Abstract
Purpose.
We determined if quantitative measurements of biogeographic ancestry (BGA) correlate with variations in optic disc area, corneal thickness (CCT), and retinal nerve fiber layer (RNFL) thickness.
Methods.
Data were obtained from 656 participants in the African Descent and Glaucoma Evaluation Study (ADAGES) cohort who consented to BGA testing. Data for CCT, optic disc area, and RNFL thickness were obtained from subjects in the ADAGES study who also had participated in the current substudy. A total of 31 ancestry informative markers (AIMs) with large allele frequencies differences between populations was used to calculate admixture proportion (implemented in STRUCTURE). Correlations with BGA adjusted for diagnosis, age, and sex for CCT and optic disc area using the whole group and RNFL thickness adjusted for age and sex for the normal study participants were determined.
Results.
The mean percentage of African admixture was 79.6% in the self-described African Descent (AD) group and 3.5% in the European Descent (ED) group. Percent African ancestry was significantly correlated with CCT (ρ = −0.27, P < 0.0001) and disc area (ρ = 0.15, P < 0.0001), but only marginally associated with RNFL thickness (ρ = 0.20, P = 0.092) in adjusted models.
Conclusions.
The BGA correlates with variation in ocular features that significantly differ across racial groups and that have been associated with the development of glaucoma. While BGA can provide an objective measurement of the biologic component of self-described race for ocular research, for most nongenetic epidemiologic studies, self-described race may adequately describe the associations with these ocular characteristics. (ClinicalTrials.gov number, NCT00221923.)
Keywords: biogeographic ancestry, optic disc, retinal nerve fiber layer, corneal thickness, race
The proportion of African ancestry assessed with biogeographic genetic analysis correlates with corneal thickness and optic disc area.
Introduction
Several epidemiologic studies have demonstrated a greater susceptibility to primary open angle glaucoma (POAG) and higher rates of blindness in populations of African descent (AD) compared to European descent (ED).1–6 These racial differences in the susceptibility to glaucomatous injury prompted the initiation of the National Eye Institute-funded African Descent and Glaucoma Evaluation Study (ADAGES).7 The ADAGES enrolled healthy, glaucoma suspect, ocular hypertensive, and glaucomatous individuals of AD and ED.
As in all prior clinical studies of glaucoma, ADAGES uses self-report to define racial classification. This categorization has demonstrated differences in the risk of developing glaucoma among individuals6,8–10 and in ocular features, such as corneal thickness,11 optic nerve morphology, and retinal nerve fiber layer (RNFL) thickness.12 However, racial categorization by self-description is an imprecise measure of biologic diversity and represents an amalgam of socioeconomic, cultural, and geographic factors in addition to the biologic components of the disease process that ADAGES was designed to explore.7
Alternatively, quantitative assessment of biogeographic ancestry (BGA) can provide a genetic definition of race that is free from the imprecision of self-description. Further, it may more appropriately describe race as a continuous rather than categorical variable. Thus, shifting focus from self-described race to genetically defined BGA may allow for more accurate characterization of the population substructure associated with the biologic component of what is termed “race.” The purpose of this study is to describe the proportion of African versus European ancestry in the ADAGES cohort using BGA genetics information and to determine if this proportion is associated with variation in ocular phenotype seen with self-reported race, specifically optic disc size, corneal thickness, and RNFL thickness.
Methods
Study Design
The methodology for ADAGES has been described in the baseline study design.13 The relevant methodology to this particular report is reviewed below. Baseline data from participants in the National Eye Institute (NEI; Bethesda, MD, USA)–funded Diagnostic Innovations in Glaucoma Study (DIGS) and the ADAGES without ocular disease (defined below) were used for all the analyses in the current study. The methodologies followed in DIGS and ADAGES are identical. All participants provided written informed consent. The institutional review boards at all three sites approved the study methods. Methods adhere to the tenets of the Declaration of Helsinki and to the Health Insurance Portability and Accountability Act (HIPAA). Healthy control subjects for ADAGES and DIGS were recruited to join the study by advertisement, from family members of patients, and from primary eye care clinics, whereas glaucoma patients, suspects, and ocular hypertensive patients were obtained from glaucoma subspecialty clinics. Each participant was asked to identify his or her ancestry by self-report using the NEI Inclusion/Enrollment system describing ethnicity and race (available in the public domain at http://orwh.od.nih.gov/pubs/outreach.pdf).
All participants were over 18 years of age. Eligible participants had open angles, best-corrected acuity of 20/40 or better, and refractive error no greater than 5.0 diopters (D) sphere and 3.0 D cylinder at study entry. Diabetic participants with no evidence of retinal involvement were included. A family history of glaucoma was allowed. Glaucoma patients and normal subjects were excluded if they had a history of intraocular surgery (except for uncomplicated laser or cataract surgery), other intraocular eye disease, or other diseases known to affect the visual field (e.g., central nervous system disease, demyelinating diseases, human immunodeficiency virus [HIV]–positive, or AIDS).
