Abstract
The diverse receptor families of the innate immune system activate signal transduction pathways that are important for host defence, but common themes to explain the operation of these pathways remain undefined. In this Opinion article, we propose — on the basis of recent structural and cell biological studies — the concept of supramolecular organizing centres (SMOCs) as location-specific higher-order signalling complexes in which increased local concentrations of signalling components promote the intrinsically weak allosteric interactions that are required for enzyme activation. We suggest that SMOCs are assembled on various membrane-bound organelles or other intracellular sites, which may assist signal amplification to reach a response threshold and potentially define the specificity of cellular responses that are induced in response to infectious and non-infectious insults.
Perhaps no area of immunology has benefitted more from the sequencing of the human (and mouse) genome than that of innate immunity. Modern studies of innate immunity received widespread attention with the discovery in the late 1990s that Toll-like receptors (TLRs) link microbial detection with the induction of adaptive immunity1. Because TLRs and their associated families of signalling proteins have sequence homology, surveying the human and mouse genomes for uncharacterized orthologous proteins became a common approach to study these biological processes. Thus, within a few years of the discovery of cell-surface and endosomal TLRs1-3, more than 100 genes had been identified that regulate the signalling pathways induced by these receptors4-8, as well as the functionally related, cytosolic NOD-like receptors (NLRs), RIG-I-like receptors (RLRs) and others9 (FIG. 1; TABLE 1). Individual members of these pattern recognition receptor (PRR) families detect conserved pathogen-associated molecular patterns (PAMPs) that are present on bacteria, viruses and fungi, or recognize intrinsic damage-associated molecular patterns (DAMPs) that are elicited by cellular injury. Upon ligand binding, these receptors activate numerous cellular responses to fight infection and restore homeostasis.
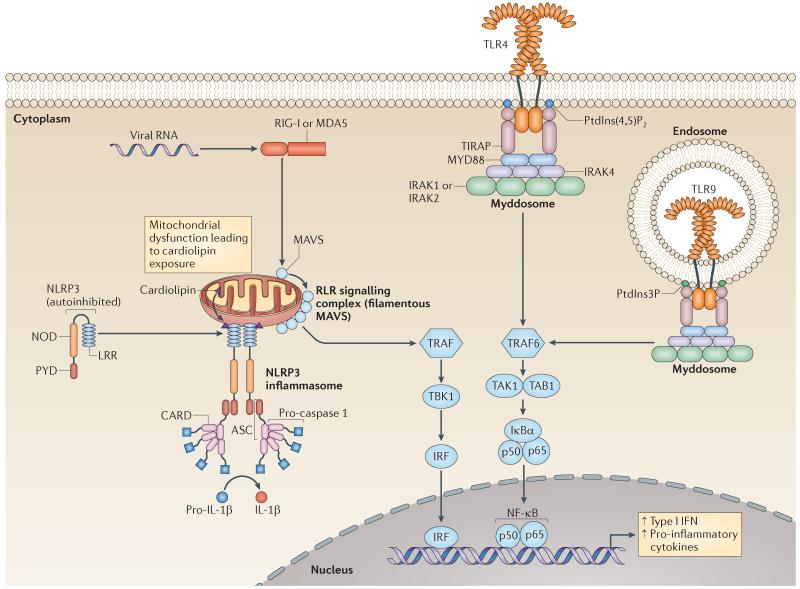
Figure 1. SMOC formation for TLRs, RLRs and NLRs.
