources and improving environmental quality. Annual Review of Environment and Resources 28: 315–358. [Google Scholar]
- Catalán P, Müller J, Hasterok R, et al. 2012. Evolution and taxonomic split of the model grass Brachypodium distachyon. Annals of Botany 109: 385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Obanor F, Westecott R, Abeywickrama K. 2010. Wheat crown rot pathogens Fusarium graminearum and F. pseudograminearum lack specialization. Phytopathology 100: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Charity JA, Hughes P, Anderson MA, et al. 2005. Pest and disease protection conferred by expression of barley β-hordothionin and Nicotiana alata proteinase inhibitor genes in transgenic tobacco. Functional Plant Biology 32: 35–44. [DOI] [PubMed] [Google Scholar]
- Cook RJ, Schillinger WF, Christensen NW. 2002. Rhizoctonia root rot and take-all of wheat in diverse direct-seed spring cropping systems. Canadian Journal of Plant Pathology 24: 349–358. [Google Scholar]
- Couch BC, Kohn LM. 2002. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94: 683–693. [DOI] [PubMed] [Google Scholar]
- Crouch J, Beirn L. 2009. Anthracnose of cereals and grasses. Fungal Diversity 39: 19. [Google Scholar]
- Cui Y, Lee MY, Huo N, et al. 2012. Fine mapping of the Bsr1 barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS ONE 7: e38333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, et al. 2013. A TILLING platform for functional genomics in Brachypodium distachyon. PLoS ONE 8: e65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels A, Lucas JA, Peberdy JF. 1991. Morphology and ultrastructure of W and R pathotypes of Pseudocercosporella herpotrichoides on wheat seedlings. Mycological Research 95: 385–397. [Google Scholar]
- Demircan T, Akkaya M. 2010. Virus induced gene silencing in Brachypodium distachyon, a model organism for cereals. Plant Cell, Tissue and Organ Culture 100: 91–96. [Google Scholar]
- Desjardins AE. 2006. Fusarium mycotoxins: chemistry, genetics, and biology. St. Paul: American Phytopathological Society (APS Press). [Google Scholar]
- Desmond OJ, Manners JM, Stephens AE, et al. 2008. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Molecular Plant Pathology 9: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drader T, Kleinhofs A. 2010. A synteny map and disease resistance gene comparison between barley and the model monocot Brachypodium distachyon. Genome 53: 406–417. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LAJ, Jenkins G, et al. 2001. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology 127: 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. 2010. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Current Opinion in Plant Biology 13: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Gill B. 1996. The deletion stocks of common wheat. Journal of Heredity 87: 295–307. [Google Scholar]
- Falter C, Voigt C. 2014. Comparative cellular analysis of pathogenic fungi with a disease incidence in Brachypodium distachyon and Miscanthus × giganteus. BioEnergy Research 7: 958–973. [Google Scholar]
- Faris J, Liu Z, Xu S. 2013. Genetics of tan spot resistance in wheat. Theoretical and Applied Genetics 126: 2197–2217. [DOI] [PubMed] [Google Scholar]
- Figueroa M, Alderman S, Garvin DF, Pfender WF. 2013. Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host-pathogen incompatibility. PLoS ONE 8: e56857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiz E, Ozdemir BS, Budak F, Vogel JP, Tuna M, Budak H. 2009. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52: 876–890. [DOI] [PubMed] [Google Scholar]
- Forster R, Schaad N. 1988. Control of black chaff of wheat with seed treatment and a foundation seed health program. Plant Disease 72: 935–938. [Google Scholar]
- Freeman J, Ward E. 2004. Gaeumannomyces graminis, the take-all fungus and its relatives. Molecular Plant Pathology 5: 235–252. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Upadhyaya NM, Stiller J, et al. 2014. Genomic analysis of Xanthomonas translucens pathogenic on wheat and barley reveals cross-kingdom gene transfer events and diverse protein delivery systems. PLoS ONE 9: e84995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin DF, Gu Y-Q, Hasterok R, et al. 2008. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Science 48: S-69–S-84. [Google Scholar]
