Abstract
Influenza is a continuing world-wide public health problem that causes significant morbidity and mortality during seasonal epidemics and sporadic pandemics. The existing vaccination program is variably effective from year to year, and drug resistance to available antivirals is a growing problem, making the development of additional antivirals an important challenge. Influenza virus non-structural protein 1 (NS1) is the centerpiece of the viral response to the host interferon (IFN) system. NS1 was demonstrated previously to be a potential therapeutic target for antiviral therapy by the identification of specific small-molecule inhibitors. One inhibitory compound, NSC125044, was subjected to chemical evaluation. Initial synthetic work comprised simplifying the core structure by removing unwanted functionality and determination of key features important for activity. Several subclasses of molecules were designed and synthesized to further probe activity and develop the basis for a structure activity relationship. Apparent potency, as judged by activity in virus replication assays, increased dramatically for some analogs, without cytotoxicity. Results suggest that the target binding site tolerates hydrophobic bulk as well as having a preference for weakly basic substituents.
1. Introduction
Influenza is a continuing worldwide public health problem. The World Health Organization estimate of annual mortality due to seasonal influenza is 250,000 to 500,000, and there are approximately 30,000 influenza associated deaths and 200,000 hospitalizations in the United States per year 1. Severe infections are most commonly seen in the elderly, the very young, and the chronically ill 2–4.
In addition to seasonal epidemics, there have been four documented influenza pandemics during this and the previous century, the most serious occurring in 1918. During an eight month period the 1918 Spanish influenza claimed 20–40 million lives 5. Two subsequent pandemics in 1957 and 1968 were less severe but shared the characteristic of having avian origin 6. There is concern that another devastating pandemic is inevitable, perpetuated by the spread of the avian H5N1 virus through multiple bird populations in Asia, Africa and Europe, which periodically causes human infections 7–10. Although avian H5N1 has not acquired the ability to transmit from person to person, those who are infected through direct contact with birds are at a very high risk for mortality, approximately 60% 11, 12. The H1N1 swine influenza pandemic of 2009 has added weight to concerns regarding the pandemic threat 13–15.
The most common method for combating influenza virus is vaccination. However, viral antigenic drift dictates that a new vaccine be developed each year, and the effectiveness of the vaccine is variable 16. In addition there are obstacles to worldwide vaccine distribution, and reluctance on the part of some to vaccine administration 17. Two classes of drugs have been used to treat influenza infections, including one that targets the viral M2 ion channel and one that inhibits the viral neuraminidase protein. Due to the emergence of viral strains that are resistant to M2 and neuraminidase inhibitors, the variable effectiveness of seasonal vaccines, and the high probability of future pandemics, it is important to identify additional viable viral targets to treat the influenza infections 18.
Previously we identified non-structural protein 1 (NS1) of influenza virus as a novel drugable antiviral target 1, 19. NS1 is encoded by all strains of influenza A and is highly conserved 20–22. It is a multifunctional protein with essential roles in viral replication and evasion of the cellular innate immune response. The central role of NS1 in virus propagation and spread make it an attractive drug target 20, 21. Numerous studies have shown that NS1 participates in a wide range of functions. NS1 binds to double stranded RNA in a non-sequence specific manner 23, 24. In doing so NS1 is able to shield dsRNA from detection by the 2’-5’ oligo(A) synthetase (OAS)/RNase L pathway, which is responsible for degradation of viral RNA 25. NS1 also binds to the 30 kDa subunit of cleavage and polyadenylation specificity factor (CPSF30), and poly (A)-binding protein II 26, 27. In doing this the 3’-end processing of cellular mRNAs is inhibited, impeding the metabolism of essential RNAs. Among these are mRNAs for interferons (IFNs), which are produced in response to viral infection 26–29. Additionally NS1 associates with cellular protein kinase R (PKR), thereby preventing it from phosphorylating translation elongation factor eIF-2α and allowing viral protein synthesis to continue 30, 31. Recently, it has been demonstrated that NS1 interacts with TRIM25. This interaction prevents the activation of retinoic acid-inducible gene I (RIG-I), thereby suppressing the production of cellular IFN 32–34.
In a recent study we identified several compounds that specifically inhibit NS1 function in cells 1. Importantly, four of the compounds with anti-NS1 activity also demonstrated significant antiviral activity in cell culture assays and reversed NS1-dependent blockade of the cellular interferon synthesis pathway 1. In this report we describe the design, synthesis and biological testing of a series of compounds based on one of the compounds reported previously, NSC125044. These studies have resulted in several highly potent antiviral compounds.
2. Results
2.1 Method validation
Efficacy of an antiviral compound in cell culture is a complex function of its activity, permeability, and other factors. In addition, virus replication is a multi-dimensional non-linear process, so the overall effect of chemical inhibition of a viral target may be complex as well. In the case of inhibition of NS1 function there is the added complexity of the interplay between NS1 and the host cell innate immune response, which involves modulation of cellular interferon production and the consequences of it downstream antiviral effects 20.
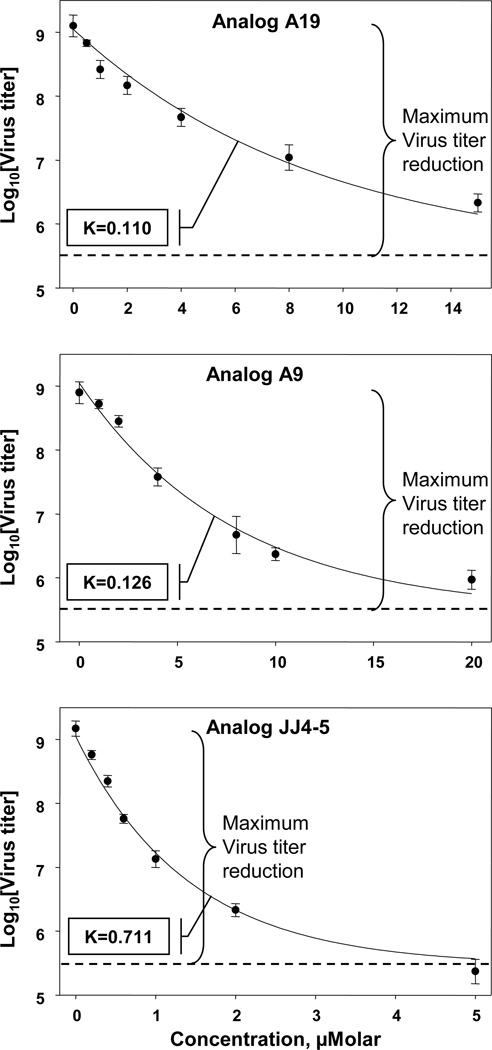
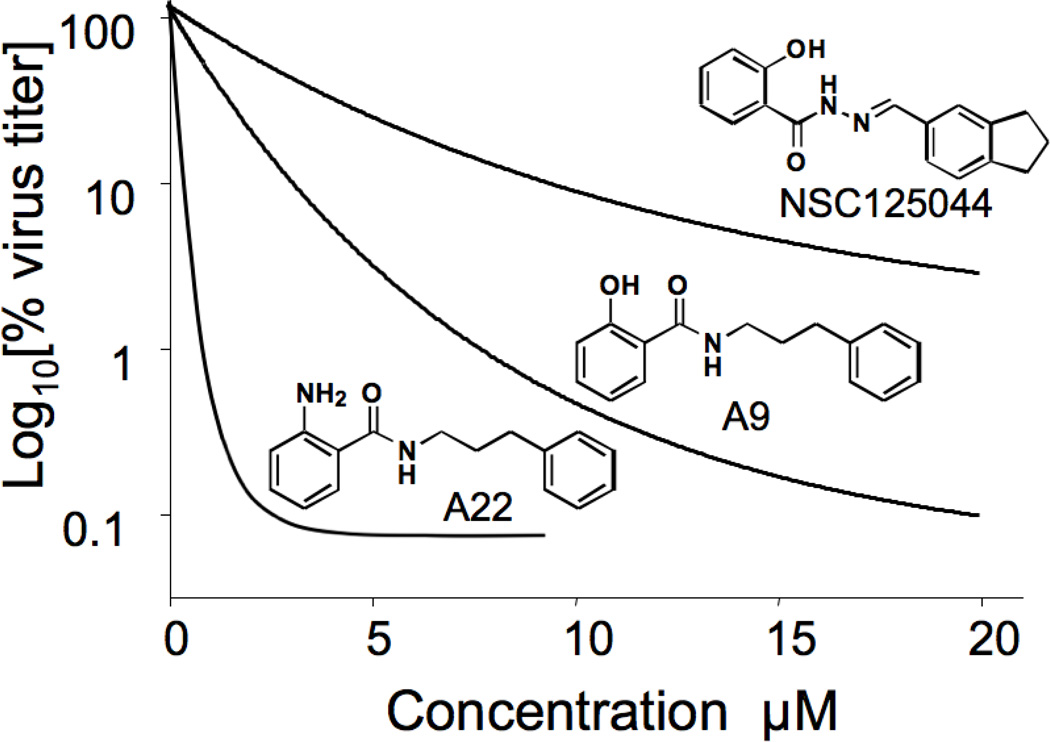
As described in Methods a live cell virus replication assay was used throughout this study. Interestingly, in comparing the efficacy of compounds we noticed that we could discriminate between them based two parameters, (1) apparent potency and (2) maximum efficacy. Medicinal chemistry decisions took both factors into account. To fully present our data we have included graphs depicting representative concentration curves for some compounds so that these can be inspected visually (in Figures or Supplemental Data). Furthermore, examination of the general shape of the relationship between Log10(virus titer) and concentration suggested an exponential decay that approached a residual virus replication level at high compound concentrations. This analysis yielded two parameters of interest: (1) a constant K, indicative of apparent potency and (2) a maximum inhibition of virus replication (maximum efficacy), being the difference between the no-compound value and that observed at high concentrations. To establish that this relationship held for compounds of low and high activity three analogs were chosen and assayed in triplicate. As can be seen from Figure 1, in each case the data were an excellent fit to an exponential decay relationship with readily determined values of K and maximum efficacy. For some of our early compound series, data were collected to determine the maximum inhibition of virus replication at a single, high concentration (e.g. Table 1), which provided sufficient information to make initial medicinal chemistry decisions.