Depicted are the best-studied supramolecular organizing centres (SMOCs), including the ligands and regulatory proteins that promote their assembly, and the downstream biological activities induced by these protein complexes. The figure does not show the exact stoichiometry of the protein components in each signalling complex. Binding of lipopolysaccharide (not shown) activates Toll-like receptor 4 (TLR4), leading to assembly of a Myddosome on the plasma membrane. By contrast, unmethylated CpG-containing DNA oligonucleotides (not shown) promote TLR9 to assemble an endosomal Myddosome. The TLR-specific sorting adaptor Toll/IL-1R domain-containing adaptor protein (TIRAP) facilitates Myddosome assembly. TIRAP has an amino-terminal lipid-binding domain that interacts promiscuously with acidic phosphoinositides and phosphatidylserine. For example, TIRAP is depicted as binding phosphatidylinositol-3-phosphate (PtdIns3P) and phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) on the endosomal membrane and the plasma membrane, respectively. Binding of cardiolipin, which translocates to the outer mitochondrial membrane upon mitochondrial dysfunction, relieves the autoinhibited state of NOD-, LRR- and pyrin domain-containing 3 (NLRP3). This in turn may promote NLRP3 inflammasome assembly through downstream pyrin domain (PYD)–PYD and caspase activation and recruitment domain (CARD)–CARD interactions. Activation of RIG-I-like receptors — retinoic acid-inducible gene I (RIG-I) or melanoma differentiation-associated protein 5 (MDA5) — leads to the formation of higher-order oligomers of mitochondrial antiviral signalling protein (MAVS). IFN, interferon; IκBα, NF-κB inhibitor-α; IL, interleukin; IRAK, IL-1 receptor-associated kinase; IRF, IFN-regulatory factor; LRR, leucine-rich repeat; MYD88, myeloid differentiation primary response protein 88; NOD, nucleotide-binding oligomerization domain; NF-κB, nuclear factor-κB; TAB1, TAK1-binding protein 1; TAK1, TGFβ-associated kinase 1; TRAF, tumour necrosis factor receptor-associated factor.
Table 1. SMOCs of the innate immune response.
| SMOC | Triggers or ligands |
Receptors or sensors |
Adaptors | Effectors | Functions |
|---|---|---|---|---|---|
| FAS DISC | FAS ligand | FAS | FADD | Caspase 8 | Apoptosis |
| PIDDosome | DNA damage | PIDD | RAIDD | Caspase 2 | Apoptosis |
| Myddosome | PAMPs | TLRs | TIRAP and MYD88 |
IRAKs | NF-κB activation |
| RLR complex | Viral RNAs | RIG-I and MDA5 |
MAVS | TRAFs | NF-κB activation and interferon response |
| Inflammasome | PAMPs and DAMPs |
NLRs and ALRs |
ASC | Caspase 1 | Pyroptosis and IL-1β maturation |
ALR, AIM2-like receptor; DAMP, damage-associated molecular pattern; DISC, death-inducing signalling complex; FADD, FAS-associated death domain protein; IL-1β, interleukin-1β; IRAK, IL-1 receptor-associated kinase; MAVS, mitochondrial antiviral signalling protein; MDA5, melanoma differentiation-associated protein 5; MYD88, myeloid differentiation primary response protein 88; NF-κB, nuclear factor-κB; NLR, NOD-like receptor; PAMP, pathogen-associated molecular pattern; PIDD, p53-inducible protein with a death domain; RAIDD, RIP-associated ICH1/CED3-homologous protein with a death domain; RIG-I, retinoic acid-inducible gene I; RLR, RIG-I-like receptor; SMOC, supramolecular organizing centre; TIRAP, Toll/IL-1R domain-containing adaptor protein; TLR, Toll-like receptor; TRAF, tumour necrosis factor receptor-associated factor.
The success of using bioinformatics and reverse genetics to study innate immune signalling pathways came at the expense of alternative strategies to address these areas. As such, studies of the biochemistry, cell biology and dynamics of these signalling pathways have been much less common. In fact, most early studies of TLRs and their associated signalling proteins did not include any analysis of the subcellular localization of the newly identified protein(s). Thus, although we know the identity of many genes that are involved in innate immunity, the functional mechanisms of the proteins encoded by these genes, and how they interact in space and time, are poorly understood.
The lack of knowledge on the specific activities of the proteins that control innate immunity has given rise to biological models that do not address many aspects of the signalling process, such as the subcellular site where a given signalling event occurs, or the dynamics of putative protein–protein interactions. Current models of TLR, NLR or RLR signalling rather depict a series of arrows connecting receptors with downstream signalling proteins, yet we have little understanding of what these arrows actually represent. Do they represent direct protein–protein interactions? If so, are these interactions constitutive or are they induced upon microbial encounter? How are these interactions regulated and where in the cell do they occur? As described below, recent biochemical and cell biological studies have provided important insight into these questions. These new studies indicate that numerous protein regulators of innate immunity are organized into higher-order signalling complexes that define the subcellular sites and specificity of innate immune signal transduction.
