Abstract
MicroRNAs (miRNAs) are endogenously expressed small non-coding RNAs that act as post-transcriptional regulators of gene expression. Dysregulation of these molecules has been indicated in the development of many cancers. Altered expression levels of several miRNAs were identified also in glioblastoma. It was repeatedly found that miRNAs are involved in important signalling pathways, which play roles in crucial cellular processes, such as proliferation, apoptosis, cell cycle regulation, invasion, angiogenesis and stem cell behaviour. Therefore, miRNAs represent promising therapeutic targets in glioblastoma. In this review, we summarize the current knowledge about miRNAs significance in glioblastoma, with special focus on their involvement in core signalling pathways, their roles in drug resistance and potential clinical implications.
Keywords: glioblastoma, microRNAs, prognosis, prediction, therapeutic targets
Introduction
Glioblastoma is the most frequently occurring primary malignant brain tumour, with an incidence of 355 new cases per 100,000 Caucasians per year. Despite introduction of modern therapeutic approaches, this cancer remains associated with very poor prognosis characterized by median overall survival less than 1 year [1, 2]. For glioblastoma, rapid diffuse and infiltrative growth with high level of cellular heterogeneity associated with therapeutic resistance is typical. There are also multiple genetic alterations characteristic for glioblastoma. Primary glioblastoma arising de novo is often characterized by EGFR amplification and PTEN mutations, whereas TP53 mutations are typical for secondary glioblastoma developing from lower-grade astrocytomas. Nevertheless, this distinction is not absolute and both glioblastoma types may harbour other genetic and chromosomal changes, for example, the loss of heterozygosity (LOH) 10q is the most frequent aberration in both primary and secondary glioblastomas [3, 4].
miRNAs are small non-coding RNAs, 22-nt in length that guide post-transcriptional gene silencing of their mRNA targets. miRNAs are encoded by genes that are presumably transcribed into long primary miRNAs (pri-miRNAs) by RNA polymerases II/III [5]. Afterwards, RNase III enzyme converts pri-miRNAs into pre-miRNAs hairpin transcripts. At last, pre-miRNAs are processed into mature miRNAs that are incorporated into a ribonucleoprotein complex RISC. Within RISC, miRNAs act as specific determinants, whereas protein components achieve target mRNA silencing [6]. Deregulation of miRNAs can affect carcinogenesis if their mRNA targets are encoded by oncogenes or tumour suppressor genes; overexpression, silencing or switching off specific miRNAs have been described in carcinogenesis of glioblastoma. Up-regulation of mature miRNA may occur as a consequence of transcriptional activation or amplification of the miRNA encoding gene, whereas silencing or reduced expression may result from deletion of a particular chromosomal region, epigenetic silencing, or defects in their biogenesis [7]. In this review, we summarize recent work on miRNAs, with emphasis on their alterations and roles in glioblastoma pathogenesis and their potential used as disease biomarkers or novel therapeutic targets.
miRNAs signatures of glioblastoma tissue and cell lines
Global analysis of miRNA expression profiles of both glioblastoma tissues and glioblastoma cell lines allowed to identify a group of miRNAs with significantly altered expression in this tumour. When primary glioblastoma tissue was compared to non-malignant brain tissue, expression levels of nine miRNAs were significantly increased, whereas levels of four miRNAs were decreased (Table 1). The most significant results indicated strongly overexpressed miR-221, and down-regulated, miR-128a, miR-181a, miR-181b and miR-181c from a set of brain-enriched miRNAs [8]. The same authors performed miRNA expression analysis of 10 glioblastoma cell lines using identical methodic approach and data analysis. Interestingly, miRNAs underexpressed in glioblastoma cell lines generally confirmed primary tumour data, whereas only miR-21 and miR-221 that were overexpressed in tumours were deregulated also in the cell lines [8].
