Abstract
Every surface of the human body is colonized by a diverse microbial community called the microbiota, yet the impact of microbiota on viruses is unclear. Recent research has advanced our understanding of how microbiota influence viral infection. Microbiota inhibit infection of some viruses and promote infection of other viruses. These effects can occur through direct and/or indirect effects on the host and/or virus. This review examines the known effects and mechanisms by which the microbiota influence mammalian virus infections. Furthermore, we suggest strategies for future research on how microbiota impact viruses. Overall, microbiota may influence a wide array of viruses through diverse mechanisms, making the study of virus-microbiota interactions a fertile area for future investigation.
Keywords: virus, microbiota, bacteria, innate immunity
INTRODUCTION
Every surface of the human body exposed to the environment is colonized by a diverse microbial community called the microbiota. The microbiota include bacteria, fungi, and viruses and these microorganisms are thought to outnumber human cells (1–3). The complexity of the microbiota is only now beginning to be appreciated. A majority of the microbiota reside in the gastrointestinal tract, but distinct populations are also found on the skin, mouth, and genitourinary tract (4–8). Interestingly, the body site, not the individual, appears to be the primary determinant for diversity in these microbiota communities (9, 10).
Human health is integrally tied to the microbiota. Microbiota are required for optimal human development and help protect the host from various pathogens (11, 12). The microbiota compete with pathogens for colonization sites and resources. Moreover, the microbiota supply important signals for immune priming and therefore aid development of the immune system (13–16). Yet what constitutes a healthy microbiota remains to be determined. Recently microbiota imbalances have been linked to many human diseases including inflammatory bowel diseases, type 2 diabetes, and obesity (17–22). Thus, the presence of this community is vital for human health.
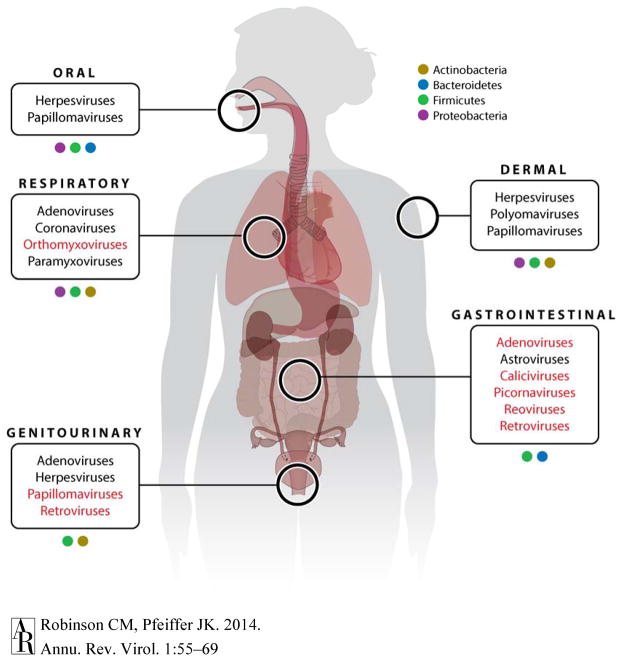
Based on the location and abundance of microbiota, unquestionably viruses have interactions with the microbiota (Figure 1). Recent progress has led to tremendous insight into the interaction of microbiota and the host. However, many microbiota-viral interactions remain unaddressed. Understandably, viruses that colonize the gastrointestinal tract have been the most studied due to site of the initial infection and the vast microbial community at that site. Enteric viruses encounter approximately 1014 bacteria in the mammalian gastrointestinal tract (2). Even though the density and composition of the microbiota differs at different body sites, nearly every virus must initiate infection at a location populated by microorganisms.
Figure 1.
Anatomical locations of potential microbiota-viral interactions. Distinct populations of the microbiota are found across the human body. Colored boxes represent the diversity in the bacterial taxa. Predominating bacterial taxa at each location are shown, although other taxa are also present (9, 10). At each of these sites, viruses initiate infection and potentially interact with the local microbiota community. Common viruses are shown. Red text indicates viruses that have been studied in the context of the microbiota.
Microbiota-virus interactions have been studied using several laboratory models, most commonly germ-free mice or antibiotic-treated mice. Germ-free mice are microbiologically sterile from birth and are maintained in a sterile environment. Illustrating the importance of microbiota on nutrition, germ-free animals consume 30% more calories per day to maintain body weight compared with conventional animals (23). While generally healthy and fertile, germ-free mice have underdeveloped immune systems, which can complicate experimental interpretation (24–29). That said, germ-free mice provide a defined environment devoid of any microbes and they can be colonized with one bacterial strain or several bacterial strains to examine virus-microbiota interactions. Therefore, germ-free mice are a very important model for virus-microbiota studies. Because germ-free mice require specialized facilities and are expensive to generate and maintain, antibiotic-treated mice have also been used to study virus-microbiota interactions. In this model, mice are treated with a combination of several antibiotics and knockdown is confirmed by culture-based methods (30). The caveats to the antibiotic treatment approach include potential alteration of host physiology, unknowns created by unculturable organisms, incomplete knockdown, and antibiotic resistance. However, the antibiotic treatment approach has the benefit that it is relatively inexpensive, can be used with any mouse strain as well as other animal models, and animals have physiological and immune development in the presence of the microbiota prior to treatment. Because both germ-free models and antibiotic-treatment models have pros and cons, utilization of both models for the study of virus-microbiota interactions is ideal when possible. Finally, it is critical to use the natural route of infection for the specific virus under investigation. Enteric viruses should be inoculated orally, respiratory viruses should be inoculated intranasally, and so forth. In fact, bypassing that natural route of infection, and therefore the relevant microbiota community, can significantly alter results (31).
