Abstract
Measurements of the abundance of common metabolites in cultured embryonic stem (ES) cells revealed an unusual state with respect to one-carbon metabolism. These findings led to the discovery of copious expression of the gene encoding threonine dehydrogenase (TDH) in ES cells. TDH-mediated catabolism of threonine takes place in mitochondria to generate glycine and acetyl–coenzyme A (CoA), with glycine facilitating one-carbon metabolism via the glycine cleavage system and acetyl-CoA feeding the tricarboxylic acid cycle. Culture media individually deprived of each of the 20 amino acids were applied to ES cells, leading to the discovery that ES cells are critically dependent on one amino acid—threonine. These observations show that ES cells exist in a high-flux backbone metabolic state comparable to that of rapidly growing bacterial cells.
Embryonic stem (ES) cells are small, rapidly dividing cells with the pluripotent capacity to form any and all cells present in the adult body. Cultured mouse ES cells measure only several micrometers in diameter, as compared to the 10- to 30-μm range typical of cultured somatic cells (1). The cell division cycle of ES cells under the standard culture conditions used in our studies lasts only 5 hours. This rate of cell division matches that measured for ICM (inner cell mass) cells of the early mouse embryo (2). This doubling time is shorter than that of even the most rapidly dividing cancer cells and approaches that of single-celled microbial organisms. When deprived of leukemia inhibiting factor (LIF), mouse ES cells slow their growth rate, increase in size, form embyroid bodies, and enter pathways leading to differentiation (3).
Mouse ES cells exist in a high-flux metabolic state
To investigate whether ES cells might exist in a metabolic state that facilitates rapid growth, we surveyed the abundance of numerous common metabolites. Mouse ES cells (line E14) grown without feeder cells were exposed to organic solvents that allow soluble extracts to be subjected to liquid chromatography–mass spectrometry (LC-MS/MS), enabling multiple reaction monitoring of scores of metabolites (4). Parameters necessary for the detection of the two most abundant daughter ions for each metabolite upon collision-induced fragmentation were optimized so that metabolite identity and abundance could be simultaneously assessed.
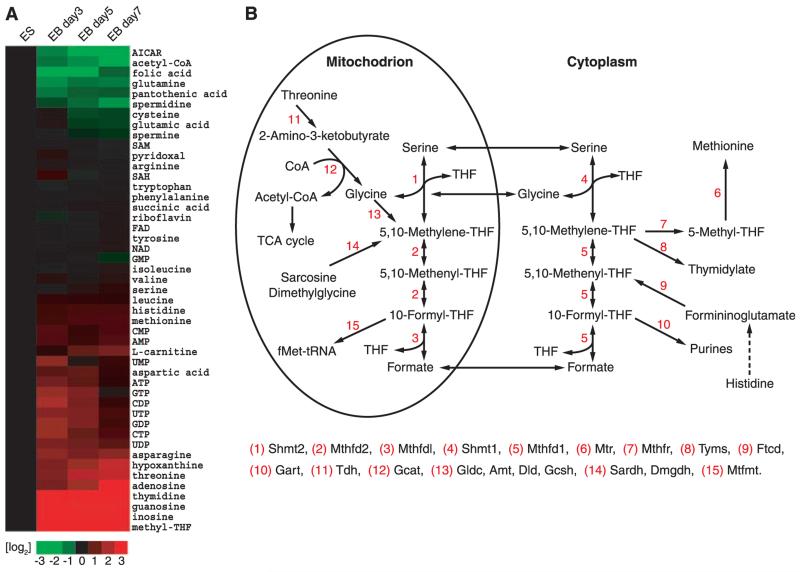
The relative amounts of individual metabolites prepared from ES cells were compared with those of embryoid body cells deprived of LIF for 3, 5, or 7 days (Fig. 1A and fig. S1). Three patterns of fluctuation in metabolite abundance were observed. One class consisted of metabolites that changed little as a function of the transition of ES cells into the differentiating cells present in embryoid bodies. Included in this class were many essential and nonessential amino acids. The second class consisted of metabolites present at a higher abundance in ES cells relative to differentiating cells. 5-Aminoimidazolecarboxamide-R (AICAR), an intermediate in purine biosynthesis, was the most abundant member of this class, followed by acetyl–coenzyme A (CoA) and folic acid. The third class included metabolites that increased in abundance as a function of differentiation. Methyl-tetrahydrofolate (mTHF) was the most prominent member of this class, being nearly undetectable in ES cells, yet normalizing considerably in differentiating cells. A similar pattern of differentiation-associated increase in abundance was observed for guanosine, adenosine, inosine, and threonine.
