Abstract
Hypoxia is an integral part of tumorigenesis and contributes extensively to the neoplastic phenotype including drug resistance and genomic instability. It has also been reported that hypoxia results in global demethylation. Because a majority of the cytosine-phosphate-guanine (CpG) islands are found within the repeat elements of DNA, and are usually methylated under normoxic conditions, we suggested that retrotransposable Alu or short interspersed nuclear elements (SINEs) which show altered methylation and associated changes of gene expression during hypoxia, could be associated with genomic instability. U87MG glioblastoma cells were cultured in 0.1% O2 for 6 weeks and compared with cells cultured in 21% O2 for the same duration. Real-time PCR analysis showed a significant increase in SINE and reverse transcriptase coding long interspersed nuclear element (LINE) transcripts during hypoxia. Sequencing of bisulphite treated DNA as well as the Combined Bisulfite Restriction Analysis (COBRA) assay showed that the SINE loci studied underwent significant hypomethylation though there was patchy hypermethylation at a few sites. The inter-alu PCR profile of DNA from cells cultured under 6-week hypoxia, its 4-week revert back to normoxia and 6-week normoxia showed several changes in the band pattern indicating increased alu mediated genomic alteration. Our results show that aberrant methylation leading to increased transcription of SINE and reverse transcriptase associated LINE elements could lead to increased genomic instability in hypoxia. This might be a cause of genetic heterogeneity in tumours especially in variegated hypoxic environment and lead to a development of foci of more aggressive tumour cells.
Keywords: DNA methylation, CpG islands, SINE, U87MG, bisulphite sequencing, genetic instability, long-term hypoxia
Introduction
An unstable tumour genome is likely to generate a significant amount of genetic heterogeneity in solid tumours such as gliomas [1] with heterogeneous microenvironment. Active repeat elements are one of the many factors contributing to genomic instability. Repeat elements such as short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs) consist of 35% of human genome [2]. In mammals the low basal level of SINE expression has been attributed to extensive methylation resulting in transcriptionally inactive heterochromatin [3]. Also, hypomethylation of LINE1 and SINEs has been associated with the clinicopathological behaviour of the tumours [4, 5]. SINEs are dependent on the reverse transcriptase activity of LINE 1 for retrotransposition and insertional mutagenesis [6]. SINE expression responds to external factors such as heat shock and stress [7]. We have previously shown that alu mediated recombination increases when cells are cultured in low doses of chemotherapeutic drug, cisplatin [8]. A recent study by Bennett et al.[9] suggests that Alu elements pose the largest transposon-based mutagenic threat to the human genome and play an important role in genetic variations in diseases.
Of the 70–80% of methylated CpG sites [10], the repetitive elements and areas of low CpG density are preferential targets of aberrant methylation [11]. DNA methylation is an important epigenetic phenomenon that has a significant role in long-term regulation of gene expression [12]. Although classical models propose that DNA methylation patterns are rigid [13], recent evidence suggests that site-specific DNA methylation is a dynamic process that depends on the interplay of several factors of the methylation machinery [14, 15]. Alterations in methylation have been linked to tumorigenesis [16] and can further induce phenotypic and gene expression changes associated with targeted de novo epigenetic alterations important in tumour progression [17]. There is a general trend of hypomethylation in tumours [18] and the extent of genome wide hypomethylation in tumours parallels the degree of malignancy [19].
As tumour cells proliferate, inadequate blood supply and aberrant neovascularization result in hypoxic tumour interiors [20]. Decreased penetration of drugs and paucity of free radical generation under hypoxic conditions often lead to resistance of the tumour to radiotherapy and chemotherapy [21]. The change in the microenvironment due to hypoxia and other factors like lactic acidosis, decreased supply of nutrients, etc may result in local changes in the epigenetic pattern of the cell genome. Increased genomic instability and mutations under hypoxic conditions and genotoxic stress have also been demonstrated by Rice et al.[22], where a transient hypoxia followed by methotrexate exposure in normoxia led to a significant increase in methotrexate resistant cells with DHFR gene amplification.
Yuan et al.[23] have attributed the increased genetic instability observed in hypoxic cells to the compromised DNA repair gene activity particularly the MMR gene. They have observed a 5-fold increase in the mutation frequency in hypoxic cells using a λ phage shuttle vector with a reporter gene for identifying mutations [24]. The down-regulation of MMR and increased genetic instability has also been recently observed in stem cells, where reduced transcription activation of MMR genes has been attributed to specific epigenetic events [25]. Hypoxia and its associated factors are also linked to the process of aging [26].
We suggested that long-term hypoxic stress may also cause repeat element mediated genetic instability. We have studied changes in the methylation status of the CpG islands within and around the repetitive SINEs of a glial tumour cell line (U87MG) and an osteosarcoma cell line (SaOS2) after exposure to prolonged hypoxic conditions. Transcription of SINEs and LINEs was studied by real-time PCR and genomic instability was determined by inter-alu PCR.
