Abstract
Nonstructural protein 1 (NS1) of influenza A virus plays a central role in virus replication and blockade of the host innate immune response, and is therefore being considered as a potential therapeutic target. The primary function of NS1 is to dampen the host interferon (IFN) response through several distinct molecular mechanisms that are triggered by interactions with dsRNA or specific cellular proteins. Sequestration of dsRNA by NS1 results in inhibition of the 2’-5’ oligoadenylate synthetase/RNase L antiviral pathway, and also inhibition of dsRNA-dependent signaling required for new IFN production. Binding of NS1 to the E3 ubiquitin ligase TRIM25 prevents activation of RIG-I signaling and subsequent IFN induction. Cellular RNA processing is also targeted by NS1, through recognition of cleavage and polyadenylation specificity factor 30 (CPSF30), leading to inhibition of IFN- mRNA processing as well as that of other cellular mRNAs. In addition NS1 binds to and inhibits cellular protein kinase R (PKR), thus blocking an important arm of the IFN system. Many additional proteins have been reported to interact with NS1, either directly or indirectly, which may serve its anti-IFN and additional functions, including the regulation of viral and host gene expression, signaling pathways and viral pathogenesis. Many of these interactions are potential targets for small-molecule intervention. Structural, biochemical and functional studies have resulted in hypotheses for drug discovery approaches that are beginning to bear experimental fruit, such as targeting the dsRNA-NS1 interaction, which could lead to restoration of innate immune function and inhibition of virus replication. This review describes biochemical, cell-based and nucleic acid-based approaches to identifying NS1 antagonists.
1. NS1 biology in the context of drug discovery
Non-structural protein 1 (NS1) of influenza A virus has attracted much attention for its role in modifying the host innate immune response and controlling virus replication. NS1 is encoded by viral segment 8, which also encodes the viral nuclear export protein, NEP. NS1 has come under scrutiny as a potential target for antiviral drug discovery based on its structure, activities, genetics, and overall importance in virus replication and pathogenesis. It is a highly conserved protein of 230-237 amino acids that is produced in abundant levels throughout infection. Structurally, NS1 consists of two distinct domains, each of which contributes to homodimer formation and function. The RNA binding domain (RBD) encompasses amino acids 1-73. It binds nonspecifically to RNA and is also required for interaction with specific cellular proteins. The C-terminal “effector” domain (ED) includes amino acids 86–230/237 and also interacts with a variety of cellular proteins. Together both domains contribute to the highly multifunctional nature of NS1 (Das et al., 2010; Garcia-Sastre, 2011; Hale et al., 2008b; Krug and Aramini, 2009).
The number of cellular proteins reported to associate with NS1 has grown very large (Table 1), although not all interactions have been proven to be direct, and there are (and are likely to be) strain-specific differences for some interactions. Primary among the functions of NS1 is inhibition of the host interferon (IFN) system, which is accomplished through several molecular mechanisms. Additional effects include regulation of viral RNA and protein synthesis and viral mRNA splicing, and activation of the PI3K pathway (Ayllon et al., 2012; Ehrhardt and Ludwig, 2009; Garcia-Sastre, 2011; Hale et al., 2008b). Therefore, it is thought that chemical inhibition of NS1 might exert pleiotropic effects that enhance innate immunity and significantly limit virus replication mechanisms in humans.
Table 1.
Host-cell proteins that interact with the influenza A virus NS1 protein.
| *interacting protein | Process affected by NS1 | Interaction domain | Reference |
|---|---|---|---|
| NS1-I | Unknown | Unknown | (Wolff et al., 1996) |
| CPSF30 | RNA processing | ED | (Nemeroff et al., 1998) |
| NS1-BP | **RNA splicing | Unknown | (Wolff et al., 1998) |
| PABII | RNA processing | ED | (Chen et al., 1999) |
| Staufen | Unknown | Unknown | (Falcon et al., 1999) |
| eIF4GI | Viral translation | aa 1-113 | (Aragon et al., 2000) |
| PABPI | Viral translation | RBD | (Burgui et al., 2003) |
| p85- | PI3K activation | ED (aa Y89) | (Hale et al., 2006) |
| PKR | Protein synthesis | ED (aa 123-127) | (Li et al., 2006) |
| PACT | Unknown | Unknown | (Li et al., 2006) |
| NXF1, Rael, E1B-AP5 | RNA export | RBD and ED | (Satterly et al., 2007) |
| p15 | RNA export | ED | (Satterly et al., 2007) |
| Importin | NS1 nuclear targeting | RBD and NLS2 | (Melen et al., 2007) |
| Crk and CrkL | PI3K activation | SH3 binding domain | (Heikkinen et al., 2008) |
| RIG-I | IFN induction | Likely indirect through TRIM25 | (Mibayashi et al., 2007) |
| (Gack et al., 2009) | |||
| nucleolin, B23 and fibrillarin | Nucleolar targeting | RBD | (Murayama et al., 2007) |
| NLS1,NLS2/NoLS | (Melen et al., 2012) | ||
| TRIM25, Riplet | RIG-I activation/IFN induction | RBD (aa R38/K41) and ED (aa E96/E97) | (Gack et al., 2009) |
| (Rajsbaum et al., 2012) | |||
| Viral Pol complex | **Viral RNA replication | Unknown | (Kuo and Krug, 2009) |
| Gas8 | Unknown | Unknown | (Zhao et al., 2009) |
| Akt | Akt pathway activation | RBD and ED | (Matsuda et al., 2010) |
| p53 | Apoptosis | Unknown | (Wang et al., 2010) |
| Scribble, Dlg1, MAGI-1, MAGI-2, MAGI-3 | Tight junctions | PDZ-binding motif | (Golebiewski et al., 2011) |
| PARP10 | **Cell cycle control | Unknown | (Yu et al., 2011) |
| RIL, c-Src | c-Src activation | PDZ-binding motif SHB domain | (Bavagnoli et al., 2011) |
| Hsp90 | Apoptosis | Unknown | (Zhang et al., 2011) |
| Viral nucleoprotein (NP) | Viral RNA replication | RBD (aa R38/K41) | (Robb et al., 2011) |
| (Kuo and Krug, 2009) | |||
| PDZ-containing proteins | Viral pathogenesis | PDZ-binding motif | (Javier and Rice, 2011) |
| NOLC1 | Nucleolar/coiled body phosphoprotein | ED | (Zhu et al., 2012) |
| RAP55 | P-bodies, stress granules | RBD (aa R38/R41) ED (aa I123, M124, K126, and N127) |
(Mok et al., 2012) |
| IKK | NF- B pathway | ED | (Gao et al., 2012) |
| hPAF1C | hPAF1C-dependent transcriptional elongation | H3N2 C-term ARSK | (Marazzi et al., 2012) |
| RNA helicase A | RNA replication | Unknown | (Lin et al., 2012) |
| -tubulin | **Apoptosis | RBD | (Han et al., 2012) |
| hGBP1 | anti-viral state | ED (aa123-144) | (Zhu et al., 2013) |
Direct and indirect interactions are listed
Proposed process affected by NS1 through interaction with this factor; (aa): Amino acid residues required for interaction; ED: Effector domain; RBD: RNA-binding domain; NLS/NoLS: nuclear localization signal/nucleolar localization signal; SHB domain: Src homology binding domain.