Table 1.
miRNAs significantly de-regulated in human glioblastoma tissues and glioblastoma cell lines
| Ciafre et al. (2005) | Slaby et al. (2010) | ||||
|---|---|---|---|---|---|
| miRNA | C/P ratio | L/B ratio (P) | miRNA | Fold change (P) | |
| Up-regulated | miR-9-2 | 1.88–10.16 | |||
| miR-10b | 1.97–13.6 | ||||
| miR-21 | 1.81–9.3 | 1.61 (0.008) | miR-21 | 8.35 | |
| (<0.001) | |||||
| miR-23a | 6.22 (<0.001) | ||||
| miR-23-b | 3.28 (0.043) | ||||
| miR-24-1 | 1.83 (<0.001) | ||||
| miR-24-2 | 1.88 (<0.001) | ||||
| miR-25 | 1.99–3.6 | ||||
| miR-123 | 1.9–2.45 | ||||
| miR-125b-1 | 2.19–2.73 | miR-125b | 1.45 | ||
| (0.502) | |||||
| miR-125b-2 | 1.95–2.88 | ||||
| miR-130a | 2.11–5.3 | ||||
| miR-191 | 1.92 (0.008) | ||||
| miR-220 | 1.68 (0.020) | ||||
| miR-221 | 1.84–4.8 | 5.34 (<0.001) | |||
| miR-222-prec | 2.43 (0.039) | ||||
| Down-regulated | miR-125b-1 | 0.31 (0.043) | |||
| miR-125b-2 | 0.39 (0.047) | ||||
| miR-128a | 0.34–0.56 | miR-128a | 0.03 | ||
| (<0.001) | |||||
| miR-128b | 0.53 (0.008) | ||||
| miR-181a | 0.082–0.56 | 0.35 (<0.001) | miR-181a | 0.4 | |
| (0.073) | |||||
| miR-181b | 0.098–0.56 | 0.34 (0.005) | miR-181b | 0.28 | |
| (0.036) | |||||
| miR-181c | 0.096–0.56 | 0.49 (0.040) | miR-181c | 0.29 | |
| (0.043) | |||||
| miR-197 | 0.33 (0.040) | ||||
| miR-221 | 0.25 | ||||
| miR-222 | 0.22 | ||||
C/P ratio represents the range of ratio between tumour samples values (C: centre of the tumour) and the control samples values (P: peripheral brain area from the same patient). L/B ratio represents the ratio between averaged cell line samples values (L) and the control sample values (B); P value is presented (t-test [8], Mann-Whitney U-test [9]).
In another study, Slaby et al. evaluated expression profiles of eight miRNAs in glioblastoma tissues compared to non-malignant brain tissues from areas surrounding arteriovenous malformation (AVM). In glioblastoma tissue, only miR-21 and miR-125b were overexpressed, whereas six miRNAs were down-regulated (Table 1) [9]. In contrast to previous study, approximately four-fold lower levels of miR-221/222 were observed in glioblastomas in comparison to the adult ‘normal-like’ brain tissue. Authors discussed that it is likely that the brain tissue, although excised from the margin of resection material, contained traces of micro-capillaries from around the AVM. It is generally known that very high levels of miR-221/222 are found in endothelial cells. This could be responsible for the apparently low levels of miR-221/222 in glioblastomas despite their absolute levels being comparable to previously published reports [9].
Taken together, only miR-21 and miRNA-181 family were significantly and consistently altered in all three studies (Table 1).
Involvement of miRNAs in core glioblastoma signalling pathways
Glioblastoma development has been linked to progressive acquisition of mutations in genes with a crucial role in cell growth, proliferation and programmed cell death. As shown in many different studies, miRNAs might perfectly fit and integrate model of glioblastoma pathogenesis by controlling its core signalling pathways [3].
EGFR and PI3K/AKT signalling pathways
The epidermal growth factor receptor (EGFR) signalling network contributes to promotion and progression of a broad spectrum of solid tumours; it is a promising and, at least for some tumours, a validated target for anticancer therapy. Stimulation of the EGFR and, subsequently, KRAS signalling, lead to activation of numerous signal transduction molecules initiating a cascade of downstream effectors that mediate tumour growth, survival, angiogenesis and metastasis [10].
Several recent reports have also identified up-regulation of miR-21 in glioblastoma tissue [8, 9]. Consequently, mechanistic studies identified mRNA targets of miR-21 among important components of the EGFR signalling pathway. Glioblastoma cell lines U251 (mutant PTEN) and LN229 (wild-type PTEN) showed a decreased expression of EGFR, activated AKT, Cyclin D and Bcl-2 after treatment by miR-21–specific antisense oligonucleotide [11]. Although miR-21 is known to regulate PTEN and down-regulation of miR-21 led to increased PTEN expression, the glioblastoma suppressor effect of antisense-miR-21 is most likely independent of PTEN status because U251 has mutated PTEN [11, 12]. Oncogenic phenotype indicates also miR-26a, which is highly up-regulated in glioblastoma tissue [8]. Phenomenon of PTEN down-regulation followed by AKT activation was described after transfection of glioblastoma cells with the primary transcript of miR-26a-2. Similarly, the miR-26a mimics decreased PTEN protein levels and increased AKT phosphorylation [13, 14]. Modulation of AKT signalling cascade using miRNAs in glioblastoma cell lines was described also in Nan et al. In this study, transfection of miR-451 mimicked reduced expression levels of Akt1, Cyclin D1, MMP-2, MMP-9 and Bcl-2. By contrast, miR-451 down-regulation led to increase in p27 levels. According to phenotypic experiments, miR-451 inhibited invasive ability, induced cell cycle arrest in the G0/G1 phase, delayed the progression of cell cycle, inhibited cell proliferation and induced apoptosis in glioblastoma cells in vitro. To conclude, it seems that miR-451 affects glioblastoma cells via regulation of the PI3K/AKT signalling pathway [15].
