Abstract
Background and Aims Phenolic compounds contribute to food quality and have potential health benefits. Consequently, they are an important target of selection for Citrus species. Numerous studies on this subject have revealed new molecules, potential biosynthetic pathways and linkage between species. Although polyphenol profiles are correlated with gene expression, which is responsive to developmental and environmental cues, these factors are not monitored in most studies. A better understanding of the biosynthetic pathway and its regulation requires more information about environmental conditions, tissue specificity and connections between competing sub-pathways. This study proposes a rapid method, from sampling to analysis, that allows the quantitation of multiclass phenolic compounds across contrasting tissues and cultivars.
Methods Leaves and fruits of 11 cultivated citrus of commercial interest were collected from adult trees grown in an experimental orchard. Sixty-four phenolic compounds were simultaneously quantified by ultra-high-performance liquid chromatography coupled with mass spectrometry.
Key Results Combining data from vegetative tissues with data from fruit tissues improved cultivar classification based on polyphenols. The analysis of metabolite distribution highlighted the massive accumulation of specific phenolic compounds in leaves and the external part of the fruit pericarp, which reflects their involvement in plant defence. The overview of the biosynthetic pathway obtained confirmed some regulatory steps, for example those catalysed by rhamnosyltransferases. The results suggest that three other steps are responsible for the different metabolite profiles in ‘Clementine’ and ‘Star Ruby’ grapefruit.
Conclusions The method described provides a high-throughput method to study the distribution of phenolic compounds across contrasting tissues and cultivars in Citrus, and offers the opportunity to investigate their regulation and physiological roles. The method was validated in four different tissues and allowed the identification and quantitation of 64 phenolic compounds in 20 min, which represents an improvement over existing methods of analysing multiclass polyphenols.
Keywords: Phenolic compounds, flavonoids, coumarins, Citrus, UPLC–MS, high-throughput method, phylogenetic relationships, tissue diversity, biosynthetic pathway.
INTRODUCTION
Phenolic compounds are one of the most important groups of plant secondary metabolites; they are widely distributed in plants and encompass more than 8000 molecules. Numerous studies have focused on polyphenols because they have been reported to have multiple biological effects. Polyphenols are involved in many plant processes, such as development (participating in plant hormone signalling or pollen germination), reproduction (pigments attracting pollinators) and plant defence (protecting from UV, pathogens and predators) (Winkel-Shirley, 2002; Treutter, 2006; Agati et al., 2012). Polyphenols also play important roles in food quality, contributing to colour and taste and potentially participating in the prevention of chronic diseases in relation to their antioxidant properties (Cheynier, 2005; Widelski et al., 2009).
Citrus fruits, whose production reached 131 million tons in 2012 (http://faostat.fao.org), represent an important source of polyphenols in the human diet. The predominant soluble phenolic compounds in Citrus are flavonoids, such as flavones and flavanones, and coumarins, such as furanocoumarins. In Citrus species, the analysis of phenolic compounds is challenging due to the diversity of metabolites and their wide range of solubilities and contents. In addition, some metabolites are specific to some Citrus species. For example, hesperidin and naringin are flavanone-7-O-glycosides that accumulate specifically in mandarins and oranges and in pomelos and grapefruits respectively (Gattuso et al., 2007). Many flavonoids are present in Citrus as their glycosyl derivatives, which increases their solubility in water. Polymethoxyflavones, which are more lipophilic, are abundant in the essential oil fraction of the fruit pericarp and less abundant in fruit juices (Gattuso et al., 2007). The wide range of solubilities also renders the preparation of plant tissue and the extraction process very tricky. Nogata et al. (2003) showed that the process used to extract fruit juices can significantly modify the profiles of polymetoxylflavones. This may be related to the fraction of flavedo that is extracted with the juice, or to the filtration and centrifugation steps.
In line with this, although there has been considerable research on Citrus phenolic metabolites, numerous studies have focused on a single class of polyphenol, a single plant tissue or a single cultivar (Kawaii et al., 1999, 2000b; Gattuso et al., 2007; Barreca et al., 2011c, 2013; Abad-García et al., 2012). For example, Kawaii et al. (2000b) analysed 23 flavonoids from the leaves of 68 Citrus cultivars and near-Citrus relatives. High-throughput methods have recently been developed to analyse flavonoids on the one hand (Di Donna et al., 2013) and coumarins and furanocoumarins on the other hand (Dugrand et al., 2013). Only a few studies have focused simultaneously on Citrus flavonoids and coumarins. Gardana et al. (2008) analysed 18 flavonoids and two furanocoumarins in the same run using a 50-min gradient. These methods consist of a multiplex approach using HPLC–DAD–ESI–MS (where DAD is diode array detection and ESI is electrospray ionization) for the identification and quantitation of polyphenols in Citrus. This approach allows qualitative and quantitative analysis of phenolic compounds. This system has been optimized, as described by Chen et al. (2008), using ultra-high-performance liquid chromatography (UPLC). Thanks to the combination of high optimal flow rates and shorter column lengths, UPLC instrumentation and columns hold advantages in terms of speed of analysis, peak separation and better quantitation efficiency. Prokudina et al. (2012) analysed 26 compounds from five chemical families in Vigna radiata. Alarcón-Flores et al. (2013) went further and analysed more than 30 phytochemicals from five different chemical families in five different species. These methods are particularly suited to the investigation of the phenotypic effects of abiotic or biotic stresses (Brunetti et al., 2013).
Many phenolic compounds help the plant to adapt to fluctuating environments (Pollastri and Tattini, 2011; Agati et al., 2012). Accordingly, phenolic compound biosynthesis and accumulation vary greatly with developmental and environmental conditions (Ryan et al., 2002; Kaffarnik et al., 2006; Mellway et al., 2009; Daniel et al., 2011; Koyama et al., 2012). However, these factors were usually not monitored in the numerous studies aiming to quantify polyphenols in Citrus. Further analyses allowing the quantitation of several classes of metabolites in different tissues and cultivars are required for a better understanding of the phenolic pathway in Citrus, i.e. for the identification of flux-determining steps, the determination of regulation points in relation to developmental or environmental cues, and the connections with other pathways. It is important to develop rapid methods in order to analyse the responses of single organs to a given external condition. What is at stake is our capacity to compare several organs/tissues and several external conditions. From these comparisons, we should be able to infer the strategies of different organs and species in these environmental conditions and the roles of specific metabolites.
In this context, the objectives of this work were (1) to develop and validate a rapid method that allows the separation and quantitation of a wide range of phenolic compounds and can be applied to contrasting tissues and species, and (2) to draw a map of the distribution of these metabolites across 11 Citrus cultivars growing in similar environmental conditions and in four tissues with similar developmental stages. We propose an optimization of the UPLC–MS system for the detection and quantitation of the main flavones, flavanones, flavonols, anthocyanins, coumarins and furanocoumarins in Citrus leaves and fruits (albedo, flavedo and pulp tissues) extracted with hydro-methanolic solvents. Sixty-four phenolic compounds were simultaneously analysed in a single run of 20 min. We also discuss the potential regulatory nodes of the biosynthetic pathway on the basis of results of mapping the genetic and tissue diversity of these metabolites.
MATERIALS AND METHODS
Plant materials
Leaves and fruits of 11 cultivated Citrus of commercial interest were collected from adult trees grown in an experimental orchard near San Giuliano in Corsica (42°18′ 55″ N, 9°29′ 29″ E; 51 m a.s.l.). The trees were submitted to standard cultural practices: water was supplied every day on the basis of 100 % replacement of actual evapotranspiration estimated from the Penman–Monteith equation (Monteith, 1965) and fertilizers were supplied according to the recommendations of the local agriculture department. Insects and diseases were also controlled according to the agriculture department’s recommendations. Four cultivars of orange [Citrus sinensis (L.) Obs.], three cultivars of grapefruit (Citrus paradisi Macf.) and four cultivars of mandarin or mandarin types [Citrus deliciosa Ten., Citrus clementina Hort. ex Tan., Citrus reticulata Blanco × Citrus sinensis (L.) Obs. and Citrus clementina Hort. ex Tan. × Citrus tangerina Hort ex Tan.] were analysed. Cultivar and rootstock combinations are detailed in Table 1. The experiments were conducted in open fields and standard procedures were applied to minimize variations due to potentially different environmental conditions. Around 275 non-fruiting branches (11 cultivars × 25 branches) and 275 fruiting branches (11 cultivars × 25 branches) were first selected as follows. All branches had the same east orientation and experienced similar exposure to light. These branches were a similar height above ground (about 1·5 m). The leaves were adult leaves about 1 year old from shoots of the spring flush of the previous season. Then, for each cultivar, five independent leaf and fruit samples/observations were randomly collected among the previously selected branches. The leaves were only collected from non-fruiting branches to avoid leaf–fruit interactions.
Table 1.
Cultivar–rootstock combinations used in the trial. Cultivars are identified with Tanaka’s classification system
| Species | Tanaka’s system | Variety/cultivar | Rootstock |
|---|---|---|---|
| Mandarin or mandarin type | Citrus deliciosa Ten. | Willowleaf | Carrizo citrange |
| Citrus clementina Hort. ex Tan | Clementine | Carrizo citrange | |
| Citrus reticulata Blanco × Citrus sinensis L. Osb | Murcott | Carrizo citrange | |
| Citrus clementina Hort. ex Tan. × Citrus tangerina Hort. ex Tan | Fortune | Poncirus trifoliata | |
| Orange | Citrus sinensis L. Osb | Hamelin | Poncirus trifoliata |
| Washington Navel | Poncirus trifoliata | ||
| Sanguinelli | Poncirus trifoliata | ||
| Navelina | Poncirus trifoliata | ||
| Grapefruit | Citrus paradisi Macf. | Marsh | Poncirus trifoliata |
| Star Ruby | Carrizo citrange | ||
| Duncan | Poncirus trifoliata |
The five independent leaf samples were composed of leaves with similar bud break ranking on the selected shoots. Leaf size and leaf fresh and dry weights are detailed in Supplementary Data Table S1. The leaf surface area was determined by image analysis with ImageJ 1.46 software. After collection, leaves were weighed and immediately frozen in liquid nitrogen. They were freeze-dried in the dark to protect coumarins and flavonoids from the light, and weighed for dry weight determination. Then, they were ground into a fine powder with a marble grinder (Retsch, Haan, Germany) for 5 min at 30 rotations s–1 immediately after freeze-drying; the samples were kept frozen during this process.
Details of fruit diameter, tissue thicknesses, fruit fresh weight and maturity index (ratio of soluble solid content to titratable acidity) are given in Supplementary Data Table S2. The fruits were randomly selected among the fruiting branches. Five independent samples, which represent five observations per cultivar, were analysed. The flavedo (exocarp), albedo (mesocarp) and pulp (endocarp) of each fruit were separately weighed and immediately frozen in liquid nitrogen. They were freeze-dried, weighed and ground according to the method used for the leaf samples. Fruit aliquots were used to extract juices and to determine the maturity index according to the official method detailed by Helrich (1990). Titratable acidity was determined by titration to pH 8·2 with 0·1 m sodium hydroxide (NaOH) using an automated titration system (Mettler DL 25; Mettler-Toledo, Viroflay, France), and was expressed as a percentage of anhydrous citric acid. The soluble solids content was determined with a refractometer (Atago model, 0–32 % mas Sacch; VWR, West Chester, PA, USA). The maturity index was evaluated as the soluble solids content/titratable acidity ratio.
Standards and reagents
Methanol of HPLC grade was purchased from Sigma-Aldrich (St Louis, MO, USA) and HPLC-grade acetonitrile from VWR International (Radnor, PA, USA).