Importantly, genetic analyses of NS1 in the context of infected cells and animals have demonstrated that virus replication, spread, and pathogenesis are very dependent on the function of this protein. As part of efforts to develop a live, attenuated influenza vaccine carrying deletions of NS1 coding sequences, it has been demonstrated that viruses lacking NS1 function have highly diminished replication and pathogenic capacity in a variety of animal models. (Falcon et al., 2005; Garcia-Sastre et al., 1998; Richt and Garcia-Sastre, 2009; Talon et al., 2000b; Zhou et al., 2010). These findings satisfy an important criterion for an anti-influenza virus target, since drugs that inhibit the action of the target must be able to slow virus production and/or pathogenesis as a consequence. They also suggest that treatment of humans with NS1 inhibitors would not result in toxicity due to dysregulation of the innate immune response.
2. Targeting the IFN-antagonist functions of NS1
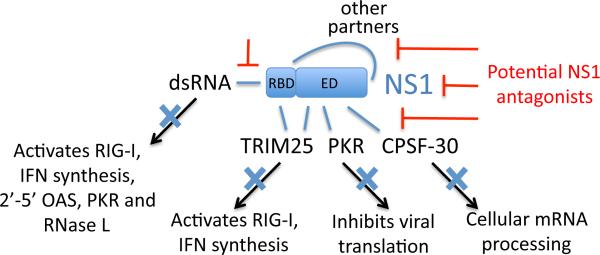
Several fascinating functions for NS1 have been described that form the basis for its anti-IFN activity, and most therapeutic targeting strategies include disruption of these functions in order to relieve viral inhibition of the innate immune response. As an RNA-binding protein, NS1 can interact with a variety of RNA species, including double-stranded RNA (dsRNA), the 3’ poly-A tail of mRNAs, the U6 snRNA and negative-sense viral RNA (Hatada and Fukuda, 1992; Hatada et al., 1997; Hatada et al., 1992; Qiu and Krug, 1994; Qiu et al., 1995). Binding to dsRNA inhibits the 2’-5’ oligoadenylate synthetase/RNase L pathway for degradation of viral RNAs, thereby limiting a major arm of the IFN system. It also dampens the activity of transcription factor pathways that depend on dsRNA, thus reducing the expression of IFN- in infected cells (Ludwig et al., 2002; Min and Krug, 2006; Talon et al., 2000a; Wang et al., 2000). NS1 also inhibits activation of RIG-I, a pattern-recognition receptor that senses viral RNA and in turn triggers the IFN response (Gack et al., 2009; Guo et al., 2007; Inn et al., 2011; Opitz et al., 2007; Pichlmair et al., 2006). NS1 inhibits RIG-I by binding to the ubiquitin ligases TRIM25 and Riplet that are required for RIG-I activation (Gack et al., 2009; Inn et al., 2011; Rajsbaum et al., 2012).
NS1 also modifies cellular pre-mRNA processing, including 3′-end formation, by binding to the 30-kDa subunit of cleavage and polyadenylation specificity factor (CPSF30), which results in inhibition of IFN- mRNA processing, among other cellular mRNAs (Krug et al., 2003; Nemeroff et al., 1998; Noah et al., 2003; Twu et al., 2006). NS1 also binds directly to and inhibits activation of cellular PKR, an important effector of the IFN system (Bergmann et al., 2000; Min et al., 2007; Pindel and Sadler, 2011), and it inhibits nuclear non-viral RNA export by associating with several cellular proteins that mediate RNA export, including NXF1/TAP, p15/NXT, Rae1/mrnp41, and E1B-AP5 (Fortes et al., 1994; Qiu and Krug, 1994; Satterly et al., 2007).
3. Structural basis for small-molecule inhibition of NS1
X-ray crystallographic and NMR studies of NS1 have revealed structures of the individual RBD and ED homodimers, as well as the RBD homodimer in association with dsRNA (Das et al., 2010). A crystallographic study of full-length NS1 from the highly pathogenic avian H5N1 strain has also been reported (Bornholdt and Prasad, 2008). The RBD exists in a unique six-helical symmetric head-to-tail homodimer structure (Cheng et al., 2009; Chien et al., 1997; Liu et al., 1997; Yin et al., 2007). Arginine 38 within this domain interacts directly with dsRNA, and both members of the dimer contribute to the RNA-binding architecture. Accordingly, mutation of Arg38 results in a highly attenuated virus, and NS1 protein carrying this mutation is defective in dsRNA binding in vitro. Dimerization itself is also required for dsRNA binding activity (Min and Krug, 2006; Wang et al., 1999). Thus, the dsRNA-NS1 interaction is a potential target for small-molecule inhibition, either by disruption of the dsRNA-NS1 complex or by interfering with homodimer stability (Krug and Aramini, 2009). Such inhibitors would be expected to restore dsRNA-dependent antiviral functions such as activation of the 2’-5’ oligoadenylate synthetase/RNase L and PKR pathways, and RIG-I mediated activation of the IFN response. As new interactions between the RBD and specific cellular proteins are explored, additional opportunities for small-molecule intervention may become apparent through structural analysis.