Patients enrolled in ADAGES were categorized as glaucoma, defined as a glaucomatous visual field defect and optic nerve consistent with glaucomatous optic neuropathy (defined below), glaucoma suspect (normal visual field, but a glaucomatous optic nerve), ocular hypertension (documented IOP > 22 mm Hg with a normal visual field and optic nerve), or normal.
The ADAGES required two reliable tests on 24-2 standard automated perimetry (SAP; Carl Zeiss Meditec, Dublin, CA, USA) using the Swedish Interactive Thresholding Strategy (SITA) at baseline for inclusion. Reliability was defined as <33% false-positives, false-negatives, and fixation losses. All fields were reviewed by the University of California, San Diego (UCSD), Visual Field Assessment Center (VisFACT). The SAP visual fields were considered abnormal if the pattern standard deviation was triggered at the 5%, 2%, 1%, or 0.5% levels or the Glaucoma Hemifield Test (GHT) was “outside normal limits.” Abnormality had to be confirmed with an additional field.
Each baseline stereoscopic optic disc photograph was graded by two independent graders according to a standard protocol using the standard photographs as reference. All photographs were graded for quality and evidence of glaucoma damage. Each grader was masked to the participant's identity, diagnostic status, study, race, and other results. A third senior grader adjudicated all cases of disagreement. Glaucomatous optic disc damage was defined as evidence of excavation, neuroretinal rim thinning or notching, localized or diffuse RNFL defect, or a between eye asymmetry of the vertical cup–disc ratio > 0.2.
For the correlations between the proportion of African admixture, central corneal thickness (CCT), and optic disc area, all subjects were included regardless of diagnosis. For the correlation with RNFL thickness, only normal subjects were included as this parameter is affected by glaucomatous injury. The average values for CCT, disc area, and RNFL thickness for both eyes were used for this analysis when both eyes were eligible for inclusion. Corneal thickness was measured using a Pachette 2 ultrasonic pachymeter (DGH Technology, Inc., Exton, PA, USA). A mean of three measurements taken at the center of the cornea was used.
The protocol for image acquisition, including the assessment of quality by the UCSD Imaging Data Evaluation and Analysis Center (IDEA) center has been described previously.14 Optic disc area for this study was defined using the Heidelberg Retina Tomograph (HRT; Heidelberg Engineering, Heidelberg, Germany) obtained at the baseline visit. The HRT II (software version 3.0) uses a diode laser and confocal imaging to produce three-dimensional measurements of optic disc topography. Image acquisition, processing, and reproducibility of HRT measurements has been described in detail previously (Zangwill L, et al. IOVS 2014;40:ARVO E-Abstract 983 and Refs. 15–19). An experienced IDEA center operator evaluated image quality and outlined the disc margin (i.e., the contour line that defines the reference plane used for some analyses) with the aid of available stereoscopic photographs of the optic disc.
The RNFL thickness used for this study was determined using high resolution Spectralis optical coherence tomography (OCT) circle scans (software version 5.4.7.0; Heidelberg Engineering). The operation and IDEA Center review of the Spectralis OCT scans has been described in detail previously.20 To be considered good quality, the scans had to be clear, have a good signal strength and quality score (>15), and be centered vertically and horizontally in the scan window. The scan circle also had to be well centered around the optic disc. The average global RNFL thickness for both eyes was used for this analysis. Only the normal participant groups were used for the correlation between BGA and RNFL thickness due to the effect of glaucoma on RNFL thickness.
After separate informed consent for BGA analysis, blood was drawn from study participants in heparinized and EDTA tubes. Whole heparinized blood was used to create immortalized human B cell lines. The DNA was extracted from Epstein-Barr transformed lymphoblastoid cell lines for the University of Alabama at Birmingham site and blood for the other sites using the Gentra Puregene Blood Kit (Qiagen, Valencia, CA, USA), following the manufacturer's protocol. Only samples that had an absorbance score (A260/280) > 1.7 were genotyped.
The SNP genotyping was completed at Washington University using the Sequenom MassARRAY iPLEX (Sequenom, Inc., San Diego, CA, USA). A total of 53 SNPs was genotyped, with an average genotyping call rate of 85.3% to 87.9%. Of these SNPs, 31 ancestry informative markers (AIMs), demonstrating large allele frequency differences between populations were used to estimate admixture proportion in our samples using previously described methods.21 This approach estimates admixture proportions using 3517 subjects from 107 worldwide reference populations, including subjects collected by the Human Genome Diversity Project (HGDP).22 The reference populations were from five distinct continental regions (K5), including Africa, Eurasia, East Asia, Americas, and Oceania, using 20,000 burn-in cycles and 20,000 MCMC replications under the admixture model. The “infer α” option with the same, uniform α for all populations was used under the λ = 1 option. Allele frequencies were updated using only individuals with population information (i.e., reference subjects) at a migration prior of 0.05. All other parameters were set at default. The program STRUCTURE version 2.3 was used to determine admixture proportions.23,24
Statistical Analysis
Differences in the proportion of African ancestry, CCT, disc area, and RNFL thickness were compared across self-described racial groups using t-tests. Differences in sex across self-described racial groups were compared using a χ2 test. The proportion of African ancestry was correlated with CCT and disc area in the whole group and RNFL thickness in normal subjects using Spearman's Rho for nonparametric data. For CCT and disc area, general linear models were used to adjust for age, sex, and diagnostic category, whereas for RNFL thickness, the model was adjusted for age and sex. Similar general linear models were used to determine the correlations between the proportion of African ancestry and these three ocular parameters within each self-described racial group.