Another miRNA involved in the EGFR signalling pathway is miR-7. Kefas et al. published that miR-7 directly inhibited EGFR expression via its 3′-UTR and independently suppressed the AKT pathway via targeting upstream regulators, such as IRS-1 and IRS-2. Moreover, transfection with miR-7 oligonucleotides decreased viability and invasiveness of primary glioblastoma cell lines [16]. Webster et al. confirmed that miR-7 down-regulates EGFR mRNA and protein expression in glioblastoma cell lines via two of the three predicted sites, and induces cell cycle arrest and apoptosis. Furthermore, these authors also described Raf1, another member of the EGFR signalling pathway, as a direct target of miR-7 in cancer cells [17].
Godlewski et al. published that miR-128 expression significantly reduced glioma cell proliferation in vitro and correspondingly glioma xenograft growth in vivo. This effect was explained by direct regulation of the Bmi-1 mRNA 3′-UTR, through a single miR-128 binding site. Bmi-1 expression was significantly up-regulated and miR-128 was down-regulated compared to normal brain. In addition, miR-128 expression leads to a decrease in H3K27 methylation and modulation of cellular pathways, especially p21CIP1 and Akt, involved in cell cycle arrest and survival [18]. Some of the investigations supported by revelation that Bmi-1 transcriptionally down-regulates expression of the tumour suppressor PTEN in tumour cells through direct association with the PTEN locus [19].
Finally, miR-221 and miR-222 were revealed using bioinformatics analysis as potential regulators of many target genes involved in AKT signalling pathway. Up-regulation of miR-221/222 resulted in remarkable increase of p-Akt and significant changes in expression of Akt-related genes in glioma cells. Consequently, miR-221/222 overexpression increased glioma cell proliferation and invasion in vitro and induced glioma growth in a subcutaneous mouse model. These results suggest that miR-221/222 enhance glioma malignant phenotype via activation of the AKT signalling pathway mediated by regulation of common gene expression (Fig. 1) [20].
Fig 1.
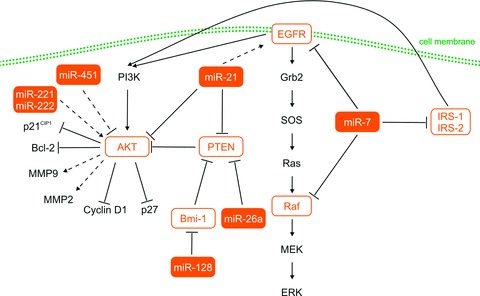
MiRNAs involved in EGFR and PI3K/AKT signalling pathways. EGFR: epidermal growth factor receptor; AKT: serine/threonine protein kinase Akt; PTEN: phosphatase and tensin homologue; Bmi-1: polycomb ring finger oncogene; Raf: raf kinase, effector of Ras; IRS1/2: insulin receptor substrate 1/2; PI3K: Phosphotidylinositol 3 kinase; MMP9/2: matrix metallopeptidase 9/2; p27: cyclin-dependent kinase inhibitor 1B (p27, Kip1); p21: cyclin-dependent kinase inhibitor 1A (p21, Cip1); Bcl-2: B-cell CLL/lymphoma 2; Grb2: growth factor receptor-bound protein 2; SOS: son of sevenless homologue 1; MEK: mitogen-activated protein kinase kinase 1; ERK: extracellular signal-regulated kinase. Dashed lines indicated indirect regulation, solid lines indicate direct regulation.