SCOPE AND INTENT
Our goal in this article is to provide a broad review of what is currently known about how microbiota influence mammalian virus infections. There are many other interesting interactions between viruses and microbiota that are not discussed here: For example, bacteriophage interactions with microbiota, plant virus interactions with plant and soil microbiota, and virus interactions with protozoa. While the effect of microbiota on mammalian virus infection has been studied since the 1960s, there is not a deep understanding of how microbiota influence any single virus. Instead, we have a relatively broad and shallow knowledge of general effects and “phenomenology” for a wide variety of viruses. For any given viral system, there are only a handful of publications. This may be due to several factors including the cross-disciplinary nature of the research and technical hurdles. However, some of the technical hurdles have been at least partially overcome, such as the more common availability of germ-free mice and deep sequencing. Therefore, the 50-year old field of mammalian virus-microbiota interactions is poised for a period of great productivity. The intent here is to summarize current knowledge of microbiota-viral interactions in a broad approach covering many viral systems and to suggest areas of future investigation. As shown below, microbiota can be detrimental or beneficial to viral infection. Furthermore, these consequences can occur through direct effects by interactions between the microbiota and virus particles, indirect effects through microbiota-mediated immune priming, and other effects. As we explore this new crossroads between microbiology and virology, future work using techniques that are now feasible will help identify important mechanisms that influence viral infections.
NEGATIVE EFFECTS OF THE MICROBIOTA ON VIRUSES
Rotavirus
For many years evidence has suggested a connection between the microbiota and rotavirus. Rotavirus, a nonenveloped, double-stranded RNA virus from the Reoviridae family, is significant cause of viral diarrhea worldwide. Probiotics have been shown to reduce the duration of viral diarrhea and administration of Lactobacillus rhamnosus GG reduces rotavirus shedding (32–34). Recently it was determined that soluble factors from commensal bacteria block rotavirus infection in intestinal epithelial cells in vitro (35). Varyukina et al. hypothesize that these soluble factors modify the intestinal epithelial cell-surface glycans and prevent rotavirus attachment. It remains to be determined how these factors may limit rotavirus attachment, but highlights the ability of the commensal bacteria to potentially protect the host from viral infections by altering the host environment.
Influenza
Stimulation of the host immune system by the microbiota has been shown to affect influenza virus disease. Influenza virus, an enveloped, negative-strand RNA virus from the Orthomyxoviridae family, is spread by the respiratory route. In the 1960s Dolowy et al. examined influenza virus pathogenesis in conventional versus germ-free mice. They determined that germ-free mice are more susceptible to influenza A virus compared to conventional mice (36). Recently, three groups have shed light on the mechanisms behind this observation. Ichinohe et al. demonstrated higher pulmonary influenza virus titers in antibiotic-treated mice compared with conventional mice (37). Not all commensal bacteria within conventional mice were responsible for host protection, as neomycin-sensitive bacteria were associated with protective immune responses in the lung. Interestingly, stimulation with toll-like receptor (TLR) agonists was sufficient to restore immune responses in antibiotic-treated mice, suggesting that certain gut bacteria may prime the immune system for influenza virus protection. Similarly, Abt et al. demonstrated increased influenza virus titers and pathogenesis in antibiotic-treated mice compared with conventional mice (38). The enhanced viral replication and disease in antibiotic-treated mice correlated with reduced virus-specific CD8+ T cell responses and IgG and IgM antibody levels, suggesting impaired adaptive immune responses in mice with depleted microbiota. Furthermore, antibiotic treatment impaired antiviral immune responses in alveolar macrophages (38). Recently, Wang et. al. found that M2 alveolar macrophages were downstream mediators of viral clearance following priming with a upper respiratory tract commensal bacterium, Staphylococcus aureus (39). These macrophages were shown to reduce influenza pathogenesis by limiting inflammation in the lung. Priming was TLR2 dependent, confirming the role of pattern recognition receptors in stimulating influenza immune responses. Overall, these findings suggest that commensal bacteria from both the intestinal and upper respiratory tracts may play important roles in limiting influenza virus infections by providing a tonic signal that calibrates the immune system.