Additional linear regression models were developed to determine the associations between self-reported race with CCT and disc area, adjusted for diagnostic category, age, and sex, in additional to RNFL thickness, adjusted for age and sex. This allowed for a subjective comparison of the performance of self-described race and BGA similar to that described by Halder et al.25 The optic disc area was log transformed in each of these models. The JMP software (SAS Institute, Inc., Cary, NC, USA) was used for all analysis and a P value < 0.05 was considered statistically significant.
Results
Table 1 illustrates the demographic and ocular characteristics for ED and AD groups. The ED group was significantly older and had a higher proportion of males than the AD group. As expected, the AD group had a larger disc area, thinner CCT, and thicker RNFL than the ED group.
Table 1.
Demographic and Clinical Characteristics by Self-Described Racial Groups
|
African Descent, N= 297 |
European Descent, N= 359 |
PValue |
|
| Age, y | 60.5 ± 13 | 65.6 ± 13 | <0.0001 |
| Sex | |||
| Male | 97 | 161 | 0.0015 |
| Female | 200 | 198 | |
| % of African ancestry | 79.6 ± 25.9 | 3.5 ± 13.4 | <0.0001 |
| Central corneal thickness, mm | 533.1 ± 36.1 | 554.8 ± 38.1 | <0.0001 |
| Optic disc area, mm2 | 2.15 ± 0.5 | 1.96 ± 0.4 | <0.0001 |
| Retinal nerve fiber layer thickness, μm | 88.7 ± 15.9 | 85.3 ± 15.2 | 0.0072 |
| Diagnostic category: | |||
| Normal subjects | 77 | 52 | |
| Glaucoma patients and suspects | 220 | 307 | |
Numbers are means ± SD or n (%).
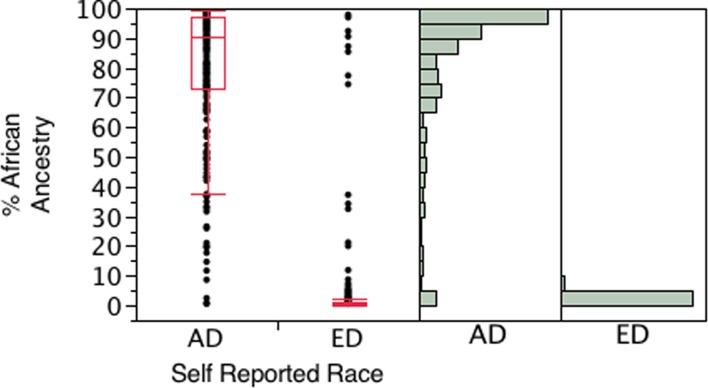
Figure 1 presents the distribution of the percentage of African admixture across racial groups. The mean percentage of African admixture in the AD group was 79.6% with a range from 0.3% to 99.3%. In the ED group, the mean percentage was 3.5% with a range from 0.22 to 98%. A total of 18 individuals of self-described African ancestry had less than 20% African ancestry with genetic ancestry testing, whereas six subjects of self-described European ancestry had greater than 80% African ancestry with BGA testing.
Figure 1.
Box and whisker plot and histograms showing the distribution of proportion of African ancestry determined by biogeographic testing for the AD (n = 297) and ED (n = 359) groups used in the analysis.
Table 2 presents the crude (Spearman's ρ) and adjusted correlations between the proportion of African ancestry and CCT, disc area, and RNFL thickness for the entire group and within each self-described racial group. An increase in the proportion of African ancestry was negatively correlated with cornea thickness (ρ = −0.27, P < 0.0001) and positively associated with a larger disc area (ρ = 0.15, P < 0.0001) in the entire cohort following adjustment for the differences in sex, age, and diagnostic category between self-described racial groups. There was a marginal association with a thicker RNFL and higher proportion of African ancestry within the normal group (ρ = 0.20, P = 0.091) adjusted for age and sex. The associations between BGA and ocular parameters also were evaluated within each racial group. However, none of these associations reached statistical significance.
Table 2.