Materials and methods
Cell line and culture
U87MG (glioblastoma cell line, wild-type p53) and SaOS2 (p53 null osteosarcoma cell line) lines have been used for all the experiments (Supporting Information). Experiments on normoxia and hypoxia were initiated simultaneously with similar passage number (12–15). Cells were cultured in 75 cm2 and 25 cm2 vented tissue culture flasks.
In vitro exposure to hypoxic condition
The vented cap flasks were kept in Anoxomat chambers (Mart® Microbiology, and the Anoxomat™ system) with appropriate O2 concentrations for hypoxia (0.1%) and normoxia (21.0%). These atmospheres were created in the anaerobic chambers after vacuuming and then flooding the chambers with pure CO2 and N2 according to the set partial pressures. We frequently measured the partial pressure of oxygen with the help of dissolved oxygen (DO) meter in one of the control flask after 6 hrs to confirm the level of hypoxia. Culture media were pre-exposed to hypoxia for 2 hrs before feeding the cells. Cells were routinely passaged on attaining 75–80% confluency at a split ratio of 1:3. At the time of passaging, minimum possible time (5–10 min.) was spent for trypsinization and replating, and then the cells were immediately shifted to the hypoxic conditions. With the help of the DO meter we have also observed that atmospheric O2 is not significantly miscible in media upto 10–15 min. As the duration of culture under hypoxia progressed, the requirement for passaging the cells decreased in frequency as a result of decrease in proliferation, but the media were replenished every 3 days. As a measure of cellular response to hypoxia, we have observed an increase in the induction of HIF-1 protein starting from 4 hrs and in 24 hrs of hypoxia indicating that cells respond to the hypoxic stress given (Fig. S1). Cells were in three groups (i) 6-week hypoxia (H) (ii) 6-week normoxia (N) and (iii) 6-week hypoxia followed by further 4-week reversion to normoxia (HR). DNA and RNA were extracted from cells after appropriate time-points. The experiments were repeated twice.
Cell viability studies
U87MG cells from same passage number were cultured for 6 weeks in hypoxia (0.1% O2) and normoxia (21% O2) conditions, respectively. A flask of hypoxic cells was reverted back to normoxia for 4 weeks. The cells from hypoxic, normoxic and hypoxia revertent conditions were plated in a 96-well plate at a density of 5 × 103 cells / 100 μl / well and subsequently cultured in their respective conditions and counted at 2, 4, 6 and 8 days of culture. Viability was measured by Trypan blue dye exclusion.
Morphology and caspase-3 cleavage for apoptosis
Morphological analysis was done for identifying features of apoptosis in U87MG and SaOS2 cells grown in hypoxia. The microphotographs were taken at various magnifications in cells grown under normoxia and hypoxia.U87MG cells subjected to hypoxia for various time-points and normoxia were lysed in a triple detergent buffer (sodium deoxycholate, NP-40 and SDS) and protein inhibitor cocktail (Amresco, Solon, OH, USA). Total protein was quantified using the BCA protein assay kit (Pierce, Rockford, IL, USA) and 20 μg protein was run on a 15% SDS-PAGE and electroblotted on a PVDF membrane (Schleicher and Schuell, Germany) with the Mini-Protean system from Bio-Rad (Hercules, CA, USA). The blots were checked for transfer with Ponceau S (Sigma-Aldrich, St. Louis, MO, USA) and blocked in 4% non-fat milk in TBS (Tris buffered saline). The blots were probed with the monoclonal anti-caspase-3 antibody (BD Pharmingen, San Diego, CA, USA) overnight, goat antimouse alkaline phosphatase linked IgG secondary antibody and developed with BCIP/NBT system (Promega, Madison, WI, USA).
Isolation of RNA and real-time RT-PCR for SINE (alu) and LINE expression
Total RNA was isolated from U87MG and SaOS2 cells using TRIZOL® (Invitrogen, Carlsbad, CA, USA) and reverse transcribed with RETROscript® reverse transcriptase kit (Ambion, Foster City, CA, USA). Real-time PCR was done using intra-alu primers for SINE expression; L1-F and L1-R for LINE expression and b-actin primers (IDT, Coralville, IA, USA) as normalization control (Table 1) in RotorGene cycler (Corbett Research, Sydney, Australia) in two biological replicates.
Table 1.