Lymphocytic Choriomeningitis Virus (LCMV)
As with influenza virus, stimulation of the host immune system by the microbiota influences LCMV infection. LCMV, an enveloped, negative-strand RNA virus from the Arenaviridae family, can undergo acute or persistent infection in mice depending upon the viral strain. Abt et al. demonstrated that LCMV clearance was delayed in antibiotic-treated mice, indicating that microbiota promote antiviral responses (38). The impaired viral control correlated with reduced LCMV-specific CD8+ T cell responses and IgG antibody titers. In fact, CD8+ T cells from antibiotic-treated mice demonstrated increased inhibitory receptors and decreased production of effector molecules, pointing toward T cell exhaustion in the absence of conventional microbiota. While macrophage recruitment was not impaired in antibiotic-treated mice, macrophages from conventional mice expressed higher antiviral response genes, suggesting an impaired innate immune response in antibiotic-treated mice (38). These results suggest that the altered environment in antibiotic-treated mice diminishes innate and adaptive immune responses to LCMV infection.
Dengue Virus
Microbiota also influence viral infection in nonhuman hosts such as insects. Dengue virus, an enveloped, single-stranded RNA virus from the Flaviviridae family, is transmitted from mosquitoes to humans. When a mosquito feeds and ingests a virus-containing blood meal, the virus encounters the mosquito midgut and undergoes replication. Like the human gastrointestinal tract, the mosquito midgut is colonized by microbiota. Xi et al. treated Aedes aegypti mosquitoes with antibiotics to deplete the midgut microbiota and examined effects on dengue virus replication. They found higher dengue virus loads in the midgut of antibiotic-treated mosquitoes, suggesting that microbiota limit viral replication (40). Knockdown of the toll-like receptor adapter protein MyD88 also increased viral loads in the midgut, suggesting that the toll pathway is involved in limiting dengue virus replication. The microbiota are likely to be the source for toll pathway stimulation culminating in antiviral defense. In fact, antidengue virus immune responses elicited by some bacterial strains were more effective than immune responses elicited by other bacterial strains (41). Interestingly, one member of the insect microbiota, Wolbachia, is being explored as a method to limit transmission of dengue virus and other mosquito-borne infectious agents. Wolbachia species are symbiotic parasites that are maternally transmitted within insect populations. Infection of mosquitoes with Wolbachia species confers dengue virus resistance and is being explored as a potential tool to control mosquito-born diseases (42, 43). Recent evidence suggests that Wolbachia induce oxidative stress within the host mosquito that activates the antiviral toll pathway (44). Future work is needed to provide the precise mechanisms underlying Wolbachia effects on dengue virus replication, but offers promising studies for future control of dengue virus transmission and mosquito-borne diseases.
POSITIVE EFFECTS OF THE MICROBIOTA ON VIRUSES
Theiler’s Murine Encephalomyelitis Virus (TMEV)
Long used as a model virus, TMEV is an enteric virus and common contaminant of laboratory rodent colonies due to fecal-oral spread. TMEV is a nonenveloped, single-stranded RNA virus from the Picornaviridae family and can replicate in neurons and induce multiple sclerosis-like disease in mice. Pullen et al. demonstrated that TMEV replication and disease was enhanced by treatment with lipopolysaccharide (LPS) (45). It is likely that these effects occur through increased inflammation in the central nervous system, which enhances viral replication.
Poliovirus
Although poliovirus is an enteric virus spread by the fecal-oral route, it can rarely invade the human central nervous system and cause paralysis. Poliovirus is a nonenveloped, single-stranded RNA virus from the Picornaviridae family. Our laboratory demonstrated that poliovirus replication and pathogenesis was reduced in antibiotic-treated mice, suggesting that microbiota promote poliovirus infection (31). We recently proposed two different mechanisms by which bacteria enhance poliovirus infectivity. First, bacterial polysaccharides from both Gram-positive and Gram-negative bacteria bind to poliovirus and stabilize the virion to prevent premature RNA release (46). N-acetylglucosamine-containing bacterial polysaccharides, including peptidoglycan and LPS, bind poliovirus and limit thermal inactivation. Second, using LPS as a model bacterial polysaccharide, we found that LPS aids poliovirus attachment to host cells by enhancing viral binding to its cellular receptor, the poliovirus receptor (46). To get mechanistic insight into virus-polysaccharide interactions, we identified a poliovirus mutant with diminished LPS binding. A single amino acid mutation in the VP1 capsid protein, T99K, reduced LPS binding and virion stabilization by LPS (46). While replication and pathogenesis of this mutant virus was not altered in vivo, the mutant virus was unstable in mouse feces. These data suggest that binding of polysaccharides to poliovirus may stabilize the capsid for transmission to a new host. The mechanism by which the microbiota enhance poliovirus pathogenesis in mice remains to be determined.
Reovirus
It is likely that all humans are infected with mammalian reovirus by age five through fecal-oral transmisison. Reovirus is a nonenveloped, double-stranded RNA virus from the Reoviridae family and reovirus symptoms are generally mild or absent. Using orally inoculated conventional or antibiotic-treated mice, we demonstrated that reovirus replication and pathogenesis was reduced in antibiotic-treated mice (31). These results suggest that microbiota promote reovirus replication and pathogenesis in vivo, although the mechanisms remain unknown.