Correlations Between the Proportion of African Ancestry and CCT, Disc Area, and RNFL Thickness
|
CCT |
ONH |
RNFL |
||||||||||
|
n |
ρ |
PValue |
PValue* |
n |
ρ |
PValue |
PValue* |
n |
ρ |
PValue |
PValue† |
|
| Total | 650 | −0.2741 | <0.0001 | <0.0001 | 648 | 0.1499 | <0.0001 | <0.0001 | 126 | 0.2020 | 0.0233 | 0.09183 |
| AD | 295 | −0.0787 | 0.1778 | 0.210 | 297 | 0.0081 | 0.8897 | 0.808 | 74 | 0.1349 | 0.2518 | 0.4192 |
| ED | 355 | −0.0937 | 0.0780 | 0.518 | 351 | −0.0019 | 0.9714 | 0.531 | 52 | 0.2051 | 0.1446 | 0.5592 |
Adjusted for age, sex, and diagnosis.
Adjusted for age and sex.
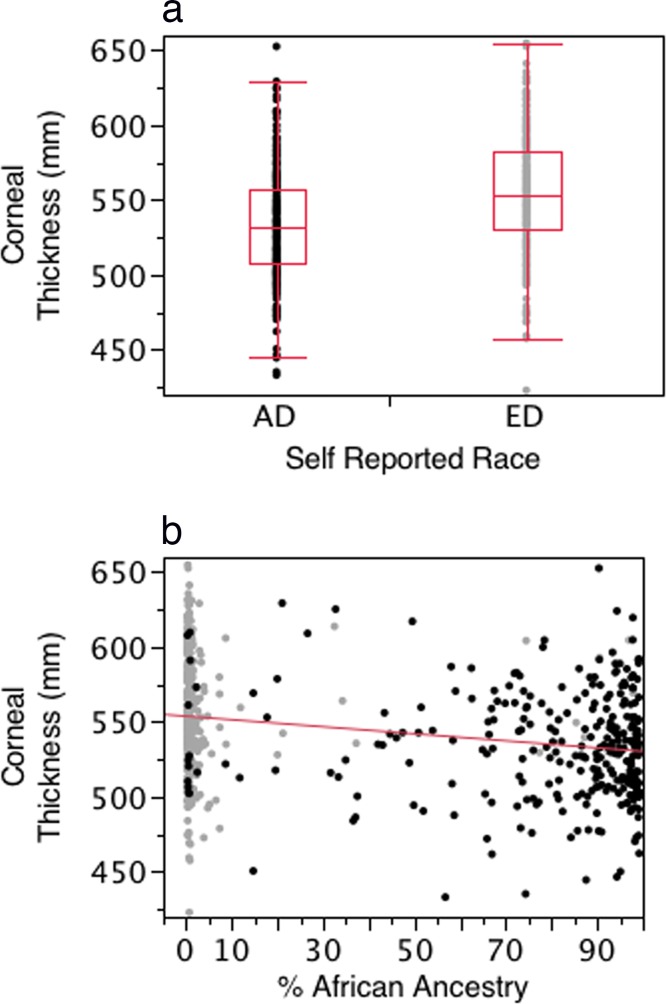
Figures 2a and 2b present scatterplots illustrating the association between self-reported race (Fig. 2a) and BGA (Fig. 2b) with CCT. The CCT was thinner in the self-described AD group and showed increased thinning with increasing proportion of African ancestry as defined by BGA. In models adjusting for age, sex, and diagnostic category, self-reported race and BGA provided similarly highly significant associations with CCT (P < 0.0001 for both self-report and BGA). Within each self-described racial group, BGA was not significantly associated with CCT (AD, P = 0.21; ED, P = 0.52).
Figure 2.
(a) Box and whisker plot of the distribution of corneal thickness for the AD and ED groups. (b) Scatterplot illustrating the association between the proportion of African ancestry and CCT for the entire cohort. Dark dots indicate ED group, light dots indicate AD group. The associations for self-report and BGA were highly significant (P < 0.0001 for both).