Primers used in the study
| Primers | Sequence (5′–3′) | Annealing temperature (°C) |
|---|---|---|
| Bisulphite sequencing primers | ||
| At chromosome 16 | ||
| 16 outer F | TTTATAGGGTAGTAGTTTTAGTGGG | 50 |
| 16 outer R | TATTTACAAAAAACCAAAAATTCAC | |
| 16 inner F | AGTATAGGTGGGGAGGAAGTAGAG | 50 |
| 16 inner R | TTTACAAAAAACCAAAAATTCACTATC | |
| At chromosome 7 | ||
| 7 outer F | TGAATAGAATTTTAGAAATAGGA | 46 |
| 7 outer R | CTAACAATTTTCATTCCAAC | |
| 7 inner F | TATTAAGATTTATAGAGGTGG | 52 |
| 7 inner R | ATTATTTAACTTTCAATAATCA | |
| LINE-COBRA primers | ||
| LINE-3 | GYGTAAGGGGTTAGGGAGTTTTT | 50 |
| LINE-4 | AACRTAAAACCCTCCRAACCAAATATAAA | |
| Primers for qRT/RT | ||
| Intra-alu F | CACGCCTGTAATCCCAGCAC | 52 |
| Intra-alu R | GGAGTCTCGCTCTGTCG | |
| L1 F | GCTGGATATGAAATTCTGGGTTGA | 60 |
| L1 R | AGGAAATACAGAGAACGCCACAA | |
| β-actin F | TCATGAAGTGTGACGTTGACATCCGT | 52/60 |
| b-actin R | CCTAGAAGCATTTGCGGTGCACGATG | |
| Inter-alu primers | ||
| alu264 | CAGAGCGAGACTCC | 54 |
| alu267 | AGCGAGACTCCG | 56 |
Bisulphite treatment
Genomic DNA was extracted from experimental U87MG and SaOS2 cells. Bisulphite treatment of genomic DNA was done using EpiTect kit (Qiagen GmBH, Hilden, Germany). The DNA after bisulphite treatment was purified with Wizard™ DNA Clean-up System (Promega).
Primers targeting CpG sites of SINE
SINE sequence at chromosome 16 nucleotide 12755020 to 12754683 (NW_926528.1) and chromosome 7 nucleotide 4397659 to 439838 (NT_079595.2) were chosen for their highest homology with the consensus SINE sequence using NCBI BLAST. CpG islands were identified within 500 bp of the above genomic sequence. Nested primers and M13 tagged inner primers were designed from the flanking regions of the sequences of interest (Table 1).
DNA methylation analysis by PCR sequencing
The PCR products were sequenced directly using primers designed specifically against one of the bisulphite-converted strand. The sequencing chromatograms were analysed for change in methylation status of the CpG islands by comparing height of peaks of T (indicating unmethylated cytosine) and the peaks of C (indicating methylated cytosine) at a CpG island (Supporting Information). Twenty-three sites on chromosome 16 and 25 sites on chromosome 7 could be reproducibly analysed.
Global methylation
Global methylation of the bisulphite treated DNA samples was assessed by Line COBRA using LINE 3 and LINE 4 primers (Table 1) as described by Matsuzaki et al. in two biological replicates [27]. The IDV of digested (methylated) and undigested (unmethylated) bands was determined by Chemi-Imager gel documentation system (Alpha Innotech Corp., San Leandro, CA, USA) to obtain the percent fractional methylation at the LINE TaqI site (Supporting Information).
HIF-1α and DNMT1 protein expression
U87MG cells exposed to normoxia and hypoxia (various time-points) were lysed in a triple detergent buffer (as described previously), 30 μg of total protein was resolved on a 7.5% SDS-PAGE. The gel was electroblotted onto a PVDF membrane (Schneider and Schuell) and blocked in 4% BSA overnight at 4°C. It was further probed with rabbit polyclonal anti-DNMT1 (Abcam, Cambridge, MA, USA) or mouse monoclonal anti-HIF-1α (BD Biosciences, San Jose, CA, USA), goat anti-rabbit or antimouse alkaline phosphatase linked IgG (Santa Cruz Biotech. Inc., Franklin Lake, NJ, USA) and developed with BCIP/NBT (Promega) as per manufacturers protocol.
Statistical analysis
The change in methylation at site specific CpG at SINE loci on chromosome 16 and 7 was calculated for hypoxia (N – H = percentage methylation in N minus percentage methylation in H), for reverts with respect to normoxia (percentage methylation in N minus percentage methylation in HR) and for revert with respect to hypoxia (percentage methylation in H minus percentage methylation in HR). Statistical differences between test and control for each group were evaluated by paired two-tailed t-test for significance.
Inter-Alu PCR
Inter-alu primers were taken from Krajinovic et al.[28]. The conditions were followed as detailed in Srivastava et al.[8] (Table 1). The amplified products were resolved in native 8% PAGE on Midi protean apparatus (Bio-Rad). Amplification patterns of the samples were obtained and altered bands were identified as either loss/gain or changes in intensity in H and HR samples as compared to N. Each PCR was performed on two biological replicates, repeated at least three times and scoring was done by two independent observers.
PAGE elution, cloning and sequencing
Altered bands were selected and carefully excised from H samples for elution (Supporting Information). They were cloned in PGEMTeasy (Promega) and sequenced in an ABI prism sequencer (Applied Biosystems, Foster City, CA, USA). The sequence analysis was done using MegaBLAST and repeatBLAST and homologies were further analysed for flanking alu sequences.