Mouse Mammary Tumor Virus (MMTV)
Retroviruses, such as MMTV, are commonly transmitted across mucosal surfaces that are rich in microbiota. MMTV is an enveloped virus of the Retroviridae family and has been used as a model system for decades. MMTV is spread from mouse mother to mouse pup through milk, and therefore infection is initiated in the gut. The microbiota have been hypothesized to play an important role in MMTV and other retrovirus infections. C3H/HeJ mice, which lack TLR4, have delayed mammary tumor development when infected with MMTV, implicating a role for TLR4 in productive MMTV infection (47). In C3H/HeN mice, which have functional TLR4, MMTV can establish persistent infection. Jude et al. demonstrated that the immunoregulatory cytokine interleukin 10 (IL-10) was produced in a TLR4-dependent manner to establish MMTV persistence (48). Yet for many years the mechanism by which TLR4 signaling was induced and promoted MMTV transmission was unknown. Recently, Kane et al. provided a mechanism for the TLR4/IL-10 stimulation and MMTV persistence: The link between the TLR signaling and viral persistence was found to be microbiota-dependent. Kane et al. found that antibiotic-treated and germ-free mice fail to transmit MMTV to their offspring through maternal milk (49). LPS from Gram-negative bacteria bound to MMTV virions culminating in TLR4 signaling and IL-10 production. This work suggests that MMTV may take direct advantage of the microbiota using bacterial LPS to enhance viral tolerance through host IL-10 production.
UNCLEAR EFFECTS OF THE MICROBIOTA ON VIRUSES
Adenovirus
Adenoviruses infect humans by respiratory and enteric routes. Human adenovirus is a nonenveloped, double-stranded DNA virus of the Adenoviridae family. Replication of some adenoviruses is inhibited by defensins, which are antimicrobial peptides produced by host cells in response to microbiota. A subset of defensins, including alpha defensin 5, bind human adenovirus virions and limit replication in cultured cells by preventing uncoating in endosomes (50–54). Because defensin production is stimulated by microbiota and defensins have antiviral activity against adenovirus, microbiota may inhibit adenovirus infection. Due to the lack of a complete defensin knockout animal model, the antiviral role of defensins in vivo is uncertain. However, future studies in this area are critical, since defensins impact several different viruses including herpesviruses, human papillomaviruses, polyomaviruses, orthomyxoviruses, and retroviruses (55).
Coxsackievirus B3 (CVB3)
While microbiota and/or LPS enhance infection with poliovirus and TMEV, effects of the microbiota on the closely related CVB3 are unclear. CVB3 is a nonenveloped, single-strand RNA virus in the Picornaviridae family and is very closely related to poliovirus. In the 1960s Schaffer et al. demonstrated that germ-free mice were more susceptible to CVB3 compared with colonized mice (56). These results suggest that microbiota inhibit CVB3 infection. However, CVB3 was inoculated by intraperitoneal injection, bypassing the natural oral route of infection. Further studies will be required to determine whether microbiota effects on picornaviruses differ by virus type.
Norovirus and Murine Norovirus (MNV)
Norovirus is transmitted by through the fecal-oral route and is a common cause of viral gastroenteritis. Norovirus is a nonenveloped, single-stranded RNA virus from the Caliciviridae family. Infection with norovirus significantly alters the gut flora in humans (57). Interestingly MNV infection does not alter the gut microbiota in mice, yet MNV-induced pathologies have been shown to be microbiota-dependent (58). Genetically susceptible mice (ATG16L1HM) infected with persistent MNV induce intestinal inflammation similar to Crohn’s disease (59). In this model, antibiotic-treated mice have reduced pathology suggesting that the inflammation is microbiota-dependent. While it remains to be determined if these interactions are direct or indirect, recently it was demonstrated that strains of norovirus bind to extracellular polymeric glycans from certain strains of intestinal bacteria (60). Since this binding may play a role in viral attachment to intestinal epithelial cells, Miura et al. hypothesize that this event may alter transmission and infection of the virus.
Murine Leukemia Virus (MLV)
MLV has been used as a model virus for decades. MLV is an enveloped virus of the Retroviridae family and transmission can occur through mucosal and other routes. Two studies have shown that germ-free mice are relatively resistant to MLV-induced leukemia compared with conventional mice (61, 62). These results suggest that microbiota may promote MLV infection and disease progression. It is possible that microbial products stimulate division of lymphoid cells, facilitating viral replication (62). However, an earlier study found that germ-free mice were more sensitive to MLV-induced leukemia (63). The discrepancies among these studies include use of different MLV strains (64).
Human Immunodeficiency Virus (HIV)
Apart from direct blood-mediated transmission, HIV is transmitted through mucosal routes in microbiota-rich areas. HIV is an enveloped virus of the Retroviridae family and there is great interest in identifying host factors that influence infection. Although the impact of the microbiota on HIV infection has not been directly examined, several studies suggest that the microbiota influence HIV infection (65). First, Brenchley et al. demonstrated that HIV pathogenesis is enhanced by LPS-mediated immune activation from microbial translocation through the intestinal barrier (66). The resulting chronic immune activation contributes to development of AIDS. These results suggest that microbiota enhance HIV pathogenesis. Second, Majerle et al. demonstrated that the HIV envelope protein gp120 binds bacterial LPS and that LPS-bound gp120 has reduced binding to target cells (67). These results suggest that microbial products may reduce HIV infection. Third, bacterial metabolic products may influence HIV infection. The vaginal microbiota of healthy women is dominated by Lactobacillus species that produce lactic acid, which has been shown to be a potent microbicide (8, 68)(69). Lai et al. demonstrated that HIV-1 diffused more slowly in lactic acid-acidified cervicovaginal mucus compared with neutralized mucus (70), indicating that the presence of Lactobacilli may limit viral infection. Furthermore, Aldunate et al. demonstrated that physiological concentrations of lactic acid can inactivate HIV-1 and HIV-2 in vitro (71). These results suggest that Lactobacilli may reduce HIV infection.