Figure 1.
Determination of decay constant K constant for virus replication in cells treated with NS1 inhibitors. Cells were infected with influenza virus A/PR/8/34, treated with the indicated compounds and analyzed for virus production as described in Methods. Determination of K was based on a fit to a first order exponential relationship.
Table 1.
| Compound |
aLogTCID50 (viral titer) |
bFold decrease in replication |
|---|---|---|
| Control (DMSO) | 8.5 | - |
| A1 | 7.1 | 25 |
| A8 | 7.1 | 25 |
| A9 | 5.7 | 630 |
| A11 | 6.8 | 50 |
| A12 | 6.2 | 200 |
| A13 | 6.5 | 100 |
| A14 | 6.3 | 158 |
| A15 | 5.8 | 500 |
| A16 | 6.5 | 100 |
Viral titer, expressed as logTCID50, was determined from supernatants of MDCK cells infected for 48 hours and treated for the entire period with 50 µM of the indicated compounds.
Fold decrease in virus replication was determined by subtracting the logTCID50 value for each compound from 8.5 (control value, see Table) and calculating the inverse log.
2.2 Replacement of compound A1 indane with simple benzene ring retains activity
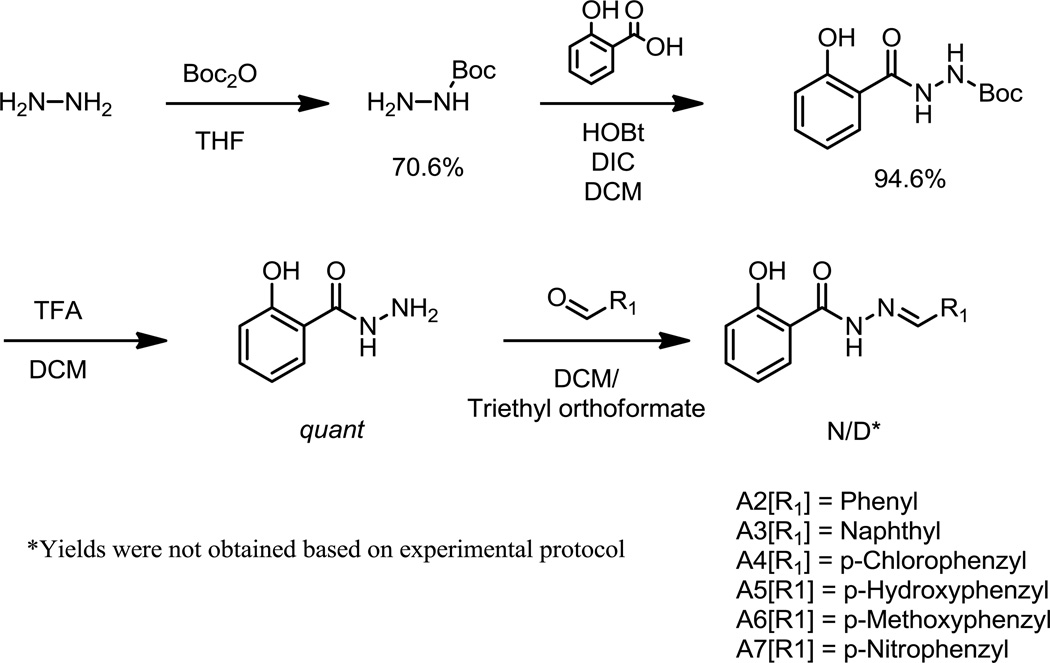
As previously reported compound A1 (NIH compound NSC125044) was identified from a screen for functional inhibitors of NS1 1. Examination of A1 in terms of developing a structure activity relationship (SAR) suggested that the right hand side indane was the least amenable to chemical manipulation, and commercial availability of building blocks was judged an impediment. In a first series of compounds (Figure 2; A2–A7) we therefore examined the necessity of the indane ring to the inhibition of virus replication. Analogs introducing a variety of alternative functionalities to the indane were synthesized through the use of para-substituted benzaldehyde derivatives (Scheme 1). These compounds were assessed using a single high concentration (50 µM) to compare their maximum efficacies. None of the resulting compounds (A2–A7) were significantly different in activity from compound A1 (data not shown). Modifications included incorporation into the benzene ring of either electron donating or withdrawing substituents, as with hydrogen bond donating and accepting functional groups. Additionally, replacement of the indane with naphthalene (A3) was accomplished. Of the analogs synthesized, compound A2 contained a simple benzene ring to replace the indane. Given the difficulty of pursuing an SAR with retention of the indane ring system it was decided to use analog A2 as the basis for further development.
Figure 2.
Hydrazide analogs
Scheme 1.
2.3 Removal of hydrazine, and carbon spacer optimization
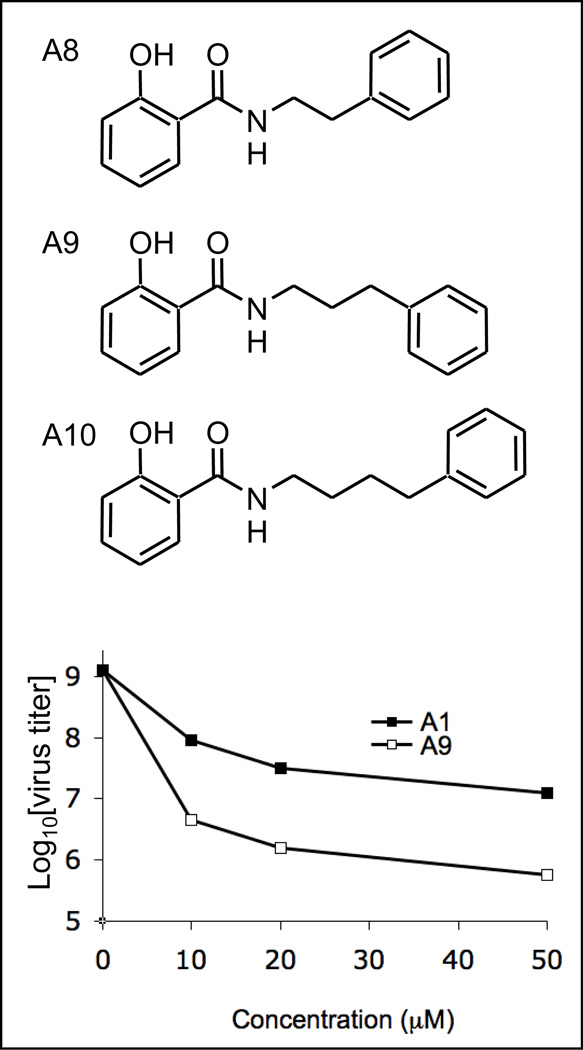
The hydrazine functionality of compounds A1 and A2 is an unattractive feature with potentially poor pharmacokinetic properties. Three molecules were synthesized that retained the amide bond nitrogen, but replaced the other with an alkyl chain (Figure 3). A8 was synthesized using 2-phenylethylamine as one of the building blocks with retention of the bond distance between the two rings. This substitution exhibited similar inhibitory activity to the compound A1 hydrazine parent (Table 1), and indicated that the hydrazine is not an essential motif required for activity. Further studies explored the chain length between the amine and the phenyl ring. Compound A9 with a three carbon spacer resulted in a substantial increase in activity over a wide concentration range (Table 1, Figure 3 and Figure 1). Also the maximum effects of A1 and A9 at high concentrations were substantially different as illustrated in Figure 3. Compounds A1 and A9 were found to be non-toxic to MDCK cells up at up to 100 µM concentration1, 19. It was observed that the three and four carbon chain spacers (A9 and A10) displayed approximately the same activity (not shown). With the better commercial availability of building block sets, the three carbon analog (A9) was chosen over the four carbon A10 as the new lead molecule. These positive results confirmed the previous findings regarding substitution of the phenyl ring on the right hand side, and also simplified the synthetic protocols from four steps down to one (see Scheme 2).
Figure 3.
Ortho hydroxybenzamide analogs
Scheme 2.
2.4 Position of the left hand side hydroxyl
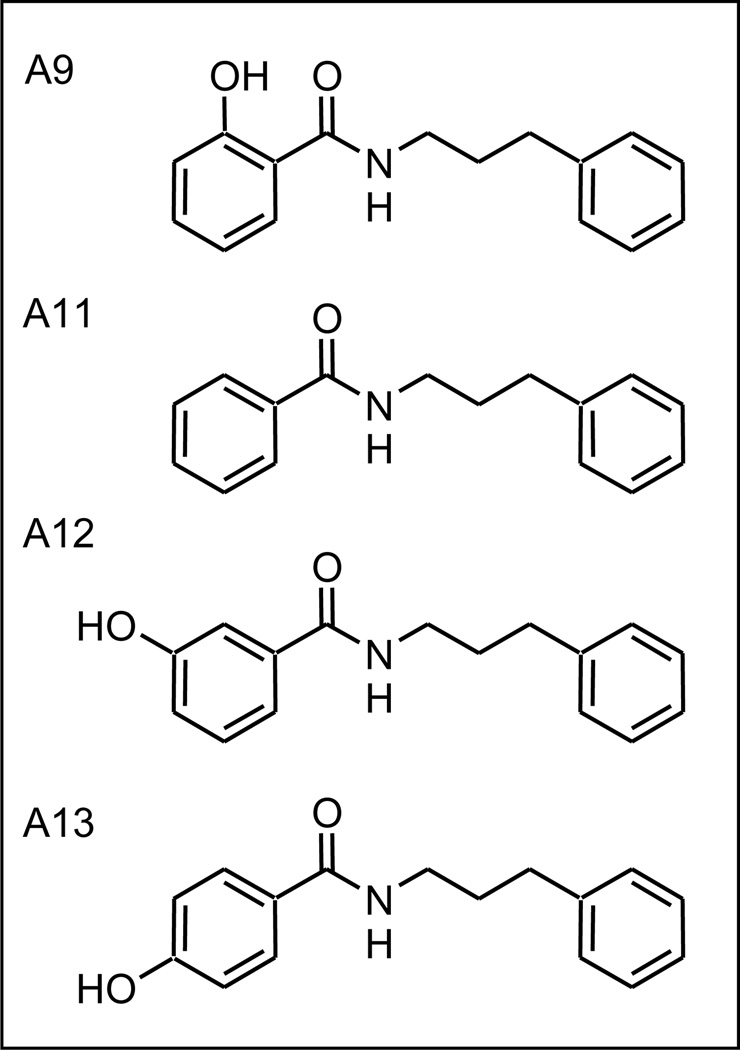
In A9, the phenol hydroxyl is ortho with respect to the amide linking group. As shown in Table 1 and Figure 4, analogs with either the hydroxy in the meta or para position (A12 and A13), resulted in decreased in activity. Furthermore, the incorporation of a simple benzene ring (A11) also led to a decrease in activity. These results indicated a preference for an ortho substituted phenyl ring as the preferred left hand side (LHS) functionality.