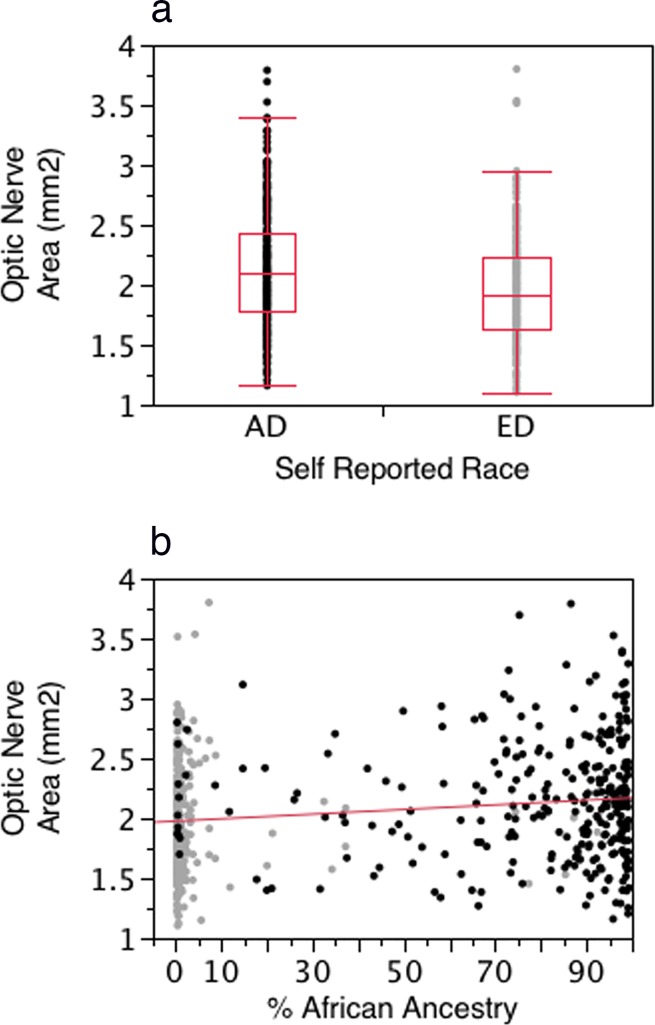
Figures 3a and 3b present scatterplots illustrating the association between self-reported race (Fig. 3a) and BGA (Fig. 3b) with disc area. Disc area was larger in the self-described AD group and increased in size with increasing proportion of African ancestry as defined by BGA. In models adjusting for age, sex, and diagnostic category, self-reported race and BGA provided similarly highly significant associations with disc area (P < 0.0001 for both self-report and BGA). Within each self-described racial group, BGA was not significantly associated with disc area (AD, P = 0.81; ED, P = 0.53).
Figure 3.
(a) Box and whisker plot of the distribution of optic disc area for the AD and ED groups. (b) Scatterplot illustrating the association between the proportion of African ancestry and optic disc area for the entire cohort. Dark dots indicate ED group, light dots indicate AD group. The associations for self-report and BGA were highly significant (P < 0.0001 for both).
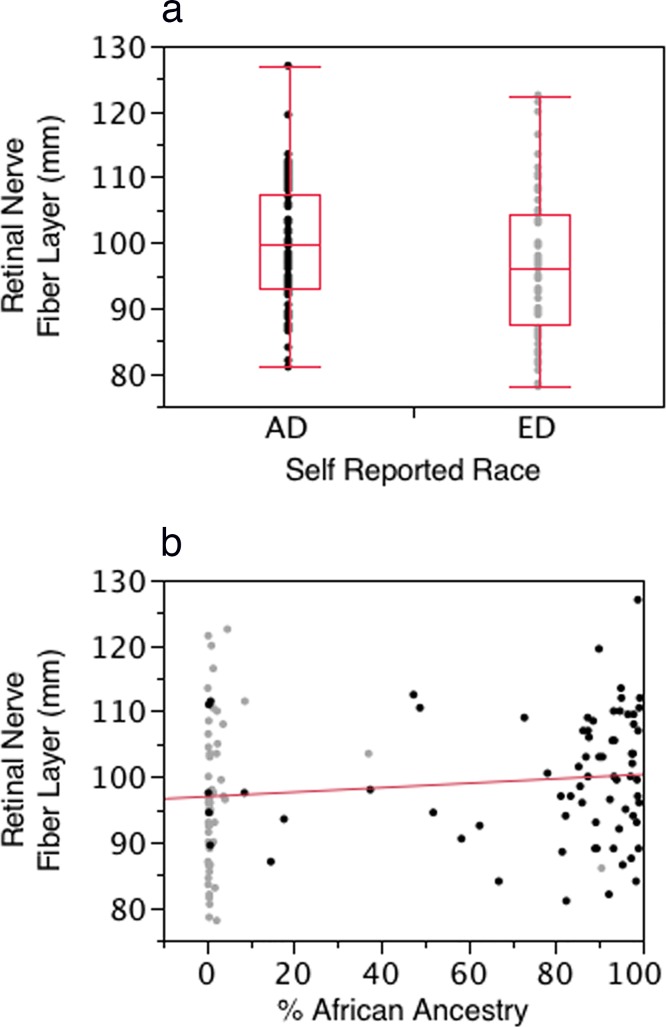
Figures 4a and 4b present scatterplots illustrating the association between self-reported race (Fig. 4a) and BGA (Fig. 4b) with RNFL thickness in the normal group. The RNFL was thicker in the self-described AD group and increased in thickness with increasing proportion of African ancestry as defined by BGA. However, in models adjusting for age and sex, self-reported race and BGA did not reach statistical significance (BGA, P = 0.092; self-report, P = 0.095). Within each self-described racial group, BGA was not significantly associated with RNFL thickness (AD, P = 0.41; ED, P = 0.56).