In this Opinion article, we propose that these higher-order signalling complexes function as ‘supramolecular organizing centres’ (SMOCs) that control cellular responses induced by specific families of upstream receptors. We discuss how SMOCs can operate from various locations within the cell, and describe how they consist of proteins that either sense the activation of upstream receptors or elicit specific downstream effector responses. We further propose that these complexes include context-dependent components, which may be cell type-specific or organelle-specific regulators, such that a given SMOC can elicit diverse cellular responses depending on the stimulus. A benefit of coordinating innate immune signalling pathways around a set of organizing centres may be the modularity of the system, whereby numerous upstream stimuli can be directed into a common downstream module. Indeed, this is the case when considering the operation of other non-membranous organizing centres in mammalian cells, such as the microtubule organizing centre (MTOC) and the proteasome. In these examples, a large protein complex coordinates an entire biological process that may be needed to address diverse cellular needs.
SMOCs for PRRs
The classical view of signal transduction — involving a serial reaction in which ligands induce conformational changes in receptors followed by the activation of enzymes and the generation of second messengers — required serious modifications in the case of PRRs. The emerging concept for PRRs supports the formation of higher-order signalling complexes as the mode of signal transduction and amplification. Indeed, with the exception of the signalling pathway involving cyclic GMP–AMP synthase (cGAS) and stimulator of interferon genes protein (STING) described below, there is little or no role of second messengers in the earliest events associated with PRR signal transduction.
Early studies
Cellular studies using light microscopy imaging provided the first evidence for the existence of higher-order signalling complexes in innate immune pathways. The tumour necrosis factor (TNF) receptor superfamily comprises some of the earliest discovered members of the innate immune system, such as TNF receptor 1 (TNFR1) and FAS (also known as CD95 and TNFRSF6). These receptors do not have intrinsic enzymatic activity, but aggregate upon stimulation by trimeric ligands of the corresponding TNF superfamily. As an example, FAS — as part of the death-inducing signalling complex (DISC) — forms aggregated clusters or puncta of hundreds of nanometres to micrometres in diameter10,11, which are much larger than ligand–receptor trimers. The formation of sizable clusters that are visible by light microscopy has become a recurrent observation for activated innate immune receptors, including TLRs12-14, RLRs15,16, and NLR or absent in melanoma 2 (AIM2) inflammasomes17,18 (FIG. 1; TABLE 1).
Biochemical and structural studies of innate immune signalling complexes indicated that such protein clusters are not random aggregates but instead have a defined molecular basis of assembly. An almost ubiquitous feature of innate immune pathways is the participation of signal transduction proteins with death domains, or the related death effector domains (DEDs), caspase activation and recruitment domains (CARDs) and pyrin domains (PYDs)19. Crystal structures of oligo meric death domain complexes revealed an ordered helical assembly mechanism that underlies the oligomerization of several proteins19 including the 5:7 complex of p53-inducible protein with a death domain (PIDD) and RIP-associated ICH1/CED3-homologous protein with a death domain (RAIDD; also known as CRADD) in the core of the PIDDosome for caspase 2 activation20; the 5:5 complex of FAS and FAS-associated death domain protein (FADD) in the DISC for caspase 8 activation21; and the 6:4:4 complex of myeloid differentiation primary response protein 88 (MYD88), interleukin-1 receptor (IL-1R)-associated kinase 4 (IRAK4) and IRAK2 in the Myddosome for kinase activation in the TLR pathway22 (TABLE 1). The common helical symmetry indicates that death domains and their related domains might be able to form large helical filaments and even larger filamentous signalling complexes to generate microscopically visible SMOCs.