Results
Growth characteristics after exposure to long-term hypoxia
U87MG cells cultured in 0.1% O2 demonstrated evidence of HIF-1α protein expression (Fig. S1) by 4 hrs. The cells also showed characteristic signs of apoptosis like cellular shrinkage, nuclear condensation and membrane blebbing in the form of apoptotic bodies even at 6 weeks (Fig. 1A). Similar characteristic changes were also observed in SaOS2 cells. The apoptotic feature of cleaved caspase was also seen in U87MG cell lysates at 1, 2 and 3 days of hypoxia and was found to persist till at least 6 weeks of hypoxic treatment (Fig. 1C).
Fig 1.
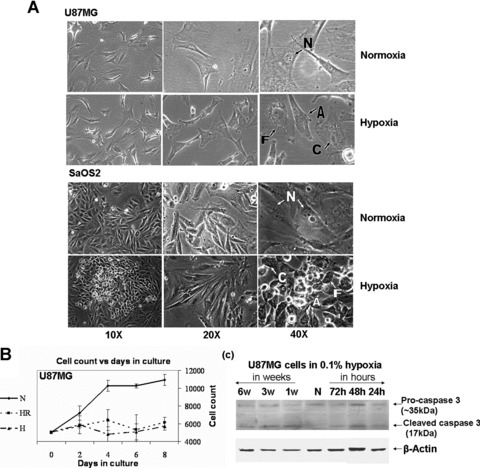
(A) Morphological analysis: Effects of prolonged (6 weeks) normoxia (21% O2) and hypoxia (0.1% O2) on the morphology of the U87MG and SaOS2 cells. Characteristic signs of apoptosis like cellular shrinkage, nuclear condensation, blebbing of cell membrane in the form of apoptotic bodies are marked in hypoxic cells. N – Normal, A – Apoptotic bodies, F – Fragmentation, C – Condensation. (B) Cell viability assay: Trypan blue stained cell counting was done for measuring survival in hypoxia and normoxia treated U87MG cells with equal number of seeding (5 × 103 cells) per well. The cells cultured initially in hypoxia and normoxia conditions were continued in the same condition (N or H) or reverted back to normoxia (HR). (C) Caspase-3 cleavage for apoptosis: Western blot for pro-caspase 3 and cleaved caspase-3 in cell extracts. Cleaved caspase was observed in U87MG cells exposed to 0.1% hypoxia at various time-points including 6-week hypoxia.
The growth rate of the U87MG cells was determined on cells cultured for 4 weeks under hypoxic and normoxic conditions and replated in 96-well plates. Maximum growth was noted in cells grown continuously in 21% O2 (N) (Fig. 1B). When cells were cultured in hypoxia (H), the growth rate was less than in N, but more than the cells reverted to normoxia for 4 weeks after initial culture in hypoxia for 6 weeks (HR). The cells cultured in hypoxia (H) and the revertent HR showed very slow growth rate and remained less than N throughout the assay duration.
By day 8, cell proliferation of N and H were significantly different, demonstrating a decrease in cell growth after long-term hypoxia. HR had a significantly lower growth rate compared to N but not H (Fig. 1B) indicating that the decrease in growth rate may be an irreversible phenomenon.
Expression of SINE and LINE under hypoxia
Real-time PCR and RT-PCR was done with cDNA prepared from total RNA extracted from cells cultured in hypoxia, normoxia and hypoxia of 6 weeks reverted to 4-week normoxia. Average values of two biological replicates are shown. After 6-week hypoxia, there was a 9.57-fold increase in relative expression of SINE in U87MG cells as compared to normoxia (P= 0.0008). In case of SaOS2 the increase in the expression level is approximately 6.30-fold in hypoxia (P= 0.0004). SINE transcripts levels increased only by 1.29-fold in U87MG and 1.36-fold in SaOS2 revertents (Fig. 2A).
Fig 2.
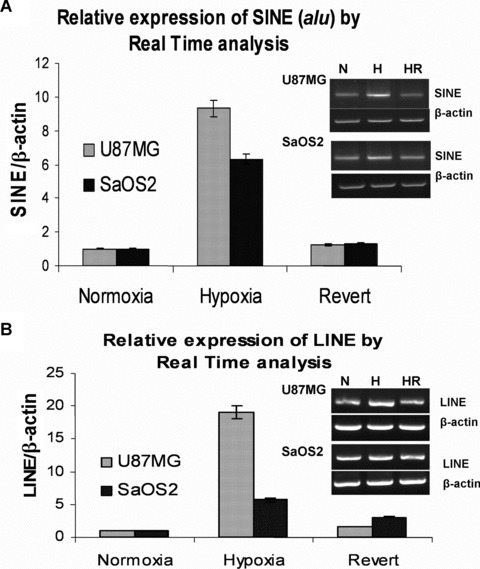
SINE (alu) and LINE transcript levels: Relative expression of (A) SINE and (B) LINE in U87MG and SaOS2 cell lines under normoxia, hypoxia and 4-week revert to normoxia by real-time PCR. The SINE and LINE expression are markedly increased in 6-week hypoxia as shown by average values from two biological replicates. Representative photographs of RT-PCR are shown in inset. The values are normalized with respect to b-actin.