The isolated ED of NS1 also forms a homodimer in solution, with each subunit containing a novel -helix -crescent fold. However, structural studies of the ED from different influenza strains have yielded conflicting results regarding the architecture of the dimer interface (Bornholdt and Prasad, 2006; Bornholdt and Prasad, 2008; Hale et al., 2008a; Kerry et al., 2011; Xia et al., 2009). Tryptophan 187 (W187) in the ED is required for dimer formation, and mutation at this position resulted in exclusively monomeric species (Aramini et al., 2011; Hale et al., 2008a; Xia and Robertus, 2010). Interestingly, the interface responsible for ED dimer formation includes amino acid residues that help form a hydrophobic pocket for binding to CPSF30. Cellular expression of a small fragment of CPSF30 sufficient to bind NS1 was also shown to inhibit virus replication and increase production of IFN-β mRNA, presumably through a dominant negative mechanism (Aramini et al., 2011; Das et al., 2008; Twu et al., 2006). It was therefore proposed that the hydrophobic CPSF30-binding pocket in NS1 is an attractive target for drug discovery (Das et al., 2010; Krug and Aramini, 2009; Twu et al., 2006).
An NS1 protein with a W187Y mutation in the ED also retained the ability to bind CPSF30, and the structure of its CPSF30 binding pocket was almost identical to that of wild-type ED, suggesting that this non-dimerized mutant could provide an efficient platform for drug discovery targeting the NS1-CPSF30 interaction (Xia and Robertus, 2010). Structural and biochemical definition of the dimer interface and the CPSF30 binding pocket of NS1 are therefore likely to lead to new NS1 inhibitors through small-molecule screening or rational design. The ED has also been co-crystallized with the inter-SH2 (coiled-coil) domain of p85beta subunit of PI3K, which may lead to proposals for inhibitor design (Hale et al., 2010).
Based on structural data from the H5N1 A/Vietnam/1203/2004 strain (Bornholdt and Prasad, 2008) and the large number of virus isolates whose sequence has been determined, Darapaneni et al. analyzed NS1 for its most highly conserved amino acid residues, then used the Q-SiteFinder binding site prediction algorithm (Laurie and Jackson, 2005) to identify conserved pockets that might function as protein-protein interaction sites. Several pockets were identified and proposed as potential sites for small-molecule inhibition of NS1 function (Darapaneni et al., 2009).
4. In vitro platforms for NS1-targeted drug discovery
Cho et al. reported the development of a high-throughput screen (HTS) assay to identify NS1 inhibitors, in which the RNA-binding activity of NS1 from A/Udorn/72 was targeted using a sensitive fluorescence polarization (FP) assay (Cho et al., 2012). A filter-binding assay previously reported by Chien et al. (Chien et al., 2004) was adapted to detect displacement of prebound, labeled dsRNA. The assay measured FP changes in the presence of small-molecules that inhibit the interaction between NS1 and a fluorescein-labeled 16-mer dsRNA, resulting in a decrease in FP that correlated with the efficiency of displacement. Importantly, a mutant NS1 containing an Arg38 to Ala substitution, which abolishes binding to dsRNA, failed to yield an FP signal with the probe dsRNA, confirming the specificity of the screening assay for the dsRNA-NS1 interaction.
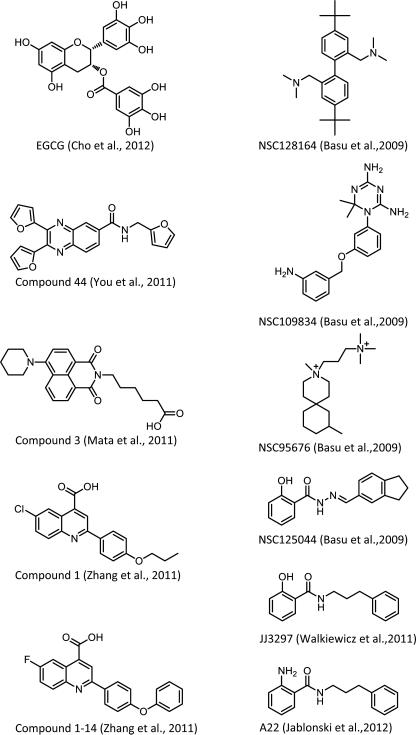
A pilot screen using 466 compounds from the NIH clinical collection demonstrated high signal to noise ratios and Z’ scores in the range of 0.71 – 0.82. Hits were validated by re-screening, followed by counterscreening to rule out direct effects on the dsRNA probe. Four of 6 hit compounds showed concentration-dependent inhibition of binding. One of these, epigallocatechin gallate (EGCG, see Figure 1), potently inhibited binding (IC50 0.29 M). Interestingly EGCG had previously been reported as an inhibitor of influenza virus infection, but without identification of its target (Furuta et al., 2007; Nakayama et al., 1993; Song et al., 2005). Based on its structure, a second study explored a quinoxaline scaffold, which shares structural similarities with EGCG (You et al., 2011). A library of 46 compounds was synthesized that retained the quinoxaline core and several of these showed significant inhibition of RNA-NS1 binding. One compound was tested for antiviral activity against A/Udorn/72 in an MDCK cell assay at 30 and 60 M and inhibited replication by 10-fold over a time course of infection (see Figure 1, Compound 44), although cytotoxicity data were not reported.
Figure 1.