Figure 4.
(a) Box and whisker plot of the distribution of RNFL thickness for the AD and ED groups. (b) Scatterplot illustrating the association between the proportion of African ancestry and RNFL thickness for the entire normal group RNFL. Dark dots indicate ED group, light dots indicate AD group. In models adjusting for age and sex neither BGA (P = 0.092) nor self-report (P = 0.095) reached statistical significance.
Discussion
The current study demonstrated that objective measurements of BGA separated racial groups in the ADAGES across participants of self-described ED and AD. Moreover, these measurements correlated well with phenotypic variations of ocular features, such as CCT11 and optic nerve head area,12 that are known to exist across these racial groups. However, within each self-described racial group, there were no significant correlations. Since self-describe race also demonstrated highly significant association with these phenotypic characteristics, it is unclear what added value BGA provides over standard self-report in this setting. To our knowledge, this is the first study to correlate ocular phenotype with genetic measurement of ancestry.
Quantitative measurements of BGA have been determined using large numbers of single nucleotide polymorphisms (SNPs) to define ancestry accurately in a number of admixed populations26–36 and have been applied to the study of a variety of conditions that have exhibited racial variation, such as hypertension,26 obesity,33 lupus erythematosis,31 and diabetes.28,36 Although the genetic variation among the various human populations is only a small percentage (~10%–15%)29 of the total genetic variation between individuals, some genetic markers exhibit large allele frequency differences between ancestral groups or continents and are termed AIMs. By determining the polymorphisms within a small number of AIMs developed from geographically distinct populations, it is possible to determine the proportional genetic ancestry of an individual related to these ancestral populations.21,27,34,35,37 These markers also have demonstrated significant correlations with phenotypic variations in human populations in ocular features, such as iris38,39 and skin pigmentation,30,40 and provide useful adjuncts to forensic medicine, pharmocogenetic studies, and in the study of diseases, like POAG, which exhibit significant differences in outcome based on ancestral affllications.41 Our study population had wide variation in the percentage of African ancestry within each racial group and is in agreement with recent studies examining the distribution of African ancestry in the United States.42
Substantial prior research has demonstrated a strong heritability component to the characteristics of the optic disc, including optic disc size, cup size, and cup–disc ratio, with minimal contribution from environmental factors. Armaly43 first described this observation in 1967 and subsequent twin studies and populations-based cohort studies have confirmed a strong genetic component to the structure of the optic disc44 and RNFL thickness.45 The Beaver Dam Study examined correlations between siblings, parents, children, and cousins and showed high heritability estimates for optic disc, cup size and cup-to-disc ratio.46 Similarly, multiple prior studies have demonstrated strong heritability for CCT.47–52 Therefore, using genetically defined BGA could better reflect the biologic diversity as it relates to differences in optic nerve structure and corneal thickness compared to current imprecise measure based on self-described race.
The current study has some limitations. First, the degree of admixture seen in the ADAGES cohort may have limited the ability to detect a correlation of the ocular phenotypes with BGA within the AD groups. It is possible that other, more admixed cohorts may be better suited to test these associations. A larger sample size might also reveal more subtle associations. Further, the general use of BGA may be limited as it requires skilled laboratories and increases study expense.
In addition, ancestry estimates were derived from a relatively small SNP panel including 31 AIMs. Although other studies have used similar numbers of AIMs to determine BGA28 and our genetic results corresponded well with self-identified ancestry, larger marker panels will increase the accuracy of individual ancestry estimates. Future work will test if more accurate ancestry estimates will result in significant correlations of our phenotypes within admixed populations.
In summary, BGA correlates well to the variation in features of various ocular structures that significantly differ between racial groups and that have been associated with the development of glaucoma. While quantitative BGA can provide an objective measurement of the biologic component of self-described race it does not appear to provide any significant information within the AD and ED racial groups in regards to the three occular phenotypes investigated in the present study. The relationship between glaucomatous disease progression and BGA deserves further study to fully determine its usefulness in comparison to standard methods of self-report.
Acknowledgments
The authors thank Alcon Laboratories, Allergan, Pfizer, Merck, and Santen for supplying glaucoma medication.
Supported by NEI Grants P30EY022589, EY11008, U10EY14267, and EY019869; unrestricted grants from Research to Prevent Blindness (New York, NY, USA) to the University of Alabama and UCSD; the Eyesight Foundation of Alabama; the Edith C. Blum Foundation Fund of the New York Glaucoma Research Institute (New York, NY, USA); and Grants R01MH093500 and P30 DK079337 (CMN, AXM).