Figure 4.
Benzamide analogs
2.5 Substitution at the left hand side ortho position
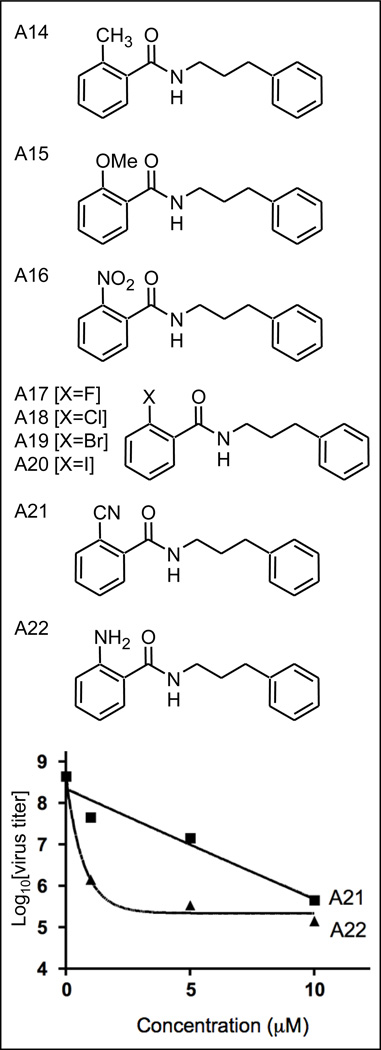
The optimum substitution pattern for the molecule was determined to be the ortho position. To examine the requirement for a hydroxyl group, as in A9, various ortho substituted benzoic acids were selected to produce analogs A14–A22 (Figure 5) and to further explore electronic effects at this position on virus inhibition.
Figure 5.
Phenylpropylamide analogs
No clear trend was observed. Methyl (A14) and methoxy (A15), both weaker electron donating groups than hydroxy led to weaker virus inhibition (Table 1). Compounds A16 (Table 1) and A21 (Figure 5) also showed weak activity. In contrast a substantial increase in virus inhibition was observed for the ortho substituted amino (A22 K=1.40 compared to A9 K=.126; see also Figure 5). Like compound A9, compound A22 was found to be non-toxic in MDCK cells up to 100 µM (data not shown). Some halogen analogs (A18 and 20) but not others (A17 and A19) also displayed greater activity than compound A9; see Supplemental Figure S1). These results would seem to rule out hydrogen bonding, either as an acceptor or donator, as a role for the hydroxy functionality in interacting with its target. An additional consideration is the possible intramolecular hydrogen bonding between the ortho hydroxyl group and the carbonyl group through a six-membered ring. This can partially lock the molecule in a conformation which may affect compound cell permeability. Also of interest was the finding that analog A20 (I-substituted) at high concentrations gave a lower overall reduction in virus replication than the parent hydroxy containing analog despite a greater apparent potency at lower concentrations (see Supplemental Figure S1).
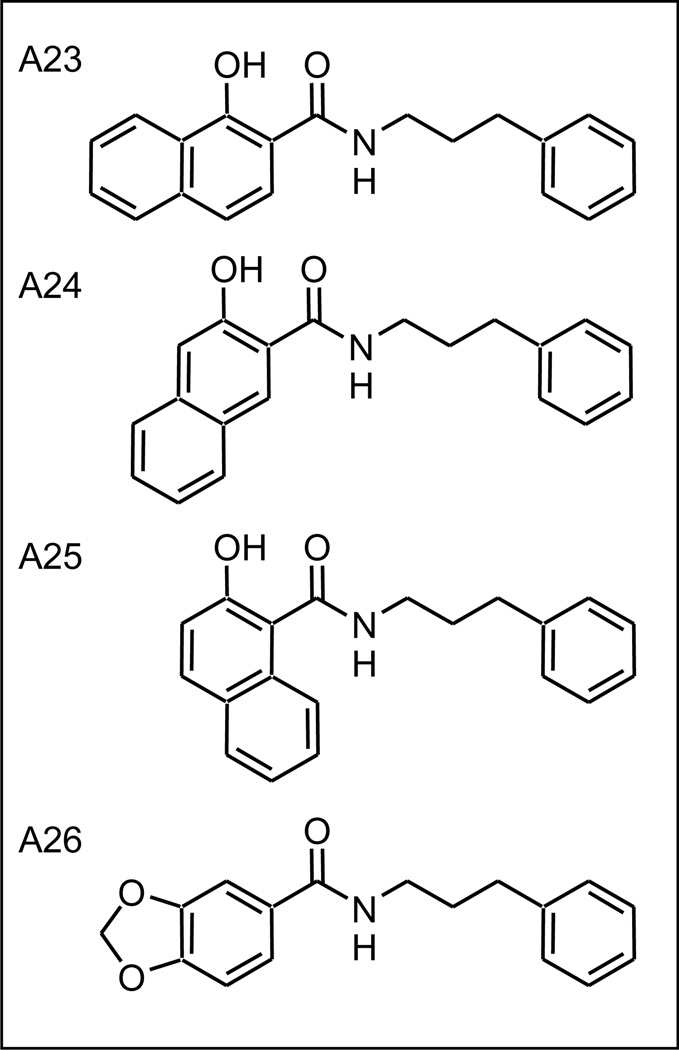
2.6 Increased hydrophobic bulk on the left hand side increases activity
While retaining a hydroxy functionality ortho to the linking amide bond, possible size restrictions associated with the LHS of the molecule were examined. Three, hyroxy-naphthoic acid derivatives were used in the synthesis of new compounds (Figure 6). Each of these analogs (A23–A25) project the second aromatic ring in a different direction probing both the size and shape of the target binding site. The observed virus inhibition of these analogs was similar (A23 K=657; A24 K=546; A25 K=658; see also Supplemental Figure S2) but significantly better than for the parent (A9, K=0.126), suggesting a large hydrophobic LHS binding pocket in the binding site. The activity of analog A26 (K=0.15), another non-aromatic fused ring system but without the presence of the ortho hydroxy function, is consistent with this hypothesis but has overall weaker activity due to its unsubstituted benzene LHS, as also observed for A11 (Table 1).
Figure 6.
Naphthoic acid analogs
2.7 Pyridyl and Quinolyl analogs
As mentioned above it was demonstrated that replacing the α-hydroxy group by an amine led to greater virus inhibition for compound A22 (K=1.40). Using picolinic acid (Figure 7) for the LHS (A27), with a weakly basic ring nitrogen, was less active (K= 0.771). Combination of the ring nitrogen in the context of the naphthalene ring system, analogs A28–A30, were synthesized to take advantage of the suggested larger LHS binding site (Figure 7). These three quinolyl derivatives, similar to the naphthalene fused ring systems series (A23–A25), showed an improvement in virus inhibition activity over compound A27, with compound A28 being somewhat more active (Supplemental Figure S3).
Figure 7.
Pyridyl analog A27 and Quinolyl analogs A28 – A30
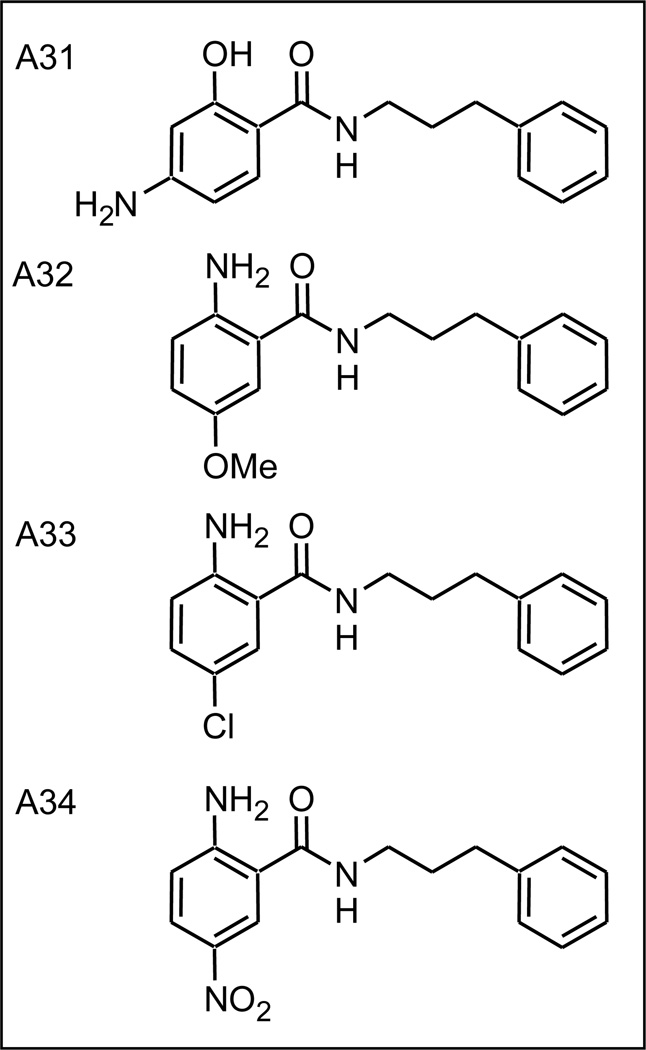
2.8 Left hand side substitution leads to highly potent compound A32
Having determined that both hydroxy and amine ortho substituted benzamides, and the dual ring system analogs, showed potent virus inhibition activity, a series of analogs with more than one functionality in the LHS were synthesized (Figure 8; compounds A31–A34). For compound A31, addition of an amino group para to the amide group increased activity a small amount compared with A9 (A9 K=0.126; A31 K=0.22). Similarly the addition of a para-substituent, chloro (A33, K=0.27), methoxy (A32, K=0.73) or nitro (A34, K=0.86) gave analogs with increased virus inhibitory activity (Supplemental Figure S4).