Major cellular antiviral pathways affected by the influenza virus NS1 protein, and potential points of intervention by NS1 antagonists. The roles of dsRNA, TRIM25, PKR and CPSF-30 are described in the text. Blue Xs indicate inhibition of cellular antiviral pathways by NS1. NS1 antagonists (red) could act by binding directly to NS1 or by interfering with interactions between NS1 and its cellular targets, or by other mechanisms. RBD: NS1 RNA binding domain; ED: NS1 effector domain.
Maroto et al. also reported the development of a HTS assay that targeted the RNA-NS1 interaction. Microplate wells were precoated with His-tagged NS1, and the binding of a 35S-labeled RNA probe was measured in the presence or absence of small-molecules (Maroto et al., 2008). A 240-nucleotide negative-polarity RNA corresponding to a portion of influenza segment 8 was used to form the target complex. It had been shown previously that this RNA interacts with NS1 in a manner that is not competed using a 1000-fold excess of nonspecific RNA (Marion et al., 1997). Assay optimization resulted in a signal-to-noise ratio of 16 and a Z’ score of 0.78. A screen was carried out with a Merck collection of small-molecules composed of 27,520 mixtures, each containing 10 compounds, using a combination of 96-well and 384-well formats. A cut-off of 40% inhibition of binding was chosen, and a counterscreen was conducted using an RNA-Staufen complex as the target, to rule out nonspecific inhibitors of RNA-protein interactions. This resulted in 13 NS1-specific hits that were subsequently deconvoluted, but their structures were not reported. Five of the hits were reported to have IC50s in the range of 0.12-0.85 M. The antiviral activity of three compounds was qualitatively demonstrated using MDCK cells and virus A/Victoria/3/75, but the SI appeared to be quite low (see Maroto et al., Figure 7).
5. Cell-based platforms
Nuclear NS1 directly inhibits mRNA processing and export by interacting with specific host factors, resulting in changes in cellular gene expression (Nemeroff et al., 1998; Qian et al., 1994; Qiu and Krug, 1994; Satterly et al., 2007; Wolff et al., 1998). An important consequence of this regulation is a decrease in mature cytoplasmic IFN mRNA and other mRNAs encoding antiviral factors. Accordingly, Mata et al. targeted the ability of NS1 to inhibit host gene expression. A CMV promoter-driven luciferase reporter was used in cells co-transfected with an NS1 expression construct, which triggered a 95% reduction in luciferase expression. Cells were screened with 5 M of 200,000 compounds assembled from ChemDiv, ChemBridge, ComGenex and TimTek libraries. Those that restored luciferase expression were evaluated for antiviral activity against A/WSN/1933 in immortalized human bronchial epithelial cells (HBECs). Eight structural classes of compounds were identified among the hits, and one of the most active was a naphthalimide compound that was explored further, including analysis of analogs (Mata et al., 2011).
Non-cytotoxic concentrations of one analog (see Figure 1, Mata et al. Compound 3) reduced viral titers from 103-106-fold, depending on the influenza virus strain and dose, and also significantly relieved virus-induced retention of poly(A) RNA in the nucleus. A/WSN/1933, A/Texas/36/91 and A/Brevig/Mission/1/1918 were used as challenge virus in these studies, and it will be interesting to determine if the observed differences in compound sensitivity are due to genetic differences in NS1 between the different strains. The selective index (SI) for this compound was 31 for MDCK cells infected with A/WSN/1933. Interestingly, compound 3 also had activity against vesicular stomatitis virus (VSV), suggesting a common, cellular target. To explore this possibility, gene expression profiles were determined in the presence and absence of the compound, and this analysis suggested the possible involvement of the mTORC1 pathway. REDD1, an inhibitor of this pathway, was found to be induced at the mRNA and protein levels in cells treated with compound 3. Significantly, REDD1−/− mouse embryonic fibroblasts were observed to be hypersensitive to influenza virus and VSV infection compared to REDD1+/+ cells, and the inhibitor had no effect on virus replication in REDD−/− cells. Likewise, the inhibitor had no effect on viral protein expression in TSC2-knockout cells, further solidifying the chemical-genetic interactions between the mTORC1 pathway and compound 3. Together these data defined REDD1 as a host defense factor and demonstrated that chemical induction of REDD1 can overcome the effects of NS1 on host gene expression.
A second hit (see Figure 1, Compound 1, Zhang et al) from the screen conducted by Mata et al. is structurally related to brequinar, a quinolone carboxylic acid known to inhibit dihydroorotate dehydrogenase (DHODH) (Vyas and Ghate, 2011; Zhang et al., 2012). Inhibitors of DHODH have been shown to have activity against a variety of DNA and RNA viruses including influenza (Hoffmann et al., 2011). Compound 1 at 10 M inhibited A/WSN/1933 in MDCK cells by a factor of 104 with no cytotoxicity. Indeed, novel analogs of the compound 1 inhibited DHODH in vitro, (Zhang et al., 2012). Analog 1-14 (Figure 1) also prevented the NS1-mediated blockade of host cell RNA export usually seen in influenza virus-infected cells. This result is consistent with the idea that inhibition of DHODH causes reduced viral RNA synthesis through its effects on pyrimidine synthesis, leading to decreased NS1 protein production and increased export of host cell nuclear mRNAs. Interestingly however, inhibition of DHODH also reversed NS1-dependent effects on mRNA export in uninfected cells containing an NS1 expression construct. This finding indicated a novel relationship between the pyrimidine biosynthetic pathway and the regulation of mRNA export by NS1. Accordingly, RNAs encoding members of the cellular HIF1-pathway, part of the antiviral response, were found enriched among those released from the mRNA export block by DHODH inhibition. In addition, inhibition of DHODH triggered accumulation of the mRNA export protein NXF1 in cells expressing NS1.