p53, TGF-β and apoptotic signalling pathways
Papagiannakopoulos et al. reported that p53, TGF-β and mitochondrial apoptotic networks are de-repressed in response to miR-21 knockdown. They published a panel of genes involved in particular pathways and simultaneously modulated by miR-21 treatment. From this panel, p63, JMY, TP53BP2, HNRPK, TOPORS, IGFB3, APAF1, PPIF, TGFBR2/3, DAXX, HNRNPK were predicted to be direct targets of miR-21 that can stabilize p53 protein levels by interfering with MDM2 and/or act as p53 transcriptional cofactors [21]. Inhibition of miR-21 increased also endogenous levels of PDCD4 in human glioma cell lines and activated caspases 9 and 3, which may be mediated by modulating multiple potential target genes, such as TIMP3 [22, 23]. Protein PDCD4 inhibits translation by its interaction with the factor that initiates translation of eIF4A and eIF4G. PDCD4 also inhibits proliferation via activation of p21CIP1[6]. In addition, specific inhibition of miR-21 led to elevated levels of RECK and TIMP3 and therefore reduced MMP activities in vitro and in model of gliomas in nude mice. Consequently, down-regulation of miR-21 decreased migratory and invasive abilities in glioma cells (Fig. 2) [24].
Fig 2.
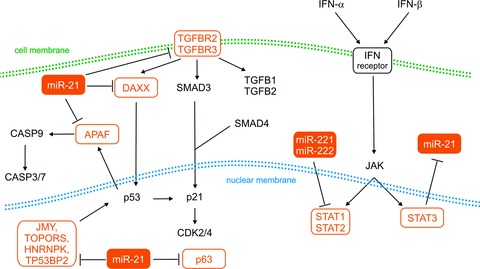
MiRNAs involved in TGF-β and IFN-α/IFN-β signalling pathways. TGFBR2/3: transforming growth factor β receptor 2/3; TGFB1/2: transforming growth factor β 1/2; DAXX: death-domain associated protein; SMAD3/4: SMAD family member 3/4; APAF: apoptotic peptidase activating factor; CASP3/7/9: caspase 3/7/9; p53: tumour protein p53; p21: cyclin-dependent kinase inhibitor 1A (p21, Cip1); p63: tumour protein p63; CDK2/4: cyclin-dependent kinase 2/4; JMY: junction mediating and regulatory protein, p53 cofactor; TOPORS: topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase; HNRNPK: heterogeneous nuclear ribonucleoprotein K; TP53BP2: tumour protein p53 binding protein, 2; IFN: interferon; STAT1/2/3: signal transducer and activator of transcription 1/2/3; JAK: Janus kinase. Dashed lines indicated indirect regulation, solid lines indicate direct regulation. EGFR: epidermal growth factor receptor.
IFN-α/IFN-β signalling pathways
Interferons (IFNs) are cytokines released by lymphocytes that have antiviral, antiproliferative and immunomodulatory effects. They are connected with the JAK-STAT (Janus kinase-Signal Transducer and Activator of Transcription) signalling cascade and allow communication between cells to trigger protective defences of the immune system leading to eradication of affected cells [25].
Insight into transcriptional regulation of miRNAs through both intracellular and extracellular mechanisms is one of the fundamental ideas leading to understanding oncogenesis. Ohno et al. investigated the possibility that IFN-β may induce or down-regulate cellular miRNAs in human gliomas. They analysed the effect of IFN-β treatment on miR-21 expression in glioma cells and intracranial glioma xenografts. Systematic delivery of IFN-β markedly reduced the level of miR-21 in all glioma cells. The pri-miR-21 transcript levels decreased 6 hrs after the addition of IFN-β and began to recover after 48 hrs. These results indicate that decrease in the levels of miR-21 is the result of transcriptional suppression. In contrast, the addition of the STAT3-specific inhibitor increased the level of miR-21 and inhibited IFN-β–mediated suppression of miR-21, suggesting that miR-21 expression is negatively regulated by STAT3 [5].
Another study revealed miR-221 and miR-222 as possible regulators of IFN pathways. Using the KEGG pathway databases and BioCarta, Zhang et al. found that the IFN-α signalling pathway was the most significant pathway modulated by genes with the most different expression after knockdown of miR-221 and miR-222. The authors showed that STAT1 and STAT2 expression and phosphorylation were up-regulated in U251 cells with silenced miR-221/222. Tyrosine phosphorylation of STAT1 and STAT2 was present in the nucleus after repression of the same miRNAs. These data illustrate a mechanism of STAT1/2 up-regulation under the transcriptional control of IFN-α signalling after knockdown of miR-221/222 cluster in U251 glioma cells (Fig. 2) [26].