Figure 8.
Disubstituted benzylamide analogs
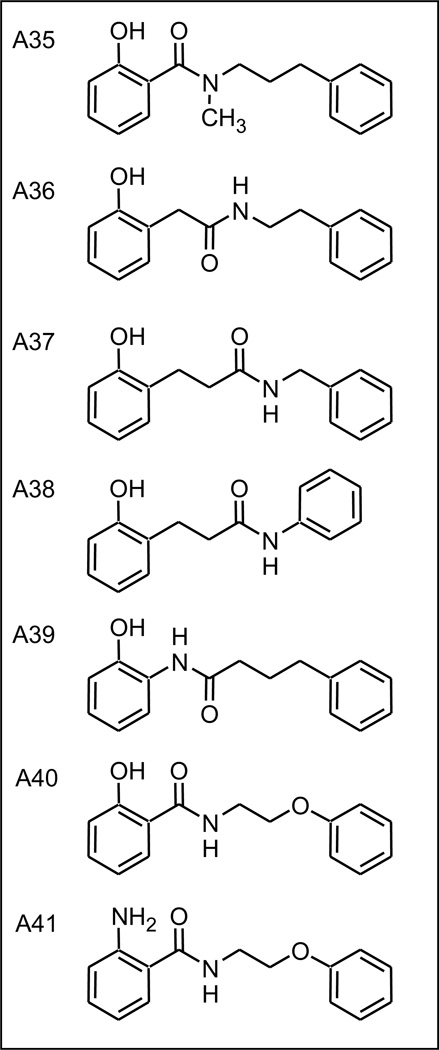
2.9 Examination of the linking region
In a last set of analogs (Figure 9) we explored within the linking chain between the LHS and RHS, the requirement of a secondary amide (A35), the position of the amide bond (A36 - A38), and the direction of the amide bond (A39). In all cases these analogs were less active than the corresponding ortho hydroxy parent A9 (data not shown). Of more interest were analogs A40 and A41, two compounds in which the RHS-most methylene was replaced by an ether oxygen. This mode of assembly was chosen to allow future development of the RHS SAR, via incorporation of substituted phenols, of which many versions are commercially available. In the case of the ortho-amino analog virus inhibitory activity was unchanged, (A22, K=1.40 to A41, K=1.46), whereas for the ortho-hydroxy analog a substantial increase was observed, (A9, K=0.126 to A40, K=1.15). Data for compounds A40 and A41 can be found in Supplemental Figure S5.
Figure 9.
Examination of the linking region
3. Discussion
Starting from hit NSC125044 (compound A1), obtained from a screen as reported peviously 1, a systematic analoging approach has yielded a simple molecule that inhibits virus replication in cells by greater than three orders of magnitude (Figure 5, Supplemental Figure S5, and Figure 10). This is virtually the same magnitude of effect on replication observed with a virus carrying a deletion of the NS1 gene (“delNS1”) in the A/PR/8/34 background, the same used in this study 35. This strongly suggests that members of the compound series reported here are capable of nearly complete abolishment of NS1 function in the cellular context. Interestingly, 100% of mice inoculated with the delNS1 virus survived infection, indicating that ablation of NS1 function is strongly protective in vivo 35. In vivo studies with several of the compounds reported here are ongoing.
Figure 10.
Improvement in apparent potency and maximum efficacy in cell culture
Not surprising is the finding that the many of the features of the originally identified molecule NSC125044 were not essential for activity; with only ~ 2000 compounds library used in the initial screen the likelihood of obtaining a near optimal molecule is very small. As was shown in these studies, optimally the LHS requires a large hydrophobic core with a weakly basic function, the RHS was adequately represented as a simple aromatic ring, and the linking region comprised a left justified amide bond followed by a 3 or 4 methylene chain.
The SAR as developed herein relied on a resource intensive cell culture virus replication assay. In this context it is recognized that the SAR is at a minimum the combination of both the cell penetration properties of the molecule as well as its ability to bind its molecular target. As shown in Figure 5, compound A22 triggered a greater than 2 log effect on replication at 1 µM, indicating that the KD for its target is quite low. Nonetheless, establishment of a direct binding assay will allow direct comparison of analogs in terms of binding constants (KD values), and further chemical optimization based on affinity. Experiments of this type are being pursued.
4. Experimental
4.1 Biological Methods
4.1.1 Mammalian cells and viruses
MDCK cells were maintained in Iscove medium supplemented with 10% fetal bovine serum and 2 mM L-glutamine. Medium and serum were from Invitrogen. For infections, viral stocks were diluted in growth medium supplemented with 0.3% bovine serum albumin, 0.22% sodium bicarbonate, and 0.25 U of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Invitrogen)/ml. Influenza virus A/PR/8/34 was propagated in 10-day-old embryonated chicken eggs at 37°C. Titers of influenza virus were determined by 50% tissue culture infective dose (TCID50) analysis on MDCK cells using the hemagglutination assay protocol of Reed and Muench 36.
4.1.2 Virus replication assays
Confluent cell monolayers were infected at a multiplicity of infection (MOI) of 0.1 for 48 h in the presence or absence of compound or DMSO. Compounds were added at the beginning of infection and were present throughout the infection. After 48 h virus titers were determined by TCID50 analysis as described previously 36.
4.2 Chemistry
Reagents and solvents used for synthesis were reagent grade and used as received. HOBt and DIC were purchased from Advanced ChemTech/Creosalus. HPLC grade H2O and MeCN were purchased from VWR and were used as received. All other reagents and solvents were purchased from Acros Organics, Fisher Scientific, or Sigma-Aldrich.
1H NMR spectra were obtained using a Varian UnityInova 300/54 at 300K unless otherwise noted. Chemical shifts were reported in parts per million (δ) relative to the solvent as follows: CDCl3 (δ 7.24) and DMSO-d6 (δ 2.50). NMR spectra data are abbreviated as follows: s, singlet; d, doublet; t, triplet; q, quartet; p, pentet; m, multiplet.
High-resolution mass spectrometry data was obtained from the Mass Spectrometry Laboratory in the School of Chemical Sciences at the University of Illinois Urbana-Champagne (Urbana, IL).All analytical High Performance Liquid Chromatography analysis was performed using Gilson Unipoint software and analytical equipment. This includes Gilson 215 Liquid Handler, Gilson 306 pumps, Gilson 811c Dynamic Mixer, Gilson 806 Manometric Module, and a Gilson 170 Diode Array Detector with deuterium lamp. The column used was a Thomson Instrument Inc. Advantage C18, 60A, 5u, 250 × 4.6 mm column. In conjunction with the HPLC analysis, mass spectrometry data was collected simultaneously using a single quad PE SCIEX API 150EX mass spectrometer utilizing Analyst software. A two-solvent mobile phase gradient was used for all LCMS analysis, starting at 90% water/0.01% TFA and 10% acetonitrile/0.01% TFA. The mobile phase changed linearly to 100% acetonitrile/0.01% TFA over 10 minutes. The 100% acetonitrile/0.01% TFA phase was held for an additional 10 minutes. A constant flow rate of 1.0 mL/min was maintained for the duration of analysis. Retention times are reported in minutes and abbreviated as tR; mass to charge ratios are reported as m/z.
Preperative HPLC was performed using a Gilson 215 Liquid Handler, Gilson 306 pumps, Gilson 811c Dynamic Mixer, Gilson 805 Manometric Module, and a Gilson UV/VIS-156 detector. A Thomson Instrument Inc. Advantage C18, 60A, 5u, 250 × 10.0 mm column and a Thomson Instrument Inc. Advantage C18, 60A, 5u, 250×21.2mm column were used in the purification of samples. A two-solvent mobile phase gradient was used which started at 90% water/0.01% TFA, 10% acetonitrile/0.01% TFA and changed linearly to 100% acetonitrile/0.01% TFA over 10 minutes. The 100% acetonitrile/.01% TFA phase was held for an additional 10 minutes. When the 250 × 10.0 mm column was used, a constant flow rate of 5.0 mL/min was used. When the 250 × 21.2 mm column was used, a constant flow rate of 18.0 mL/min was used for the duration of the method.
4.2.1 General procedure A for the final imine synthesis
The benzhydrazide was dissolved in a 1:1 mixture of triethyl orthoformate in DCM. To this, 1.1 equivalents of the benzaldehyde derivative were added andthe reactions were mixed overnight at room temperature. LCMS analysis of the reaction mixture indicated the formation of the desired product. The reaction mixture was concentrated under reduced pressure and preceded directly to preparative HPLC purification without further work-up.