- Glawe DA. 2008. The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annual Review of Phytopathology 46: 27–51. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. 2001. Genes controlling expression of defence responses in Arabidopsis – 2001 status. Current Opinion in Plant Biology 4: 301–308. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. 1997. Use of Arabidopsis for genetic dissection of plant defence responses. Annual Review of Genetics 31: 547–569. [DOI] [PubMed] [Google Scholar]
- Goddard R, Peraldi A, Ridout C, Nicholson P. 2014. Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Molecular Plant-Microbe Interactions 27: 1095–1106. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Priest H, Des Marais DL, et al. 2014. Genome diversity in Brachypodium distachyon: deep sequencing of highly diverse inbred lines. Plant Journal 79: 361–374 doi: 10.1111/tpj.12569 [DOI] [PubMed] [Google Scholar]
- Großkinsky DK, van der Graaff E, Roitsch T. 2012. Phytoalexin transgenics in crop protection—fairy tale with a happy end? Plant Science 195: 54–70. [DOI] [PubMed] [Google Scholar]
- Halbritter AH, Carroll GC, Güsewell S, Roy BA. 2012. Testing assumptions of the enemy release hypothesis: generalist versus specialist enemies of the grass Brachypodium sylvaticum. Mycologia 104: 34–44. [DOI] [PubMed] [Google Scholar]
- Hall D, Tegström C, Ingvarsson PK. 2010. Using association mapping to dissect the genetic basis of complex traits in plants. Briefings in Functional Genomics 9: 157–165. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. 1999. Phytoalexins: what have we learned after 60 years? Annual Review of Phytopathology 37: 285–306. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. 1997. Plant disease resistance genes. Annual Review of Plant Physiology and Plant Molecular Biology 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Harwood WA. 2012. Advances and remaining challenges in the transformation of barley and wheat. Journal of Experimental Botany 63: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S. 2009. Host–pathogen warfare at the plant cell wall. Current Opinion in Plant Biology 12: 406–413. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, et al. 2012. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics 44: 32–39. [DOI] [PubMed] [Google Scholar]
- Huang X-Q, Röder M. 2004. Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137: 203–223. [Google Scholar]
- The International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Jackson AO, Lim H-S, Bragg J, Ganesan U, Lee MY. 2009. Hordeivirus replication, movement, and pathogenesis. Annual Review of Phytopathology 47: 385–422. [DOI] [PubMed] [Google Scholar]
- Jansen C, von Wettstein D, Schäfer W, Kogel K-H, Felk A, Maier FJ. 2005. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proceedings of the National Academy of Sciences of the United States of America 102: 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Huang X, Zhi H, et al. 2013. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nature Genetics 45: 957–961. [DOI] [PubMed] [Google Scholar]
- Jung K-H, An G, Ronald PC. 2008. Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nature Reviews: Genetics 9: 91–101. [DOI] [PubMed] [Google Scholar]
- Keller B, Feuillet C, Yahiaoui N. 2005. Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize genome. Genetics Research 85: 93–100. [DOI] [PubMed] [Google Scholar]
- Kollers S, Rodemann B, Ling J, et al. 2014. Genome-wide association mapping of tan spot resistance (Pyrenophora tritici-repentis) in European winter wheat. Molecular Breeding 34: 363–371. [Google Scholar]
- Kollers S, Rodemann B, Ling J, et al. 2013. Whole genome association mapping of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS ONE 8: e57500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, et al. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kumar J, Schäfer P, Hückelhoven R, et al. 2002. Bipolaris sorokiniana, a cereal pathogen of global concern: cytological and molecular approaches towards better control. Molecular Plant Pathology 3: 185–195. [DOI] [PubMed] [Google Scholar]
- Lee MY, Yan L, Gorter FA, et al. 2012. Brachypodium distachyon line Bd3-1 resistance is elicited by the barley stripe mosaic virus triple gene block 1 movement protein. Journal of General Virology 93: 2729–2739. [DOI] [PubMed] [Google Scholar]
- Leonard KJ, Bushnell WR. 2003. Fusarium head blight of wheat and barley. St. Paul: American Phytopathological Society (APS Press). [Google Scholar]