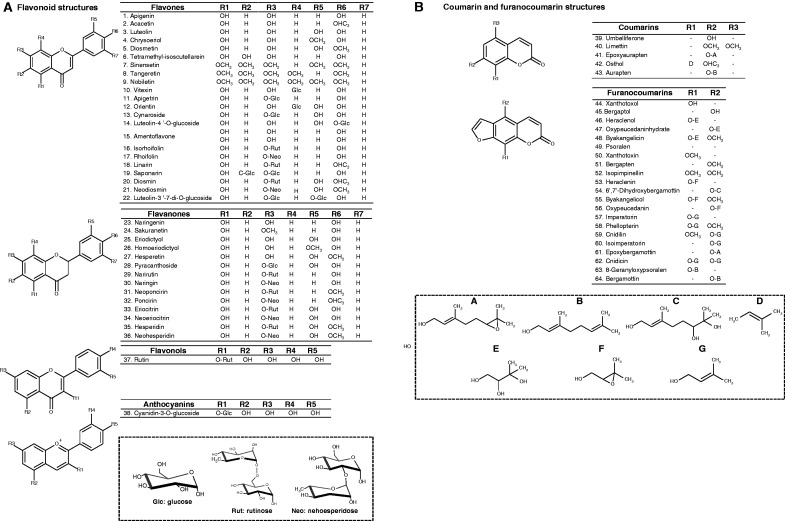
Sixty-six standards were tested: 38 flavonoids, comprising 22 flavones, 14 flavanones, 1 flavonol and 1 anthocyanin; 7 coumarins; and 21 furanocoumarins (see Fig. 1 detailing the structure of 64 compounds; note that the structures of the two metabolites used as internal standards are not presented here). Flavonoid standards were purchased from Extrasynthese (Genay, France), except for rutin, which was purchased from Sigma-Aldrich (St Louis, MO, USA). Coumarins and furanocoumarins were purchased from Herboreal Ltd (Edinburgh, UK) except for umbelliferone, psoralen, xanthotoxol, bergaptol, xanthotoxin and bergapten, which were purchased from Sigma-Aldrich (St Louis, MO, USA) and angelicin, osthol and isopimpinellin, which were provided by Extrasynthese (Genay, France).
Fig. 1.
List and structure of the flavonoids, coumarins and furanocoumarins identified in the four Citrus tissues of 11 cultivars. Metabolites are grouped by class: (A) flavones, flavanones, flavonols and anthocyanins; and (B) coumarins and furanocoumarins. The structure of each class is presented and ramifications are detailed for each metabolite. Note that in (A), compound no. 15 is a biflavonoid which is why there are two lines representing this metabolite and showing the repetition of the first structure. Abbreviations for sugars: Glc, glucose; Rut, rutinose; Neo, neohesperidose.
Phenolic compound extraction
The leaves and the three fruit tissues were analysed separately.
Flavonoids, coumarins and furanocoumarins were isolated according to a modified method from Tanaka and Brugliera (2013). The metabolites were simultaneously extracted from 20 mg of leaf or fruit powder with 400 µL of MeOH–H2O (80 : 20, v : v) acidified with 2 mm of hydrochloric acid and supplemented with 5-methoxyflavone and angelicin as internal standards at a concentration of 5 µm. Samples were sonicated for 10 min in a water bath sonicator. Finally, they were centrifuged for 1 h at 20 000 g at 4 °C. Samples were stored at −20 °C until analysis.
Preparation of standard solutions
A stock solution of each compound was prepared by dissolving 2 mg of compound powder in DMSO–MeOH (1 : 1, v : v) to obtain a final concentration of 10 mm. Two blending solutions of standards were used to characterize and quantify phenolic compounds. The first calibration solution contained 37 flavonoids and two internal standards (5-methoxyflavone and angelicin) and the second contained 27 coumarins and furanocoumarins with the same two internal standards. The two solutions were diluted to obtain five concentration points: 1, 7·5, 15, 22·5 and 30 µm. These ranges of standard solutions were injected at the beginning and end of each run before and after sample injections.
UPLC–MS analysis
Flavonoid, coumarin and furanocoumarin detection and quantitation were performed with a Nexera UPLC® (Ultra-High Performance Liquid Chromatographic – LC-30AD) system (Shimadzu Corp., Kyoto, Japan). This system was equipped with a binary pump, an autosampler cooler kept at 10 °C, a column heater kept at 60 °C, a degasser using nitrogen and a diode array detector monitoring absorbance between 210 and 800 nm (SPDM20A; Shimadzu). The diode array detector was connected in series with a mass spectrometer (single quadrupole; LC-MS-2020; Shimadzu).
Flavonoids, coumarins and furanocoumarins were separated on a ZORBAX RRHD Eclipse Plus C18 reverse-phase column (2·1 mm × 150 mm, 1·8 µm; Agilent Technologies Inc., Santa Clara, CA, USA), protected with an Agilent 1290 Infinity pre-column and thermostated at 60 °C. The gradient elution programme was arranged with two eluents: eluent A, pure water–formic acid (99·9 : 0·1, v : v); eluent B, acetonitrile–formic acid (99·9 : 0·1, v : v). The flow rate was set at 0·4 mL min–1 and the injection volume at 2 µL. Details of this 20-min programme are given in Table 2.
Table 2.
Details of the UPLC gradient
| Time (min) | Acetonitrile/formic acid (%) |
|---|---|
| 0·01 | 20 |
| 4·00 | 24 |
| 5·50 | 33 |
| 8·00 | 35 |
| 9·00 | 37 |
| 10·50 | 52 |
| 13·30 | 54 |
| 16·30 | 60 |
| 16·70 | 80 |
| 17·00 | 97 |
| 17·50 | 100 |
| 19·00 | 100 |
| 19·05 | 20 |
| 20·00 | 20 |
The UPLC system was connected to the mass spectrometer by a dual ion source (DUIS), a combination of ESI, here operating in positive mode (ESI+), and atmospheric pressure chemical ionization (APCI). Inlet, desolvation line (DL) and heating block temperatures were set at 350, 250 and 300 °C respectively. The capillary voltage was set at 4·5 kV. For each compound, the voltages of the DL and Q-array were optimized in order to increase the sensitivity of detection (Table 3). For this purpose, each standard was directly introduced into the mass spectrometer at the concentration of 1 mm. Detection was performed in single-ion monitoring (SIM) mode to increase selectivity. Analytical time was split into four segments in order to increase the sensitivity of the detection.
Table 3.
Chromatographic and MS parameters of 64 metabolites, including linearity and accuracy indicators
| Compound | Ion species | m/z | Retention time (Rt) |
Detection |
Linearity |
Accuracy |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Valuea (min) | Window | DL (V) | Q-array (V) | Equation | r2 | Mean recovery (µm) | s.d. | |||
| Flavones | ||||||||||
| Apigenin | [M + H]+ | 271 | 6·987 | 2 | 0 | 0 | y = 0·0152x + 0 | 0·9997 | 4·154 | 0·416 |
| Acacetin | [M + H]+ | 285 | 11·196 | 3 | 0 | 0 | y = 0·1056x + 0 | 0·9988 | 4·039 | 0·394 |
| Luteolin | [M + H]+ | 287 | 5·796 | 2 | 0 | 0 | y = 0·0137x + 0 | 0·9945 | 5·983 | 0·287 |
| Chrysoeriol | [M + H]+ | 301 | 7·558 | 2 | 80 | 0 | y = 0·0585x + 0 | 0·9992 | 4·103 | 0·464 |
| Diosmetin | [M + H]+ | 301 | 7·621 | 2 | 80 | 0 | y = 0·0691x + 0 | 0·9965 | 4·090 | 0·447 |
| Tetramethyl isoscutellarein | [M + H]+ | 343 | 11·683 | 3 | 0 | 0 | y = 1·0326x + 0 | 0·9925 | 5·999 | 0·420 |
| Sinensetin | [M + H]+ | 373 | 10·900 | 3 | 40 | 0 | y = 0·3330x + 0 | 0·9976 | 5·512 | 0·285 |
| Tangeretin | [M + H]+ | 373 | 12·400 | 3 | 40 | 0 | y = 0·5164x + 0 | 0·9901 | 4·962 | 0·391 |
| Nobiletin | [M + H]+ | 403 | 11·700 | 3 | 40 | 0 | y = 0·4735x + 0 | 0·9990 | 4·692 | 0·222 |
| Vitexin | [M + H]+ | 433 | 2·329 | 1 | 0 | 0 | y = 0·0398x + 0 | 0·9993 | 4·931 | 0·275 |
| Apigetrin | [M + H]+ | 433 | 3·325 | 1 | 0 | 0 | y = 0·0288x + 0 | 0·9958 | 4·661 | 0·182 |
| Orientin | [M + H]+ | 449 | 1·929 | 1 | 40 | 0 | y = 0·0433x + 0 | 0·9998 | 4·318 | 0·485 |
| Cynaroside | [M + H]+ | 449 | 2·462 | 1 | 40 | 0 | y = 0·0188x + 0 | 0·9941 | 4·347 | 0·309 |
| Luteolin-4′-O-glucoside | [M + H]+ | 449 | 3·175 | 1 | 40 | 0 | y = 0·0324x + 0 | 0·9938 | 4·727 | 0·347 |
| Amentoflavone | [M + H]+ | 539 | 5·150 | 2 | 40 | 0 | y = 0·0318x + 0 | 0·9973 | 4·913 | 0·353 |
| Isorhoifolin | [M + H]+ | 579 | 2·438 | 1 | 0 | 0 | y = 0·0349x + 0 | 0·9933 | 4·609 | 0·105 |
| Rhoifolin | [M + H]+ | 579 | 3·117 | 1 | 0 | 0 | y = 0·0265x + 0 | 0·9999 | 4·881 | 0·187 |
| Linarin | [M + H]+ | 593 | 6·217 | 2 | 0 | 0 | y = 0·0419x + 0 | 0·9932 | 4·771 | 0·322 |
| Saponarin | [M + H]+ | 595 | 1·688 | 1 | 0 | 0 | y = 0·0404x + 0 | 0·9945 | 4·680 | 0·182 |
| Diosmin | [M + H]+ | 609 | 3·347 | 1 | 0 | 0 | y = 0·0065x + 0 | 0·9994 | 5·172 | 0·166 |
| Neodiosmin | [M + H]+ | 609 | 3·917 | 1 | 0 | 0 | y = 0·0261x + 0 | 0·9981 | 4·851 | 0·140 |
| Luteolin-3′-7-di-O-glucoside | [M + H]+ | 611 | 1·712 | 1 | 0 | 0 | y = 0·0230x + 0 | 0·9993 | 5·079 | 0·213 |
| Flavanones | ||||||||||
| Naringenin | [M + H]+ | 273 | 6·821 | 2 | 40 | 0 | y = 0·0126x + 0 | 0·9950 | 4·519 | 0·224 |
| Sakuranetin | [M + H]+ | 287 | 11·083 | 3 | 0 | 0 | y = 0·0259x + 0 | 0·9912 | 4·048 | 0·204 |
| Eriodictyol | [M + H]+ | 289 | 5·304 | 2 | 0 | 0 | y = 0·0032x + 0 | 0·9999 | 5·055 | 0·116 |
| Homoeriodictyol | [M + H]+ | 303 | 7·267 | 2 | -40 | 0 | y = 0·0163x + 0 | 0·9955 | 4·299 | 0·382 |
| Hesperetin | [M + H]+ | 303 | 7·588 | 2 | -40 | 0 | y = 0·0183x + 0 | 0·9999 | 4·583 | 0·274 |
| Pyracanthoside | [M + H]+ | 451 | 2·379 | 1 | 0 | 0 | y = 0·0051x + 0 | 0·9999 | 4·815 | 0·314 |
| Narirutin | [M + H]+ | 581 | 2·375 | 1 | 40 | 0 | y = 0·0162x + 0 | 0·9984 | 4·729 | 0·229 |