Fig. 1.
Coordinated changes in metabolite abundance during ES cell differentiation. Metabolites extracted from cells in 50% aqueous methanol were subjected to LC-MS/MS analysis (4). (A) Heat map showing the relative levels of metabolites in ES cells as a function of 3, 5, and 7 days after withdrawal of LIF and differentiation into embryoid bodies. Metabolite levels are expressed as log2 transformation of the fold change at days 3, 5, and 7 relative to the same measurement in undifferentiated ES cells; green indicates metabolites that decrease in abundance as a function of differentiation and red, metabolites that increase as ES cells differentiate. (B) Schematic diagram of the pathways facilitating folate-mediated, one-carbon metabolism.
Knowing that ES cells must replicate the genome at a prolific rate, it was not surprising to observe that six of the metabolites changing most markedly in abundance as a function of ES cell differentiation are involved in the purine biosynthetic pathway (folic acid, mTHF, AICAR, guanosine, adenosine, and inosine). The de novo synthesis of purine nucleotides requires that each purine base receive carbon atoms at two different steps along the pathway. The source of one-carbon metabolism for purine synthesis is formyl-tetrahydrofolate (5). This and the other two “charged” forms of tetrahydrofolate, methylene-tetrahydrofolate and methyl-tetrahydrofolate, are the sole sources of one-carbon metabolism (6, 7). Notably, the AICAR intermediate in purine synthesis is located immediately downstream of one of the two single-carbon donating steps in the pathway. Because AICAR was present at increased abundance in rapidly growing ES cells, and because folic acid and mTHF levels fluctuated substantially as a function of conversion of ES cells into embryoid bodies, we hypothesized that the demand for metabolic flux via one-carbon metabolism might be unusually high in undifferentiated ES cells.
The threonine dehydrogenase gene and enzyme are expressed copiously in mouse ES cells
Quantitative polymerase chain reaction (qPCR) assays were performed on samples prepared from ES cells and seven tissues of the adult mouse to measure the levels of mRNAs specifying the 15 enzymes most proximal to the pathway of one-carbon metabolism (Fig. 1B). Such measurements revealed unusually accentuated, ES cell–specific expression of the gene encoding threonine dehydrogenase (TDH). The level of mRNA specifying synthesis of the TDH enzyme in ES cells was roughly 1000 times as high as that in any of the seven tissues that were assayed (fig. S2). In only one other case did the ES mRNA level of a folate-associated metabolic enzyme exceed that of other tissues (the level of thymidylate synthase mRNA in ES cells was roughly twice as high as the levels observed in lung, intestine, and testis).
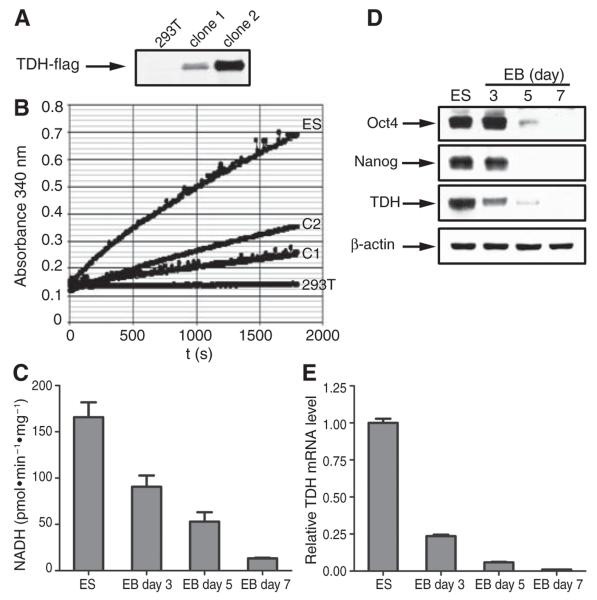
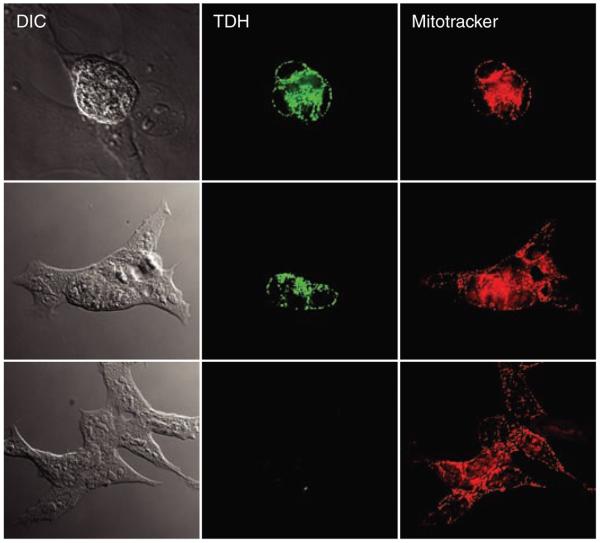
The TDH enzyme performs the initial, rate-limiting step in an atypical form of threonine catabolism, wherein threonine is converted to glycine and acetyl-CoA. In eukaryotic cells, the TDH enzyme is located within mitochondria. There it supplies acetyl-CoA for direct entry into the tricarboxylic (TCA) cycle, and likewise passes glycine to tetrahydrofolate charging via the glycine cleavage system (8). If the TDH enzyme is selectively active in undifferentiated ES cells, this might explain why threonine abundance was reduced in ES cells compared with differentiating cells associated with embryoid bodies (Fig. 1A). High TDH enzyme activity