Basu et al. employed a yeast-based phenotypic assay to identify specific NS1 antagonists (Basu et al., 2009). Expression of NS1 from a galactose-inducible promoter in S. cerevisiae produced a pronounced slow growth phenotype, as had been reported previously (Ward et al., 1994). This was exploited to screen for small-molecules that could restore growth by direct or indirect inhibition of NS1 function. Nine positive compounds were identified from the National Cancer Institute Diversity Set library of ~2,000 compounds, four of which also inhibited influenza virus replication in MDCK cells, but not respiratory syncytial virus (RSV) replication (Figure 1, see NSC compounds). Depending on the influenza strain (A/Hong Kong/19/68, A/WSN/1933, or A/PR/8/34) and the inhibitor used, EC50s were found in the range of 2 – 20 M, with SI values ranging from 12 -- 200. It will be important to determine if the variations in strain sensitivity are due to differences in NS1 sequence. Interestingly, in cells infected with WT A/PR/8/34, all four inhibitors restored levels of IFN- mRNA, comparable to those seen when cells were infected with a viral NS1 deletion mutant, indicating that the compounds significantly reversed the anti-IFN effects of NS1. In addition, the compounds had no effect on IFN mRNA levels in uninfected cells, demonstrating that they are not direct inducers of IFN, and that virus infection was required for the activity of the compounds. To confirm the role of NS1 in compound sensitivity, cells were co-transfected with an NS1 expression plasmid plus an IFN- reporter construct, and the four compounds were able to efficiently reverse the effects of NS1 on dsRNA-dependent IFN- promoter activation.
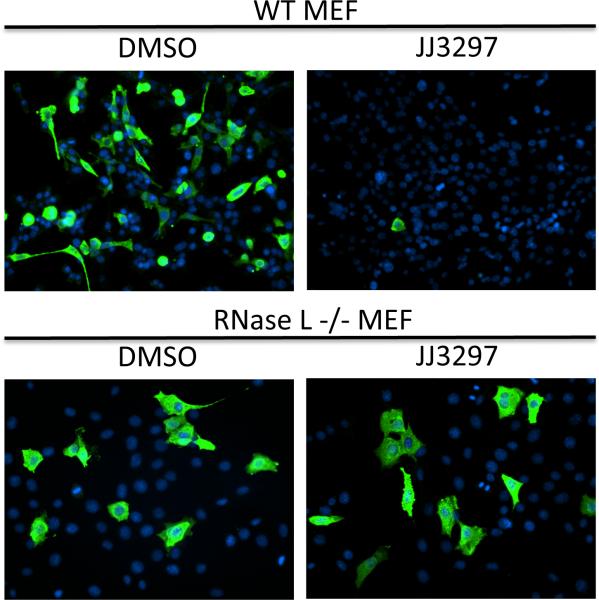
In subsequent studies, compound NSC125044 from Basu et al. was analyzed further and an initial structure-activity relationship study was conducted. Non-toxic concentrations of analog JJ3297 inhibited A/PR/8/34 replication with an EC50 of 0.8 M and a maximal inhibition of >1000-fold. This compound retained the ability of the parental NSC125044 to restore IFN- expression in infected cells. Moreover, JJ3297 facilitated the induction of an IFN-like antiviral state in cells infected at a low MOI, resulting in increased resistance to subsequent challenge with vesicular stomatitis virus (VSV). The activity of JJ3297 absolutely required the function of cellular RNase L, indicating that an intact IFN system is essential for function of the compound (see Figure 2) (Walkiewicz et al., 2011). Additional medicinal chemistry development of JJ3297 resulted in compound A22 (Figure 1), with an EC50 of ~50 nM (Jablonski et al., 2012). Together, these findings indicate that this class of compounds acts as an NS1 antagonist. It will be interesting to determine if the compounds act directly on the NS1 protein or perturb an aspect of NS1-related pathways.
Figure 2.
Compounds that antagonize the influenza virus NS1 protein in biochemical or cell-based assays. Biochemical assays were used identify inhibitors of the dsRNA-NS1 interaction (Cho et al., 2012; You et al., 2011). Cell-based assays identified compounds that bypass NS1 function and relieve the block to cellular RNA export (Mata et al., 2011; Zhang et al., 2012). An NS1-dependent assay in yeast identified compounds that specifically reverse NS1 activity in mammalian cells (Basu et al., 2009; Jablonski et al., 2012; Walkiewicz et al., 2011).
6. Nucleic acid-based NS1 inhibitors
Nucleic acid-based approaches are also being to be used to target NS1. Wu et al. reported antisense RNA targeting of the avian H5N1 NS1 sequence using three selected oligomers of 18 nucleotides (Wu et al., 2008), building on previous reports of siRNA or antisense approaches that effectively controlled influenza A replication in vitro or in vivo by targeting other viral genes (DeVincenzo, 2012; Wong et al., 2010). In chicken embryo fibroblast cells, 2 M of the transfected NS1-directed oligonucleotides reduced viral replication to 2.4–7.3 HA units, compared with titers of 52.4 and 55.9 in control transfections. Importantly, these oligonucleotides were effective in protecting SPF chickens from H5N1-induced clinical symptoms and mortality. FITC-labeled oligonucleotides delivered intranasally were shown to be efficiently taken up in the lung and observed 24 hours after administration, and a mixture of three unlabeled oligonucleotides protected 87.5% of infected birds up to 14 days post-infection. This correlated with a 5- to 7-fold reduction in lung viral titer at 24 hours post infection. However, gastrointestinal involvement in these H5N1 infections was not addressed. It is interesting to note that a substantial protection rate was associated with this modest decrease in lung viral titer (although lung titers at later time points were not measured). This could be due to an amplified innate immune response when decreased amounts of NS1 are present in the animal.