4.2.2 General procedure B - amide bond formations which lead to final products
1 mmol of the carboxylic acid was dissolved in DCM. To this, 1.1 equivalents of HOBt and the desired primary amine were added. Following this, 1 eq of DIC was added to the mixture and the reaction was mixed overnight. Upon completion of the reaction, the diisopropylurea was filtered away and the DCM layer was washed with saturated bicarbonate solution (1×) and 2M HCl (1×). The organic layer was dried over anhydrous sodium sulfate and the solvent was then removed under reduced pressure. Typical yields for all amide bond formations were ≥ 80%. A small portion of the resulting compounds were carried through and used for HPLC purification.
4.2.3 tert-butyl hydrazinecarboxylate
To a round bottom flask equipped with a stir bar, 25mmol of hydrazine monohydrate was dissolved in THF at 0°C. Added drop-wise to this was a solution of Boc2O in THF. The reaction was allowed to mix at 0°C for an additional hour. The reaction was then diluted with ethyl acetate and washed with water (2x). The organic layer was dried over Na2SO4 and the solvent was removed under reduced pressure. 3.53 mmol of desired product was collected (70% yield). LCMS: tR = 7.23 minutes, m/z = 133.2 (M+H)
4.2.4 tert-butyl 2-(2-hydroxybenzoyl)hydrazinecarboxylate
The 3.53 mmol of tert-butyl hydrazinecarboxylate was dissolved in DCM in a round bottom flask with a stir bar. To this, 1.1 equivalents of HOBt and salicylic acid were added at room temperature. Following this, 1 equivalent of DIC was added to the flask. The reaction was mixed overnight at room temperature. Upon completion of the reaction, the DIU was filtered away and the resulting DCM was washed with saturated bicarbonate solution (1×) and 2M HCl (1×). The organic layer was dried over Na2SO4 and pumped down under reduced pressure to produce a white solid. This produced 3.34 mmol of the desired product (94% yield). LCMS: tR = 9.68 minutes, m/z = 253.2(M+H).
4.2.5 2-hydroxybenzohydrazide
The 3.34 mmol of tert-butyl hydrazinecarboxylate which was collected was dissolved in a 1:1 mixture of TFA in DCM in a round bottom flask equipped with a stir bar. The reaction was mixed for one hour and then the excess TFA and DCM was removed under vacuum for 1 hour producing a clear oil (quant). The crude reaction material was carried on to the next step without further purification steps.
4.2.6 N'-benzylidene-2-hydroxybenzohydrazide TFA salt (A2)
General procedure A was used to form the imine bond using benzaldehyde to produce a white solid. 1H NMR (300 MHz, DMSO) δ 11.96 - 11.85 (m, 2H), 8.45 (s, 1H), 7.87 (d, J = 6.4 Hz, 1H), 7.76 – 7.70 (m, 2H), 7.49 – 7.37 (m, 4H), 7.00 – 6.87 (m, 2H), 6.54 (s, 1H). LCMS: tR = 10.45 minutes. m/z = = 241.3 (M+H).
4.2.7 2-hydroxy-N'-(naphthalen-2-ylmethylene)benzohydrazide TFA salt (A3)
General procedure A was used to form the imine bond using 2-naphthalaldehyde to produce an off-white solid. 1H NMR (300 MHz, DMSO) δ 10.17 (s, 1H), 9.34 (s, 1H), 8.59 (s, 1H), 8.21 – 7.83 (m, 5H), 7.62 – 7.50 (m, 2H), 7.47 – 7.38 (m, 1H), 7.08 – 6.88 (m, 3H), 6.54 (s, 1H). LCMS: tR = 11.68 minutes. m/z = 291.0 (M+H).
4.2.8 N'-(4-chlorobenzylidene)-2-hydroxybenzohydrazide TFA salt (A4)
General procedure A was used to form the imine bond using 4-chlorobenzaldehyde and produced an off-white solid. 1H NMR (300 MHz, DMSO) δ 12.00 – 11.69 (m, 1H), 10.18 (s, 1H), 9.51 – 9.24 (m, 1H), 8.16 – 8.07 (m, 1H), 8.07 – 7.99 (m, 1H), 7.85 – 7.70 (m, 1H), 7.64 – 7.53 (m, 3H), 7.48 – 7.34 (m, 1H), 7.10 - 6.99 (m, 1H), 6.54 (s, 1H). LCMS: tR = 11.45 minutes. m/z = 274.9 (M+H).
4.2.9 2-hydroxy-N'-(4-hydroxybenzylidene)benzohydrazide TFA salt (A5)
General procedure A was used to form the imine bond using 4-hydroxybenzaldehyde. An off-white solid was produced after HPLC purification. 1H NMR (300 MHz, DMSO) δ 11.98 (s, 1H), 11.68 (s, 1H), 9.97 (s, 1H), 8.33 (s, 1H), 7.87 (d, J = 8.2 Hz, 1H), 7.56 (d, J = 8.4 Hz, 2H), 7.48 – 7.35 (m, 1H), 6.94 (d, J = 7.7 Hz, 2H), 6.90 (s, 1H), 6.83 (d, J = 8.5 Hz, 2H). LCMS: tR = 8.90 minutes. m/z= 257.1 (M+H).
4.2.10 2-hydroxy-N'-(4-methoxybenzylidene)benzohydrazide TFA salt (A6)
General procedure A was used to form the imine bond using 4-methoxybenzaldehyde. An off-white solid was produced after purification. 1H NMR (300 MHz, DMSO) δ 11.92 (s, 1H), 11.73 (s, 1H), 8.38 (s, 1H), 7.87 (d, J = 7.2 Hz, 1H), 7.68 (d, J = 8.9 Hz, 1H), 7.45 – 7.39 (m, 1H), 7.29 – 7.12 (m, 1H), 7.02 (d, J = 8.7 Hz, 2H), 6.94 (t, J = 7.8 Hz, 2H), 6.53 (s, 1H), 3.80 (s, 3H). LCMS: tR = 10.34 minutes. m/z = 271.2 (M+H).
4.2.11 2-hydroxy-N'-(4-nitrobenzylidene)benzohydrazide TFA salt (A7)
General procedure A was used to form the imine bond using 4-nitrobenzaldehyde and after purification, an off-white solid was produced. 1H NMR (300 MHz, DMSO) δ 12.30 (s, 1H), 11.69 (s, 1H), 10.96 (s, 1H), 8.03 – 7.90 (m, 1H), 7.90 – 7.82 (m, 1H), 7.71 – 7.61 (m, 1H), 7.49 – 7.43 (m, 1H), 7.28 – 7.17 (m, 2H), 7.02 – 6.91 (m, 1H), 6.53 (s, 1H), 6.38 (d, J = 9.7 Hz, 1H). LCMS: tR = 10.68 minutes. m/z = 285.9 (M+H).
4.2.12 2-hydroxy-N-phenethylbenzamide (A8)
General Procedure B was used to couple salicylic acid to phenethylamine. The reaction was carried out at room temperature overnight and after HPLC purification, a white solid was produced. 1H NMR (300 MHz, CDCl3) δ 12.37 (s, 1H), 7.46 – 7.16 (m, 7H), 7.07 – 6.98 (m, 1H), 6.89 – 6.79 (m, 1H), 6.33 (s, 1H), 3.81 – 3.71 (m, 2H), 2.98 (t, J = 6.8 Hz, 2H). LCMS: tR = 12.12 minutes. m/z = 242.3 (M+H).
4.2.13 2-hydroxy-N-(3-phenylpropyl)benzamide (A9)
General Procedure B was used to couple salicylic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. A white solid was produced after purification. 1H NMR (300 MHz, CDCl3) δ 12.42 (s, 1H), 7.44 – 7.17 (m, 6H), 7.12 (d, J = 7.9 Hz, 1H), 7.00 (d, J = 8.4 Hz, 1H), 6.82 (t, J = 7.6 Hz, 1H), 6.29 (s, 1H), 3.53 (dd, J = 13.1, 6.4 Hz, 2H), 2.77 (t, J = 7.4 Hz, 2H), 2.02 (p, J = 7.2 Hz, 2H). LCMS: tR = 13.90 minutes. m/z = 256.3 (M+H).
4.2.14 2-hydroxy-N-(4-phenylbutyl)benzamide (A10)
General Procedure B was used to couple salicylic acid to 4-phenylbutylamine. The reaction was carried out at room temperature overnight. An off-white solid was produced following HPLC purification. 1H NMR (300 MHz, CDCl3) δ 7.39 (ddd, J = 8.7, 7.5, 1.5 Hz, 1H), 7.34 – 7.24 (m, 3H), 7.20 (t, J = 6.7 Hz, 3H), 6.98 (d, J = 8.4 Hz, 1H), 6.87 – 6.78 (m, 1H), 6.27 (s, 1H), 3.47 (dd, J = 12.8, 6.8 Hz, 2H), 2.68 (t, J = 7.1 Hz, 2H), 1.79 – 1.62 (m, 4H). LCMS: tR = 13.90 minutes. m/z = 270.4 (M+H).
4.2.15 N-(3-phenylpropyl)benzamide (A11)
General Procedure B was used to couple benzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced an off-white solid after preparative HPLC. 1H NMR (300 MHz, CDCl3) δ 7.75 – 7.67 (m, 2H), 7.58 – 7.40 (m, 3H), 7.38 – 7.29 (m, 2H), 7.29 – 7.20 (m, 3H), 6.17 (s, 1H), 3.55 (dd, J = 13.4, 6.5 Hz, 2H), 2.77 (t, J = 7.5 Hz, 2H), 2.08 – 1.94 (m, 2H). LCMS: tR = 11.34 minutes. m/z = 240.3 (M+H).
4.2.16 3-hydroxy-N-(3-phenylpropyl)benzamide (A12)
General Procedure B was used to couple 3-hydroxybenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid. 1H NMR (300 MHz, CDCl3) δ 7.45 (s, 1H), 7.35 – 7.26 (m, 2H), 7.22 (d, J = 6.3 Hz, 4H), 7.08 – 6.95 (m, 2H), 6.51 (s, 1H), 6.10 (s, 1H), 3.51 (dd, J = 13.3, 6.7 Hz, 2H), 2.73 (t, J = 7.6 Hz, 2H), 2.03 – 1.91 (m, 2H). LCMS: tR = 9.90 minutes. m/z = 256.2 (M+H).