| Naringin | [M + H]+ | 581 | 3·046 | 1 | 40 | 0 | y = 0·0050x + 0 | 0·9980 | 4·899 | 0·160 |
| Neoponcirin | [M + H]+ | 595 | 6·458 | 2 | 0 | 0 | y = 0·0261x + 0 | 0·9937 | 4·660 | 0·123 |
| Poncirin | [M + H]+ | 595 | 6·500 | 2 | 0 | 0 | y = 0·0261x + 0 | 0·9966 | 4·893 | 0·098 |
| Eriocitrin | [M + H]+ | 597 | 6·004 | 2 | 0 | 0 | y = 0·0114x + 0 | 0·9940 | 4·668 | 0·127 |
| Neoeriocitrin | [M + H]+ | 597 | 2·283 | 1 | 0 | 0 | y = 0·0117x + 0 | 0·9997 | 4·872 | 0·263 |
| Hesperidin | [M + H]+ | 611 | 3·258 | 1 | 0 | 0 | y = 0·0162x + 0 | 0·9976 | 4·779 | 0·205 |
| Neohesperidin | [M + H]+ | 611 | 3·556 | 1 | 0 | 0 | y = 0·0117x + 0 | 0·9939 | 5·222 | 0·264 |
| Flavonols | ||||||||||
| Rutin | [M + H]+ | 611 | 1·957 | 1 | 0 | 0 | y = 0·0207x + 0 | 0·9926 | 5·535 | 0·330 |
| Anthocyanins | ||||||||||
| Cyanidin-3-O-glucoside | [M + H]+ | 449 | 1·162 | 1 | 0 | 0 | y = 0·0264x + 0 | 0·9936 | 4·178 | 0·338 |
| Coumarins | ||||||||||
| Umbelliferone | [M + H]+ | 163 | 2·750 | 1 | 80 | 0 | y = 0·0330x + 0 | 0·9916 | 6·370 | 0·839 |
| Limettin | [M + H]+ | 207 | 8·387 | 2 | 0 | 0 | y = 1·2774x + 0 | 0·9960 | 4·303 | 0·393 |
| Epoxyaurapten | [M + H]+ | 315 | 13·500 | 3 | 0 | 0 | y = 0·7909x + 0 | 0·9927 | 5·471 | 0·799 |
| Osthol | [M + H]+ | 245 | 13·842 | 3 | 0 | 0 | y = 3·0618x + 0 | 0·9960 | 3·686 | 0·323 |
| Aurapten | [M + H]+ | 299 | 18·108 | 4 | 80 | 0 | y = 0·7222x + 0 | 0·9911 | 3·092 | 0·284 |
| Furanocoumarins | ||||||||||
| Xanthotoxol | [M + H]+ | 203 | 4·400 | 1 | 80 | 0 | y = 0·2504x + 0 | 0·9946 | 8·274 | 1·498 |
| Bergaptol | [M + H]+ | 203 | 5·608 | 2 | 80 | 0 | y = 0·1620x + 0 | 0·9924 | 4·336 | 0·914 |
| Heraclenol | [M + H]+ | 305 | 5·825 | 2 | 0 | 0 | y = 0·3284x + 0 | 0·9907 | 4·437 | 0·491 |
| Oxypeucedanin hydrate | [M + H]+ | 305 | 6·475 | 2 | 0 | 0 | y = 1·0039x + 0 | 0·9981 | 5·270 | 0·632 |
| Byakangelicin | [M + H]+ | 335 | 6·825 | 2 | 80 | 30 | y = 0·1256x + 0 | 0·9955 | 5·694 | 0·373 |
| Psoralen | [M + H]+ | 187 | 7·058 | 2 | 0 | 0 | y = 0·3516x + 0 | 0·9991 | 5·299 | 0·349 |
| Xanthotoxin | [M + H]+ | 217 | 7·583 | 2 | 0 | 0 | y = 0·8156x + 0 | 0·9995 | 4·802 | 0·299 |
| Bergapten | [M + H]+ | 217 | 8·800 | 2 | 0 | 0 | y = 0·9985x + 0 | 0·9983 | 2·884 | 0·243 |
| Isopimpinellin | [M + H]+ | 247 | 9·033 | 2 | 80 | 0 | y = 1·3823x + 0 | 0·9983 | 5·735 | 0·600 |
| Heraclenin | [M + H]+ | 287 | 10·208 | 3 | 0 | 30 | y = 0·7295x + 0 | 0·9907 | 3·907 | 1·016 |
| 6′,7′-Dihydroxybergamottin | [M + H]+ | 373 | 11·192 | 3 | 0 | 0 | y = 0·8056x + 0 | 0·9966 | 4·740 | 0·385 |
| Byakangelicol | [M + H]+ | 317 | 11·221 | 3 | 80 | 0 | y = 1·4634x + 0 | 0·9947 | 4·737 | 0·276 |
| Oxypeucedanin | [M + H]+ | 287 | 11·225 | 3 | 0 | 30 | y = 0·8076x + 0 | 0·9925 | 5·742 | 0·648 |
| Imperatorin | [M + H]+ | 271 | 12·967 | 3 | 80 | 30 | y = 1·2433x + 0 | 0·9979 | 4·842 | 0·651 |
| Phellopterin | [M + H]+ | 301 | 13·692 | 3 | 80 | 30 | y = 2·3466x + 0 | 0·9930 | 4·369 | 0·640 |
| Cnidilin | [M + H]+ | 301 | 18·021 | 4 | 80 | 30 | y = 3·1095x + 0 | 0·9981 | 4·148 | 0·269 |
| Isoimperatorin | [M + H]+ | 271 | 14·325 | 3 | 80 | 30 | y = 0·3661x + 0 | 0·9939 | 3·888 | 0·283 |
| Epoxybergamottin | [M + H]+ | 355 | 14·929 | 3 | 0 | 30 | y = 0·2798x + 0 | 0·9968 | 2·977 | 0·384 |
| Cnidicin | [M + H]+ | 355 | 17·938 | 4 | 0 | 30 | y = 3·7462x + 0 | 0·9901 | 4·424 | 0·417 |
| 8-Geranyloxypsoralen | [M + H]+ | 339 | 18·080 | 4 | 80 | 0 | y = 1·3745x + 0 | 0·9952 | 2·767 | 0·266 |
| Bergamottin | [M + H]+ | 339 | 18·254 | 4 | 80 | 0 | y = 0·3705x + 0 | 0·9943 | 4·390 | 0·612 |
| Internal standards | ||||||||||
| 5-Methoxyflavone | [M + H]+ | 253 | 10·871 | 3 | 0 | 0 | ||||
| Angelicin | [M + H]+ | 187 | 7·463 | 2 | 0 | 0 | ||||
aExtracted from ion chromatograms.
Data were acquired and analysed using LabSolution software version 5.52 sp2 (Shimadzu).
Peak identification and quantitation
Flavonoid, coumarin and furanocoumarin standards were individually injected into the UPLC–MS system to increment the software database (retention time and m/z ratio), which was used further to perform a semi-automatic identification of each molecule in the samples.
Quantitation of molecules was based on the MS signal on the addition of two internal standards to evaluate recoveries and on the determination of calibration curves of all the standards. 5-Methoxyflavone and angelicin, which were absent from Citrus extracts, were used as internal standard for flavonoids and coumarins and furanocoumarins, respectively. Standard solutions at five concentrations were used to draw calibration curves for each molecule. All curves were linear and forced to pass through 0 (Table 3).
Method validation
The method was validated in terms of linearity, sensitivity (limits of detection and quantitation), specificity, precision, accuracy and robustness.
Calibration curves were constructed by preparing five different concentrations of all standards, ranging from 1 to 30 µm. All calibration curves were plotted based on a linear regression analysis of the integrated peak area (y) versus concentration (x, µm).
Accuracy was evaluated according to recommendations for bioanalytical method validation (Green, 1996; Gustavo González and Ángeles Herrador, 2007). This study was performed by spiking metabolites in tomato matrix, which was free of flavanones, flavones, coumarins and furanocoumarins. Five tomato leaf extracts (Solanum lycopersicum var. Micro-Tom) were therefore injected. They were extracted according to the protocol described above. All standards were added to the tomato extracts at a concentration of 5 µm before injection. Accuracy was determined by calculating the recovery of each standard. The results are expressed in Table 3 as the mean of n = 5 five replicates ± s.d.
For each matrix, the limits of detection (LOD) and quantitation (LOQ) were calculated by analysing the signal-to-noise (S/N) ratio of each compound, in order to establish the lowest concentration detectable (LOD, S/N = 3) and the lowest concentration quantifiable (LOQ, S/N = 10) (Table 4).
Table 4.
Limit of detection (LOD) and limit of quantitation (LOQ) of the 64 phenolic compounds determined in the four Citrus tissues
| Compound | Flavedo |
Albedo |
Pulp |
Leaf |
||||
|---|---|---|---|---|---|---|---|---|
| LOD (mg kg–1) | LOQ (mg kg–1) | LOD (mg kg–1) | LOQ (mg kg–1) | LOD (mg kg–1) | LOQ (mg kg–1) | LOD (mg kg–1) | LOQ (mg kg–1) | |
| Flavones | ||||||||
| Apigenin | 0·028 | 0·093 | 0·035 | 0·117 | 0·066 | 0·219 | 0·075 | 0·251 |
| Acacetin | 0·009 | 0·031 | 0·008 | 0·026 | 0·008 | 0·027 | 0·013 | 0·045 |
| Luteolin | 0·038 | 0·127 | 0·018 | 0·060 | 0·072 | 0·241 | 0·078 | 0·259 |
| Chrysoeriol | 0·083 | 0·275 | 0·080 | 0·268 | 0·017 | 0·058 | 0·020 | 0·066 |
| Diosmetin | 0·082 | 0·272 | 0·083 | 0·276 | 0·019 | 0·064 | 0·021 | 0·068 |
| Tetramethyl isoscutellarein | 0·001 | 0·004 | 0·002 | 0·006 | 0·006 | 0·019 | 0·005 | 0·017 |
| Sinensetin | 0·005 | 0·016 | 0·004 | 0·015 | 0·004 | 0·013 | 0·011 | 0·036 |
| Tangeretin | 0·006 | 0·019 | 0·003 | 0·010 | 0·003 | 0·009 | 0·009 | 0·030 |
| Nobiletin | 0·032 | 0·106 | 0·004 | 0·015 | 0·004 | 0·013 | 0·015 | 0·049 |
| Vitexin | 0·018 | 0·061 | 0·033 | 0·110 | 0·031 | 0·104 | 0·081 | 0·269 |
| Apigetrin | 0·011 | 0·036 | 0·014 | 0·047 | 0·069 | 0·231 | 0·118 | 0·392 |
| Orientin | 0·014 | 0·048 | 0·010 | 0·033 | 0·023 | 0·077 | 0·024 | 0·081 |
| Cynaroside | 0·011 | 0·036 | 0·013 | 0·045 | 0·023 | 0·075 | 0·018 | 0·060 |
| Luteolin-4′-O-glucoside | 0·068 | 0·226 | 0·011 | 0·037 | 0·024 | 0·081 | 0·015 | 0·049 |
| Amentoflavone | 0·269 | 0·896 | 0·003 | 0·011 | 0·068 | 0·227 | 0·140 | 0·466 |
| Isorhoifolin | 0·062 | 0·205 | 0·176 | 0·586 | 0·055 | 0·182 | 0·360 | 1·199 |
| Rhoifolin | 0·092 | 0·307 | 0·263 | 0·875 | 0·066 | 0·219 | 0·493 | 1·644 |
| Linarin | 0·086 | 0·286 | 0·034 | 0·113 | 0·026 | 0·086 | 0·029 | 0·097 |
| Saponarin | 0·006 | 0·021 | 0·007 | 0·023 | 0·005 | 0·018 | 0·015 | 0·051 |
| Diosmin | 0·956 | 3·187 | 0·473 | 1·578 | 0·572 | 1·905 | 0·668 | 2·226 |
| Neodiosmin | 0·265 | 0·884 | 0·113 | 0·377 | 0·135 | 0·450 | 0·155 | 0·518 |
| Luteolin-3′-7-di-O-glucoside | 0·024 | 0·080 | 0·218 | 0·727 | 0·026 | 0·086 | 0·041 | 0·138 |