Disclosure: C.A. Girkin, None; C.M. Nievergelt, None; J.Z. Kuo, None; A.X. Maihofer, None; C. Huisingh, None; J.M. Liebmann, None; R. Ayyagari, None; R.N. Weinreb, None; R. Ritch, None; L.M. Zangwill, None
References
- 1. Seddon JM. The differential burden of blindness in the United States. N Engl J Med. 1991; 325: 1440–1442. [DOI] [PubMed] [Google Scholar]
- 2. Sommer A. Glaucoma risk factors observed in the Baltimore Eye Survey. Curr Opin Ophthalmol. 1996; 7: 93–98. [DOI] [PubMed] [Google Scholar]
- 3. Sommer A. Epidemiology, ethnicity, race, and risk. Arch Ophthalmol. 2003; 121: 1194. [DOI] [PubMed] [Google Scholar]
- 4. Tielsch JM, Katz J, Quigley HA, Javitt JC, Sommer A. Diabetes, intraocular pressure, and primary open-angle glaucoma in the Baltimore Eye Survey. Ophthalmology. 1995; 102: 48–53. [DOI] [PubMed] [Google Scholar]
- 5. Munoz B, West SK, Rubin GS, et al. Causes of blindness and visual impairment in a population of older Americans: The Salisbury Eye Evaluation Study. Arch Ophthalmol. 2000; 118: 819–825. [DOI] [PubMed] [Google Scholar]
- 6. Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. JAMA. 1991; 266: 369–374. [PubMed] [Google Scholar]
- 7. Burchard EG, Ziv E, Coyle N, et al. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003; 348: 1170–1175. [DOI] [PubMed] [Google Scholar]
- 8. Sommer A, Tielsch JM, Katz J, et al. Relationship between intraocular pressure and primary open angle glaucoma among white and black Americans. The Baltimore Eye Survey. Arch Ophthalmol. 1991; 109: 1090–1095. [DOI] [PubMed] [Google Scholar]
- 9. Gordon MO, Beiser JA, Brandt JD, et al. The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002; 120: 714–720, discussion 829–830. [DOI] [PubMed] [Google Scholar]
- 10. Investigators AGIS. The Advanced Glaucoma Intervention Study (AGIS): 9. Comparison of glaucoma outcomes in black and white patients within treatment groups. Am J Ophthalmol. 2001; 132: 311–320. [DOI] [PubMed] [Google Scholar]
- 11. La Rosa FA, Gross RL, Orengo-Nania S. Central corneal thickness of Caucasians and African Americans in glaucomatous and nonglaucomatous populations. Arch Ophthalmol. 2001; 119: 23–27. [PubMed] [Google Scholar]
- 12. Girkin CA, Sample PA, Liebmann JM, et al. African Descent and Glaucoma Evaluation Study (ADAGES): II. Ancestry differences in optic disc, retinal nerve fiber layer, and macular structure in healthy subjects. Arch Ophthalmol. 2010; 128: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sample PA, Girkin CA, Zangwill LM, et al. The African Descent and Glaucoma Evaluation Study (ADAGES): design and baseline data. Arch Ophthalmol. 2009; 127: 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Medeiros FA, Zangwill LM, Bowd C, Vessani RM, Susanna R Jr, Weinreb RN. Evaluation of retinal nerve fiber layer, optic nerve head, and macular thickness measurements for glaucoma detection using optical coherence tomography. Am J Ophthalmol. 2005; 139: 44–55. [DOI] [PubMed] [Google Scholar]
- 15. Artes PH, Chauhan BC. Longitudinal changes in the visual field and optic disc in glaucoma. Prog Retin Eye Res. 2005; 24: 333–354. [DOI] [PubMed] [Google Scholar]
- 16. Bathija R, Zangwill L, Berry CC, Sample PA, Weinreb RN. Detection of early glaucomatous structural damage with confocal scanning laser tomography. J Glaucoma. 1998; 7: 121–127. [PubMed] [Google Scholar]
- 17. Chauhan BC, Blanchard JW, Hamilton DC, LeBlanc RP. Technique for detecting serial topographic changes in the optic disc and peripapillary retina using scanning laser tomography. Invest Ophthalmol Vis Sci. 2000; 41: 775–782. [PubMed] [Google Scholar]
- 18. Mardin CY, Horn FK. Influence of optic disc size on the sensitivity of the Heidelberg Retina Tomograph. Graefes Arch Clin Exp Ophthalmol. 1998; 236: 641–645. [DOI] [PubMed] [Google Scholar]
- 19. Strouthidis NG, White ET, Owen VM, Ho TA, Hammond CJ, Garway-Heath DF. Factors affecting the test-retest variability of Heidelberg retina tomograph and Heidelberg retina tomograph II measurements. Br J Ophthalmol. 2005; 89: 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miki A, Medeiros FA, Weinreb RN, et al. Rates of retinal nerve fiber layer thinning in glaucoma suspect eyes. Ophthalmology. 2014; 121: 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nievergelt CM, Maihofer AX, Shekhtman T, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Invest Genet. 2013; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002; 298: 2381–2385. [DOI] [PubMed] [Google Scholar]