Notch signalling pathway
Notch signalling is critical in stem cell maintenance and cell survival, as well as in cell fate decisions such as neuronal versus glial fate in the developing nervous system. Therefore, it is not surprising that this pathway plays a key role in brain tumours, including glioblastoma [27].
miR-326 is associated with Notch signalling pathway in glioblastomas. This miRNA was first identified among a set of miRNAs expressed in neurons and further noted on a list of miRNAs elevated in zebrafish embryos treated with a Notch inhibitor [28, 29]. To find potential miRNA mediators of Notch effect in glioma, Kefas et al. performed miRNA microarray analysis of glioma tumour stem cells transfected with Notch-1 siRNA. In these Notch-1 knockdown cells, miR-326 was one of the miRNAs significantly increased when compared to control transfected cells. Therefore, it was indicated that miR-326 is suppressed by Notch activity. However, pre-miR-326 transfection caused substantial decrease in both Notch-1 and Notch-2 protein as was shown by immunoblotting. This paper showed that another Notch pathway components are inhibited by miR-326. It was observed that miR-326 induces apoptosis and decreases glioma cells proliferation, viability and invasiveness of glioblastoma stem cell-like lines. Furthermore, miR-326 transfection also reduced glioma cell tumourigenicity in vivo [27]. Considering that the expression of miR-326 down-regulated the Hedgehog stem cell pathway in medulloblastoma cells, authors tested the effects of this miRNA on Hedgehog activity in a glioma line using the Gli-1 promoter reporter plasmid. Unfortunately, expected effects were not observed in this case; this may reflect distinct roles for miR-326 in these pathways in different cancers [30].
Computational target gene prediction identified pyruvate kinase type M2 (PKM2) as another target of miRNA-326. PKM2 has recently been shown to play a key role in cancer cell metabolism. It is crucial for aerobic glycolysis and provides a growth advantage for tumour cells [31]. To investigate whether PKM2 might be a functionally important target of miR-326, Kefas et al. used RNA interference to knockdown PKM2 expression in glioma cells. Transfection of established glioma and glioma stem cells with PKM2 siRNA reduced their growth, cellular invasion, metabolic activity, ATP and glutathione levels and activated cAMP-activated protein kinase. Levels of PKM2 negatively correlated with levels of miR-326, suggesting regulatory relationship of PKM2 and miR-326 [32]. Among others, all of these results showed that efficient delivery of miR-326 has therapeutic potential against both glioma stem-like cells and established glioma lines.
Li et al. studied the role of miR-34a in human brain tumours with a special focus on glioblastomas. They found that miR-34a inhibits Notch-1 and Notch-2 protein expression and 3′-UTR reporter activities as well as CDK6 and c-Met protein expression in glioma cells. They observed for the first time that average pre-miR-34a expression is down-regulated in human glioblastoma tissues when compared to normal human brain [33]. Other studies showed that miR-34a expression was higher in wild-type p53 glioblastoma tissues compared to mutant p53 glioblastoma; miR-34a acts as a tumour suppressor in p53-mutant glioma cells U251, partially through regulating SIRT1 [34]. Transfection of miR-34a into tested glioblastoma cell lines strongly inhibited cell proliferation, cell cycle, cell survival, cell invasion and in vivo glioblastoma xenograft growth; however, the treatment did not affect human astrocyte cell survival and cell cycle. Forced c-Met and Notch-1/2 expression partially rescued the effects of miR-34a on the cell cycle and cell death in gliomas, respectively [35] (summarized in Fig. 3).
Fig 3.
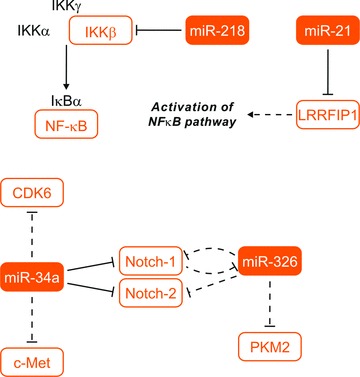
MiRNAs involved in notch and NF-κB signalling pathways. IKK-α/β/γ: inhibitor of κB kinase α/β/γ; NF-κB: nuclear factor of κB; IκB: inhibitor I κB; LRRFIP1: leucine rich repeat (in FLII) interacting protein 1; CDK6: cyclin-dependent kinase 6; c-MET: met proto-oncogene (hepatocyte growth factor receptor); PKM2: pyruvate kinase, muscle. Dashed lines indicated indirect regulation, solid lines indicate direct regulation. EGFR: epidermal growth factor receptor.