Rajput et al. observed a 60% inhibition of NS1 RNA with either of two NS1-specific siRNAs in co-transfection experiments using an NS1 expression construct in MDCK cells (Rajput et al., 2012). Remarkably, intravenous administration of increasing doses of NS1-specific siRNA in BALB/c mice challenged with 2×106 p.f.u. of A/PR/8/34 resulted in a greater than 6-log decrease in lung viral titer, assayed two days postinfection. This corresponded to a 92% decrease in NS1 RNA and a 90% decrease in NS1 protein in lung lysates. Bronchoalveolar lavage fluid showed siRNA dose-dependent increases in IL-1 , IFN- 1 and IFN- compared with untreated controls, and 15- to 20-fold decreases in IFN- and TNF- . The large decreases in IFN- and TNF- are likely a reflection of a decreased cytokine response due to significantly lower viral replication in the lungs. In a survival study, the siRNA protected 100% of mice under conditions that were lethal to 60% of the mice over a 22-day period following virus challenge; however, the time course of the lethal response was unusually long in the control animals.
7. Conclusions and ongoing challenges
Considering the central role of NS1 in influenza virus replication and pathogenesis, potent inhibitors of this protein may prove to have clinical utility, either alone or in combination with other antiviral drugs such as the neuraminidase inhibitors (Govorkova and Webster, 2010). The field of anti-NS1 inhibitors is still in its infancy, but the methodological, structural and biochemical underpinnings required to drive further discoveries toward drug development are in place and continue to expand. More biochemical small-molecule screens are needed to add lead compounds to the pipeline. As additional structural data reveal details of novel NS1-protein interactions, new potential ligand-binding pockets can be targeted for drug discovery. Cell-based screens are also likely to lead to novel chemical-genetic interactions that can be exploited for therapeutic development. Use of animal models to demonstrate antiviral efficacy will be an important next step to establish in vivo proof-of-concept for targeting NS1
Highlights.
The influenza virus NS1 protein dampens the host IFN response and facilitates viral replication.
The central role of NS1 in replication and spread make it an attractive antiviral target.
Several anti-IFN mechanisms have been elucidated, as well as the 3-dimensional structure of NS1.
Interactions between NS1 and dsRNA or a variety of protein partners could be targeted for small-molecule intervention.
Biochemical, cell-based and nucleic acid-based approaches are being applied to identify compounds that inhibit NS1.
Figure 3.
JJ3297 activity depends on an intact interferon system. The antiviral activity of JJ3297 was tested against influenza virus A/PR/8/34 in wild-type and RNase L−/− murine embryo fibroblasts (MEFs). Wild-type (A) or RNase L−/− (B) MEFs were infected at an MOI of 0.1 and treated with 5 μM JJ3297 or 1% DMSO. After 48 h the cells were fixed and stained for the viral nucleoprotein NP, a marker for virus replication. DAPI was used to visualize cell nuclei. There was a drastic effect on virus replication in WT cells, but no effect on RNase L−/− cells was observed, indicating a strong requirement for RNase L or an upstream component of the IFN system to mediate the effects of this compound (Walkiewicz et al., 2011).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J, Nieto A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol. 2000;20:6259–68. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, Xiao R, Krug RM, Montelione GT. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J Biol Chem. 2011;286:26050–60. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllon J, Garcia-Sastre A, Hale BG. Influenza A viruses and PI3K: are there time, place and manner restrictions? Virulence. 2012;3:411–4. doi: 10.4161/viru.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DA. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol. 2009;83:1881–91. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavagnoli L, Dundon WG, Garbelli A, Zecchin B, Milani A, Parakkal G, Baldanti F, Paolucci S, Volmer R, Tu Y, Wu C, Capua I, Maga G. The PDZ-ligand and Src homology type 3 domains of epidemic avian influenza virus NS1 protein modulate human Src kinase activity during viral infection. PLoS One. 2011;6:e27789. doi: 10.1371/journal.pone.0027789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. Journal of virology. 2000;74:6203–6. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–60. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–8. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgui I, Aragon T, Ortin J, Nieto A. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. The Journal of general virology. 2003;84:3263–74. doi: 10.1099/vir.0.19487-0. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. The EMBO journal. 1999;18:2273–83. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–95. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–5. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–62. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Xia S, Ma LC, Robertus J, Krug RM, Anslyn EV, Montelione GT, Ellington AD. Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay. Journal of biomolecular screening. 2012;17:448–59. doi: 10.1177/1087057111431488. [DOI] [PubMed] [Google Scholar]
- Darapaneni V, Prabhaker VK, Kukol A. Large-scale analysis of influenza A virus sequences reveals potential drug target sites of non-structural proteins. J Gen Virol. 2009;90:2124–33. doi: 10.1099/vir.0.011270-0. [DOI] [PubMed] [Google Scholar]
- Das K, Aramini JM, Ma LC, Krug RM, Arnold E. Structures of influenza A proteins and insights into antiviral drug targets. Nat Struct Mol Biol. 2010;17:530–8. doi: 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, Krug RM, Montelione GT. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A. 2008;105:13093–8. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVincenzo JP. The promise, pitfalls and progress of RNA-interference-based antiviral therapy for respiratory viruses. Antivir Ther. 2012;17:213–25. doi: 10.3851/IMP2064. [DOI] [PubMed] [Google Scholar]
- Ehrhardt C, Ludwig S. A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cellular microbiology. 2009;11:863–71. doi: 10.1111/j.1462-5822.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon AM, Fernandez-Sesma A, Nakaya Y, Moran TM, Ortin J, Garcia-Sastre A. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J Gen Virol. 2005;86:2817–21. doi: 10.1099/vir.0.80991-0. [DOI] [PubMed] [Google Scholar]