Human Papillomavirus (HPV)
HPV is spread by mucosal and skin routes and may also be affected by microbiota (72). HPV is a nonenveloped, double-stranded DNA virus in the Papovaviridae family. In analysis of the vaginal microbiota from 70 healthy women, Gao et al. reported that Lactobacillus gasseri is found at a significantly higher frequency in HPV-positive women (73). Future work is needed to determine whether this correlation implicates Lactobacilli as pro-HPV factors and to further define microbiota-viral interactions in the genitourinary tract.
Kilham Rat Virus (KRV)
KRV has been used as a model virus to induce type 1 diabetes (T1D) in the LEW1.WR1 rat (74). KRV is a nonenveloped, single-stranded DNA virus in the Parvoviridae family. How T1D is triggered remains unknown, but viruses are commonly associated with the onset of T1D (75). Emerging data also suggest that intestinal microbiota play a role in the course of T1D (76). Hara et al. demonstrated that KRV-induced T1D in LEW1.WR1 rats was abrogated with antibiotic treatment (77). Following infection with KRV the gut microbiota in the rat is also altered. This suggests that the intestinal microbiota may promote virally-induced T1D. However, Wen et al. demonstrated that microbiota colonized MyD88-deficient mice do not develop T1D, whereas germ-free MyD88-deficient mice develop robust T1D. This work suggests that the intestinal microbiota may play a role in prevention of diabetes (78). Species differences between the rat and mouse models may be responsible for the different outcomes.
CONCLUSIONS AND FUTURE DIRECTIONS
We are only just beginning to “scratch the surface” of the intricacies that exist during a viral infection, particularly how viruses are impacted by microbiota. As described above, microbiota may limit viral infection, promote viral infection, or have no effect. Microbiota may also have direct or indirect effects on viral infection. For example, microbiota (or their products) may directly interact with viral particles to alter infectivity or responses (31, 46, 49, 60). Conversely, microbiota may indirectly impact viral infection by inducing immune priming (37–40, 44, 49). In fact, it appears that microbiota-mediated immune priming through TLR signaling is operative for all viruses that are negatively impacted by the microbiota (37–40, 44). It is possible and perhaps likely that microbiota will impact each virus by multiple mechanisms.
While we typically view viruses as pathogens, it has become apparent that healthy individuals are also colonized by an immense number of viruses. These viruses make up the ‘virome’ and remain an understudied member of microbiota community. The virome is comprised largely of viruses that infect bacteria (bacteriophage) and plants, but eukaryotic viruses are also common. The presence of bacteriophage and plant viruses is likely due to host commensal bacteria and diet (79–83). It remains to be determined if detection of these viruses through metagenomics represents a “snapshot in time” or represents truly persistent viruses. Additionally, the extent to which humans harbor mammalian viruses and the effect of mammalian viruses on human biology are unclear.
Clearly, more studies are needed to understand how microbiota impact viral infections. But what viruses should be examined and what experimental systems should be used? We think that a combination of model viruses and pathogenic human viruses, examined using several experimental systems such as germ-free mice and antibiotic-treated mice, will provide significant insight. Model viruses, including the natural mouse viruses MMTV, MLV, TMEV, MNV, LCMV, and mouse adenovirus, have many benefits for these studies. For example, they are capable of infecting many different mouse strains, they can undergo robust replication, natural inoculation routes can be used, and they are adapted to the mouse host. Furthermore, they offer a more natural infection process, where disease is often rare. The downside to using these mouse viruses is that they may not accurately recapitulate processes in humans. Some human viruses, such as influenza, replicate reasonably well in mice following inoculation by the natural route. Other human viruses, including CVB3, HIV, dengue, and HPV, are clinically important but can be more difficult to study using mouse models. For example, HIV and dengue virus require unique and/or transgenic mouse models and lessons learned from these mouse models may not translate directly to what occurs in humans (84–86). Furthermore, gaining mechanistic insight following initial observations can be difficult if mice carrying multiple gene knockouts and/or transgenes must be constructed. Because microbiota-virus studies are inherently complex, we suggest using human viruses that replicate well in standard mouse strains or using rodent viruses. Beyond mouse models, there is also great potential for other experimental systems such as Drosophila (87), mosquitoes (40, 44), and primates. In addition, retrospective and prospective studies of viral infections in humans treated with antibiotics may provide some insight, although interpretation may be difficult.