NF-κB signalling pathway
Nuclear factor-kappa B (NF-κB) is the transcription factor with pleiotropic activity owing to its central roles in various biological processes. Aberrant activation of NF-κB signalling pathway has been proved to be important for invasiveness and metastatic capacity of tumours through up-regulation of matrix metalloproteinases (MMPs) and transcription factors regulating E-cadherin, such as Snail, Twist or Slug. A critical component in NF-κB regulation is the IκB kinase (IKK-β) complex [36, 37].
Song et al. identified miR-218 expression in glioma cells lines and in human primary glioma tissues was substantially down-regulated, when compared to miR-218 expression in normal human astrocytes and normal brain tissues. Forced up-regulation of miR-218 dramatically reduced the migratory speed and invasive ability of analysed cells. Ectopic expression of miR-218 down-regulated matrix MMP-9 and reduced NF-κB transactivity at transcriptional level, whereas inhibition of miR-218 enhanced the expression of MMP-9 and transcriptional activity of NF-κB. Authors demonstrated that miR-218 could inactivate NF-κB/MMP-9 signalling by directly targeting the 3′-UTR of the IKK-β[37].
miR-21 was revealed as another post-transcriptional regulator involved in NF-κB signalling pathway in glioblastoma. Combining target prediction by bioinformatics with expression profiling, Li et al. identified LRRFIP1 gene, which was remarkably up-regulated in miR-21-knockdown cells, as a candidate target gene of miR-21. Further, through sequence analysis, they found that LRRFIP1 mRNA carried a putative miR-21 binding site. Further analyses confirmed LRRFIP1 as a direct target of miR-21. Moreover, their data suggest that miR-21 likely contributes to vepesid resistance through depression of LRRFIP1 expression, leading to the reduction of cytotoxicity of chemotherapeutic drugs through activation of the NF-κB pathway [38] (summarized in Fig. 3).
Single nucleotide polymorphisms and miRNAs: risk factors for glioblastoma
In gliomas, only one polymorphism found in mature miRNA sequence, specifically a polymorphism of miR-196a (rs11614913), has been studied so far. Published data suggest that the CC genotype of miR-196a (rs11614913) polymorphism is associated with decreased risk of glioma in the Chinese population (OR = 0.74, 95% CI: 0.56–0.98). Significant association was observed also between these genotypes and risk of particular glioma subgroups: patients over 18 years (OR = 0.73, 95% CI: 0.55–0.98), male glioma patients (OR = 0.69, 95% CI: 0.48–0.99) and patients with high-grade glioma-glioblastoma (OR = 0.58, 95% CI: 0.37–0.91). In contrast to other tumours, such as lung cancer and breast cancer [39, 40], data in glioblastoma showed opposite association between miR-196a genotype and cancer risk. This may be related to the diversity on the tissues origin and characteristic molecular alterations in different cancers [41].
miRNAs in glioblastoma prognosis and prediction of therapeutic response
Clinical significance of miRNA expression profiles in glioblastoma has not been explored very much. Nevertheless, 16 candidate miRNAs were published to associate with malignant behaviour of gliomas (miR-196a, miR-15b, miR-105, miR-367, miR-184, miR-196b, miR-363, miR-504, miR-302b, miR-128b, miR-601, miR-21, miR-517c, miR-302d, miR-383, miR-135b). Among them, miR-196a and miR-196b indicated the highest level of significance (P = 0.0038 and 0.0371, respectively). Both miRNAs showed increased expression levels in glioblastomas relative to anaplastic astrocytomas and normal brain tissues. Higher level of miR-196 transcript significantly correlated with poorer survival as demonstrate by the Kaplan–Meier method (P = 0.0073); moreover, multivariate analysis showed that these expression levels were independent predictors of overall survival in glioblastoma patients (P = 0.021; HR, 2.81) [42]. Malzkorn et al. investigated the miRNA expression profiles in four patients with primary WHO grade II gliomas that spontaneously progressed to WHO grade IV secondary glioblastomas. They identified 12 miRNAs (miR-9, miR-15a, miR-16, miR-17, miR-19a, miR-20a, miR-21, miR-25, miR-28, miR-130b, miR-140 and miR-210) showing increased expression and two miRNAs (miR-184 and miR-328) showing reduced expression upon tumour progression. Validation experiments on an independent series of primary low-grade and secondary high-grade astrocytomas confirmed miR-17 and miR-184 as interesting candidates contributing to glioma progression [43].