4.2.17 4-hydroxy-N-(3-phenylpropyl)benzamide (A13)
General Procedure B was used to couple 4-hydroxybenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. A white solid was produced after purification. 1H NMR (300 MHz, CDCl3) δ 7.56 – 7.48 (m, 2H), 7.33 – 7.25 (m, 2H), 7.23 – 7.15 (m, 3H), 6.86 – 6.77 (m, 2H), 6.07 (s, 1H), 3.49 (dd, J = 12.9, 6.9 Hz, 2H), 2.72 (t, J = 7.5 Hz, 2H), 2.03 – 1.90 (m, 2H). LCMS: tR = 9.98 minutes. m/z = 256.3 (M+H).
4.2.18 2-methyl-N-(3-phenylpropyl)benzamide (A14)
General Procedure B was used to couple 2-methylbenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and subsequently purified using HPLC, producing an-off-white solid. 1H NMR (300 MHz, CDCl3) δ 7.36 – 7.10 (m, 9H), 5.79 (s, 1H), 3.47 (dd, J = 13.6, 6.6 Hz, 2H), 2.73 (t, J = 7.6 Hz, 2H), 2.43 (s, 3H), 2.00 – 1.89 (m, 2H). LCMS: tR = 11.68 minutes. m/z = 254.2 (M+H).
4.2.19 2-methoxy-N-(3-phenylpropyl)benzamide (A15)
General Procedure B was used to couple 2-methoxybenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid. 1H NMR (300 MHz, CDCl3) δ 8.22 (dd, J = 7.8, 1.8 Hz, 1H), 7.90 (s, 1H), 7.48 – 7.40 (m, 1H), 7.34 – 7.24 (m, 2H), 7.25 – 7.15 (m, 3H), 7.13 – 7.06 (m, 1H), 6.98 (d, J = 8.3 Hz, 1H), 3.96 (s, 3H), 3.50 (dd, J = 12.9, 7.0 Hz, 2H), 2.74 (t, J = 7.5 Hz, 2H), 1.96 (dt, J = 14.7, 7.3 Hz, 2H). LCMS: tR = 11.90 minutes. m/z = 270.3 (M+H).
4.2.20 2-nitro-N-(3-phenylpropyl)benzamide (A16)
General Procedure B was used to couple 2-nitrobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight which produced a white solid. 1H NMR (300 MHz, CDCl3) δ 8.04 (dd, J = 8.1, 1.1 Hz, 1H), 7.60 (dtd, J = 17.2, 7.5, 1.4 Hz, 2H), 7.43 (dd, J = 7.4, 1.5 Hz, 1H), 7.35 – 7.24 (m, 2H), 7.26 – 7.14 (m, 3H), 5.83 (s, 1H), 3.49 (dd, J = 13.1, 6.9 Hz, 2H), 2.75 (t, J = 7.4 Hz, 2H), 2.04 – 1.91 (m, 2H). LCMS: tR = 11.01 minutes. m/z = 285.2 (M+H).
4.2.21 2-fluoro-N-(3-phenylpropyl)benzamide (A17)
General Procedure B was used to couple 2-fluorobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight which produced a white solid after purification. 1H NMR (300 MHz, CDCl3) δ 8.10 (td, J = 7.9, 1.9 Hz, 1H), 7.52 – 7.42 (m, 1H), 7.34 – 7.27 (m, 3H), 7.24 – 7.16 (m, 3H), 7.15 – 7.06 (m, 1H), 6.75 (s, 1H), 3.52 (q, J = 5.8 Hz, 2H), 2.74 (t, J = 7.4 Hz, 2H), 1.98 (dt, J = 14.7, 7.4 Hz, 2H). LCMS: tR = 12.34 minutes. m/z = 258.3 (M+H).
4.2.22 2-chloro-N-(3-phenylpropyl)benzamide (A18)
General Procedure B was used to couple 2-chlorobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. After HPLC purification, a white solid was produced. 1H NMR (300 MHz, CDCl3) δ 7.67 – 7.54 (m, 1H), 7.42 – 7.10 (m, 8H), 6.24 (s, 1H), 3.50 (q, J = 10.0 Hz, 2H), 2.75 (t, J = 7.3 Hz, 2H), 2.05 – 1.89 (m, 2H). LCMS: tR – 11.57 minutes. m/z = 274.1 (M+H).
4.2.23 2-bromo-N-(3-phenylpropyl)benzamide (A19)
General Procedure B was used to couple 2-bromobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid 1H NMR (300 MHz, CDCl3) δ 7.58 (d, J = 7.0 Hz, 1H), 7.49 (dd, J = 7.6, 1.8 Hz, 1H), 7.39 – 7.15 (m, 7H), 6.02 (s, 1H), 3.50 (dd, J = 13.4, 6.7 Hz, 2H), 2.76 (t, J = 7.4 Hz, 2H), 1.97 (dt, J = 14.6, 7.2 Hz, 2H). LCMS: tR = 12.01 minutes. m/z = 320.1 (M+H).
4.2.24 2-iodo-N-(3-phenylpropyl)benzamide (A20)
General Procedure B was used to couple 2-iodobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid after purification. 1H NMR (300 MHz, CDCl3) δ 7.90 −7.80 (m, 1H), 7.40 – 7.15 (m, 7H), 7.13 – 7.03 (m, 1H), 5.77 (s, 1H), 3.50 (dd, J = 13.5, 6.6 Hz, 2H), 2.77 (t, J = 7.5 Hz, 2H), 1.99 (dt, J = 14.7, 7.2 Hz, 2H). LCMS: tR = 12.34 minutes. m/z = 366.1(M+H).
4.2.25 2-cyano-N-(3-phenylpropyl)benzamide (A21)
General Procedure B was used to couple 2-cyanobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. A white solid was produced after purification. 1H NMR (300 MHz, CDCl3) δ 7.81 – 7.74 (m, 2H), 7.73 – 7.56 (m, 2H), 7.37 – 7.20 (m, 5H), 6.26 (s, 1H), 3.59 (dd, J = 12.9, 7.0 Hz, 2H), 2.80 (t, J = 7.6 Hz, 2H), 2.12 – 1.99 (m, 2H).LCMS: tR = 11.90 minutes. m/z = 265.2 (M+H). HRMS m/z calcd for C17H17N2O (M+H), 265.1341; found, 265.1339.
4.2.26 2-amino-N-(3-phenylpropyl)benzamide (A22)
General Procedure B was used to couple 2-aminobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced an off-white solid. 1H NMR (300 MHz, CDCl3) δ 7.37 −7.25 (m, 2H), 7.25 – 7.18 (m, 3H), 7.15 (dd, J = 10.0, 4.7 Hz, 2H), 6.79 – 6.54 (m, 2H), 6.04 (s, 1H), 5.00 (s, 2H), 3.45 (dd, J = 12.9, 6.2 Hz, 2H), 2.72 (t, J = 7.5 Hz, 2H), 2.04 – 1.86 (m, 2H).LCMS: tR = 12.01 minutes. m/z = 255.3 (M+H).
4.2.27 1-hydroxy-N-(3-phenylpropyl)-2-naphthamide (A23)
General Procedure B was used to couple 1-hydroxy-2-naphthoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. After purification, a white solid was produced. 1H NMR (300 MHz, CDCl3) δ 8.41 (d, J = 8.1 Hz, 1H), 7.74 (d, J = 7.5 Hz, 1H), 7.61 – 7.47 (m, 2H), 7.37 – 7.27 (m, 2H), 7.27 – 7.18 (m, 4H), 7.04 (d, J = 8.8 Hz, 1H), 6.25 (s, 1H), 3.55 (dd, J = 12.8, 6.9 Hz, 2H), 2.77 (t, J = 7.4 Hz, 2H), 2.08 – 1.96 (m, 2H). LCMS: tR = 17.02 minutes. m/z = 306.2 (M+H).
4.2.28 3-hydroxy-N-(3-phenylpropyl)-2-naphthamide (A24)
General Procedure B was used to couple 3-hydroxy-2-naphthoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. HPLC purification produced a white solid. 1H NMR (300 MHz, CDCl3) δ 11.84 (s, 1H), 7.72 (t, J = 8.9 Hz, 2H), 7.60 (s, 1H), 7.54 – 7.46 (m, 1H), 7.42 – 7.34 (m, 2H), 7.34 – 7.27 (m, 5H), 6.41 (s, 1H), 3.62 (dd, J = 13.6, 5.7 Hz, 2H), 2.84 (t, J = 7.2 Hz, 2H), 2.10 (p, J = 7.1 Hz, 2H). LCMS: tR = 15.57 minutes. m/z = 306.3 (M+H).
4.2.29 2-hydroxy-N-(3-phenylpropyl)-1-naphthamide (A25)
General Procedure B was used to couple 2-hydroxy-1-naphthoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid after HPLC purification. 1H NMR (300 MHz, CDCl3) δ 11.49 (s, 1H), 8.06 (d, J = 8.5 Hz, 1H), 7.84 – 7.76 (m, 2H), 7.56 – 7.46 (m, 1H), 7.41 – 7.12 (m, 7H), 6.35 (s, 1H), 3.61 (dd, J = 13.7, 6.6 Hz, 2H), 2.78 (t, J = 7.5 Hz, 2H), 2.10 −1.97 (m, 2H). LCMS: tR = 13.12 minutes. m/z = 306.3 (M+H). HRMS m/z calcd for C20H20NO2 (M+H), 306.1494; found, 306.1495.