Recent advances
Exciting new studies have now confirmed the structural predictions, as well as further extended our mechanistic understanding of innate immune signalling. The RLR signalling adaptor mitochondrial antiviral signalling protein (MAVS), which contains an amino-terminal CARD, forms helical filaments that activate the interferon pathway16,23,24 (FIG. 2). The CARD-containing adaptor B cell lymphoma 10 (BCL-10) — which activates mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) for nuclear factor-κB (NF-κB) signalling downstream of several innate and adaptive immune receptors — assembles into similar helical filaments25. Recent studies of cytosolic inflammasomes, which activate caspase 1 to induce interleukin-1β (IL-1β) maturation and pyroptosis, show that filaments of the adaptor protein ASC (which contains both a PYD and a CARD) and caspase 1 (which contains a CARD) form star-shaped structures that mediate signal transduction and proximity-induced caspase 1 activation26-28 (FIG. 2). Electron microscopy-based structural studies showed that these filaments are assembled through three conserved types of interactions that were initially observed in the crystal structures of death domain complexes24,25,27.
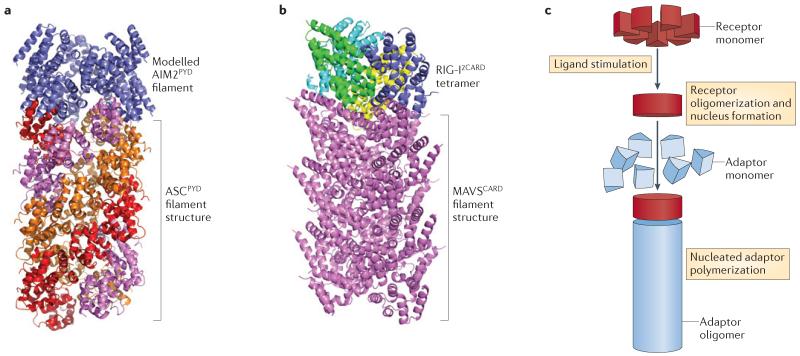
Figure 2. Structures of SMOCs that are formed by the mechanism of nucleated polymerization.
a | A ribbon diagram of the electron cryomicroscopic structure of polymerized ASC pyrin domain (ASCPYD) filaments in inflammasomes (shown in red, purple and orange for each of the three helical strands), in complex with polymerized absent in melanoma 2 PYD (AIM2PYD) filaments (shown in blue) formed upon double-stranded DNA (dsDNA) stimulation. b | A ribbon diagram of the electron cryomicroscopic structure of polymerized mitochondrial antiviral signalling protein caspase activation and recruitment domain (MAVSCARD) in the RIG-I-like receptor (RLR) pathway (shown in purple), in complex with a retinoic acid-inducible gene I (RIG-I) double CARD (RIG-I2CARD) tetramer (shown in blue, cyan, green and yellow) upon viral RNA stimulation. c | Proposed mechanism of nucleated polymerization for the formation of ASCPYD and MAVSCARD SMOCs. Receptors (for example, AIM2 and RIG-I) are shown in red wedges for monomers and in red disks for oligomerized forms. Adaptors (for example, ASC and MAVS) are shown in blue wedges for monomers and in blue cylinders for filaments.
These new advances indicate that nucleated polymerization may be a general principle in promoting the assembly of filamentous complexes upon stimulation to mediate innate immune signal transduction (FIG. 2). A sensor protein (receptor) in a pathway first becomes activated in the presence of ligands by overcoming auto-inhibition and subsequently oligomerizes. The oligomerized sensor then provides the platform to nucleate a downstream protein to polymerize into filaments. For example, recognition of cytosolic double-stranded DNA (dsDNA) by the AIM2 inflammasome or the response to various infectious and danger signals by NLR inflammasomes disrupts intra molecular domain interactions29 to enable oligomerization of the PYDs of inflamma some sensor proteins26,27. These oligomerized PYDs then function as a platform to nucleate the polymerization of the PYDs in the adaptor protein ASC. The ASCPYD filaments bring the CARDs of ASC into proximity to nucleate the polymerization of the CARDs of caspase 1, which in turn promotes caspase 1 dimerization and activation. Similarly, the polyubiquitin-stabilized double CARD of retinoic acid-inducible gene I (RIG-I) forms a helical tetramer to imprint the polymerization of MAVS through its CARDs to promote downstream signalling24,30-32.