Similarly, the relative expression of LINE 1 also increased by 19.51-fold (P= 0.0006) and 5.88-fold (P= 0.001) in U87MG and SaOS2 cell exposed to 6-week hypoxia (Fig. 2B). When cells were reverted back to normoxia for 4 weeks, the increase was only 1.68-fold in U87MG and 3.21-fold in SaOS2.
Methylation analysis by bisulphite sequencing
Methylation analysis was done in consensus SINE locus on two chromosomes Chr16 and Chr7 in both U87MG and SaOS2. Tendency for a particular CpG site to show hypo- or hypermethylation was consistent in all the biological replicates for both the cell lines.
From the consensus SINE locus of chromosome 16, 23 CpG sites were chosen for bisulphite PCR sequencing. In U87MG, there was statistically significant decrease in methylation status after exposure to long-term hypoxic conditions which did not change even after reverting back to normoxic conditions. Seventeen sites showed average decrease in methylation by 9.76% (from 91.37% to 81.63%) (P= 0.01) in hypoxia with respect to normoxia (Fig. 3A-i) (Fig. S2). Major hypomethylation was seen at two specific CpG sites (site14: from 91.6% to 47.8% and site17: from 77.1% to 34.2%). Of these two sites, site 17 showed no change in methylation on reversion to normoxia, while at site 14 there was some increase in methylation in revertent (62.8%). The average percentage of methylation of these 17 sites in revertents was 81.5%, i.e. there was almost no change (0.05%) in methylation in comparison to hypoxia (P= 0.97) (Fig. 3A-i).
Fig 3.
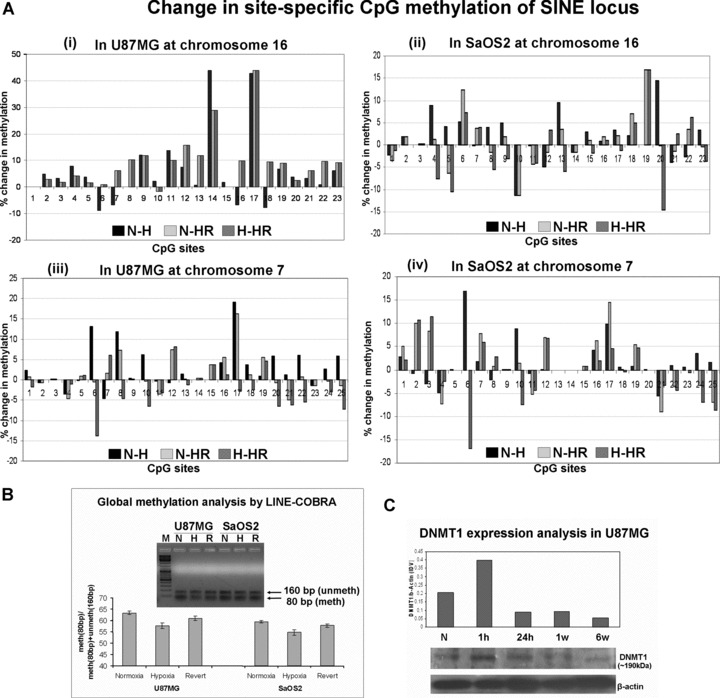
(A) Change in percentage (%) of site-specific CpG methylation at SINE locus (i) at chromosome 16 in U87MG, (ii) at chromosome 16 in SaOS2, (iii) at chromosome 7 in U87MG, (iv) at chromosome 7 in SaOS2. Change is calculated for test group as follows: N(-)H: percentage methylation in normal minus percentage methylation in hypoxia; N(-)R: percentage methylation in normal minus percentage methylation in hypoxia; H(-)HR: percentage methylation in hypoxia minus percentage methylation in revert. x-axis: CpG sites; y-axis: percentage change in methylation. (B) Global methylation assay: LINE-COBRA was done in two biological replicates of normoxia (N), hypoxia (H) and reverted samples (HR) of U87MG and SaOS2 cells and the average value is depicted. In the representative picture shown, the band of 160 bp shows the unmethylated fraction and 80 bp is of methylated fraction. Densitometry shows global hypomethylation in hypoxia samples as compared to normoxia controls in both U87MG (P= 0.033) and SaOS2 (P= 0.032) cells. (C) DNMT1 expression: Western blot of U87MG cells exposed to various durations of 0.1% hypoxia showed a marked decrease in DNMT1 levels in 6-week hypoxia (H) as compared to N. The sudden increase in DNMT1 expression in 1 hr hypoxia was followed by reduced expression in 24 hrs, 1 week and 6 weeks. The IDV values are normalized to b-actin levels.
The other six CpG sites at chromosome 16 studied in U87MG behaved differently and showed either increase or no change in the methylation status due to hypoxia in comparison to normoxia (Fig. S2). The average increase in methylation in H at these sites was 5.05% (P= 0.027) and in HR there was significant decrease of 11.16% in comparison to H (P= 0.007) (Fig. 3 A-i). Methylation of reverted cells was even lower than the original normoxic levels (P= 0.02). The results point to the reversible nature of these changes.