Treatment of malignant gliomas remains one of the greatest challenges facing oncologists today through a frequent resistance to both chemo- and radiotherapeutics and short survival [44]. Important question for management of glioblastoma patients is the possibility of predicting therapeutic outcome. The miRNA expression profiles of glioblastoma tissues have shown association of miR-181b and miR-181c with response to concomitant chemoradiotherapy with temozolomide (RT/RMZ). MiR-181b and miR-181c were significantly down-regulated in glioblastoma tissue of patients who responded to RT/TMZ (P = 0.016 and 0.047, respectively) in comparison to patients with progressive disease [9].
miRNAs involved in drug resistance of glioblastoma
Temozolomide is an oral alkylating agent, which is frequently used for the treatment of glioblastoma. To explore the mechanism of resistance to TMZ, Shi et al. found that overexpression of miR-21 in glioblastoma cells could significantly reduce TMZ-induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity [45] The miR-21 inhibitor could also enhance the chemosensitivity of human glioblastoma cells to paclitaxel via inhibition of STAT3 expression and phosphorylation. Moreover, the same treatment by miR-21 antisense oligonucleotides led to enhanced cytotoxicities of vepesid [12, 21]. The results of glioblastoma in vitro experiments showed also other miRNAs involved in the TMZ resistance. miR-195, miR-455–3p and miR-10a* were the three most up-regulated miRNAs in the drug resistant glioblastoma cell line (U251R) [46].
Multidrug resistance protein ABCG2 (ATP-binding cassette sub-family G member 2) is the target gene for miR-328. Li et al. observed that miR-328 is underexpressed in many cancers including glioblastoma and contributes to tumour chemoresistance through ABCG2, which is highly expressed in glioblastoma cells [47]. Another research group reported the possible impact on the therapeutic effect by transfection of miR-451 in combination with imatinib mesylate treatment. Up-regulation of miR-451 led to differentiation of glioblastoma stem cells [48].
miRNAs as potential therapeutic targets
The association of miRNA deregulation with pathogenesis and progression of malignant disease illustrates great potential of utilizing miRNAs as targets for therapeutic intervention. The basic strategy of current miRNA-based treatment studies is either to antagonize the expression of target miRNAs with antisense technology or to restore or strengthen the function of given miRNAs to inhibit the expression of certain protein-coding gene. There is a number of experimentally, in vitro or/and in vivo, proved miRNAs presenting potential therapeutic targets in glioblastoma, which were mentioned in the context of altered signalling pathways (e.g. miR-21, miR-451, miR-7, miR-128, miR-221/222).
Considering angiogenesis, which is critical in most solid tumours, including glioblastoma, miR-296 has been demonstrated to be up-regulated in glioblastoma-associated endothelial cells [49]. This miRNA promotes angiogenesis by down-regulating HGS (hepatocyte growth factor-regulated tyrosine kinase substrate), an inhibitor of pro-angiogenic receptors VEGFR2 and PDGFRb (vascular endothelial growth factor and platelet-derived growth factor receptor β, respectively). This study indicated the potential of anti-angiogenic therapy of glioblastoma by delivery of a miR-296 inhibitor [49].
Although siRNAs allow specific knockdown of individual gene targets, miRNAs result in a broad reduction of gene expression being affected. The ability of individual miRNAs to target multiple genes/pathways could be a major advantage, especially given studies indicating the therapeutic necessity of simultaneously targeting multiple pathways in glioblastoma [50]. Unfortunately, there are several major challenges to overcome before the application of miRNA-based treatment. First, the multitargeting nature of miRNAs gives the risk of unintended off-target effects that need to be carefully evaluated. Secondly, the expression of target gene may be controlled by several different miRNAs, which may compromise the effect of miRNA-based treatment. Finally, there is still lack of miRNA delivery system with enough specificity and efficacy.
Conclusions
The discovery of miRNAs has substantially changed the view on gene expression regulation, and new findings over the past few years have catapulted miRNAs into the centre of cancer molecular biology. It is now evident that dysregulation of miRNAs is an important step in the development of many cancers, including glioblastoma. Several studies based on expression profiling have proven there are significant changes of miRNA expression levels in glioblastomas in comparison to adult brain tissue; these expression levels identified groups of miRNAs with potential of prognostic stratification and prediction of responses to chemoradiotherapy in glioblastoma patients. To improve our knowledge of role of miRNAs in glioblastoma core signalling pathways, functional effects of particular miRNAs were successfully studied. The results of these studies suggest a great potential and relevance of miRNAs as a novel class of therapeutic targets and possibly powerful intervention tools in glioblastoma.