Oligomerization mechanisms that do not involve the death domain superfamily have also been discovered in innate immunity14,33. A common molecular mechanism of signal transduction seems to be the tight coupling between oligomerization — regardless of the type — and allostery. On the one hand, ligand-induced allosteric changes in the respective receptors promote oligomerization. On the other hand, oligomerization of effector enzymes, such as kinases and caspases, enhances the allosteric changes that are presumed to be required for enzyme activation. This oligomerization-facilitated allostery is illustrated by the recently reported crystal structure of the unphosphorylated IRAK4 kinase domain asymmetric dimer captured in the conformation of the Myddosome-induced IRAK4 trans-autophosphorylation reaction34. In the crystal structure, one IRAK4 monomer exists in the active kinase conformation despite a lack of phosphorylation and it precisely binds the activation loop phosphosite of the other IRAK4 monomer for phospho-transfer. In solution, unphosphorylated IRAK4 forms weak dimers. By markedly increasing the local concentration of IRAK4, the Myddosome promotes dimerization to drive allosteric autoactivation of IRAK4. Oligomerization-driven allosteric changes probably have crucial roles in the higher-order signalling complexes of other innate immune receptors; it is the challenge of structural biologists to characterize these often weak and transient conformations.
Mechanistic implications
The assembly of higher-order signalling complexes not only provides a phenomenological explanation for the observation of punctate structures in cells, but also implicates an elegant mechanism of signal amplification, preceding enzyme activation, that enables a response threshold to be reached. Nucleated polymerization ensures that a small number of sensor proteins can activate many downstream signalling proteins. For example, in the AIM2 inflammasome, a substoichiometric amount of AIM2 can polymerize many more ASC molecules27. In turn, activated ASC further oligomerizes many more caspase 1 molecules to amplify signal transduction. Within a single cell, it is likely that once the signalling cascade is successfully initiated, almost all caspase 1 molecules are recruited to inflammasomes so that a maximal response is generated. Signal amplification and the cooperativity in the assembly of higher-order signalling complexes may both contribute to the threshold, all-or-none host defence programmes that have been observed at the single-cell level for many innate immune pathways35,36.
In addition, signal amplification may explain the sensitivity of mammalian cells to incredibly small numbers of bacteria. For example, it has been estimated that a single Escherichia coli bacterium can activate 1,000 macrophages through TLR4 (REF. 37), suggesting that very few receptors on any individual cell are engaged during infection. We suggest that SMOC formation around the cytosolic tail of TLR4 may explain the all-or-none response that macrophages often exhibit to a wide range of lipopolysaccharide (LPS) concentrations37. In addition, the role of SMOCs in signal amplification may be fundamentally analogous to that of second messengers, such as cAMP in signal transduction by G protein-coupled receptors. However, we believe that cAMP-mediated responses may be more graded than those mediated by SMOCs, with the strength of the response being determined by the amount, half-life and deactivation kinetics of the second messengers.
If signal amplification to reach a response threshold is the mechanism by which innate immune pathways are turned on, then once formed, how are SMOCs turned off to terminate signalling? In the case of BCL-10-containing signalosomes, they are recruited to autophagosomes through an interaction between ubiquitylated BCL-10 and the autophagy adaptor p62 (also known as SQSTM1), leading to degradation38. Deficiency of ATG16L1 — a protein that is required for autophagosome formation — has been shown to enhance endotoxin-induced IL-1β and IL-18 production, probably through a TLR-mediated pathway39. Given that SMOCs may be too large to be degraded efficiently by proteasomes, autophagosome-mediated degradation seems to make sense. It is interesting to note that the size distribution of puncta can be quite different for different SMOCs. For example, TLRs form relatively small clusters, which may account for the smaller size of the Myddosome compared with inflamma some filaments. Some inflammasomes, as well as STING (which mediates the interferon response upon stimulation of cGAS by cytosolic dsDNA), have gigantic peri nuclear puncta40,41. Whether the distinct size of puncta is reflective of the threshold of activation and/or degradation mechanisms remains to be addressed.
Consistent with the ability of many death domain superfamily members to polymerize, several of these domains — including the CARD of MAVS and the PYD of ASC — have prion-like activities in yeast16,27,28. In mammalian cells, it seems that ASC-containing specks of inflamma somes are not degraded rapidly but are released upon pyroptosis into the extra cellular space, where they promote further IL-1β processing and can be engulfed by macrophages to induce inflammasome activation in the recipient cells42,43. Therefore, understanding the biophysical principles of SMOC assembly and degradation not only provides insights into unique signalling properties within each cell, but also implicates mechanisms of unexpected signal transduction between cells.