- Liu S, Yeh C-T, Tang HM, Nettleton D, Schnable PS. 2012. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ellwood SR, Oliver RP, Friesen TL. 2011. Pyrenophora teres: profile of an increasingly damaging barley pathogen. Molecular Plant Pathology 12: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JA, Dyer PS, Murray TD. 2000. Pathogenicity, host-specificity, and population biology of Tapesia spp., causal agents of eyespot disease of cereals. Advances in Botanical Research 33: 226–258. [Google Scholar]
- Lucas SJ, Baştaş K, Budak H. 2014. Exploring the interaction between small RNAs and R genes during Brachypodium response to Fusarium culmorum infection. Gene 536: 254–264. [DOI] [PubMed] [Google Scholar]
- Luo S, Zhang Y, Hu Q, et al. 2012. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiology 159: 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof K-BG. 2012. Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiology 160: 1432–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof K-BG. 2013. Plant immune responses against viruses: how does a virus cause disease? The Plant Cell Online 25: 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi KK, Pyle JD, Scholthof KB. 2014. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defences. Molecular Plant Microbe Interaction 27: 1277–1290. [DOI] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, et al. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology 13: 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. 2012. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytologist 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research 39: D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Grossniklaus U. 2010. Model organisms — a historical perspective. Journal of Proteomics 73: 2054–2063. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Allainguillaume J, Catalán P, et al. 2011. Exploiting the Brachypodium tool box in cereal and grass research. New Phytologist 191: 334–347. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Xu R, Casson SA, Stoddart WM, Routledge APM, Draper J. 2004. Characterization of a proteinase inhibitor from Brachypodium distachyon suggests the conservation of defence signalling pathways between dicotyledonous plants and grasses. Molecular Plant Pathology 5: 267–280. [DOI] [PubMed] [Google Scholar]
- Murray G, Brennan J. 2009. Estimating disease losses to the Australian wheat industry. Australasian Plant Pathology 38: 558–570. [Google Scholar]
- Murray GM, Brennan JP. 2010. Estimating disease losses to the Australian barley industry. Australasian Plant Pathology 39: 85–96. [Google Scholar]
- Mysore KS, Ryu CM. 2004. Nonhost resistance: how much do we know? Trends Plant Science 9:97–104. [DOI] [PubMed] [Google Scholar]
- Neate SM, Warcup JH. 1985. Anastomosis grouping of some isolates of Thanatephorus cucumeris from agricultural soils in South Australia. Transactions of the British Mycological Society 85: 615–620. [Google Scholar]
- Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM. 2004. Regional economic impacts of Fusarium head blight in wheat and barley. Applied Economic Perspectives and Policy 26: 332–347. [Google Scholar]
- Nordstrom KJV, Albani MC, James GV, et al. 2013. Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nature Biotechnology 31: 325–330. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology 41: 600–623. [DOI] [PubMed] [Google Scholar]
- O’Brien PA, Zamani M. 2003. Production of pectic enzymes by barepatch isolates of Rhizoctonia solani AG 8. Australasian Plant Pathology 32: 65–72. [Google Scholar]
- Oerke E-C. 2006. Crop losses to pests. The Journal of Agricultural Science 144: 31–43. [Google Scholar]
- Oliver RP, Friesen TL, Faris JD, Solomon PS. 2012. Stagonospora nodorum: from pathology to genomics and host resistance. Annual Review of Phytopathology 50: 23–43. [DOI] [PubMed] [Google Scholar]
- Oliver RP, Solomon PS. 2010. New developments in pathogenicity and virulence of necrotrophs. Current Opinion in Plant Biology 13: 415–419. [PubMed] [Google Scholar]
- Oxley S, Havis N. 2004. Development of Ramularia collo-cygni on spring barley and its impact on yield. In: Proceedings of the Dundee conference: crop protection in Northern Britain, 147–152. [Google Scholar]
- Parker D, Beckmann M, Enot DP, et al. 2008. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nature Protocols 3: 435–445. [DOI] [PubMed] [Google Scholar]