| Flavanones | ||||||||
| Naringenin | 0·476 | 1·587 | 0·459 | 1·529 | 0·397 | 1·324 | 0·327 | 1·090 |
| Sakuranetin | 0·134 | 0·448 | 0·081 | 0·271 | 0·058 | 0·193 | 0·073 | 0·243 |
| Eriodictyol | 1·000 | 3·333 | 0·214 | 0·715 | 0·896 | 2·986 | 0·577 | 1·924 |
| Homoeriodictyol | 0·346 | 1·152 | 0·226 | 0·755 | 0·259 | 0·862 | 0·320 | 1·067 |
| Hesperetin | 0·100 | 0·332 | 0·142 | 0·473 | 0·171 | 0·570 | 0·206 | 0·686 |
| Pyracanthoside | 0·187 | 0·622 | 0·092 | 0·306 | 0·443 | 1·476 | 0·479 | 1·598 |
| Narirutin | 0·347 | 1·155 | 0·877 | 2·924 | 0·850 | 2·834 | 0·360 | 1·199 |
| Naringin | 1·183 | 3·942 | 4·146 | 13·822 | 0·493 | 1·644 | 1·059 | 3·529 |
| Neoponcirin | 0·405 | 1·350 | 0·022 | 0·072 | 0·330 | 1·101 | 0·356 | 1·185 |
| Poncirin | 0·416 | 1·386 | 0·321 | 1·070 | 0·345 | 1·150 | 0·377 | 1·257 |
| Eriocitrin | 0·131 | 0·438 | 0·684 | 2·281 | 0·097 | 0·325 | 0·205 | 0·684 |
| Neoeriocitrin | 0·239 | 0·795 | 1·265 | 4·217 | 0·093 | 0·310 | 0·175 | 0·585 |
| Hesperidin | 0·368 | 1·227 | 0·377 | 1·257 | 0·136 | 0·453 | 0·222 | 0·739 |
| Neohesperidin | 0·941 | 3·136 | 0·685 | 2·284 | 0·247 | 0·823 | 0·325 | 1·082 |
| Flavonols | ||||||||
| Rutin | 0·028 | 0·094 | 0·299 | 0·998 | 0·092 | 0·307 | 0·103 | 0·344 |
| Anthocyanins | ||||||||
| Cyanidin-3-O-glucoside | 0·036 | 0·119 | 0·018 | 0·061 | 0·025 | 0·083 | 0·031 | 0·103 |
| Coumarins | ||||||||
| Umbelliferone | 1·009 | 3·363 | 0·739 | 2·463 | 0·903 | 3·010 | 4·942 | 16·474 |
| Limettin | 0·013 | 0·042 | 0·008 | 0·027 | 0·006 | 0·020 | 0·023 | 0·075 |
| Epoxyaurapten | 0·223 | 0·742 | 0·061 | 0·203 | 0·034 | 0·112 | 0·070 | 0·234 |
| Osthol | 0·008 | 0·026 | 0·006 | 0·020 | 0·004 | 0·012 | 0·013 | 0·044 |
| Aurapten | 0·007 | 0·024 | 0·002 | 0·007 | 0·003 | 0·010 | 0·011 | 0·038 |
| Furanocoumarins | ||||||||
| Xanthotoxol | 0·379 | 1·262 | 0·669 | 2·230 | 0·165 | 0·549 | 1·074 | 3·579 |
| Bergaptol | 0·389 | 1·298 | 0·115 | 0·383 | 0·116 | 0·388 | 0·267 | 0·888 |
| Heraclenol | 0·080 | 0·266 | 0·030 | 0·102 | 0·019 | 0·062 | 0·146 | 0·488 |
| Oxypeucedanin hydrate | 0·070 | 0·232 | 0·026 | 0·086 | 0·016 | 0·052 | 0·111 | 0·371 |
| Byakangelicin | 0·050 | 0·168 | 0·030 | 0·099 | 0·028 | 0·095 | 0·157 | 0·522 |
| Psoralen | 0·056 | 0·187 | 0·022 | 0·073 | 0·020 | 0·066 | 0·100 | 0·335 |
| Xanthotoxin | 0·057 | 0·191 | 0·026 | 0·087 | 0·013 | 0·043 | 0·052 | 0·174 |
| Bergapten | 0·040 | 0·133 | 0·017 | 0·056 | 0·009 | 0·030 | 0·036 | 0·120 |
| Isopimpinellin | 0·020 | 0·066 | 0·011 | 0·036 | 0·006 | 0·021 | 0·023 | 0·077 |
| Heraclenin | 0·057 | 0·190 | 0·018 | 0·061 | 0·022 | 0·072 | 0·062 | 0·206 |
| 6′,7′-Dihydroxybergamottin | 0·060 | 0·201 | 0·020 | 0·068 | 0·006 | 0·021 | 0·044 | 0·145 |
| Byakangelicol | 0·026 | 0·085 | 0·004 | 0·013 | 0·005 | 0·016 | 0·020 | 0·067 |
| Oxypeucedanin | 0·004 | 0·013 | 0·013 | 0·044 | 0·017 | 0·058 | 0·039 | 0·129 |
| Imperatorin | 0·017 | 0·057 | 0·011 | 0·037 | 0·008 | 0·026 | 0·029 | 0·096 |
| Phellopterin | 0·014 | 0·047 | 0·011 | 0·038 | 0·006 | 0·021 | 0·023 | 0·077 |
| Cnidilin | 0·008 | 0·027 | 0·007 | 0·023 | 0·004 | 0·014 | 0·014 | 0·047 |
| Isoimperatorin | 0·037 | 0·122 | 0·025 | 0·082 | 0·018 | 0·062 | 0·066 | 0·220 |
| Epoxybergamottin | 0·238 | 0·794 | 0·100 | 0·332 | 0·118 | 0·392 | 0·255 | 0·851 |
| Cnidicin | 0·009 | 0·030 | 0·003 | 0·011 | 0·004 | 0·014 | 0·010 | 0·033 |
| 8-Geranyloxypsoralen | 0·016 | 0·054 | 0·003 | 0·009 | 0·004 | 0·014 | 0·007 | 0·023 |
| Bergamottin | 0·038 | 0·128 | 0·007 | 0·023 | 0·010 | 0·032 | 0·012 | 0·039 |
The specificity of the method was assessed analysing the matrix effect. The matrix effect was determined on each tissue. First, one sample of each tissue (leaf, flavedo, albedo, pulp) of ‘Star Ruby’ grapefruit was injected into the UPLC–MS system to determine flavonoid, coumarin and furanocoumarin contents. Then, specificity was determined by repeatedly spiking the extracts of each ‘Star Ruby’ grapefruit tissue with known levels of ten standards at concentrations of 25, 50, 75 and 100 % of the expected values for each metabolite. The curves linking the concentrations of the standards to the percentage of additional standards were plotted in order to evaluate the specificity of the method (the equations of these curves are given in Table 5).
Table 5.
Precision, specificity and robustness of the method for ten metabolites in four different tissues
| Tissue/compound | Specificity |
Precision (RSD, %) |
Robustness (RSD, %) |
|||
|---|---|---|---|---|---|---|
| Equation | r2 | Repeatability | Intermediate precision | Oven temperature | Flow | |
| Flavedo | ||||||
| Flavonoids | ||||||
| Saponarin | y = 0·1164x + 0 | 0·9931 | 4·49 | 7·10 | 2·87 | 2·13 |
| Isorhoifolin | y = 0·2106x + 0 | 0·9985 | 5·69 | 10·16 | 2·55 | 4·04 |
| Poncirin | y = 0·1063x + 0 | 0·9921 | 6·29 | 10·33 | 8·91 | 8·44 |
| Rutin | y = 0·0519x + 0 | 0·9911 | 5·05 | 13·60 | 9·78 | 8·72 |
| Hesperidin | y = 0·1179x + 0 | 0·9933 | 4·13 | 5·91 | 2·45 | 7·37 |
| Coumarins | ||||||
| Limettin | y = 0·0081x + 0 | 0·9954 | 4·13 | 8·01 | 3·94 | 6·44 |
| Osthol | y = 0·0591x + 0 | 0·9966 | 4·41 | 6·10 | 4·67 | 6·91 |
| Bergapten | y = 0·0277x + 0 | 0·9946 | 4·35 | 10·61 | 4·46 | 4·25 |
| Isopimpinellin | y = 0·0053x + 0 | 0·9924 | 2·68 | 11·61 | 10·52 | 8·81 |
| Bergamottin | y = 0·1548x + 0 | 0·9930 | 7·93 | 12·58 | 6·07 | 16·53 |
| Albedo | ||||||
| Flavonoids | ||||||
| Isorhoifolin | y = 0·0077x + 0 | 0·9984 | 4·90 | 16·73 | 3·30 | 7·87 |
| Rhoifolin | y = 0·1114x + 0 | 0·9913 | 1·26 | 8·76 | 2·05 | 3·89 |
| Neodisomin | y = 0·0041x + 0 | 0·9933 | 7·16 | 15·60 | 7·46 | 6·95 |
| Saponarin | y = 0·1738x + 0 | 0·9929 | 2·65 | 19·70 | 2·90 | 9·39 |
| Hesperidin | y = 0·0391x + 0 | 0·9966 | 2·68 | 13·40 | 7·94 | 6·04 |
| Coumarins | ||||||
| Limettin | y = 0·0011x + 0 | 0·9906 | 3·48 | 8·13 | 4·90 | 6·72 |
| Osthol | y = 0·0007x + 0 | 0·9990 | 3·62 | 6·99 | 5·83 | 9·03 |
| Bergapten | y = 0·0009x + 0 | 0·9975 | 1·64 | 2·06 | 4·63 | 7·05 |
| Isopimpinellin | y = 0·0005x + 0 | 0·9964 | 5·03 | 9·45 | 2·46 | 7·43 |
| Bergamottin | y = 0·0022x + 0 | 0·9906 | 8·36 | 15·67 | 10·22 | 8·04 |
| Pulp | ||||||
| Flavonoids | ||||||
| Saponarin | y = 0·0316x + 0 | 0·9975 | 2·08 | 17·96 | 9·46 | 3·41 |
| Isorhoifolin | y = 0·1869x + 0 | 0·9948 | 2·96 | 9·52 | 9·58 | 6·83 |
| Poncirin | y = 0·0108x + 0 | 0·9970 | 2·30 | 10·22 | 2·01 | 5·62 |
| Rutin | y = 0·0139x + 0 | 0·9938 | 1·98 | 8·10 | 2·03 | 3·15 |
| Hesperidin | y = 0·0644x + 0 | 0·9942 | 2·07 | 9·80 | 9·84 | 9·33 |
| Coumarins | ||||||
| Limettin | y = 0·0005x + 0 | 0·993 | 1·37 | 4·35 | 4·72 | 3·81 |
| Osthol | y = 0·0003x + 0 | 0·9911 | 4·01 | 7·79 | 3·99 | 5·75 |
| Bergapten | y = 0·0019x + 0 | 0·9907 | 3·34 | 3·27 | 2·84 | 6·56 |
| Isopimpinellin | y = 0·0005x + 0 | 0·9901 | 3·19 | 4·30 | 4·16 | 6·27 |
| Bergamottin | y = 0·0345x + 0 | 0·9993 | 5·79 | 16·28 | 28·99 | 4·19 |
| Leaf | ||||||
| Flavonoids | ||||||
| Saponarin | y = 0·1713x + 0 | 0·9932 | 3·13 | 9·35 | 5·99 | 9·11 |
| Isorhoifolin | y = 0·2061x + 0 | 0·9950 | 2·46 | 15·23 | 7·37 | 9·08 |
| Poncirin | y = 0·0631x + 0 | 0·9990 | 3·53 | 15·20 | 5·95 | 9·83 |
| Rutin | y = 0·0191x + 0 | 0·9903 | 3·56 | 11·54 | 8·52 | 2·02 |
| Hesperidin | y = 0·2014x + 0 | 0·9924 | 1·95 | 7·32 | 8·22 | 8·18 |
| Coumarins | ||||||
| Limettin | y = 0·0985x + 0 | 0·9948 | 5·47 | 11·77 | 6·81 | 5·46 |
| Osthol | y = 0·0009x + 0 | 0·9983 | 7·80 | 11·33 | 8·48 | 4·14 |
| Bergapten | y = 0·0263x + 0 | 0·9936 | 7·09 | 12·60 | 9·03 | 9·05 |
| Isopimpinellin | y = 0·0075x + 0 | 0·9955 | 3·97 | 11·35 | 9·86 | 7·15 |
| Bergamottin | y = 0·0113x + 0 | 0·9948 | 6·59 | 8·77 | 4·17 | 3·42 |
The intra-day (repeatability) and inter-day (intermediary precision, also assimilated to the reproducibility) precisions were evaluated by injecting five technical replicates (five extracts from the same sample of flavedo, albedo, pulp and leaf) of ‘Star Ruby’ grapefruit on five different days. The intra-day and inter-day precisions were determined for ten metabolites in each tissue by dividing the s.d. (n = 5) by the mean concentration to obtain the relative standard deviation (RSD), expressed as a percentage (Table 5).