The subcellular localization of SMOCs
Cell biological and biochemical analyses of individual components of SMOCs have shown that these complexes are usually assembled on the cytosolic surface of membranous organelles of mammalian cells. This trend of SMOC assembly on membranes can be observed for TLRs, as well as for RLRs and NLRs. Because the RLRs and NLRs are not transmembrane proteins, their need to induce SMOC assembly on organelles is intriguing, and it indicates that membrane-based SMOC assembly is not simply a consequence of signal transduction being initiated by a transmembrane receptor such as a TLR.
Toll-like receptors
Although the Myddosome has been defined structurally22, there is an incomplete understanding of the composition, dynamics and regulation of the endogenous TLR-induced Myddosome in mammalian cells. Recent work has provided insight into the behaviour of the Myddosome in macrophages, through the demonstration that it can form either at the plasma membrane or on endosomes44. For example, LPS treatment of macrophages activates TLR4 to assemble a Myddosome at the cell surface, whereas unmethylated CpG-containing DNA oligonucleotides activate TLR9 to assemble an endosomal Myddosome. In both cases, an activated receptor is not sufficient to induce Myddosome assembly; rather, the TLR-specific Toll/IL-1R (TIR) domain-containing adaptor protein (TIRAP; also known as MAL) is required for Myddosome assembly44. Thus, TIRAP is the first natural regulator of TLR-induced Myddosome formation to be identified. TIRAP is a peripheral membrane protein that contains an N-terminal lipid-binding domain that interacts promiscuously with acidic phosphoinositides and phosphatidylserine45. This promiscuity of lipid binding enables TIRAP to survey several plasma membrane and endosomal subdomains for the presence of an activated (ligand-bound) TLR. When TIRAP detects a ligand-bound TLR through its carboxy-terminal TIR domain46,47, it recruits the core component of the Myddosome, MYD88, which seeds the formation of this SMOC through interactions with IRAKs22,44,48. Altering the lipid specificity of TIRAP such that it can only interact with either plasma membrane-localized or endosome-localized lipids restricts the ability of the cell to assemble Myddosomes from the location in which TIRAP resides44. Thus, although TIRAP contains a promiscuous lipid-binding domain, the individual targets of this domain enable location-specific assembly of a SMOC. TIRAP was originally thought to be a regulator that defines the specificity of signalling pathway activation induced by plasma membrane-localized TLRs, as compared with endosomal TLRs8. The observation that TIRAP regulates Myddosome formation from both subcellular locations reignites the question of how organelle-specific innate immune responses are achieved. We suggest that whereas TIRAP and the other components of the Myddosome participate in TLR signalling from multiple locations within the cell, organelle-specific regulators may exist to determine the specificity of signal transduction. Identifying these putative regulators may provide tools to understand how the composition and function of SMOCs can be modulated naturally, and perhaps therapeutically.
RIG-I-like receptors
The RLR system assembles a SMOC on membranes, despite the fact that the upstream receptors are cytosolic proteins. The core of the RLR-induced SMOC is MAVS, which is localized to membranes though a C-terminal transmembrane domain15,16. Although the localization domain of MAVS is not structurally similar to that of TIRAP, these domains are similar in that they can be targeted to multiple organelles. In the case of MAVS, its transmembrane domain enables it to localize to mitochondria, peroxisomes and the mitochondria-associated membranes (MAMs) of the endoplasmic reticulum15,49,50. Also, similar to TIRAP, MAVS can induce signalling responses from each of these organelles15,49,50, although biochemical evidence for the formation of higher-order oligomers is only available for mitochondria-localized MAVS16. It remains to be determined whether similar MAVS-containing SMOCs are assembled on peroxisomes and MAMs during viral infections. Recent studies have shown that functional TIRAP or MAVS proteins can be generated by replacing the localization domains of either of these proteins with other domains that have similar membrane-targeting activities15,44,50. These data provide strong evidence that the sole function of these domains is to direct TIRAP or MAVS to the membranes that are necessary for PRR-induced SMOC assembly. Although it is clear that MAVS must localize to specific organelles to initiate SMOC assembly and signalling, it remains unclear why mitochondria, peroxisomes and MAMs have evolved as the preferred sites of RLR signal transduction. One possibility is that the metabolic functions of these organelles control MAVS activation, a theory that is supported by recent studies indicating that dysfunctional mitochondria cannot promote efficient RLR-dependent innate immune responses51,52.