might also explain why ES cells contained increased amounts of acetyl-CoA relative to embryoid body cells. To test this hypothesis, mitochondria were isolated from ES cells, assayed for the threonine-dependent conversion of NAD (nicotinamide adenine dinucleotide) to NADH, and found to contain much higher TDH activity than human 293T cells tailored to express an exogenous form of the TDH gene (Fig. 2, A and B). Similarly, TDH enzyme activity, TDH mRNA levels, and immunoreactivity to TDH-specific antibodies as detected by Western blotting all declined sharply as a function of differentiation of ES cells into embryoid bodies (Fig. 2, C to E). The availability of TDH-specific antibodies (see Materials and Methods) allowed immunohistochemical staining of ES cell colonies as a function of differentiation. Before LIF withdrawal–induced differentiation, all cells in each ES colony stained positive for mitochondria-associated TDH (Fig. 3). One day after LIF withdrawal, TDH immunoreactivity was lost in differentiating cells located around the periphery of each colony, yet maintained in the small, centrally located cells. After prolonged LIF-withdrawal, all TDH-specific immunoreactivity was lost. Together, these observations suggest that threonine catabolism via the TDH enzyme might be important to the growth and metabolic state of mouse ES cells.
Fig. 2.
Measurements of TDH enzyme activity, mRNA abundance, and protein abundance as a function of ES cell differentiation. (A) Western blotting assays of 293T cells stably transformed with an expression vector encoding a Flag-tagged version of TDH. Western blotting revealed no TDH signal in parental 293T cells, an intermediate signal in transformed clone 1, and a higher signal in transformed clone 2. (B) TDH enzyme activity assays for ES cells (ES), untransformed 293T cells (293T), transformed 293T clone 1 cells (C1), and transformed 293T clone 2 cells (C2). Mitochondria were isolated from ES cells and the three 293T clones by differential centrifugation (see Materials and Methods). Equal amounts (10 μg) of mitochondrial protein were subjected to an assay reaction containing 100 mM tris–HCl (pH 8.0), 50 mM NaCl, 25 mM threonine, and 5 mM NAD+ at 25°C. Absorbance at 340 nm was recorded over time on a microplate reader to monitor conversion of NAD+ to NADH. (C) TDH enzyme activity present in mitochondrial extracts from undifferentiated ES cells and day 3,5, and 7 embryoid body cells as determined under the same reaction conditions as in (B). Experiments were performed in triplicate; error bars indicate ±SD. (D) Western blotting signals for Oct4, Nanog, TDH, and actin in protein samples prepared from undifferentiated ES cells and day 3, 5, and 7 embryoid body cells. (E) shows TDH mRNA levels as monitored by qPCR in ES cells and day 3, 5, and 7 embryoid body cells. Experiments were performed in triplicate; error bars indicate ±SD.
Fig. 3.
Immunohistochemical staining of TDH in mouse ES cells as a function of differentiation. Mouse ES cells were grown in chamber slides and subjected to LIF withdrawal–mediated differentiation. Shown are images of an undifferentiated ES colony (top), a partially differentiated colony (middle), and an extensively differentiated colony (bottom). Cells were fixed and stained with antibodies specific to the mouse TDH enzyme. TDH immunoreactivity was visualized with Alexa488-labeled goat antibody to rabbit secondary antibodies (green). Before fixation, cells were incubated with Mitotracker dye, allowing visual localization of mitochondria (red).