Method robustness was tested by changing two parameters. First, three different temperatures were tested on the column oven (55, 60 and 65 °C) and then three different flow rates (0·35, 0·40 and 0·45 mL min–1) were also tested. The robustness of the method was evaluated by the quantitation of ten metabolites in each ‘Star Ruby’ grapefruit tissue across three different conditions of temperature and flow rate and by the calculation of RSD (n = 3) for each metabolite (Table 5).
Data analysis
Data were analysed using R statistical software (http://www.R-project.org).
Results for metabolites across the four tissues and the 11 cultivars are expressed as means (n = 5 biological replicates) ± s.d.
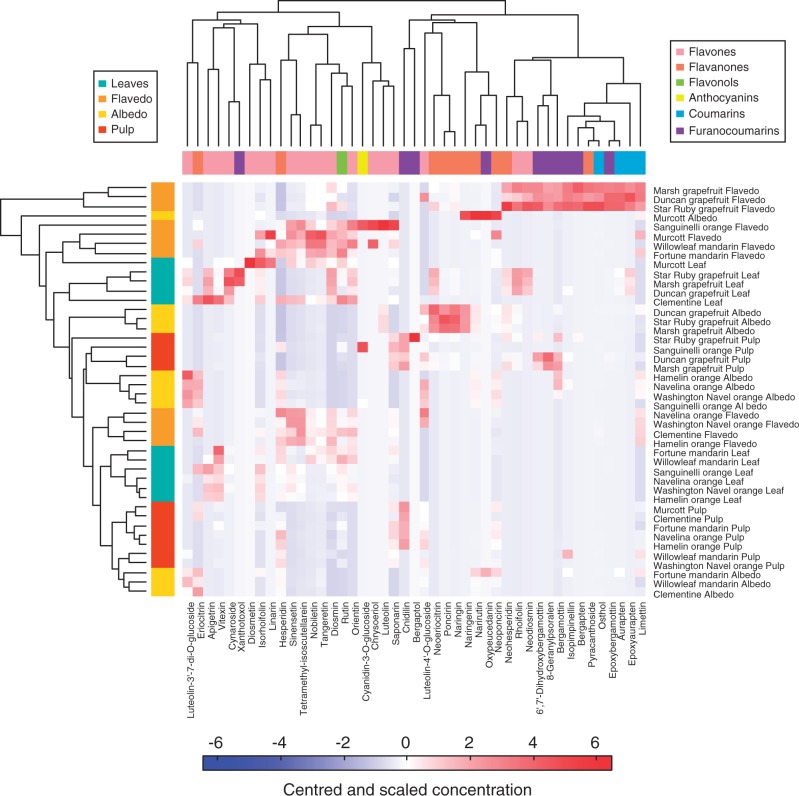
The gplots R package and the heatmap function were used to construct the heat map. Data comprised the concentrations of 45 metabolites and 44 observations (4 tissues × 11 cultivars). Data (means of five biological replicates) were centred and scaled by metabolites. The 45 metabolites and the 44 observations were classified according to Euclidean distances. Values were then associated with a colour, ranging from blue (low) to red (high) (Fig. 2).
Fig. 2.
Heat map illustrating the patterns of phenolic compound across 11 Citrus cultivars and four tissues: leaves, flavedo, albedo and pulp. Forty-five phenolic compounds were quantified in these tissues with the UPLC–MS method developed in this study, and the values shown correspond to the means of five observations. The dataset comprised centred and scaled values. Colours vary from blue for low concentrations to red for high concentrations. The two classifications of metabolites on the one hand and observations (tissues of cultivars) on the other hand were based on Euclidian distances. The colour codes of the two classifications correspond to the different tissues and to the different classes of metabolites, as indicated in the two keys. The heat map clearly illustrates the separation between tissues and between cultivars.
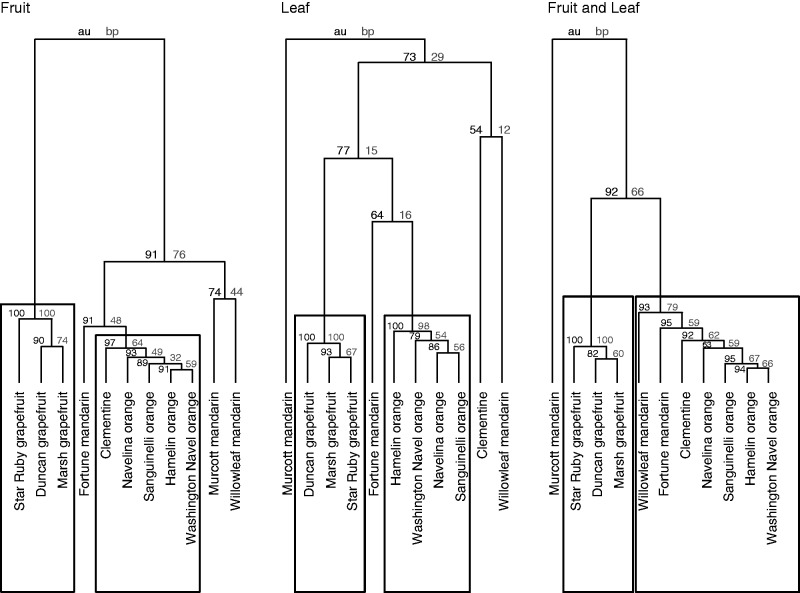
The phylogenetic trees of the 11 Citrus cultivars were constructed with the pvclust package and the pvclust function of R software, performing a hierarchical cluster analysis. Two types of P-values were calculated: approximately unbiased (AU) and bootstrap probability (BP) (Fig. 3). The AU P-value was computed by multiscale bootstrap resampling and the BP value by normal bootstrap resampling (Suzuki and Shimodaira, 2006).
Fig. 3.
Phylogenetic trees of the 11 Citrus cultivars based on the polyphenol contents calculated with the UPLC–MS procedure developed in this study and expressed as µmol g–1 dry wt. Data are means of five biological replicates. The first tree is based on concentrations determined in the three tissues of the fruit, whereas the second is based only on concentrations in the leaf and the third on concentrations calculated in all targeted tissues. The trees were constructed with the pvclust function of R software, performing a hierarchical cluster analysis. Two types of P-value are indicated for each cluster (indicating how strongly each cluster is supported by the data): numbers in black type represent approximately unbiased (AU) P-values and numbers in grey represent bootstrap probability (BP) values. The AU is more accurate than the BP value and clusters with high AU values (≥95 %) are strongly supported by the data (Suzuki and Shimodaira, 2006); these clusters are enclosed in rectangles.
The Ade4 R package and the dudi.pca function were used to perform principal components analysis (PCA) (Fig. 4). Data consisted of the concentrations of 45 metabolites and 22 observations (2 tissues × 11 cultivars). Data (means of five biological replicates) were centred and scaled by metabolites. The relative contributions of the metabolites to the two components and their coordinates on these axes are given in Supplementary Data Table S3
Fig. 4.
Principal components analysis of 45 phenolic compounds quantified in external tissues (leaves and flavedo) of 11 Citrus cultivars. The objective of this analysis was to test whether these external tissues can be differentiated on the basis of their phenolic compound contents. The dataset was composed of centre-scale values. The first principal component (PC1) explains 39 % of the total variance while the second component (PC2) accounts for 16 %. The 45 quantitative variables (metabolites) are plotted on these two components in (A). Metabolites are identified by numbers and a colour code, according to their class, if they contribute significantly to PC1 or to PC2. Observations corresponding to tissues and cultivars are presented in (B). Each point corresponds to the mean of five replicates.
Multiple mean comparisons were performed using the Kruskal–Wallis non-parametric test at α = 0·05 (Fig. 5).
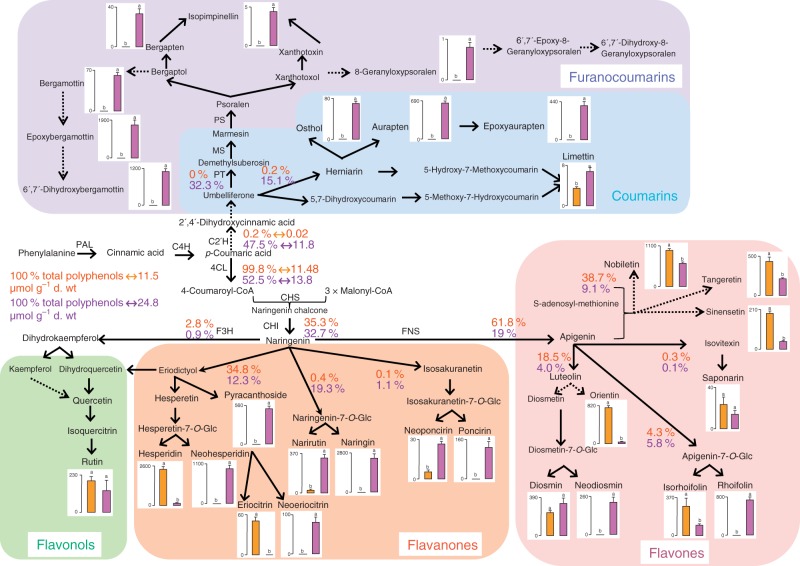
Fig. 5.
Comparison of the biosynthesis and accumulation of phenolic compounds between ‘Clementine’ and ‘Star Ruby’ grapefruit. A simplified polyphenol biosynthetic pathway, starting from phenylalanine, was constructed on the basis of ‘omics’ data from previous studies in Citrus (Bourgaud et al., 2006; Vialart et al., 2012; Frydman et al., 2013) and on the basis of the metabolite patterns found in the flavedo of ‘Clementine’ and ‘Star Ruby’ grapefruit. Accordingly, the compounds not quantifiable in these tissues were deliberately omitted. This simplified scheme does not display all possible routes between metabolites. For example, luteolin can also be formed via eriodictyol, which is not presented here. Arrows indicate one or several enzymatic steps of the pathway. Dotted arrows represent enzymes that are uncharacterized in Citrus. For the sake of clarity, details about enzymes are omitted within the classes of phenolic compounds. The contents of quantifiable end-products are presented. Concentrations of the indicated metabolites were measured in the flavedo of ripe fruits using the method developed in this study. ‘Clementine’ is represented in orange and ‘Star Ruby’ grapefruit in purple. Contents are expressed in mg kg–1 dry wt and are means (n = 5 biological replicates) ± s.d. Different letters indicate significant differences between ‘Clementine’ and ‘Star Ruby’ grapefruit at α = 0·05. The contents of phenolic compounds are also expressed in µmol phenylalanine equivalent per g dry wt. The sum is equivalent to the total amount of phenylalanine contained in soluble phenolic compounds. The proportions of each class were determined in relation to this total amount of phenolic rings and are shown close to the corresponding arrows in orange for ‘Clementine’ and in purple for ‘Star Ruby’. This information highlights the rate-limiting nodes/enzymes in the pathway. For example, in ‘Clementine’ flavones, flavanones and flavonols represent almost 100 % of the total phenolic rings (11·5 µmol g–1 dry wt), whereas in ‘Star Ruby’ grapefruit this percentage is around 50 %. Abbreviations for the main enzymes: CHS, chalcone synthase; CHI, chalcone isomerase; C2′H, p-coumaroyl CoA 2′-hydroxylase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate CoA ligase; F3H, flavanone 3-hydroxylase; FNS, flavone synthase; MS, marmesin synthase; PAL, phenylalanine ammonia lyase; PT, prenyltransferase; PS, psoralen synthase.