- Falcon AM, Fortes P, Marion RM, Beloso A, Ortin J. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 1999;27:2241–7. doi: 10.1093/nar/27.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. Embo J. 1994;13:704–12. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T, Hirooka Y, Abe A, Sugata Y, Ueda M, Murakami K, Suzuki T, Tanaka K, Kan T. Concise synthesis of dideoxy-epigallocatechin gallate (DO-EGCG) and evaluation of its anti-influenza virus activity. Bioorganic & medicinal chemistry letters. 2007;17:3095–8. doi: 10.1016/j.bmcl.2007.03.041. [DOI] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, Wang Q, Kang T, Chen S, Huang W. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol. 2012;14:1849–66. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–8. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–30. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Golebiewski L, Liu H, Javier RT, Rice AP. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J Virol. 2011;85:10639–48. doi: 10.1128/JVI.05070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova EA, Webster RG. Combination chemotherapy for influenza. Viruses. 2010;2:1510–29. doi: 10.3390/v2081510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–9. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- Hale BG, Barclay WS, Randall RE, Russell RJ. Structure of an avian influenza A virus NS1 protein effector domain. Virology. 2008a;378:1–5. doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci U S A. 2006;103:14194–9. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ, Randall RE, Russell RJ. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc Natl Acad Sci U S A. 2010;107:1954–9. doi: 10.1073/pnas.0910715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008b;89:2359–76. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Han X, Li Z, Chen H, Wang H, Mei L, Wu S, Zhang T, Liu B, Lin X. Influenza virus A/Beijing/501/2009(H1N1) NS1 interacts with beta-tubulin and induces disruption of the microtubule network and apoptosis on A549 cells. PLoS One. 2012;7:e48340. doi: 10.1371/journal.pone.0048340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73(Pt 12):3325–9. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J Gen Virol. 1997;78(Pt 5):1059–63. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- Hatada E, Takizawa T, Fukuda R. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J Gen Virol. 1992;73(Pt 1):17–25. doi: 10.1099/0022-1317-73-1-17. [DOI] [PubMed] [Google Scholar]
- Heikkinen LS, Kazlauskas A, Melen K, Wagner R, Ziegler T, Julkunen I, Saksela K. Avian and 1918 Spanish influenza a virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling. J Biol Chem. 2008;283:5719–27. doi: 10.1074/jbc.M707195200. [DOI] [PubMed] [Google Scholar]
- Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A. 2011;108:5777–82. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Molecular cell. 2011;41:354–65. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski JJ, Basu D, Engel DA, Geysen HM. Design, synthesis, and evaluation of novel small molecule inhibitors of the influenza virus protein NS1. Bioorg Med Chem. 2012;20:487–97. doi: 10.1016/j.bmc.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol. 2011;85:11544–56. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One. 2011;6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Aramini JM. Emerging antiviral targets for influenza A virus. Trends Pharmacol Sci. 2009 doi: 10.1016/j.tips.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–9. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Kuo RL, Krug RM. Influenza a virus polymerase is an integral component of the CPSF30- NS1A protein complex in infected cells. J Virol. 2009;83:1611–6. doi: 10.1128/JVI.01491-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–16. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lin L, Li Y, Pyo HM, Lu X, Raman SN, Liu Q, Brown EG, Zhou Y. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J Virol. 2012;86:1942–54. doi: 10.1128/JVI.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–9. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- Ludwig S, Wang X, Ehrhardt C, Zheng H, Donelan N, Planz O, Pleschka S, Garcia-Sastre A, Heins G, Wolff T. The influenza A virus NS1 protein inhibits activation of Jun N- terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–71. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, Garcia-Sastre A, Roeder RG, Tarakhovsky A. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–33. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Aragon T, Beloso A, Nieto A, Ortin J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–7. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Fernandez Y, Ortin J, Pelaez F, Cabello MA. Development of an HTS assay for the search of anti-influenza agents targeting the interaction of viral RNA with the NS1 protein. Journal of biomolecular screening. 2008;13:581–90. doi: 10.1177/1087057108318754. [DOI] [PubMed] [Google Scholar]
- Mata MA, Satterly N, Versteeg GA, Frantz D, Wei S, Williams N, Schmolke M, Pena-Llopis S, Brugarolas J, Forst CV, White MA, Garcia-Sastre A, Roth MG, Fontoura BM. Chemical inhibition of RNA viruses reveals REDD1 as a host defense factor. Nat Chem Biol. 2011;7:712–9. doi: 10.1038/nchembio.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Suizu F, Hirata N, Miyazaki T, Obuse C, Noguchi M. Characterization of the interaction of influenza virus NS1 with Akt. Biochemical and biophysical research communications. 2010;395:312–7. doi: 10.1016/j.bbrc.2010.03.166. [DOI] [PubMed] [Google Scholar]
- Melen K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Tynell J, Fagerlund R, Roussel P, Hernandez-Verdun D, Julkunen I. Influenza A H3N2 subtype virus NS1 protein targets into the nucleus and binds primarily via its C-terminal NLS2/NoLS to nucleolin and fibrillarin. Virol J. 2012;9:167. doi: 10.1186/1743-422X-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr., Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–24. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2'-5' oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–5. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JY, Li S, Sen GC, Krug RM. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–43. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- Mok BW, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau SY, Wu WL, Matsumoto K, Yuen KY, Chen H. The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection. J Virol. 2012;86:12695–707. doi: 10.1128/JVI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama R, Harada Y, Shibata T, Kuroda K, Hayakawa S, Shimizu K, Tanaka T. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochemical and biophysical research communications. 2007;362:880–5. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Suzuki K, Toda M, Okubo S, Hara Y, Shimamura T. Inhibition of the infectivity of influenza virus by tea polyphenols. Antiviral research. 1993;21:289–99. doi: 10.1016/0166-3542(93)90008-7. [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–95. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–8. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pindel A, Sadler A. The role of protein kinase R in the interferon response. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- Qian XY, Alonso-Caplen F, Krug RM. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J Virol. 1994;68:2433–41. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J Virol. 1994;68:2425–32. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Nemeroff M, Krug RM. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. Rna. 1995;1:304–16. [PMC free article] [PubMed] [Google Scholar]