Elegant studies of metabolic flux in rapidly growing bacterial cells have defined reaction pathways that flow at rates more than three to four orders of magnitude higher than those of other pathways of intermediary metabolism (9). The core circuitry of what has been termed the high-flux backbone (HFB) of metabolism includes the TDH enzyme. Whereas most amino acids are processed into biomass via translation during HFB metabolism, TDH converts most threonine into glycine and acetyl-CoA. Because most microbial organisms, including yeast (10), can synthesize threonine de novo, the HFB pathway allows synthesis of sufficient amounts of threonine to satisfy the demands of both threonine catabolism to glycine and acetyl-CoA, as well its requirement for protein synthesis.
Mouse ES cells critically depend on threonine
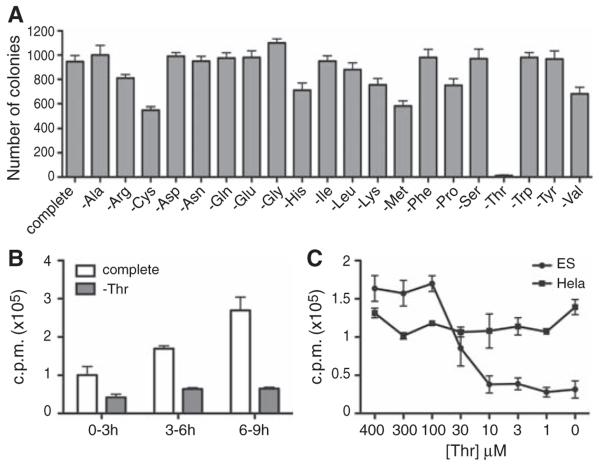
Mammalian cells, unlike bacteria or yeast, require threonine as an essential amino acid. To test whether ES cells might deploy the TDH enzyme to adopt a metabolic state comparable to the HFB of bacterial cells, we prepared culture media that were individually deprived of each of the 20 amino acids. ES cells of the E14 line were plated and exposed to each of the “drop-out” culture media. After 36 hours of exposure, normal ES colonies were observed to grow in all culture media except that deprived of threonine (Fig. 4A and fig. S3A). Colonies grown from either control media, or media individually lacking all amino acids except threonine, stained positively with the alkaline phosphatase marker prototypical of undifferentiated ES cells. Light microscopic inspection of ES cell colonies showed little or no difference in colony morphology, cell size, or cell number as a function of media type. All of the media samples tested contained 10% fetal bovine serum. The serum component provides cells with residual amounts of amino acids, as well as with serum proteins that can be hydrolyzed by cells to salvage amino acids for protein synthesis and other metabolic needs. These residual amounts of amino acids are sufficient for the growth of ES cells in all cases except threonine deprivation. Almost indistinguishable observations of acute threonine dependence were obtained on the CCE line of mouse ES cells, as well as the AOK-5P line of mouse embryonic fibroblast (MEF)–derived induced pluripotent stem (iPS) cells (fig. S3, B and C). Moreover, as with the E14 line of ES cells, the CCE and AOK-5P lines were observed by immunofluorescence staining to express copious amounts of mitochondrial TDH enzyme (fig. S4).
Fig. 4.
Inhibition of cell growth and DNA synthesis in ES cells deprived of threonine. (A) Growth dependence of ES cells on threonine supplementation to culture media. After plating at single-cell density and growth on gelatinized dishes for 6 hours, E14Tg2A mouse ES cells were exposed for 36 hours to complete culture medium or to medium missing a single amino acid. The numbers of alkaline phosphatase–positive colonies were counted and plotted. The experiments were performed in triplicate; error bars indicate±SD. (B)Time-dependent effects of threonine deprivation on DNA synthesis in cultured ES cells. After growth of undifferentiated ES cells in complete culture medium for 1 day, 2 × 105 cells were exposed to complete (white bar) or threonine-deprived (gray bar) medium for 3, 6, or 9 hours. Cells were metabolically labeled with 2 μCi of [3H]thymidine at 3-hour intervals as indicated (see Materials and Methods). Progressively increased amounts of label incorporation at 6 and 9 hours were observed for cells grown in complete culture medium, reflective of increased cell number owing to a 5-hour doubling time. Experiments were performed in triplicate; error bars indicate ±SD. (C) Dose-dependent effects of threonine deprivation on DNA synthesis in cultured ES cells. ES cells (2 × 105) (closed circle) or HeLa cells (5 × 105) (closed square) were exposed for 3 hours to culture medim containing the indicated concentrations of threonine, and then metabolically labeled for 3 hours with [3H]thymidine. DNA was purified from each sample and the amount of radioactivity incorporated was measured by scintillation counting. Experiments were performed in triplicate; error bars indicate ±SD.