The distribution of aromatic rings along the polyphenol network was analysed by (1) the construction of a network using available ‘omics’ data on the genes and enzymes of the phenolic compound pathway in Citrus (Bourgaud et al., 2006; Frydman et al., 2013; Vialart et al., 2012), (2) phenotyping of two cultivars with the developed method and the simplification map according to metabolite measurements, and (3) calculation of the distribution of aromatic rings along the network relative to the total abundance of phenolic compounds and using the information available on reaction stoichiometries. It was then possible to express metabolite contents in phenylalanine equivalents and to determine the absolute concentration of aromatic rings. This led to the calculation of the percentage of the different classes of metabolites relative to this absolute concentration.
RESULTS AND DISCUSSION
Optimizing the UPLC–MS analysis to identify and quantitate a wide range of phenolic compounds in a single run
The objective of this work was to propose a procedure (from tissue sampling to UPLC–MS analysis) to analyse phenolic metabolites in Citrus species. The metabolites were first separated and identified by UPLC–MS analysis. Sixty-four phenolic compounds were isolated and characterized in 20 min. Figure 1 and Table 3 summarize the structure, chromatographic data and MS data of the 64 metabolites, belonging to six different classes of phenolic compounds. Identification was performed by combining UV and ion chromatograms and UV and MS spectra. This sequence is detailed in Supplementary Data Fig. S1 for cyanidin-3-O-glucoside and for four compounds sharing the same m/z (611). References presenting the full MS data of these compounds are listed in Supplementary Data Table S4. The data from Citrus extracts were compared with the data obtained with 64 authentic standards. Two standard solutions were prepared, the first with 37 flavonoids and two internal standards and the second with 27 coumarins and furanocoumarins. These standard solutions were injected at the beginning and end of each run sequence to avoid mis-identification due to changes in retention times. To complete this identification process, co-elution tests were carried out by spiking sample extracts with standards.
These first results prompted us to investigate two parameters of the UPLC–MS system. Firstly, different mobile phases, a methanol–water system and an acetonitrile–water system, both acidified with 0·1 % formic acid, were tested to optimize peak separation. Responses were higher and peaks were of better shape with the acetonitrile–water system compared with the methanol–water system, allowing better separation of isomers. For example, homoeriodictyol and hesperetin could not be successfully separated with a two-head peak at 10·127 and 10·163 min in the methanol–water system. On the contrary, in an acetonitrile–water system hesperetin and homoeriodictyol could be distinguished at 7·267 and 7·588 min respectively. Accordingly, the acetonitrile–water system was selected as the mobile phase and various gradient elution profiles were investigated to optimize analytical performance. Elution was performed using four linear gradient steps (from 0·01 to 17·50 min with 20–100 % of eluent B) followed by two consecutive isocratic steps (from 17·50 to 19 min and from 19·05 to 20 min, with 100 and 20 % of eluent B respectively) in order to wash and equilibrate the column between samples. With this method all 38 flavonoids and 26 coumarins and furanocoumarins could be efficiently separated for subsequent MS detection (Table 3). Secondly, MS detection was optimized by raising the sensitivity and selectivity of the MS device. In order to increase the selectivity of MS detection, SIM mode was used. Detection sensitivity was improved using a dual ion source interface, which managed ACPI and ESI in parallel. Finally, ionization parameters (DL and Q-array) of the mass spectrometer were optimized for each compound recorded in SIM mode (Table 3). The product ion mass spectra obtained in these conditions were extracted and compared with data from the literature and data obtained with authentic standards. These spectral data, combined with UV spectra and co-elution tests, helped to distinguish between metabolites with the same m/z value as the parental ion and presenting similar retention times.
This procedure, allowing the analysis of 64 phenolic compounds in 20 min, represents an improvement over existing methods for analysing flavonoids and coumarins in Citrus (Gardana et al., 2008; Barreca et al., 2011a, b; VanderMolen et al., 2013), although only one flavonol (rutin) and only one anthocyanin (cyanidin-3-O-glucoside) are reported. On the whole, rapid methods allow the identification and quantitation of a reduced number of molecules. For example, in the work of VanderMolen et al. (2013), four flavonoids and two furanocoumarins were quantified in 4·5 min. On the contrary, Barreca et al. (2011b) analysed a larger number of metabolites, with 22 flavonoids and two furanocoumarins, using a gradient of 55 min. UPLC systems decrease the duration of analysis and solvent consumption. These systems, analysing a reduced number of metabolites (generally the major flavonoids in Citrus) in a very short time, seem particularly efficient in detecting the adulteration of standardized products such as Citrus juices (Medina-Remon et al., 2011). However, a better understanding of the biosynthesis and accumulation of phenolic compounds in Citrus in response to genetic, developmental and environmental cues requires the analysis of a larger range of molecules in numerous samples. The method developed here therefore seems promising to address this issue since the major phenolic compounds found in Citrus can be investigated. Flavonols and anthocyanins are present in much smaller amounts than flavones and flavanones in Citrus species (Kawaii et al., 1999, 2000a; Nogata et al., 2006; Gattuso et al., 2007) apart from specific cultivars, such as some representatives of Citrus aurantifolia for flavonols (Loizzo et al., 2012) or blood oranges for anthocyanins (Rapisarda et al., 2000). The investigation of flavonols (including kaempferol derivatives) or anthocyanins (including malonylglycoside anthocyanins) in Citrus would require focusing analysis on these classes or increasing the duration of the gradient. For example, Abad-Garcia et al. (2012) reported the identification in Citrus of 58 flavonoids by HPLC– DAD–ESI–CID–MS/MS (where CID is collision-induced dissociation) using a gradient programme of more than 140 min, and 11 novel flavonols were identified among these compounds.
Analysing different tissues to validate the proposed method
The linearity, accuracy, sensitivity, specificity, precision and robustness of this method were then evaluated. Linearity, accuracy and sensitivity were tested for all the metabolites found in the extracts while the other method validation parameters were determined on a selection of ten metabolites quantified in the four tissues of ‘Star Ruby’ grapefruit, which was found to be a rich source of phenolic compounds.
Calibration curves of the peak area (y) versus the concentration (x) of each standard were constructed to evaluate linearity. These curves were based on the injection of five dilution points and on the linear regression passing through 0. They showed good linearity (r2 > 0·99), as summarized in Table 3. To evaluate the accuracy of this method, five tomato leaf extracts were incremented with all standards at a concentration of 5 µm. Results of the UPLC–MS quantitation showed that the recoveries were close to 5 µm (Table 3). These results demonstrate that the accuracy of this assay is within the acceptable range of 80–120 % recovery for almost all standards. Finally, calculation of the LOD and LOQ was based on determination of the concentration and the S/N ratio. The LOD and LOQ values for the 64 phenolic compound standards analysed in this study ranged from 0·001 to 4·942 µg mL–1 and from 0·004 to 16·474 µg mL–1, respectively (Table 4).
To determine the specificity of this method, tests of what happens during the extraction process were performed. The recovery of some targeted metabolites was used as an indicator of a matrix effect. This analysis was carried out on the four previously characterized tissues of ‘Star Ruby’ grapefruit. The recovery of five flavonoids (saponarin, isorhoifolin, poncirin, rutin and hesperidin) and five furanocoumarins (limettin, osthol, bergapten, isopimpinellin and bergamottin) already quantified in the ‘Star Ruby’ grapefruit extracts were analysed. The four tissue samples were repeatedly spiked with four different concentrations of the ten metabolites: 25, 50, 75 and 100 % of the expected value. They were then submitted to the same extraction process. The equations linking the compound areas to the percentages of the initial concentration added were determined for these ten molecules. The relations were linear with optimal correlation coefficients (r2 > 0·99), as shown in Table 5. We concluded that additional compounds present in the extract such as chlorophylls and carotenoids had a negligible effect on the contents of flavonoids, coumarins and furanocoumarins.
The intra- and inter-day precisions were calculated for these ten metabolites by the injection of five replicates of the tissue extracts of ‘Star Ruby’ grapefruit for 5 d and determination RSD. The intra- and inter-day precisions (RSDs) ranged from 1·26 to 8·36 % and from 2·06 to 19·70 %, respectively. Precision depends on metabolite level (Green, 1996; Gustavo González and Ángeles Herrador, 2007). Accordingly, in most cases the RSDs were within 10 %. Higher values were found when studying intermediate precision and metabolites with low concentrations (<1 ppm). These results allowed us to conclude that this method was repeatable and precise (Table 5).
Finally, two parameters were changed to determine the robustness of the method: oven temperature and flow rate. Five flavonoids and five furanocoumarins that were quantifiable in ‘Star Ruby’ grapefruit were analysed. The RSD values were around 10 % (except for bergamottin quantified in flavedo or pulp), showing the robustness of the method (Table 5). High variability has previously been reported for bergamottin (Dugrand et al., 2013). The sensitivity of this metabolite to temperature could partly explain these variations.
Profiling the phenolic compounds of contrasting species and tissues in Citrus to challenge phylogenetic relationships and reveal metabolic shifts in different tissues
The proposed high-throughput method allows the accurate identification and quantitation of 64 phenolic compounds. To go further into this analysis, this procedure was applied to four tissues of 11 commercial Citrus varieties. Four orange, three grapefruit and four mandarin or mandarin-type cultivars were analysed. Details of the cultivars and rootstocks are given in Table 1. Because the phenolic compound pathway is tightly regulated by developmental and environmental factors, the sampling procedure was adapted to minimize these two sources of variation in open fields. Data on the developmental characteristics of leaves and fruits are given in Supplementary Data Tables S1 and S2, respectively. Climatic conditions are summarized in Supplementary Data Table S5. The leaves were adult leaves from the spring flush of the previous season. The fruits were dissected to obtain the three tissues: flavedo (coloured part of the exocarp), albedo (white spongy mesocarp) and pulp (endocarp).
The heat map in Fig. 2 summarizes quantitative data on metabolite distribution in four Citrus tissues. Sixty-four metabolites were analysed and 45 were quantified across the four targeted tissues and 11 cultivars. A colour was associated with the amount of metabolites: from blue for low concentrations to red for high concentrations. Two classifications were obtained: the first showed relations between metabolites and the second showed similarities between observations. A heat map with metabolites grouped by chemical classes is presented in Supplementary Data Fig. S2. The 45 metabolites belonged to six classes: flavones, flavanones, flavonols, anthocyanins, coumarins and furanocoumarins (Fig. 2). Flavones, flavonols and anthocyanins clustered in the same group, whereas coumarins, furanocoumarins and most of the flavanones were linked in a second group (Fig. 2). One furanocoumarin, xanthotoxol, was included in the flavone–flavonol–anthocyanin group. This metabolite was only found in the leaves of ‘Star Ruby’ and ‘Marsh’ grapefruits. Two flavanones, eriocitrin and hesperidin, were also found in this first cluster. Eriocitrin and hesperidin were present in all tissues of mandarins, oranges and ‘Clementine’. The second classification showed that samples were first clustered by tissues and then by species. The flavedos from grapefruits were grouped according to their coumarin and furanocoumarin profiles. Very large amounts of coumarins and furanocoumarins were found in this tissue [the total content was up to 4000 mgkg–1 of dry weight (dry wt)] compared with the other tissues. Interestingly, flavedo and leaves appeared to be the richest sources of coumarins, furanocoumarins and flavonoids. Few metabolites seemed to be specific to internal tissues: naringenin, oxypeucedanin, cnidilin and bergaptol. On the contrary, many metabolites accumulated in high amounts only in flavedo or leaves. It should also be pointed out that the ‘Murcott’ albedo showed a unique profile, rich in naringenin, narirutin, neoponcirin and oxypeucedanin
Genetic effects on the build-up of phenolic compounds have been studied to a much greater extent than tissue effects. The tissue specificity of some metabolites prompted us to investigate whether the classification of cultivars could change on a restricted dataset. The phylogenetic trees in Fig. 3 were constructed on the basis of the metabolite contents calculated (1) in the three tissues of the fruit (first tree), (2) in the leaf only (second tree) and (3) in all tissues (third tree). In all cases, the three grapefruits constituted a specific cluster and the high P-values indicated a strong involvement of the data in this cluster (Fig. 3). The classifications of the four oranges did not present significant differences between the trees. Note that ‘Fortune’ mandarin was always associated with the orange cluster. However, the results showed that the classification of other mandarins and mandarin derivatives varied according to the dataset used to perform the clustering analysis. ‘Clementine’ clustered with oranges in the first and third dendrogram but constituted a new branch associated with ‘Willowleaf’ mandarin in the second one, based on the leaf only. ‘Murcott’ mandarin appeared linked to ‘Willowleaf’ mandarin only in the first clustering, based on fruit tissues (Fig. 3).