Bisulphite sequencing of chromosome 16 in SaOS2 showed 13 sites with significant average decrease of methylation in hypoxia by 4.98% (P= 0.0005) (Fig. 3A-ii) from 92.23% to 87.26% (Fig. S3). Ten sites (which showed either increase or no change in methylation) showed significant (P= 0.03) average increase of 2.71% in methylation (from 89.61% to 92.32%) as compared to normoxia (Fig. 3A-ii).
Similarly, the locus on chromosome 7 showed significant average decrease by 5.35% (Fig. 3A-iii) from 94.43% to 89.08% in a major group of 16 CpG sites (Fig. S4) in hypoxic U87MG cells (P= 0.001). The other minor group of nine sites showed average increase by 1.24% from 94.03 to 95.28% (P= 0.052) (Fig. 3A-iii).
The corresponding locus in SaOS2 also showed a major group of 16 sites undergoing average decrease of methylation by 3.33% from 97.42% to 94.08% (P= 0.012) in hypoxia as compared to normoxia (Fig. 3A-iv). The other group of nine sites showed average increase by 1.89% from 93.42 to 95.32% (P= 0.03) (Fig. 3A-iv) (Fig. S5).
Global methylation assay
Normoxic (N), hypoxic (H) and reverted (HR) U87MG cells showed an average global methylation from two biological replicates of 63.35%, 57.7% and 60.95%, respectively (H < N, P= 0.033), while the methylation of SaOS2 cells showed 59.45%, 54.7% and 57.75%, respectively (H< N, P= 0.032) (Fig. 3B). Representative photograph of a LINE-COBRA is shown in Fig. 3B[23].
DNMT1 protein expression
The DNMT1 expression was markedly reduced in U87MG cells exposed to 6-week hypoxia. Although the DNMT1 expression of U87MG cells showed a steep rise in expression immediately on exposure to hypoxia, the levels reduced in 24 hrs and subsequent time-points. This phenomenon was observed in two biological replicates and a representative Western blot is shown (Fig. 3C).
Inter-alu PCR for genetic instability
Inter-alu PCR is a modified fingerprinting technique to study genome instability [8, 28–30]. PCR product amplified from DNA extracted from H and HR were compared with N. A number of changes in intensity were observed, while clear gains or losses of bands were few and difficult to analyse. We were able to carefully extract and sequence four alterations which were gain of intensity (marked a, b, c and d in Fig. 4).
Fig 4.
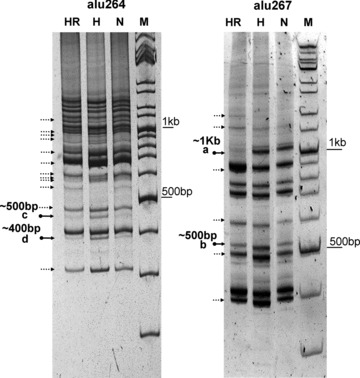
Inter-alu PCR for detecting genomic instability: Representative inter-Alu PCR profile generated for normoxia, hypoxia and revert with primers alu264 and alu267 in U87MG cells. Photographs have been inverted for better visualization. A number of alterations are observed, mostly as changes in intensity  . The alterations which were cloned and sequenced are marked differently
. The alterations which were cloned and sequenced are marked differently  , and these changes were consistent among biological replicates.
, and these changes were consistent among biological replicates.
Although three of the sequenced bands showed homology to sequences flanked by inverted alu elements, one of the smaller bands (400 bp, ‘d’) showed homology to chr 14 (AL355834.4) and alu at only one end indicating that a single priming event had taken place. Two altered bands of size ∼1 kb (‘a’) and 500 bp (‘c’) showed a change in intensity. The 1 kb (loss of intensity in revert) band showed homology to 963 bp on chromosome 8 (nt5375089–5376042) and is flanked by Zn finger motif and BTB domain 214048 bp on its 5′ side and by a hypothetical protein 41,263 bp at 3 side. The ∼500 bp band (‘c’) showed homology to chromosome 9 (NT_024000.16) and encodes a pseudogene similar to part of translocation protein (myeloid leukaemia associated protein, SET), the 5 end of the gene codes for euchromatic histone methyltransferase1 and it houses two CpG islands. One of the bands visualized as gain of intensity ∼500 bp (marked ‘b’) showed homology to homo sapiens F-box protein 27 (FBXO27), mRNA (NM_178820) which forms a part of the SCF complexes acting as protein-ubiquitin ligases. F-box proteins interact with S-phase kinase protein (SKP-1), through the F box.
Discussion
In solid tumours; microenvironment, hypoxia, increased genomic instability and mutation have been demonstrated to be associated in numerous studies [31]. Cross-talk between tumorigenesis, hypoxia and methylation pattern may determine the pathophysiology of oncogenesis and tumour metastasis [32–34]. Increase in radio and chemo resistance with increase in intra-tumour hypoxia may also be correlated with the change in methylation pattern [35, 36]. However not much is known about the transcription and methylation status of repeat elements under long-term hypoxia and consequent effects on genomic instability.