- Parker D, Beckmann M, Zubair H, et al. 2009. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. The Plant Journal 59: 723–737. [DOI] [PubMed] [Google Scholar]
- Pasquet J-C, Chaouch S, Macadré C, et al. 2014. Differential gene expression and metabolomic analyses of Brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of Fusarium graminearum. BMC genomics 15: 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitz TC, Okubara PA, Schroeder KL. 2009. Integrated control of soilborne pathogens of wheat. In: Gisi U, Chet I, Gullino ML, eds. Recent Developments in Management of Plant Diseases. Dordrecht: Springer, 229–245. [Google Scholar]
- Peraldi A, Beccari G, Steed A, Nicholson P. 2011. Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biology 11: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraldi A, Griffe LL, Burt C, McGrann GRD, Nicholson P. 2014. Brachypodium distachyon exhibits compatible interactions with Oculimacula spp. and Ramularia collo-cygni providing the first pathosystem model to study eyespot and Ramularia leaf spot diseases. Plant Pathology 63: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Cnudde F, Gerats T. 2003. Forward genetics and map-based cloning approaches. Trends in Plant Science 8: 484–491. [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Lionetti V, Fursova OV, et al. 2013. Arabidopsis and Brachypodium transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiology 162: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, Temanni M-R, van Zanten M, et al. 2012. Illumina sequencing technology as a method of identifying T-DNA insertion loci in activation-tagged Arabidopsis thaliana plants. Molecular Plant 5: 948–950. [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, et al. 2003. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. Journal of Biological Chemistry 278: 47905–47914. [DOI] [PubMed] [Google Scholar]
- Qin B, Cao A, Wang H, et al. 2011. Collinearity-based marker mining for the fine mapping of Pm6, a powdery mildew resistance gene in wheat. Theoretical and Applied Genetics 123: 207–218. [DOI] [PubMed] [Google Scholar]
- Richardson T, Thistleton J, Higgins TJ, Howitt C, Ayliffe M. 2014. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture doi: 10.1007/s11240-014-0564-7. [Google Scholar]
- Rocha O, Ansari K, Doohan FM. 2005. Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Additives and Contaminants 22: 369–378. [DOI] [PubMed] [Google Scholar]
- Routledge APM, Shelley G, Smith JV, Talbot NJ, Draper J, Mur LAJ. 2004. Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Molecular Plant Pathology 5: 253–265. [DOI] [PubMed] [Google Scholar]
- Sandoya GV, Buanafina MMO. 2014. Differential responses of Brachypodium distachyon genotypes to insect and fungal pathogens. Physiological and Molecular Plant Pathology 85: 53–64. [Google Scholar]
- Sastry KS. 2013. Economic significance of seed-transmitted plant virus diseases. Seed-borne plant virus diseases. Berlin: Springer. [Google Scholar]
- Schneebeli K, Mathesius U, Watt M. 2014. Brachypodium distachyon is a pathosystem model for the study of the wheat disease Rhizoctonia root rot. Plant Pathology doi: 10.1111/ppa.12227. [Google Scholar]
- Schneeberger K, Weigel D. 2011. Fast-forward genetics enabled by new sequencing technologies. Trends in Plant Science 16: 282–288. [DOI] [PubMed] [Google Scholar]
- Scholthof K-BG. 1999. A synergism induced by satellite panicum mosaic virus. Molecular Plant-Microbe Interactions 12: 163–166. [Google Scholar]
- Schroeder H, Christensen J. 1963. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 53: 831–838. [Google Scholar]
- Schroeder K, Paulitz T. 2008. Effect of inoculum density and soil tillage on the development and severity of Rhizoctonia root rot. Phytopathology 98: 304–314. [DOI] [PubMed] [Google Scholar]
- Schweiger W, Boddu J, Shin S, et al. 2010. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Molecular Plant-Microbe Interactions 23: 977–986. [DOI] [PubMed] [Google Scholar]
- Schweiger W, Pasquet J-C, Nussbaumer T, et al. 2013. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Molecular Plant-Microbe Interactions 7: 781–792. [DOI] [PubMed] [Google Scholar]
- Silvar C, Perovic D, Scholz U, Casas A, Igartua E, Ordon F. 2012. Fine mapping and comparative genomics integration of two quantitative trait loci controlling resistance to powdery mildew in a Spanish barley landrace. Theoretical and Applied Genetics 124: 49–62. [DOI] [PubMed] [Google Scholar]