Plants of the Citrus genus are rich sources of a wide range of phytochemicals. The complexity of Citrus taxonomy arises from the diversity of phenotypic, traits such as the secondary metabolite composition, sexual compatibility between species and apomixis. Several classifications have been made on the basis of secondary metabolite contents and the contents of flavonoids in particular (Kawaii et al., 1999, 2000b; Fanciullino et al., 2006). These classifications and further phylogenetic analyses, based on molecular markers, converged to demonstrate that the phenotypic diversity is related to the initial differentiation of the four wild ancestral species C. maxima (Burm.) (pomelo), C. medica L. (citron), C. reticulata Blanco (mandarin) and C. micrantha, which are likely to be the ancestors of the cultivated Citrus species (Froelicher et al., 2011; Garcia-Lor et al., 2013; Wu et al., 2014). For instance, ‘Clementine’ has been found to be a hybrid between ‘Willowleaf’ mandarin and a sweet orange (Wu et al., 2014). Garcia-Lor et al. (2013) concluded that the contribution of the mandarin to the gene pool is higher than that of the pomelo. Our procedure seems to be a powerful tool to analyse interspecific relationships, as illustrated by the example of ‘Clementine’, whose polyphenol profile is very close to that of ‘Willowleaf’ mandarin in the case of leaf tissue and closer to oranges in the case of fruit tissues (especially flavedo and pulp) (Figs 2 and 3). In the same way, ‘Murcott’ mandarin is believed to be a hybrid of unknown parentage; it was proposed that sweet orange is a grandparent, whereas the other grandparent and parent are not known (Wu et al., 2014). Cluster analysis based on the four tissues seems to confirm this complex parentage. Nogata et al. (2006) analysed 17 flavonoids in the fruit tissues of 42 species. They came to the conclusion that the polyphenol profiles within a section defined by Tanaka’s system of taxonomy were very similar, with few exceptions. Our data support this conclusion. Our analysis also demonstrate the role of the combination of datasets obtained from different tissues, such as leaf, flavedo, albedo and pulp, to analyse the linkage between Citrus cultivars. This conclusion is consistent with the existence of multigene families for many enzymes of the polyphenol biosynthetic pathway and the differential expression of these members according to developmental cues or in response to environmental conditions (Daniel et al., 2011).
The specificity of the leaf tissue was further studied using data on external tissues exposed to similar environmental conditions. The PCA in Fig. 4 presents the similarities and differences between leaves and flavedo in more detail. The variables correspond to the 45 phenolic compounds quantified across the 11 Citrus cultivars. The first two components, principal component 1 (PC1) and principal component 2 (PC2), explained 55 % of the total variance. The 45 metabolites are plotted on the first two principle components in (A) while (B) shows the projection of the observations. The relative contributions and the coordinates of the metabolites on the two axes are presented in Supplementary Data Table S3. Regarding PC1, one flavone (orientin) and one flavanone (hesperidin) correlated positively with PC1, and two flavones (rhoifolin and neodiosmin), four flavanones (pyracanthoside, naringin, poncirin and neohesperidin), three coumarins (osthol, aurapten and epoxyaurapten) and six furanocoumarins correlated negatively with PC1. Fewer metabolites were well represented on PC2: four flavones (tetramethyl isoscutellarin, sinensetin, tangeretin and nobiletin) correlated with the positive values of PC2, while three flavones (apigetrin, cynaroside, and luteolin-3′-7-diglucoside), one flavanone (neoeriocitrin) and one furanocoumarin (xanthotoxol) correlated with the negative values. Interestingly, cultivars were separated on PC1 whereas PC2 explained differences between tissues (Fig. 4). Grapefruit tissues, especially flavedo, can be related to high contents of flavone or flavanone neohesperidosides and of coumarins and furanocoumarins.
Nogata et al. (2006) showed that flavonoid neohesperidosides (bitter metabolites) were largely responsible for cultivar classification. In line with this, we also demonstrated that most flavonoid neohesperidosides clustered together (Fig. 4). Surprisingly, these metabolites were also positively related to coumarins and coumarin derivatives, although there is no direct link between the biosynthetic steps leading to flavonoid neohesperidosides and the steps responsible for the formation of coumarins and their derivatives in the pathway. We also found that apigetrin, cynaroside and luteolin-3′-7-diglucoside positively correlated and strongly contributed to component 2, which allowed separation of the leaf from the flavedo (Fig. 4). More precisely, adult leaves preferentially accumulated apigenin glucosides and luteolin glucosides, while the flavedo of ripe fruits concentrated higher amounts of polymethoxyflavones such as nobiletin, tangeretin and sinensetin. Thus, it is likely that this distribution is triggered by developmental or environmental cues. For example, leaf chloroplasts are major sources of reactive oxygen species (ROS) in plants. In line with this, according to Agati et al. (2012), luteolin-7-O-glycosides accumulate in response to UV-B and salinity and this accumulation should be interpreted in terms of their involvement as antioxidants in plants more than in terms of their role in preventing the formation of ROS.
Taken together, our findings demonstrate the importance of analysing the distribution of multiclass phenolic compounds across tissues and cultivars for a better understanding of the organization of the biosynthetic pathway and its regulation.
Analysing a wide range of phenolic compounds to understand the metabolic network
We then focused on two contrasting cultivars: ‘Clementine’ and ‘Star Ruby’ grapefruit, which accumulated high contents of phenolic compounds in the flavedo. We investigated the distribution of aromatic rings along the phenolic compound network relative to the total abundance. Phenolic compounds such as coumarins or flavonoids derive from the aromatic amino acid phenylalanine, provided by the shikimate pathway. Figure 5 shows polyphenol biosynthesis for each cultivar. The values plotted correspond to the concentration of each metabolite expressed in mg per kg dry wt. The contents were also expressed as µmol phenylalanine equivalent (PE) per g dry wt using the metabolite contents measured in each tissue and reaction stoichiometries. We cumulated the contents at the main branches in the pathway and determined the percentage of each class of metabolites relative to the total concentration of phenolic compounds (Fig. 5). Coumarins and furanocoumarins appeared specific to ‘Star Ruby’ grapefruit. ‘Star Ruby’ grapefruit produced 11·8 µmol PE g–1 dry wt of coumarins and furanocoumarins and 13·8 µmol PE g–1 dry wt of flavanones, flavones and flavonols. Consequently, almost 50 % of the total aromatic rings were invested in the synthesis of coumarins and coumarin derivatives and 50 % were used to form flavonoids. In the case of ‘Clementine’, almost 100 % of the total aromatic rings were used to synthesize flavonoids. Despite these differences, ‘Clementine’ and ‘Star Ruby’ grapefruit concentrated similar amounts of flavonoids (11·5 versus 13·8 µmol PE g–1 dry wt). Interestingly, flavones accounted for 61·8 % (i.e. 7·1 µmol PE g–1 dry wt) and 19 % (i.e. 4·7 µmol PE g–1 dry wt) of the total aromatic rings in ‘Clementine’ and ‘Star Ruby’ respectively. In addition, ‘Clementine’ accumulated large amounts of flavanone and flavone-7-O-rutinosides such as hesperidin (4·9 µmol PE g–1 dry wt). ‘Star Ruby’ grapefruit synthesized flavonoid-7-O-rutinosides such as neoponcirin or diosmin but preferentially accumulated flavonoid-7-O-neohesperidosides such as naringin, neohesperidin, neoeriocitrin and neodiosmin (Fig. 5). The total content of flavonoid-7-O-neohesperidoside was 7·8 µmol PE g–1 dry wt in the flavedo of ‘Star Ruby’ grapefruit while the total content of flavonoid-7-O-rutinoside was 1·5 µmol PE g–1 dry wt.
The measurements of multiclass metabolites led to the construction of a map showing the distribution of aromatic rings along the network. This distribution map allowed comparisons between contrasting cultivars (Fig. 5) and highlighted potential regulatory nodes involved in cultivar variability. For example, conversions of flavonoid-7-O-glucosides into flavanoid-7-O-rutinosides or flavanoid-7-O-neohesperidosides catalysed by rhamnosyltransferases are already known regulatory steps. Frydman et al. (2013) showed that 1,2-rhamnosyltransferase, found in pomelos but not in mandarin-type cultivars, produced bitter flavanone-7-O-hesperidosides, whereas 1,6-rhamnosyltransferase, isolated in oranges, was responsible for the formation of tasteless flavanone rutinosides. They also demonstrated that 1,6-rhamnosyltransferase, encoded by Cs1,6RahT, could use flavanones glucosylated at positions 3 or 7 as well as flavones, flavonols and anthocyanins as substrates (Frydman et al., 2013). Our analysis of multiclass metabolites in ‘Clementine’ and ‘Star Ruby’ grapefruit has confirmed that the steps catalysed by rhamnosyltransferases constitute branching nodes. Our results also suggest that these enzymes are ubiquitous, acting at several nodes of the phenolic compound pathway, since similar ratios of rutinosides to rhamnosides were found in different classes of flavonoids. In addition, results presented in Fig. 5 shed some light on the step catalysed by flavone synthase. In ‘Clementine’, more than 60 % of the total phenolic content was represented by flavones, whereas this percentage was only 19 in ‘Star Ruby’ grapefruit. This difference was not due to variation in the formation of flavonoids, since the two cultivars had the same level, around 12 µmol g–1 dry wt. Competition between flavones and flavonols should be excluded. Rutin (quercetin-3-O-rutinoside) was the only flavonol quantified in our study. Kaempferol derivatives were below the limit of detection, which is in agreement with previously published data on Citrus, showing that these compounds are barely quantifiable in several Citrus species except for C. aurantifolia (Loizzo et al., 2012). We propose that the steps leading to flavonols in ‘Clementine’ or ‘Star Ruby’ grapefruit are not regulatory nodes in our climatic conditions. This leads us to propose that the gene encoding flavone synthase (FNS) is differentially expressed in the flavedo of ‘Clementine’ and ‘Star Ruby’ grapefruit. A study comparing black dahlia cultivars with purple ones demonstrated that the competition between anthocyanidins and flavones for naringenin can be suppressed by the silencing of DvFNS, encoding a flavone synthase, which seems to be in agreement with our proposition (Deguchi et al., 2013). Post-transcriptional mechanisms affecting FNS cannot be excluded (Deguchi et al., 2013). Finally, ‘Clementine’ was barely able to produce coumarins while ‘StarRuby’ accumulated equal contents of coumarins and flavonoids. In addition, the total content of phenolic compounds expressed as PE was double in the grapefruit flavedo. These results prompt us to suggest that two other steps, the step forming 2′,4′-dihydrocinnamic acid from p-coumaric acid and probably catalysed by a p-coumaroyl CoA 2′-hydroxylase (C2′H) (Vialart et al., 2012) and the step catalysed by phenylalanine ammonia-lyase (PAL) are important regulatory nodes. The gene encoding C2′H, which directs the biosynthesis of coumarins and coumarin derivatives, and PAL might be upregulated in ‘Star Ruby’ flavedo compared with ‘Clementine’ flavedo. It could not be ruled out that the higher level of phenolic compounds in grapefruit flavedo was due to a higher flux of the precursors that enter the phenolic compound pathway (Tohge et al., 2013).