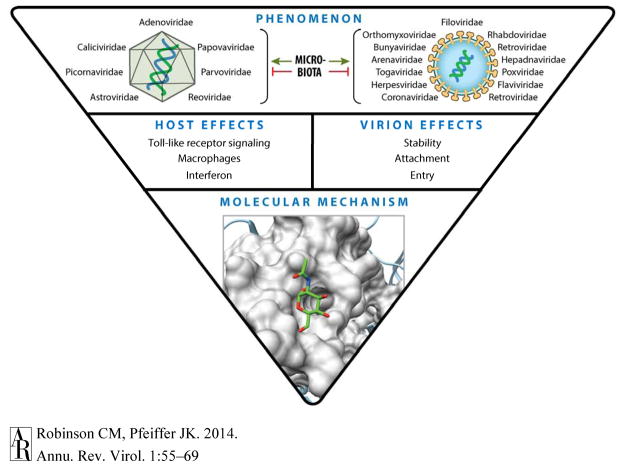
New studies of microbiota effects on viruses should be aimed at both examining the “phenomenology” and examining mechanisms underlying effects (Figure 2) (64, 88, 89). The ideal first step for any given virus is to determine whether the microbiota promote infection, limit infection, or have no effect. These experiments are relatively straightforward if an animal model is available, as described above. If an animal model is not available, ex vivo and in vitro experiments may be useful. The second step, identifying mechanisms by which microbiota impact viral infection, is much more difficult. Microbiota can influence viral infection through direct and/or indirect mechanisms on the host and/or virus, making a mechanistic search akin to “finding a needle in a haystack.” However, previous studies may inform future studies and help limit the search. There are two common themes that have emerged: 1) Microbial products initiate innate immune signaling to limit or promote viral infection (37–40, 44, 49), and 2) Microbial products interact with viral particles to alter infectivity or host responses (31, 46, 49, 60, 67). Interestingly, bacterial LPS binds poliovirus (31, 46), MMTV (49), and HIV (67), suggesting that LPS binding may be a property shared by several viruses. Moving forward, it may be helpful for investigators to examine innate immune signaling and virion interactions with bacterial products as starting points for mechanistic studies of microbiota-virus interactions. Some of these types of experiments can be performed ex vivo or in vitro, which will be especially useful for viruses lacking good animal models.
Figure 2.
Future work on microbiota-virus interactions: From phenomenology to molecular mechanism. Top: Ideally, initial studies with a given virus would start with examining the effect of the microbiota on viral infection using an animal model. Several viral families are shown (left, nonenveloped virus families; right, enveloped virus families). Middle: If animal studies indicate that microbiota promote or inhibit viral infection, then follow-up experiments to determine whether microbiota influence the host and/or viral particle should be performed. These experiments will provide clues regarding more specific mechanisms and are typically a combination of in vivo and in vitro experiments. Bottom: Once microbiota effects on the host or virion are confirmed, additional experiments may lead to identification of specific molecular mechanisms by which microbiota promote or inhibit infection. These experiments may identify specific microbial products conferring effects, binding sites on viral particles, specific immune cascades with proviral or antiviral effects, and so forth.
Although our understanding of virus-microbiota interactions is not complete, there is the potential for using existing knowledge for therapeutic design. For example, for viruses that benefit from the microbiota, it is tempting to speculate that depleting the host microbiota may confer antiviral activities. However, this is not likely to be a viable approach in most cases. Antiviral effects in mice can require massive depletion of host microbiota using several antibiotics (31), which is not feasible in humans. Furthermore, alterations to host microbiota may lead to side effects with far greater consequence than the viral infection itself (13, 90). Therefore, targeted therapeutics, rather than a broad-spectrum antimicrobial sledgehammer, will be needed and will require understanding specific mechanisms.
Future studies may reveal multiple consequences of microbiota on viruses. It is imperative as the virology field moves forward that we evaluate the complete environment that underlies a viral infection. Factors in the environment, including the microbiota, likely influence the outcomes of many infections. We anticipate that future work will help establish new paradigms in how we view individual viral infections within the complex environments within the human body.
Acknowledgments
We thank our colleagues and collaborators for many helpful and stimulating discussions. Funding sources include grants from NIH (NIAID R01 AI74668 and T32 AI007520), the Hartwell Foundation, the World Health Organization Global Polio Eradication Initiative, and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award.