- Singh RP, Hodson DP, Huerta-Espino J, et al. 2011. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annual Review of Phytopathology 49: 465–481. [DOI] [PubMed] [Google Scholar]
- Skamnioti P, Gurr SJ. 2009. Against the grain: safeguarding rice from rice blast disease. Trends in Biotechnology 27: 141–150. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Singh RP, McFadden H, et al. 2008. Fine scale genetic and physical mapping using interstitial deletion mutants of Lr34 /Yr18: a disease resistance locus effective against multiple pathogens in wheat. Theoretical and Applied Genetics 116: 481–490. [DOI] [PubMed] [Google Scholar]
- St.Clair DA. 2010. Quantitative disease resistance and quantitative resistance loci in breeding. Annual Review of Phytopathology 48: 247–268. [DOI] [PubMed] [Google Scholar]
- Steinwand MA, Young HA, Bragg JN, Tobias CM, Vogel JP. 2013. Brachypodium sylvaticum, a model for perennial grasses: transformation and inbred line development. PLoS ONE 8: e75180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Abe A, Yoshida K, et al. 2013. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant Journal 74: 174–183. [DOI] [PubMed] [Google Scholar]
- Talbot NJ. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annual Review of Microbiology 57: 177–202. [DOI] [PubMed] [Google Scholar]
- Tan S, Wu S. 2012. Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comparative and Functional Genomics 2012: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Worland B, Wright J, Bevan MW, Vain P. 2010. Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnology Journal 8: 734–747. [DOI] [PubMed] [Google Scholar]
- Thottappilly G. 1992. Plant virus diseases of importance to African agriculture. Journal of Phytopathology 134: 265–288. [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, et al. 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genetics 43: 159–162. [DOI] [PubMed] [Google Scholar]
- Tripathi P, Rabara R, Langum T, et al. 2012. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics 13: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain P. 2011. Brachypodium as a model system for grass research. Journal of Cereal Science 54: 1–7. [Google Scholar]
- Vogel J, Bragg J. 2009. Brachypodium distachyon, a new model for the Triticeae. In: Muehlbauer GJ, Feuillet C, eds. Genetics and genomics of the Triticeae. New York: Springer, 427–449. [Google Scholar]
- Vogel J, Tuna M, Budak H, Huo N, Gu Y, Steinwand M. 2009. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biology 9: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DR, Havis ND, Oxley SJP. 2008. Ramularia collo-cygni: the biology of an emerging pathogen of barley. FEMS Microbiology Letters 279: 1–7. [DOI] [PubMed] [Google Scholar]
- Wang J-Y, Wang X-Y, Li L, et al. 2012. Pathogenicity of rice blast fungus Magnaporthe oryzae on Brachypodium distachyon. Rice Science 19: 252–258. [Google Scholar]
- Watt M, Schneebeli K, Dong P, Wilson IW. 2009. The shoot and root growth of Brachypodium and its potential as a model for wheat and other cereal crops. Functional Plant Biology 36: 960–969. [DOI] [PubMed] [Google Scholar]
- Wei L, Muranty H, Zhang H. 2011. Advances and prospects in wheat eyespot research: contributions from genetics and molecular tools. Journal of Phytopathology 159: 457–470. [Google Scholar]
- Xue F, Ji W, Wang C, Zhang H, Yang B. 2012. High-density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theoretical and Applied Genetics 124: 1549–1560. [DOI] [PubMed] [Google Scholar]
- Yang X, Ma J, Li H, Ma H, Yao J, Liu C. 2010. Different genes can be responsible for crown rot resistance at different developmental stages of wheat and barley. European Journal of Plant Pathology 128: 495–502. [Google Scholar]
- Zambino PJ, Szabo L.J. 1993. Phylogenetic relationship of selected cereal rusts based on rRNA sequence analysis. Mycologia 85: 401–414. [Google Scholar]
- Zhang H, Guan H, Li J, et al. 2010. Genetic and comparative genomics mapping reveals that a powdery mildew resistance gene Ml3D232 originating from wild emmer co-segregates with an NBS-LRR analog in common wheat (Triticum aestivum L.). Theoretical and Applied Genetics 121: 1613–1621. [DOI] [PubMed] [Google Scholar]
- Zhang X, Han D, Zeng Q, et al. 2013. Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and rice. PLoS ONE 8: e57885. [DOI] [PMC free article] [PubMed] [Google Scholar]