Conclusions
We developed a high-throughput method to study the distribution of phenolic compounds across contrasting tissues and cultivars. This rapid method allowed the identification and quantitation of 64 phenolic compounds in 20 min, which represents an improvement over existing methods of analysing multiclass polyphenols. The procedure was validated in four tissues: leaves, flavedo, albedo and pulp. We were able to draw a map of the distribution of phenolic compounds along fruiting branches in relation to developmental cues and under characterized environmental conditions. We highlighted that these metabolites accumulated preferentially in external tissues, leaves and flavedo, possibly in relation to their involvement in plant defence. A combination of datasets from contrasting tissues was used to classify the Citrus cultivars. Analysis of the biosynthetic pathway for two contrasting cultivars highlighted regulatory nodes. In addition to the steps catalysed by rhamnosyltransferases, we proposed that the steps catalysed by phenylalanine ammonia lyase, the step leading to 2′,4′-dihydrocinnamic acid from p-coumaric acid and the step involving flavone synthase were important regulatory nodes in ‘Clementine’ and ‘Star Ruby’ grapefruit. This work paves the way for further analysis aiming to describe the control of the metabolism of phenolic compounds in Citrus in response to genetic and environmental factors.
SUPPLEMENTARY DATA
Supplementary Data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: leaf characterization. Table S2: fruit characterization. Table S3: details of the PCA analysis: relative contributions of the metabolites to principal components 1 and 2 and their coordinates on these axes. Table S4: list of references that include full mass spectral data on the 64 phenolic compounds. Table S5: details of climatic conditions during the trial. Figure S1: sequence identification of cyanidin-3-O-glucoside (449 [M+H]+, 287 [M+H-162]+) and of four compounds sharing the same m/z (611). Figure S2: heat map illustrating the phenolic compound patterns across 11 Citrus cultivars and four tissues; metabolites are grouped by chemical classes
ACKNOWLEDGEMENTS
This work was funded by the Collectivité Terrotoriale de Corse.
LITERATURE CITED
- Abad-García B, Garmón-Lobato S, Berrueta LA, Gallo B, Vicente F. 2012. On line characterization of 58 phenolic compounds in Citrus fruit juices from Spanish cultivars by high-performance liquid chromatography with photodiode-array detection coupled to electrospray ionization triple quadrupole mass spectrometry. Talanta 99: 213–224. [DOI] [PubMed] [Google Scholar]
- Agati G, Azzarello E, Pollastri S, Tattini M. 2012. Flavonoids as antioxidants in plants: location and functional significance. Plant Science 196: 67–76. [DOI] [PubMed] [Google Scholar]
- Alarcón-Flores MI, Romero-González R, Vidal JLM, Frenich AG. 2013. Multiclass determination of phytochemicals in vegetables and fruits by ultra high performance liquid chromatography coupled to tandem mass spectrometry. Food Chemistry 141: 1120–1129. [DOI] [PubMed] [Google Scholar]
- Barreca D, Bellocco E, Caristi C, Leuzzi U, Gattuso G. 2011a. Distribution of C- and O-glycosyl flavonoids, (3-hydroxy-3-methylglutaryl)glycosyl flavanones and furocoumarins in Citrus aurantium L. juice. Food Chemistry 124: 576–582. [Google Scholar]
- Barreca D, Bellocco E, Caristi C, Leuzzi U, Gattuso G. 2011b. Elucidation of the flavonoid and furocoumarin composition and radical-scavenging activity of green and ripe chinotto (Citrus myrtifolia Raf.) fruit tissues, leaves and seeds. Food Chemistry 129: 1504–1512. [Google Scholar]
- Barreca D, Bellocco E, Caristi C, Leuzzi U, Gattuso G. 2011c. Flavonoid profile and radical-scavenging activity of Mediterranean sweet lemon (Citrus limetta Risso) juice. Food Chemistry 129: 417–422. [DOI] [PubMed] [Google Scholar]
- Barreca D, Bisignano C, Ginestra G, et al. 2013. Polymethoxylated, C- and O-glycosyl flavonoids in tangelo (Citrus reticulata × Citrus paradisi) juice and their influence on antioxidant properties. Food Chemistry 141: 1481–1488. [DOI] [PubMed] [Google Scholar]
- Bourgaud F, Hehn A, Larbat R, et al. 2006. Biosynthesis of coumarins in plants: a major pathway still to be unravelled for cytochrome P450 enzymes. Phytochemistry Reviews 5: 293–308. [Google Scholar]
- Brunetti C, George RM, Tattini M, Field K, Davey MP. 2013. Metabolomics in plant environmental physiology. Journal of Experimental Botany 64: 4011–4020. [DOI] [PubMed] [Google Scholar]
- Chen X, Ji H, Zhang Q, et al. 2008. A rapid method for simultaneous determination of 15 flavonoids in Epimedium using pressurized liquid extraction and ultra-performance liquid chromatography. Journal of Pharmaceutical and Biomedical Analysis 46: 226–235. [DOI] [PubMed] [Google Scholar]
- Cheynier V. 2005. Polyphenols in foods are more complex than often thought. American Journal of Clinical Nutrition 81: 223S–229S. [DOI] [PubMed] [Google Scholar]
- Daniel JJ, Owens DK, McIntosh CA. 2011. Secondary product glucosyltransferase and putative glucosyltransferase expression during Citrus paradisi (c.v. Duncan) growth and development. Phytochemistry 72: 1732–1738. [DOI] [PubMed] [Google Scholar]
- Deguchi A, Ohno S, Hosokawa M, Tatsuzawa F, Doi M. 2013. Endogenous post-transcriptional gene silencing of flavone synthase resulting in high accumulation of anthocyanins in black dahlia cultivars. Planta 237: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Di Donna L, Taverna D, Mazzotti F, et al. 2013. Comprehensive assay of flavanones in citrus juices and beverages by UHPLC–ESI-MS/MS and derivatization chemistry. Food Chemistry 141: 2328–2333. [DOI] [PubMed] [Google Scholar]
- Dugrand A, Olry A, Duval T, Hehn A, Froelicher Y, Bourgaud F. 2013. Coumarin and furanocoumarin quantitation in citrus peel via ultraperformance liquid chromatography coupled with mass spectrometry (UPLC–MS). Journal of Agricultural and Food Chemistry 61: 10677–10684. [DOI] [PubMed] [Google Scholar]
- Fanciullino A-L, Dhuique-Mayer C, Luro F, Casanova J, Morillon R, Ollitrault P. 2006. Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. Journal of Agricultural and Food Chemistry 54: 4397–4406. [DOI] [PubMed] [Google Scholar]
- Froelicher Y, Mouhaya W, Bassene JB, et al. 2011. New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genetics & Genomes 7: 49–61. [Google Scholar]
- Frydman A, Liberman R, Huhman DV, et al. 2013. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. Plant Journal 73: 166–178. [DOI] [PubMed] [Google Scholar]
- Garcia-Lor A, Curk F, Snoussi-Trifa H, et al. 2013. A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Annals of Botany 111: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardana C, Nalin F, Simonetti P. 2008. Evaluation of flavonoids and furanocoumarins from Citrus bergamia (bergamot) juice and identification of new compounds. Molecules 13: 2220–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattuso G, Barreca D, Gargiulli C, Leuzzi U, Caristi C. 2007. Flavonoid composition of Citrus juices. Molecules 12: 1641–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JM. 1996. Peer reviewed: a practical guide to analytical method validation. Analytical Chemistry 68: 305A–309A. [Google Scholar]
- Gustavo González A, Ángeles Herrador M. 2007. A practical guide to analytical method validation, including measurement uncertainty and accuracy profiles. TrAC Trends in Analytical Chemistry 26: 227–238. [Google Scholar]
- Helrich K. 1990. Official methods of analysis of AOAC International. Gaithersburg: AOAC International. [Google Scholar]
- Kaffarnik F, Seidlitz HK, Obermaier J, Sandermann H, Heller W. 2006. Environmental and developmental effects on the biosynthesis of UV-B screening pigments in Scots pine (Pinus sylvestris L.) needles. Plant, Cell and Environment 29: 1484–1491. [DOI] [PubMed] [Google Scholar]
- Kawaii S, Tomono Y, Katase E, Ogawa K, Yano M. 1999. Quantitation of flavonoid constituents in Citrus fruits. Journal of Agricultural and Food Chemistry 47: 3565–3571. [DOI] [PubMed] [Google Scholar]
- Kawaii S, Tomono Y, Katase E, Ogawa K, Yano M. 2000a. Effect of coumarins on HL-60 cell differentiation. Anticancer Research 20: 2505–2512. [PubMed] [Google Scholar]
- Kawaii S, Tomono Y, Katase E, et al. 2000b. Quantitative study of flavonoids in leaves of Citrus plants. Journal of Agricultural and Food Chemistry 48: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Koyama K, Ikeda H, Poudel PR, Goto-Yamamoto N. 2012. Light quality affects flavonoid biosynthesis in young berries of Cabernet Sauvignon grape. Phytochemistry 78: 54–64. [DOI] [PubMed] [Google Scholar]
- Loizzo MR, Tundis R, Bonesi M, et al. 2012. Evaluation of Citrus aurantifolia peel and leaves extracts for their chemical composition, antioxidant and anti-cholinesterase activities. Journal of the Science of Food and Agriculture 92: 2960–2967. [DOI] [PubMed] [Google Scholar]
- Medina-Remon A, Tulipani S, Rotches-Ribalta M, Mata-Bilbao MD, Andres-Lacueva C, Lamuela-Raventos RM. 2011. A fast method coupling ultrahigh performance liquid chromatography with diode array detection for flavonoid quantification in citrus fruit extracts. Journal of Agricultural and Food Chemistry 59: 6353–6359. [DOI] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. 2009. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiology 150: 924–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith J. 1965. Evaporation and environment. Symposia of the Society for Experimental Biology and Medicine 19: 205–234. [PubMed] [Google Scholar]
- Nogata Y, Ohta H, Sumida T, Sekiya K. 2003. Effect of extraction method on the concentrations of selected bioactive compounds in mandarin juice. Journal of Agricultural and Food Chemistry 51: 7346–7351. [DOI] [PubMed] [Google Scholar]
- Nogata Y, Sakamoto K, Shiratsuchi H, Ishii T, Yano M, Ohta H. 2006. Flavonoid composition of fruit tissues of citrus species. Bioscience, Biotechnology, and Biochemistry 70: 178–192. [DOI] [PubMed] [Google Scholar]
- Pollastri S, Tattini M. 2011. Flavonols: old compounds for old roles. Annals of Botany 108: 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokudina E, Havlíček L, Al-Maharik N, Lapčík O, Strnad M, Gruz J. 2012. Rapid UPLC–ESI–MS/MS method for the analysis of isoflavonoids and other phenylpropanoids. Journal of Food Composition and Analysis 26: 36–42. [Google Scholar]
- Rapisarda P, Fanella F, Maccarone E. 2000. Reliability of analytical methods for determining anthocyanins in blood orange juices. Journal of Agricultural and Food Chemistry 48: 2249–2252. [DOI] [PubMed] [Google Scholar]
- Ryan KG, Swinny EE, Markham KR, Winefield C. 2002. Flavonoid gene expression and UV photoprotection in transgenic and mutant petunia leaves. Phytochemistry 59: 23–32. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Brugliera F. 2013. Flower colour and cytochromes P450. Philosophical Transactions of the Royal Society. Series B, Biological Sciences 368: 20120432 http://dx.doi.org/10.1098/rstb.2012.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR. 2013. Shikimate and phenylalanine biosynthesis in the green lineage. Frontiers in Plant Science 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D. 2006. Significance of flavonoids in plant resistance: a review. Environmental Chemistry Letters 4: 147–157. [Google Scholar]
- VanderMolen KM, Cech NB, Paine MF, Oberlies NH. 2013. Rapid quantitation of furanocoumarins and flavonoids in grapefruit juice using ultra-performance liquid chromatography. Phytochemical Analysis 24: 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialart G, Hehn A, Olry A, et al. 2012. A 2-oxoglutarate-dependent dioxygenase from Ruta graveolens L. exhibits p-coumaroyl CoA 2′-hydroxylase activity (C2′H): a missing step in the synthesis of umbelliferone in plants. Plant Journal 70: 460–470. [DOI] [PubMed] [Google Scholar]
- Widelski J, Popova M, Graikou K, Glowniak K, Chinou I. 2009. Coumarins from Angelica lucida L. – antibacterial activities. Molecules 14: 2729–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2002. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology 5: 218–223. [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature Biotechnology 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.