We have assessed the increase in transcript levels of alu (SINE) and of LINE, which is a source of reverse transcriptase. We have also observed genomic instability with the help of inter-alu PCR, possibly as a consequence of mobilization of the increased SINE transcripts. Our results have correlated this increase in transcript to aberrant methylation of the CpG islands, particularly at the SINE loci, under hypoxic conditions.
In our system, the cells lost their proliferative capability to a significant extent after being cultured in hypoxic conditions for 3 weeks, which could not be regained even after reverting back to normoxia. Hypoxia and HIF are known to modulate a number of cell cycle regulators and metabolic pathways which may result in a decrease in proliferating capability [37].
Our results show that global demethylation with site specific changes in the SINE loci were accompanied by increase in SINE expression. The additional increase in LINE1 transcript enhances the chance of SINEs being actively retrotransposed. Alu mediated recombination contributing to genetic instability in hypoxic cells was observed with the help of inter-alu PCR. Inter-alu PCR provides a good ‘non-invasive’ analysis of the cellular microsystem, which is highly responsive to stress. Cells with acquired genetic instability under hypoxic conditions may be selected over its normal counterpart and may or may not persist with same genetic signature even after reversal to normoxia depending on whether such instability gives a potential growth/survival advantage. Alteration of a band can occur if the particular fragment has duplicated and/or transposed to other chromosomal location. Inter-alu PCR also identifies specific sequences which are associated with instability. Alu elements are largely present in the non-coding part of the genome, and detecting an alteration associated with flanking alu within an mRNA is of considerable interest.
Previously ignored as junk, repetitive Alu elements are now known to modulate gene expression by alternate splicing, RNA editing and translation regulation [38–40] and may generate new enhancers, promoters and polyadenylation signals to many genes. Inverted alu are particularly unstable and are potential targets of RNA editing. Athanasiadis et al.[40] demonstrated favoured A-I editing when two inverted Alu element are present in close proximity (<2 kb). Such aberrant editing is found in several neurological disorders [41] and many such undiscovered potential transcriptomes may hold great physiological or pathological importance [42].
In this study we have shown data for four alterations, one of which is within an mRNA sequence of F-box protein 27. Sequence analysis shows that the F-box mRNA is interspersed with alu elements including two in reverse orientation. Incidentally its associate, SKP-1, is known to be regulated by hypoxia and HIF-1 [43, 44], and in light of this, the functional relevance of our observations is being studied. The mechanism of retrotransposition or the chromosomal localization of the recombined fragments flanked by SINE elements is yet to be addressed.
To the best of our knowledge, our observations on long-term hypoxia resulting in increased expression of SINE elements, possibly as a result of hypomethylation and subsequent alu mediated genetic instability, are a novel finding. Although alu mediated recombination and instability has been associated with a number of diseases, the importance of the methylation status of repeat elements and their expression in hypoxic regions of solid tumours needs to be further studied. This genomic instability may lead to evolution of a low-grade tumour to a high-grade one and hence needs to be addressed in order to counter the adverse phenotype associated with hypoxia.
Acknowledgments
This work has been funded by an intramural research grant (F.6–1/2003-Acad) from All India Institute of Medical Sciences and research grant (No. 63/12/2002-BMS) from Indian Council of Medical Research, India to P.C. T.S. was recipient of a Young Scientist fellowship from Department of Science and Technology, India. P.D. is a Senior Research Fellow of Council of Science and Industrial Research, India. We are grateful to Prof Chinmay K. Mukhopadhyay, Special Center for Molecular Medicine, Jawaharlal Nehru University, New Delhi for helpful discussions. We are thankful to Mathura Prasad, Satish and Jyoti for their technical and secretarial assistance.
Supporting Information
Fig. S1HIF-1 induction in U87MG cells exposed to 0.1% hypoxia.
Fig. S2 U87MG site specific CpG methylation at SINE locus on chromosome 16.
Fig. S3 SaOS2 site specific CpG methylation at SINE locus on chromosome 16.
Fig. S4 U87MG site specific CpG methlation at SINE locus on chromosome 7.
Fig. S5 SaoS2 site specific CpG methylation at SINE locus on chromosome 7.