Acknowledgments
This work was supported by grant IGA NT/11214-4/2010 and IGA NR 9875-4 of the Czech Ministry of Health, Project No. MZ0MOU2005 of the Czech Ministry of Health and by the project “CEITEC – Central European Institute of Technology” (CZ.1.05/1.1.00/02.0068). Infrastructural part of this project (Institute of Molecular and Translational Medicine) was supported from the Operational Programme Research and Development for Innovations (project CZ.1.05/2.1.00/01.0030).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2005;64:6892–9. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–89. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 3.Novakova J, Slaby O, Vyzula R, et al. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 4.Schwartzbaum JA, Fisher JL, Aldape KD, et al. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 5.Ohno M, Natsume A, Kondo Y, et al. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res. 2009;7:2022–30. doi: 10.1158/1541-7786.MCR-09-0319. [DOI] [PubMed] [Google Scholar]
- 6.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Science. 2010;101:2309–15. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schickel R, Boyerinas B, Park SM, et al. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–74. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 8.Ciafré SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Slaby O, Lakomy R, Fadrus P, et al. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–9. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 10.Jancík S, Drábek J, Radzioch D, et al. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;150960:1–13. doi: 10.1155/2010/150960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, Zhou X, Mei M, et al. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Huang W, Jiang X, et al. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci USA. 2010;107:2183–8. doi: 10.1073/pnas.0909896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nan Y, Han L, Zhang A, et al. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 16.Kefas B, Godlewski J, Comeau L, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 17.Webster RJ, Giles KM, Price KJ, et al. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 18.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/ stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 19.Song LB, Li J, Liao WT, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–36. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Han L, Ge Y, et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–20. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 21.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Liu W, Chao T, et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Zhang J, Jia Q, et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep. 2010;24:195–201. doi: 10.3892/or_00000846. [DOI] [PubMed] [Google Scholar]
- 24.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Han L, Zhang A, et al. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol. 2010;36:1503–12. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- 27.Kefas B, Comeau L, Floyd DH, et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–8. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–5. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thatcher EJ, Flynt AS, Li N, et al. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn. 2007;236:2172–80. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- 30.Ferretti E, De Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–27. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitosugi T, Kang S, Vander Heiden MG, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kefas B, Comeau L, Erdle N, et al. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–12. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guessous F, Zhang Y, Kofman A, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–6. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luan S, Sun L, Huang F. MicroRNA-34a: a novel tumor suppressor in p53-mutant glioma cell line U251. Arch Med Res. 2010;41:67–74. doi: 10.1016/j.arcmed.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 37.Song L, Huang Q, Chen K, et al. miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-β. Biochem Biophys Res Commun. 2010;402:135–40. doi: 10.1016/j.bbrc.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Li W, Yang Y, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–8. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 39.Hu Z, Shu Y, Chen Y, et al. Genetic Polymorphisms in the pre-MicroRNA Flanking Region and Non-Small-Cell Lung Cancer Survival. Am J Respir Crit Care Med. 2011;183:641–8. doi: 10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- 40.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 41.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136:1853–9. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan Y, Mizoguchi M, Yoshimoto K, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–97. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 43.Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–50. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler DS, Wright RD, Kesari S, et al. Resistance of human glioblastoma multiforme cells to growth factor inhibitors is overcome by blockade of inhibitor of apoptosis proteins. J Clin Invest. 2008;118:3109–22. doi: 10.1172/JCI34120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi L, Chen J, Yang J, et al. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352:255–64. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Ujifuku K, Mitsutake N, Takakura S, et al. miR-195, miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–8. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Li WQ, Li YM, Tao BB, et al. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance. Med Sci Monit. 2010;16:HY27–30. [PubMed] [Google Scholar]
- 48.Gal H, Pandi G, Kanner AA, et al. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- 49.Wurdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2010;14:382–93. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purow B. The elephant in the room: do microRNA-based therapies have a realistic chance of succeeding for brain tumors such as glioblastoma. J Neurooncol. 2010;102 doi: 10.1007/s11060-010-0449-5. doi: 10.1007/s11060-010-0449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