- Rajput R, Khanna M, Kumar P, Kumar B, Sharma S, Gupta N, Saxena L. Small interfering RNA targeting the nonstructural gene 1 transcript inhibits influenza A virus replication in experimental mice. Nucleic Acid Ther. 2012;22:414–22. doi: 10.1089/nat.2012.0359. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia- Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Garcia-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Current topics in microbiology and immunology. 2009;333:177–95. doi: 10.1007/978-3-540-92165-3_9. [DOI] [PubMed] [Google Scholar]
- Robb NC, Chase G, Bier K, Vreede FT, Shaw PC, Naffakh N, Schwemmle M, Fodor E. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J Virol. 2011;85:5228–31. doi: 10.1128/JVI.02562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–8. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JM, Lee KH, Seong BL. Antiviral effect of catechins in green tea on influenza virus. Antiviral research. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000a;74:7989–96. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc Natl Acad Sci U S A. 2000b;97:4309–14. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol. 2006;80:3957–65. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Ghate M. Recent developments in the medicinal chemistry and therapeutic potential of dihydroorotate dehydrogenase (DHODH) inhibitors. Mini reviews in medicinal chemistry. 2011;11:1039–55. doi: 10.2174/138955711797247707. [DOI] [PubMed] [Google Scholar]
- Walkiewicz MP, Basu D, Jablonski JJ, Geysen HM, Engel DA. Novel inhibitor of influenza non-structural protein 1 blocks multi-cycle replication in an RNase L-dependent manner. J Gen Virol. 2011;92:60–70. doi: 10.1099/vir.0.025015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. Rna. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–73. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shen Y, Qiu Y, Shi Z, Shao D, Chen P, Tong G, Ma Z. The non-structural (NS1) protein of influenza A virus associates with p53 and inhibits p53-mediated transcriptional activity and apoptosis. Biochemical and biophysical research communications. 2010;395:141–5. doi: 10.1016/j.bbrc.2010.03.160. [DOI] [PubMed] [Google Scholar]
- Ward AC, Azad AA, Macreadie IG. Expression and characterisation of the influenza A virus non-structural protein NS1 in yeast. Arch Virol. 1994;138:299–314. doi: 10.1007/BF01379133. [DOI] [PubMed] [Google Scholar]
- Wolff T, O'Neill RE, Palese P. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J Virol. 1996;70:5363–72. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, O'Neill RE, Palese P. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol. 1998;72:7170–80. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JP, Christopher ME, Salazar AM, Sun LQ, Viswanathan S, Wang M, Saravolac EG, Cairns MJ. Broad-spectrum and virus-specific nucleic acid-based antivirals against influenza. Front Biosci (Schol Ed) 2010;2:791–800. doi: 10.2741/s102. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang G, Li Y, Jin Y, Dale R, Sun LQ, Wang M. Inhibition of highly pathogenic avian H5N1 influenza virus replication by RNA oligonucleotides targeting NS1 gene. Biochem Biophys Res Commun. 2008;365:369–74. doi: 10.1016/j.bbrc.2007.10.196. [DOI] [PubMed] [Google Scholar]
- Xia S, Monzingo AF, Robertus JD. Structure of NS1A effector domain from the influenza A/Udorn/72 virus. Acta Crystallogr D Biol Crystallogr. 2009;65:11–7. doi: 10.1107/S0907444908032186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Robertus JD. X-ray structures of NS1 effector domain mutants. Arch Biochem Biophys. 2010;494:198–204. doi: 10.1016/j.abb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Khan JA, Swapna GV, Ertekin A, Krug RM, Tong L, Montelione GT. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J Biol Chem. 2007;282:20584–92. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- You L, Cho EJ, Leavitt J, Ma LC, Montelione GT, Anslyn EV, Krug RM, Ellington A, Robertus JD. Synthesis and evaluation of quinoxaline derivatives as potential influenza NS1A protein inhibitors. Bioorganic & medicinal chemistry letters. 2011;21:3007–11. doi: 10.1016/j.bmcl.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Zhang C, Yang Y, Yang Z, Zhao L, Xu L, Wang R, Zhou X, Huang P. The interaction between the PARP10 protein and the NS1 protein of H5N1 AIV and its effect on virus replication. Virol J. 2011;8:546. doi: 10.1186/1743-422X-8-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang Y, Zhou X, Yang Z, Liu X, Cao Z, Song H, He Y, Huang P. The NS1 protein of influenza A virus interacts with heat shock protein Hsp90 in human alveolar basal epithelial cells: implication for virus-induced apoptosis. Virol J. 2011;8:181. doi: 10.1186/1743-422X-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Das P, Schmolke M, Manicassamy B, Wang Y, Deng X, Cai L, Tu BP, Forst CV, Roth MG, Levy DE, Garcia-Sastre A, de Brabander J, Phillips MA, Fontoura BM. Inhibition of pyrimidine synthesis reverses viral virulence factor-mediated block of mRNA nuclear export. J Cell Biol. 2012;196:315–26. doi: 10.1083/jcb.201107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xu L, Zhou X, Zhu Q, Yang Z, Zhang C, Zhu X, Yu M, Zhang Y, Zhao X, Huang P. Interaction of influenza virus NS1 protein with growth arrest-specific protein 8. Virol J. 2009;6:218. doi: 10.1186/1743-422X-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Li Y, Belser JA, Pearce MB, Schmolke M, Subba AX, Shi Z, Zaki SR, Blau DM, Garcia-Sastre A, Tumpey TM, Wentworth DE. NS-based live attenuated H1N1 pandemic vaccines protect mice and ferrets. Vaccine. 2010;28:8015–25. doi: 10.1016/j.vaccine.2010.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zheng F, Sun T, Duan Y, Cao J, Feng H, Shang L, Zhu Y, Liu H. Interaction of avian influenza virus NS1 protein and nucleolar and coiled-body phosphoprotein 1. Virus Genes. 2012 doi: 10.1007/s11262-012-0849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, Wang S, Li B, Tong G, Ma Z. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One. 2013;8:e55920. doi: 10.1371/journal.pone.0055920. [DOI] [PMC free article] [PubMed] [Google Scholar]