References
- 1.Misra A, Chattopadhyay P, Dinda AK, et al. Extensive intra-tumor heterogeneity in primary human glial tumors as a result of locus non-specific genomic alterations. J Neurooncol. 2000;48:1–12. doi: 10.1023/a:1006435201961. [DOI] [PubMed] [Google Scholar]
- 2.Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–8. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 3.Arnaud P, Goubely C, Pélissier T, et al. SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol. 2000;20:3434–41. doi: 10.1128/mcb.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho N-Y, Kim B-H, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 5.Choi I-S, Estecio MRH, Nagano Y, et al. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20:802–10. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 6.Boeke JD. LINEs and Alu-the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu W-M, Chu W-M, Choudary PV, et al. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–65. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava T, Seth A, Datta K, et al. Inter-alu PCR detects high frequency of genetic alterations in glioma cells exposed to sub-lethal cisplatin. Int J Cancer. 2005;117:683–9. doi: 10.1002/ijc.21057. [DOI] [PubMed] [Google Scholar]
- 9.Bennett EA, Keller H, Mills RE, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–83. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 12.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–8. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Métivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 15.Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 16.Watson RE, Curtin GM, Doolittle DJ, et al. Progressive alterations in global and GC-Rich DNA methylation during tumorigenesis. Toxicol Sci. 2003;75:289–99. doi: 10.1093/toxsci/kfg190. [DOI] [PubMed] [Google Scholar]
- 17.Dumont N, Wilson MB, Crawford YG, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci USA. 2008;105:14867–72. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahrzad S, Bertrand K, Minhas K, et al. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2:119–25. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 19.Dunn BK. Hypomethylation: One side of a larger picture. Ann N Y Acad Sci. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 20.Brahimi-Horn C, Chiche J, Pouysségur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol. 2007;19:223–9. doi: 10.1016/j.ceb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Fanga JS, Gilliesb RD, Gatenbyb RA. Adaptation to hypoxia and acidosis in carcinogenesis and tumor progression. Semin Cancer Biol. 2008;18:330–7. doi: 10.1016/j.semcancer.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice GC, Hoy C, Schimke RT. Transient hypoxia enhances the frequency of dihydrofolate reductase gene amplification in Chinese hamster ovary cells. Proc Natl Acad Sci USA. 1986;83:5978–82. doi: 10.1073/pnas.83.16.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan J, Narayanan L, Rockwell S, et al. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 2000;60:4372–6. [PubMed] [Google Scholar]
- 24.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–7. [PubMed] [Google Scholar]
- 25.Rodríguez-Jiménez FJ, Moreno-Manzano V, Lucas-Dominguez R, et al. Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells. Stem Cells. 2008;26:2052–62. doi: 10.1634/stemcells.2007-1016. [DOI] [PubMed] [Google Scholar]
- 26.Katschinski DM. Is there a molecular connection between hypoxia and aging. Exp Gerontol. 2006;41:482–4. doi: 10.1016/j.exger.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki K, Deng G, Tanaka H, et al. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–9. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 28.Krajinovic M, Richer C, Labuda D, et al. Detection of a mutator phenotype in cancer cells by inter-alu polymerase chain reaction. Cancer Res. 1996;56:2733–7. [PubMed] [Google Scholar]
- 29.Kaku H, Ito S, Ebara S, et al. Positive correlation between allelic loss at chromosome 14q24–31 and poor prognosis of patients with renal cell carcinoma. Urology. 2004;64:176–81. doi: 10.1016/j.urology.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Bonafé M, Cardelli M, Marchegiani F, et al. Increase of homozygosity in centenarians revealed by a new inter-Alu PCR technique. Exp Gerontol. 2001;36:1063–73. doi: 10.1016/s0531-5565(01)00112-7. [DOI] [PubMed] [Google Scholar]
- 31.Bristow RG, Hill RP. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–92. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 32.Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52:2071–7. [PubMed] [Google Scholar]
- 33.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 34.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 35.Vaupel P, Schlenger K, Knoop C, et al. Oxygenation of human tumors: evaluation of tissue oxygen distribution in breast cancers by computerized O2 tension measurements. Cancer Res. 1991;51:3316–22. [PubMed] [Google Scholar]
- 36.Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–76. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 37.Gardner LB, Corn PG. Hypoxic regulation of mRNA expression. Cell Cycle. 2008;7:1916–24. doi: 10.4161/cc.7.13.6203. [DOI] [PubMed] [Google Scholar]
- 38.Stuart JJ, Egry LA, Wong GH, et al. The 3’-UTR of human MnSOD mRNA hybridizes to a small cytoplasmic RNA and inhibits gene expression. Biochem Biophys Res Commun. 2000;274:641–8. doi: 10.1006/bbrc.2000.3189. [DOI] [PubMed] [Google Scholar]
- 39.Landry JR, Medstrand P, Mager DL. Repetitive elements in the 5’-untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–6. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- 40.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurevich I, Tamir H, Arango V, et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–56. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 42.Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acid Res. 2006;34:5491–7. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan M, Gu Q, He H, et al. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1 alpha ubiquitination and degradation. Oncogene. 2008;27:1404–11. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- 44.Paltoglou S, Roberts BJ. HIF-1alpha and EPAS ubiquitination mediated by the VHL tumour suppressor involves flexibility in the ubiquitination mechanism, similar to other RING E3 ligases. Oncogene. 2007;26:604–9. doi: 10.1038/sj.onc.1209818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1HIF-1 induction in U87MG cells exposed to 0.1% hypoxia.
Fig. S2 U87MG site specific CpG methylation at SINE locus on chromosome 16.
Fig. S3 SaOS2 site specific CpG methylation at SINE locus on chromosome 16.
Fig. S4 U87MG site specific CpG methlation at SINE locus on chromosome 7.
Fig. S5 SaoS2 site specific CpG methylation at SINE locus on chromosome 7.