LITERATURE CITED
- 1.Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, et al. An estimation of the number of cells in the human body. Ann Hum Biol. 2013;40:463–71. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- 2.Hao WL, Lee YK. Microflora of the gastrointestinal tract: a review. Methods Mol Biol. 2004;268:491–502. doi: 10.1385/1-59259-766-1:491. [DOI] [PubMed] [Google Scholar]
- 3.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–20. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 6.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome assembly across multiple body sites in low-birthweight infants. MBio. 2013;4:e00782–13. doi: 10.1128/mBio.00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conly JM, Stein K, Worobetz L, Rutledge-Harding S. The contribution of vitamin K2 (menaquinones) produced by the intestinal microflora to human nutritional requirements for vitamin K. Am J Gastroenterol. 1994;89:915–23. [PubMed] [Google Scholar]
- 12.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–59. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–41. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–58. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–85. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–82. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 19.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–45. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 20.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–40. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. 1983;33:46–50. [PubMed] [Google Scholar]
- 24.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–70. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–34. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 27.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–13. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–11. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–7. [PMC free article] [PubMed] [Google Scholar]
- 30.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–52. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guandalini S, Pensabene L, Zikri MA, Dias JA, Casali LG, et al. Lactobacillus GG administered in oral rehydration solution to children with acute diarrhea: a multicenter European trial. J Pediatr Gastroenterol Nutr. 2000;30:54–60. doi: 10.1097/00005176-200001000-00018. [DOI] [PubMed] [Google Scholar]
- 33.Saavedra J. Probiotics and infectious diarrhea. Am J Gastroenterol. 2000;95:S16–8. doi: 10.1016/s0002-9270(99)00811-4. [DOI] [PubMed] [Google Scholar]
- 34.Guarino A, Canani RB, Spagnuolo MI, Albano F, Di Benedetto L. Oral bacterial therapy reduces the duration of symptoms and of viral excretion in children with mild diarrhea. J Pediatr Gastroenterol Nutr. 1997;25:516–9. doi: 10.1097/00005176-199711000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Varyukhina S, Freitas M, Bardin S, Robillard E, Tavan E, et al. Glycan-modifying bacteria-derived soluble factors from Bacteroides thetaiotaomicron and Lactobacillus casei inhibit rotavirus infection in human intestinal cells. Microbes Infect. 2012;14:273–8. doi: 10.1016/j.micinf.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Dolowy WC, Muldoon RL. Studies of Germfree Animals. I Response of Mice to Infection with Influenza a Virus. Proc Soc Exp Biol Med. 1964;116:365–71. doi: 10.3181/00379727-116-29249. [DOI] [PubMed] [Google Scholar]
- 37.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–70. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Li F, Sun R, Gao X, Wei H, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez JL, Souza-Neto J, Torres Cosme R, Rovira J, Ortiz A, et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl Trop Dis. 2012;6:e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310:326–8. doi: 10.1126/science.1117607. [DOI] [PubMed] [Google Scholar]
- 43.Iturbe-Ormaetxe I, Walker T, SLON Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12:508–18. doi: 10.1038/embor.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X, Zhou G, Wu J, Bian G, Lu P, et al. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2012;109:E23–31. doi: 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pullen LC, Park SH, Miller SD, Dal Canto MC, Kim BS. Treatment with bacterial LPS renders genetically resistant C57BL/6 mice susceptible to Theiler’s virus-induced demyelinating disease. J Immunol. 1995;155:4497–503. [PubMed] [Google Scholar]
- 46.Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Outzen HC, Corrow D, Shultz LD. Attenuation of exogenous murine mammary tumor virus virulence in the C3H/HeJ mouse substrain bearing the Lps mutation. J Natl Cancer Inst. 1985;75:917–23. doi: 10.1093/jnci/75.5.917. [DOI] [PubMed] [Google Scholar]
- 48.Jude BA, Pobezinskaya Y, Bishop J, Parke S, Medzhitov RM, et al. Subversion of the innate immune system by a retrovirus. Nat Immunol. 2003;4:573–8. doi: 10.1038/ni926. [DOI] [PubMed] [Google Scholar]
- 49.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–9. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gropp R, Frye M, Wagner TO, Bargon J. Epithelial defensins impair adenoviral infection: implication for adenovirus-mediated gene therapy. Hum Gene Ther. 1999;10:957–64. doi: 10.1089/10430349950018355. [DOI] [PubMed] [Google Scholar]
- 51.Smith JG, Nemerow GR. Mechanism of adenovirus neutralization by Human alpha-defensins. Cell Host Microbe. 2008;3:11–9. doi: 10.1016/j.chom.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010;6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen EK, Nemerow GR, Smith JG. Direct evidence from single-cell analysis that human {alpha}-defensins block adenovirus uncoating to neutralize infection. J Virol. 2010;84:4041–9. doi: 10.1128/JVI.02471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gounder AP, Wiens ME, Wilson SS, Lu W, Smith JG. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J Biol Chem. 2012;287:24554–62. doi: 10.1074/jbc.M112.354068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson SS, Wiens ME, Smith JG. Antiviral mechanisms of human defensins. J Mol Biol. 2013;425:4965–80. doi: 10.1016/j.jmb.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaffer J, Beamer PR, Trexler PC, Breidenbach G, Walcher DN. Response of germ-free animals to experimental virus monocontamination. I Observation on Coxsackie B virus. Proc Soc Exp Biol Med. 1963;112:561–4. doi: 10.3181/00379727-112-28105. [DOI] [PubMed] [Google Scholar]