4.2.30 N-(3-phenylpropyl)benzo[d][1,3]dioxole-5-carboxamide (A26)
General Procedure B was used to couple piperonylic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid. 1H NMR (300 MHz, CDCl3) δ 7.36 – 7.25 (m, 3H), 7.25 – 7.12 (m, 4H), 6.84 – 6.76 (m, 1H), 6.02 (s, 2H), 5.93 (s, 1H), 3.48 (dd, J = 12.9, 6.8 Hz, 2H), 2.73 (t, J = 7.5 Hz, 2H), 2.02 – 1.90 (m, 2H). LCMS: tR = 12.12 minutes. m/z = 284.2 (M+H).
4.2.31 N-(3-phenylpropyl)picolinamide (A27)
General Procedure B was used to couple picolinic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. A clear oil was produced after purification. 1H NMR (300 MHz, CDCl3) δ 8.54 (ddd, J = 4.8, 1.7, 0.9 Hz, 1H), 8.23 – 8.17 (m, 1H), 8.10 (s, 1H), 7.85 (td, J = 7.7, 1.7 Hz, 1H), 7.42 (ddd, J = 7.6, 4.8, 1.2 Hz, 1H), 7.33 – 7.26 (m, 2H), 7.24 – 7.14 (m, 3H), 3.52 (dd, J = 13.3, 7.0 Hz, 2H), 2.75 (t, J = 7.4 Hz, 2H), 1.98 (dt, J = 9.1, 7.4 Hz, 2H). LCMS: tR = 12.57 minutes. m/z = 241.4 (M+H).
4.2.32 N-(3-phenylpropyl)quinoline-2-carboxamide (A28)
General Procedure B was used to couple quinoline-2-carboxylic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a white solid after purification. 1H NMR (300 MHz, CDCl3) δ 8.32 (m, 3H), 8.10 (d, J = 8.6 Hz, 1H), 7.88 (d, J = 8.2 Hz, 1H), 7.77 (ddd, J = 8.5, 6.9, 1.5 Hz, 1H), 7.62 (t, J = 6.9 Hz, 1H), 7.33 – 7.27 (m, 2H), 7.26 – 7.17 (m, 3H), 3.58 (dd, J = 13.6, 6.9 Hz, 2H), 2.79 (t, J = 7.5 Hz, 2H), 2.05 (dt, J = 14.7, 7.5 Hz, 2H). LCMS: tR = 14.90 minutes. m/z = 291.3 (M+H).
4.2.33 N-(3-phenylpropyl)isoquinoline-3-carboxamide (A29)
General Procedure B was used to couple isoquinoline-3-carboxylic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. Following HPLC purification, a clear oil was produced. 1H NMR (300 MHz, CDCl3) δ 9.15 (s, 1H), 8.62 (s, 1H), 8.30 (s, 1H), 8.08 – 7.94 (m, 2H), 7.80 – 7.65 (m, 2H), 7.37 – 7.12 (m, 5H), 3.58 (dd, J = 13.6, 6.7 Hz, 2H), 2.78 (t, J = 7.5 Hz, 2H), 2.03 (dt, J = 14.5, 7.1 Hz, 2H). LCMS: tR = 14.12 minutes. m/z = 291.3 (M+H). HRMS m/z calcd for C19H19N2O (M+H), 291.1497; found, 291.1496.
4.2.34 N-(3-phenylpropyl)isoquinoline-1-carboxamide (A30)
General Procedure B was used to couple isoquinoline-1-carboxylic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a clear oil after preparative HPLC purification. 1H NMR (300 MHz, CDCl3) δ 9.62 (d, J = 7.6 Hz, 1H), 8.45 (d, J = 5.5 Hz, 1H), 8.26 (s, 1H), 7.87 – 7.76 (m, 2H), 7.76 – 7.64 (m, 2H), 7.34 – 7.14 (m, 5H), 3.56 (dd, J = 13.4, 6.9 Hz, 2H), 2.79 (t, J = 7.5 Hz, 2H), 2.03 (dt, J = 14.7, 7.4 Hz, 2H). LCMS: tR = 14.35 minutes. m/z = 291.3 (M+H). HRMS m/z calcd for C19H19N2O (M+H), 291.1497; found, 291.1496.
4.2.35 4-amino-2-hydroxy-N-(3-phenylpropyl)benzamide (A31)
General Procedure B was used to couple 4-amino-2-hydroxybenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced an orange solid after purification. 1H NMR (300 MHz, CDCl3) δ 7.34 – 7.17 (m, 5H), 6.89 (d, J = 8.5 Hz, 1H), 6.17 (d, J = 2.0 Hz, 1H), 6.08 (d, J = 8.5 Hz, 1H), 5.93 (s, 1H), 3.45 (dd, J = 13.3, 6.4 Hz, 2H), 2.72 (t, J = 7.5 Hz, 2H), 2.03 – 1.88 (m, 2H). LCMS: tR = 9.34 minutes. m/z = 271.3 (M+H). HRMS m/z calcd for C16H19N2O2 (M+H), 271.1447; found, 271.1444.
4.2.36 3,5-dihydroxy-N-(3-phenylpropyl)-2-naphthamide (A32)
General Procedure B was used to couple 3,5-dihydroxy-2-naphthoic acid to 3-phenylpropylamine. The reaction was mixed at room temperature overnight. Following HPLC purification, the product resulted in an off-white solid. 1H NMR (300 MHz, CDCl3) δ 11.78 (s, 1H), 7.59 (s, 1H), 7.53 (s, 1H), 7.38 – 7.26 (m, 6H), 7.18 – 7.11 (m, 1H), 6.82 (d, J = 7.3 Hz, 1H), 6.36 (s, 1H), 5.35 (s, 1H), 3.58 (dd, J = 12.6, 6.7 Hz, 2H), 2.80 (t, J = 7.2 Hz, 2H), 2.12 – 2.00 (m, 2H). LCMS: tR = 12.90 minutes. m/z = 322.3 (M+H). HRMS m/z calcd for C20H20NO3 (M+H), 322.1443; found, 322.1440.
4.2.37 2-amino-5-methoxy-N-(3-phenylpropyl)benzamide (A33)
General Procedure B was used to couple 2-amino-5-methoxybenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight. An off-white solid was produced after purification. 1H NMR (300 MHz, CDCl3) δ 7.34 – 7.26 (m, 2H), 7.26 – 7.18 (m, 3H), 7.01 – 6.89 (m, 2H), 6.80 (d, J = 2.6 Hz, 1H), 6.26 (s, 1H), 3.78 (s, 3H), 3.46 (dd, J = 12.9, 6.7 Hz, 2H), 2.73 (t, J = 7.5 Hz, 2H), 2.03 - 1.90 (m, 2H). LCMS: tR = 9.90 minutes. m/z = 285.3 (M+H).
4.2.38 2-amino-5-chloro-N-(3-phenylpropyl)benzamide (A34)
General Procedure B was used to couple 2-amino-5-chlorobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced an off-white solid. 1H NMR (300 MHz, CDCl3) δ 7.38 – 7.17 (m, 5H), 7.13 (ddd, J = 8.7, 2.3, 1.4 Hz, 1H), 7.04 (d, J = 2.4 Hz, 1H), 6.60 (dd, J = 8.7, 1.3 Hz, 1H), 5.91 (s, 1H), 4.79 (s, 2H), 3.45 (dd, J = 12.7, 6.9 Hz, 2H), 2.73 (t, J = 7.4 Hz, 2H), 2.02 – 1.90 (m, 2H). LCMS: tR = 13.35 minutes. m/z = 289.3 (M+H).
4.2.39 2-amino-5-nitro-N-(3-phenylpropyl)benzamide (A35)
General Procedure B was used to couple 2-amino-5-nitrobenzoic acid to 3-phenylpropylamine. The reaction was carried out at room temperature overnight and produced a yellow solid after purification. 1H NMR (300 MHz, CDCl3) δ 8.21 – 8.17 (m, 1H), 8.16 – 8.06 (m, 1H), 7.40 – 7.31 (m, 2H), 7.30 – 7.18 (m, 3H), 6.67 (dd, J = 9.1, 1.6 Hz, 1H), 6.54 (s, 2H), 6.14 (s, 1H), 3.52 (q, J = 7.3 Hz, 2H), 2.79 (t, J = 7.3 Hz, 2H), 2.04 (p, J = 6.7 Hz, 2H). LCMS: tR = 12.79 minutes. m/z = 300.4 (M+H). HRMS m/z calcd for C16H18N3O3 (M+H), 300.1348; found, 300.1347.
4.2.40 N-methyl-3-phenylpropan-1-amine
2mmol of 3-phenylpropylamine and 2.2mmol of N-methylmorpholine was dissolved in THF at room temperature in a round bottom flask. To this, iodomethane was added to the flask in one portion. The reaction was mixed overnight at room temperature. The next day, the precipitate was filtered away and the THF was evaporated away under reduced pressure. The crude mixture was carried on without any work-up. LCMS: tR = 9.12 minutes. m/z = 150.0 (M+H).
4.2.41 2-hydroxy-N-methyl-N-(3-phenylpropyl)benzamide (A36)
General Procedure B was used to couple salicylic acid to N-methyl-3-phenylpropan-1-amine. The reaction was carried out at room temperature overnight and produced a white solid. 2:1 mixture of cis:trans. Cis: 1H NMR (300 MHz, CDCl3) δ 7.59 – 7.08 (m, 6H), 7.07 – 6.69 (m, 3H), 3.49 (m, 2H), 2.87 (s, 3H), 2.55 (t, J = 7.5 Hz, 2H), 1.91 – 1.69 (m, 2H). Trans: 1H NMR (300 MHz, CDCl3) δ 7.59 – 7.08 (m, 6H), 7.07 – 6.69 (m, 3H), 3.27 – 3.15 (m, 2H), 3.00 (s, 3H), 2.45 (t, J = 8.0 Hz, 2H), 1.91 – 1.69 (m, 2H). LCMS: tR = 11.12 minutes. m/z = 270.3 (M+H). HRMS m/z calcd for C17H20NO2 (M+H), 270.1494; found, 270.1494.