- 23. Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Res. 2009; 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Halder I, Kip KE, Mulukutla SR, et al. Biogeographic ancestry, self-identified race, and admixture-phenotype associations in the Heart SCORE Study. Am J Epidemiol. 2012; 176: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu X, Luke A, Cooper RS, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005; 37: 177–181. [DOI] [PubMed] [Google Scholar]
- 27. Shriver MD, Mei R, Parra EJ, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genom. 2005; 2: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parra EJ, Hoggart CJ, Bonilla C, et al. Relation of type 2 diabetes to individual admixture and candidate gene polymorphisms in the Hispanic American population of San Luis Valley, Colorado. J Med Genet. 2004; 41: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jorde LB, Wooding SP. Genetic variation, classification and ‘race'. Nat Genet. 2004; 36: S28–S33. [DOI] [PubMed] [Google Scholar]
- 30. Shriver MD, Parra EJ, Dios S, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003; 112: 387–399. [DOI] [PubMed] [Google Scholar]
- 31. Molokhia M, Hoggart C, Patrick AL, et al. Relation of risk of systemic lupus erythematosus to west African admixture in a Caribbean population. Hum Genet. 2003; 112: 310–318. [DOI] [PubMed] [Google Scholar]
- 32. Gower BA, Fernandez JR, Beasley TM, Shriver MD, Goran MI. Using genetic admixture to explain racial differences in insulin-related phenotypes. Diabetes. 2003; 52: 1047–1051. [DOI] [PubMed] [Google Scholar]
- 33. Fernandez JR, Shriver MD, Beasley TM, et al. Association of African genetic admixture with resting metabolic rate and obesity among women. Obesity Res. 2003; 11: 904–911. [DOI] [PubMed] [Google Scholar]
- 34. Parra EJ, Kittles RA, Argyropoulos G, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001; 114: 18–29. [DOI] [PubMed] [Google Scholar]
- 35. Parra EJ, Marcini A, Akey J, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998; 63: 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986; 70: 433–441. [DOI] [PubMed] [Google Scholar]
- 37. Shriver MD, Kennedy GC, Parra EJ, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genom. 2004; 1: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frudakis T, Thomas M, Gaskin Z, et al. Sequences associated with human iris pigmentation. Genetics. 2003; 165: 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sturm RA, Frudakis TN. Eye colour: portals into pigmentation genes and ancestry. Trends Genet. 2004; 20: 327–332. [DOI] [PubMed] [Google Scholar]
- 40. Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Hum Genet. 2004; 115: 57–68. [DOI] [PubMed] [Google Scholar]
- 41. Yang N, Li H, Criswell LA, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005; 118: 382–392. [DOI] [PubMed] [Google Scholar]
- 42. Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The Genetic Ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015; 96: 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Armaly MF. Genetic determination of cup/disc ratio of the optic nerve. Arch Ophthalmol. 1967; 78: 35–43. [DOI] [PubMed] [Google Scholar]
- 44. Teikari JM, Airaksinen JP. Twin study on cup/disc ratio of the optic nerve head. Br J Ophthalmol. 1992; 76: 218–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hougaard JL, Kessel L, Sander B, Kyvik KO, Sorensen TI, Larsen M. Evaluation of heredity as a determinant of retinal nerve fiber layer thickness as measured by optical coherence tomography. Invest Ophthalmol Vis Sci. 2003; 44: 3011–3016. [DOI] [PubMed] [Google Scholar]
- 46. Klein BE, Klein R, Lee KE. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004; 45: 59–62. [DOI] [PubMed] [Google Scholar]
- 47. Freeman EE, Roy-Gagnon MH, Descovich D, Masse H, Lesk MR. The heritability of glaucoma-related traits corneal hysteresis, central corneal thickness, intraocular pressure, and choroidal blood flow pulsatility. PLoS One. 2013; 8: e55573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dimasi DP, Burdon KP, Craig JE. The genetics of central corneal thickness. Br J Ophthalmol. 2010; 94: 971–976. [DOI] [PubMed] [Google Scholar]
- 49. Landers JA, Hewitt AW, Dimasi DP, et al. Heritability of central corneal thickness in nuclear families. Invest Ophthalmol Vis Sci. 2009; 50: 4087–4090. [DOI] [PubMed] [Google Scholar]
- 50. Zheng Y, Ge J, Huang G, et al. Heritability of central corneal thickness in Chinese: the Guangzhou Twin Eye Study. Invest Ophthalmol Vis Sci. 2008; 49: 4303–4307. [DOI] [PubMed] [Google Scholar]
- 51. Toh T, Liew SH, MacKinnon JR, et al. Central corneal thickness is highly heritable: the twin eye studies. Invest Ophthalmol Vis Sci. 2005; 46: 3718–3722. [DOI] [PubMed] [Google Scholar]
- 52. Alsbirk PH. Corneal thickness. II. Environmental and genetic factors. Acta Ophthalmol. 1978; 56: 105–113. [DOI] [PubMed] [Google Scholar]