- 57.Nelson AM, Walk ST, Taube S, Taniuchi M, Houpt ER, et al. Disruption of the human gut microbiota following Norovirus infection. PLoS ONE. 2012;7:e48224. doi: 10.1371/journal.pone.0048224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson AM, Elftman MD, Pinto AK, Baldridge M, Hooper P, et al. Murine norovirus infection does not cause major disruptions in the murine intestinal microbiota. Microbiome. 2013:1. doi: 10.1186/2049-2618-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miura T, Sano D, Suenaga A, Yoshimura T, Fuzawa M, et al. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J Virol. 2013;87:9441–51. doi: 10.1128/JVI.01060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isaak DD, Bartizal KF, Caulfield MJ. Decreased pathogenicity of murine leukemia virus-Moloney in gnotobiotic mice. Leukemia. 1988;2:540–4. [PubMed] [Google Scholar]
- 62.Kouttab NM, Jutila JW. Friend leukemia virus infection in germfree mice following antigen stimulation. J Immunol. 1972;108:591–5. [PubMed] [Google Scholar]
- 63.Mirand EA, Grace JT., Jr Responses of Germ-Free Mice to Friend Virus. Nature. 1963;200:92–3. doi: 10.1038/200092a0. [DOI] [PubMed] [Google Scholar]
- 64.Wilks J, Golovkina T. Influence of microbiota on viral infections. PLoS Pathog. 2012;8:e1002681. doi: 10.1371/journal.ppat.1002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shu Z, Ma J, Tuerhong D, Yang C, Upur H. How intestinal bacteria can promote HIV replication. AIDS Rev. 2013;15:32–7. [PubMed] [Google Scholar]
- 66.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 67.Majerle A, Pristovsek P, Mancek-Keber M, Jerala R. Interaction of the HIV-1 gp120 viral protein V3 loop with bacterial lipopolysaccharide: a pattern recognition inhibition. J Biol Chem. 2011;286:26228–37. doi: 10.1074/jbc.M111.220434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Hanlon DE, Moench TR, Cone RA. Vaginal pH and Microbicidal Lactic Acid When Lactobacilli Dominate the Microbiota. PLoS ONE. 2013;8:e80074. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai SK, Hida K, Shukair S, Wang YY, Figueiredo A, et al. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervicovaginal mucus. J Virol. 2009;83:11196–200. doi: 10.1128/JVI.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aldunate M, Tyssen D, Johnson A, Zakir T, Sonza S, et al. Vaginal concentrations of lactic acid potently inactivate HIV. J Antimicrob Chemother. 2013;68:2015–25. doi: 10.1093/jac/dkt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veldhuijzen NJ, Snijders PJ, Reiss P, Meijer CJ, van de Wijgert JH. Factors affecting transmission of mucosal human papillomavirus. Lancet Infect Dis. 2010;10:862–74. doi: 10.1016/S1473-3099(10)70190-0. [DOI] [PubMed] [Google Scholar]
- 73.Gao W, Weng J, Gao Y, Chen X. Comparison of the vaginal microbiota diversity of women with and without human papillomavirus infection: a cross-sectional study. BMC Infect Dis. 2013;13:271. doi: 10.1186/1471-2334-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellerman KE, Richards CA, Guberski DL, Shek WR, Like AA. Kilham rat triggers T-cell-dependent autoimmune diabetes in multiple strains of rat. Diabetes. 1996;45:557–62. doi: 10.2337/diab.45.5.557. [DOI] [PubMed] [Google Scholar]
- 75.Ghazarian L, Diana J, Simoni Y, Beaudoin L, Lehuen A. Prevention or acceleration of type 1 diabetes by viruses. Cell Mol Life Sci. 2013;70:239–55. doi: 10.1007/s00018-012-1042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–49. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 77.Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol. 2012;189:3805–14. doi: 10.4049/jimmunol.1201257. [DOI] [PubMed] [Google Scholar]
- 78.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lecuit M, Eloit M. The human virome: new tools and concepts. Trends Microbiol. 2013;21:510–5. doi: 10.1016/j.tim.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14:654–9. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berges BK, Wheat WH, Palmer BE, Connick E, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−−gamma c−− (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koyanagi Y, Tanaka Y, Ito M, Yamamoto N. Humanized mice for human retrovirus infection. Curr Top Microbiol Immunol. 2008;324:133–48. doi: 10.1007/978-3-540-75647-7_9. [DOI] [PubMed] [Google Scholar]
- 86.Sun Z, Denton PW, Estes JD, Othieno FA, Wei BL, et al. Intrarectal transmission, systemic infection, and CD4+ T cell depletion in humanized mice infected with HIV-1. J Exp Med. 2007;204:705–14. doi: 10.1084/jem.20062411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu J, Hopkins K, Sabin L, Yasunaga A, Subramanian H, et al. ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc Natl Acad Sci USA. 2013;110:15025–30. doi: 10.1073/pnas.1303193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfeiffer JK, Sonnnenburg JL. The intestinal microbiota and viral susceptibility. Front Microbiol. 2011;2:92. doi: 10.3389/fmicb.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilks J, Beilinson H, Golovkina TV. Dual role of commensal bacteria in viral infections. Immunol Rev. 2013;255:222–9. doi: 10.1111/imr.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stewardson AJ, Huttner B, Harbarth S. At least it won’t hurt: the personal risks of antibiotic exposure. Curr Opin Pharmacol. 2011;11:446–52. doi: 10.1016/j.coph.2011.06.011. [DOI] [PubMed] [Google Scholar]