4.2.42 2-(2-hydroxyphenyl)-N-phenethylacetamide (A37)
General Procedure B was used to couple 2-(2-hydroxyphenyl)acetic acid to 2-phenethylamine. The reaction was carried out at room temperature overnight. Following purification, an off-white solid was produced. 1H NMR (300 MHz, CDCl3) δ 7.33 – 7.16 (m, 4H), 7.14 – 7.09 (m, 2H), 6.97 (d, J = 7.9 Hz, 2H), 6.83 (t, J = 7.4 Hz, 1H), 5.94 (s, 1H), 3.61 – 3.40 (m, 4H), 2.80 (t, J = 6.9 Hz, 2H). LCMS: tR = 11.23 minutes. m/z = 256.2 (M+H). HRMS m/z calcd for C16H1NO2 (M+H), 256.1338; found, 256.1336.
4.2.43 N-benzyl-3-(2-hydroxyphenyl)propanamide (A38)
General Procedure B was used to couple 3-(2-hydroxyphenyl)propanoic acid to benzylamine. The reaction was carried out at room temperature overnight. HPLC purification produced a white solid. 1H NMR (300 MHz, CDCl3) δ 8.69 (s, 1H), 7.36 – 7.22 (m, 3H), 7.22 – 7.10 (m, 3H), 7.05 (dd, J = 7.5, 1.6 Hz, 1H), 6.93 (d, J = 8.1 Hz, 1H), 6.89 – 6.79 (m, 1H), 5.76 (s, 1H), 4.42 (d, J = 5.7 Hz, 2H), 3.00 – 2.92 (m, 2H), 2.69 – 2.61 (m, 2H).LCMS: tR = 10.90 minutes. m/z = 256.3 (M+H).
4.2.44 3-(2-hydroxyphenyl)-N-phenylpropanamide (A39)
In a round bottom flask, 1mmol of dihydrocoumarin was dissolved in DCM. To this, 1.1mmol of aniline was added with .25mmol of DMAP. The reaction was mixed overnight at room temperature. Following this, the reaction was heated for 8 hrs at 70°C. Upon completion of the reaction, the DCM was washed with saturated bicarbonate solution (1×) and with 2M HCl (1×). The DCM was dried over Na2SO4 and the solvent was removed under reduced pressure. The crude reaction material was carried on to preparative HPLC without further work-up and produced a white solid. 1H NMR (300 MHz, CDCl3) δ 8.39 (s, 1H), 7.45 (d, J = 8.0 Hz, 2H), 7.37 - 7.26 (m, 3H), 7.16 - 7.07 (m, 3H), 6.95 – 6.81 (m, 2H), 3.03 – 2.96 (m, 2H), 2.85 – 2.77 (m, 2H). LCMS: tR = 10.79 minutes. m/z = 242.4 (M+H).
4.2.45 N-(2-hydroxyphenyl)-4-phenylbutanamide (A40)
General Procedure B was used to couple 4-phenylbutanoic acid to 2-aminophenol. The reaction was carried out at room temperature overnight. This produced a white solid following HPLC purification. 1H NMR (300 MHz, CDCl3) δ 8.77 (s, 1H), 7.37 – 7.27 (m, 2H), 7.25 – 7.15 (m, 3H), 7.15 – 7.09 (m, 1H), 7.02 (d, J = 8.2 Hz, 1H), 6.95 - 6.80 (m, 2H), 2.74 (t, J = 7.4 Hz, 2H), 2.45 (t, J = 7.5 Hz, 2H), 2.17 – 2.04 (m, 2H). LCMS: tR = 11.68 minutes. m/z = 256.2 (M+H).
4.2.46 2-hydroxy-N-(2-phenoxyethyl)benzamide (A41)
General Procedure B was used to couple salicylic acid to 2-phenoxyethanamine. The reaction was carried out at room temperature overnight. Following HPLC purification, a white solid was produced. 1H NMR (300 MHz, CDCl3) δ 12.24 (s, 1H), 7.46 – 7.26 (m, 4H), 7.03 – 6.96 (m, 2H), 6.93 (d, J = 7.9 Hz, 2H), 6.85 (t, J = 7.6 Hz, 1H), 6.77 (s, 1H), 4.17 (d, J = 5.0 Hz, 2H), 3.88 (dd, J = 10.4, 5.3 Hz, 2H). LCMS: t R = 13.12 minutes. m/z = 258.2 (M+H).
4.2.47 2-amino-N-(2-phenoxyethyl)benzamide (A42)
General Procedure B was used to couple 2-aminobenzoic acid to 2-phenoxyethanamine. The reaction was carried out at room temperature overnight and produced a white solid following HPLC purification. 1H NMR (300 MHz, CDCl3) δ 7.39 – 7.14 (m, 4H), 7.02 – 6.87 (m, 3H), 6.71 – 6.62 (m, 2H), 6.57 (s, 1H), 5.26 (s, 2H), 4.14 (t, J = 5.1 Hz, 2H), 3.83 (dd, J = 10.6, 5.3 Hz, 2H). LCMS: tR = 12.01 minutes. m/z = 257.2 (M+H).
Supplementary Material
Acknowledgements
This study was supported by Public Health Service grants R01AI071341 to D.A.E. and 5R43AI084244 to DB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DAJ. Virol. 2009;83:1881. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. Arch Intern Med. 2005;165:265. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 3.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. JAMA-J. Am. Med. Assoc. 2004;292:1333. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. JAMA-J. Am. Med. Assoc. 2003;289:179. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 5.Reid AH, Taubenberger JK, Fanning TG. Microb Infect. 2001;3:81. doi: 10.1016/s1286-4579(00)01351-4. [DOI] [PubMed] [Google Scholar]
- 6.Vittorio Farina JDB. Angew. Chem. Int. Edit. 2006;45:7330. doi: 10.1002/anie.200602623. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, Cheung CL, Xu KM, Duan L, Huang K, Qin K, Leung YHC, Wu WL, Lu HR, Chen Y, Xia NS, Naipospos TSP, Yuen KY, Hassan SS, Bahri S, Nguyen TD, Webster RG, Peiris JSM, Guan YP. Natl. Acad. Sci. USA. 2006;103:2845. [Google Scholar]
- 8.Ducatez MF, Olinger CM, Owoade AA, De Landtsheer S, Ammerlaan W, Niesters HGM, Osterhaus ADME, Fouchier RAM, Muller CP. Nature. 2006;442:37. doi: 10.1038/442037a. [DOI] [PubMed] [Google Scholar]
- 9.Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. Science. 2006;312:384. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 10.Webster RG, Govorkova EA. New Engl. J. Med. 2006;355:2174. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Ghafar AN, Chotpitayasundh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soerooso S, Uyeki TM. N. Engl. J. Med. 2008;358:261. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 12.Hayman A, Comely S, Lackenby A, Hartgroves LCS, Goodbourn S, McCauley JW, Barclay WS. J. Virol. 2007;81:2318. doi: 10.1128/JVI.01856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lera E, T. Wörner N, Sancosmed M, Fàbregas A, Casquero A, Melendo S, Miserachs M, Tórtola T, Borrego A, Campins M, Moraga F, Figueras C, Cebrián R. Eur. J. Pediatr. 2011:1. doi: 10.1007/s00431-011-1399-4. [DOI] [PubMed] [Google Scholar]
- 14.Pada S, Tambyah PA. Microb. Infect. 2011 doi: 10.1016/j.micinf.2011.01.009. In Press, Accepted Manuscript. [DOI] [PubMed] [Google Scholar]
- 15.Thickett DR, Griffiths M, Perkins GD, McAuley DF. Thorax. 2010;65:855. doi: 10.1136/thx.2010.149294. [DOI] [PubMed] [Google Scholar]
- 16.Nichol, Kristin L, Treanor John J. J. Infect. Dis. 2006;194:S111. doi: 10.1086/507544. [DOI] [PubMed] [Google Scholar]
- 17.Kieny MP, Costa A, Hombach J, Carrasco P, Pervikov Y, Salisbury D, Greco M, Gust I, LaForce M, Franco-Paredes C, Santos JI, D’Hondt E, Rimmelzwaan G, Karron R, Fukuda K. Vaccine. 2006;24:6367. doi: 10.1016/j.vaccine.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Krug RM, Aramini JM. Trends Pharmacol. Sci. 2009;30:269. doi: 10.1016/j.tips.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walkiewicz MP, Basu D, Jablonski JJ, Geysen HM, Engel DA. J. Gen. Virol. 2011;92:60. doi: 10.1099/vir.0.025015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale BG, Barclay WS, Randall RE, Russell RJ. Virology. 2008;378:1. doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Krug RM, Yuan W, Noah DL, Latham AG. Virology. 2003;309:181. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 22.Yin C, Khan JA, Swapna GVT, Ertekin A, Krug RM, Tong L, Montelione GT. J. Biol. Chem. 2007;282:20584. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- 23.Hatada E, Fukuda R. J. Gen. Virol. 1992;73:3325. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 24.Min J-Y, Krug RM. P. Natl. Acad. Sci. 2006;103:7100. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman RH. J. Virol. 2007;81:12720. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. J. Virol. 2007;81:7011. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemeroff ME, Barabino SML, Li Y, Keller W, Krug RM. Mol. Cell. 1998;1:991. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Li Y, Krug RM. EMBO J. 1999;18:2273. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noah DL, Twu KY, Krug RM. Virology. 2003;307:386. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 30.Gale M, Katze MG. Pharmacol. Ther. 1998;78:29. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 31.Min J-Y, Li S, Sen GC, Krug RM. Virology. 2007;363:236. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Gack MU, Albrecht RA, Urano T, Inn K-S, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, García-Sastre A. Cell Host Microbe. 2009;5:439. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z, Chen L-m, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. Am. J. Respir. Cell Mol. Biol. 2007;36:263. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig S, Wolff T. Cell Host Microbe. 2009;5:420. doi: 10.1016/j.chom.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Virology. 1998;252:324. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 36.Reed L, Muench H. Am. J. Hyg. 1938;27:943. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.