NOD-like receptors
The NLR system provides yet another example of SMOC assembly on membranes, at least in the case of NOD-, LRR- and PYD-containing 3 (NLRP3). Although NLRP3 is generally thought to be a cytosolic protein, it trans locates to mitochondria when cells are stimulated with several types of inflammasome activators53,54. The translocation of NLRP3 to mitochondria has been reported to occur through interactions with the lipid cardio lipin — which is only displayed on the outer membrane of damaged mitochondria55 — or with MAVS56. In this regard, NLRP3 is similar to TIRAP in that its ability to promote SMOC assembly is linked to its ability to interact with both membrane-bound lipids (phosphoinositides) and proteins (TLRs) in a specific region of the cell. It is unclear whether other NLR family members also interact with cardiolipin or any other lipid to promote SMOC assembly, but these studies provide a strong mandate to consider this possibility.
Conclusions and future perspectives
Examples now exist for the three major families of PRRs — TLRs, RLRs and NLRs — that SMOC assembly occurs on membranes, even when the upstream receptors are cytosolic proteins. In each documented example, a membrane protein seeds the formation of a higher-order signalling complex that activates specific innate immune responses. A biophysical explanation for the apparent membrane localization of SMOCs may be the marked energetic enhancement of protein–protein interactions on a two-dimensional membrane surface compared with those in a three-dimensional cellular milieu57. For this reason, we propose that future studies of the biochemical mechanisms of SMOC assembly and function would benefit from a greater consideration of the subcellular sites where this assembly occurs. This analysis may also help to address the question of why SMOCs have evolved to operate from specific organelles. Additional studies of the relationship between organelle function and SMOC assembly should address the possibility that organelles are not solely needed as a scaffold for SMOC assembly, but rather that a metabolic or biochemical activity of the organelle may contribute as well. This cell biological analysis may reveal important means of controlling and manipulating SMOC assembly and subsequent inflammatory responses.
Acknowledgements
J.C.K. is supported by the US National Institutes of Health (NIH; grants AI093589, AI072955 and AI113141-01) and an unrestricted gift from Mead Johnson & Company. J.C.K. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. H.W. is supported by NIH and the Asa and Patricia Springer Professorship of the Harvard Medical School.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 3.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nature Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 6.Oshiumi H, et al. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to Toll-like receptor 4 TICAM-1 that induces interferon-β. J. Biol. Chem. 2003;278:49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 7.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nature Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 8.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 9.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 10.Algeciras-Schimnich A, et al. Molecular ordering of the initial signaling events of CD95. Mol. Cell. Biol. 2002;22:207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RM, et al. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J. Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latz E, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nature Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 13.Visintin A, Latz E, Monks BG, Espevik T, Golenbock DT. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to Toll-like receptor-4 aggregation and signal transduction. J. Biol. Chem. 2003;278:48313–48320. doi: 10.1074/jbc.M306802200. [DOI] [PubMed] [Google Scholar]
- 14.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nature Struct. Mol. Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, et al. The Fas–FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nature Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu B, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao Q, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly. Mol. Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J. Mol. Biol. 2014;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 30.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrao R, et al. IRAK4 dimerization and trans-autophosphorylation are induced by myddosome assembly. Mol. Cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay S, et al. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol. Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 38.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franklin BS, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nature Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baroja-Mazo A, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nature Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 44.Bonham KS, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of Toll-like receptor signal transduction. Cell. 2014;156:705–716. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 46.Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nature Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 47.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 48.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl Acad. Sci. USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koshiba T, Yasukawa K, Yanagi Y, Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal. 2011;4:ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 52.Odendall C, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nature Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 54.Misawa T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nature Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 55.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]