When the same samples of drop-out media were tested for colony formation assays using human cancer (HeLa) cells, only leucine-deficient culture medium impeded cell growth (fig. S5A). Threonine-deficient medium supported HeLa colony formation as well as fully supplemented culture medium. Similar studies on MEF cells and mouse NIH 3T3 cells also failed to reveal differential sensitivity to threonine deprivation. MEF and 3T3 cell growth was instead exceptionally sensitive to cysteine deprivation (fig. S5, B and C). MEF cells grew indistinguishably in the presence of the 19 other drop-out media, including that lacking threonine. 3T3 cells grew at a slower rate in culture medium deprived of either arginine or leucine. Mildly reduced growth was also observed for culture medium lacking methionine, glutamine, lysine, threonine, tryptophan, phenylalanine, or valine. Finally, media samples deprived of serine, alanine, glycine, asparagine, histidine, tyrosine, aspartic acid, isoleucine, or proline supported 3T3 cell growth to a level equivalent to that of fully supplemented culture medium. The unusual sensitivity to cysteine deprivation may indicate that some aspect of sulfur metabolism, perhaps relating to the production of glutathione, is critical to the growth or survival of these partially differentiated, embryo-derived cell types. It can be concluded, however, that the growth properties of HeLa, MEF, and 3T3 cells are not uniquely sensitive to threonine deprivation, as was observed for mouse ES cells.
If TDH-mediated threonine breakdown in ES cells indeed supplies glycine to fuel one-carbon metabolism for enhanced purine biosynthesis, one might anticipate that deprivation of threonine would impede DNA synthesis. To test this hypothesis, we cultured ES cells in the presence of varying levels of threonine and exposed the cells to brief labeling with [3H]thymidine. The incorporation of [3H]thymidine into DNA was substantially reduced as a function of threonine concentration in the culture medium (Fig. 4C). Reduction of threonine supplementation to 30 μM partially inhibited [3H]thymidine incorporation into DNA. When threonine concentrations were reduced to 10 μM, DNA synthesis was impeded to a level similar to that observed by complete elimination of threonine from culture medium (except for residual amounts present in serum). By contrast, when HeLa cells were grown in culture medium supplemented with varying concentrations of threonine, DNA synthesis was unimpeded even under conditions of complete threonine deprivation (Fig. 4C).
Assuming that the TDH enzyme might allow ES cells to convert threonine into glycine as an alternative and theoretically enhanced mode of one-carbon metabolism, we supplemented culture medium deprived of threonine by increased concentrations of glycine. Instead of the normal, 400 μM concentration of glycine, threonine-deficient medium was supplemented with 4 mM glycine. ES cells subjected to this threonine-deficient, glycine-enriched medium grew no better than those subjected to the threonine drop-out medium itself. It is possible that ES cells cannot deliver the supplemented glycine to mitochondria to feed the glycine cleavage system and folate charging. Alternatively, TDH-mediated production of acetyl-CoA could be equally important to the HFB-like metabolic state. This alternative interpretation is consistent with the observation that among all metabolites that decline in abundance most precipitously as a function of the conversion of ES cells into differentiating embryoid bodies, acetyl-CoA was second only to the purine intermediate AICAR (Fig. 1). To investigate these alternatives, we used 3-hydroxynorvaline (3-HNV), a synthetic variant of threonine containing an extra carbon atom. The TDH enzyme can hydrolyze this threonine analog, yet instead of producing glycine and acetyl-CoA, catabolism of 3-HNVyields glycine and propionyl-CoA (11). Compared with the 3.56 mM Km (Michaelis constant) for threonine, the TDH enzyme catabolizes 3-HNV with a Km of 11.48 mM. ES cells were cultured in either normal or threonine-deficient medium supplemented with varying amounts of 3-HNV. Not only did the analog fail to complement the absence of threonine, but it instead inhibited ES cell colony formation in the presence of normal culture medium (fig. S6A). The inhibitory effect of 3-HNV was not observed on the growth of HeLa, MEF, or 3T3 cells (fig. S6B). These data are consistent with the interpretation that mouse ES cells critically rely on TDH-mediated production of both glycine and acetyl-CoA. Because 3-HNV is a known TDH enzyme substrate, we hypothesize that it competes with the TDH enzyme in mitochondria of ES cells, yielding only one of the two metabolites required of HFB metabolism.
Growth of mouse embryos is critically dependent on threonine
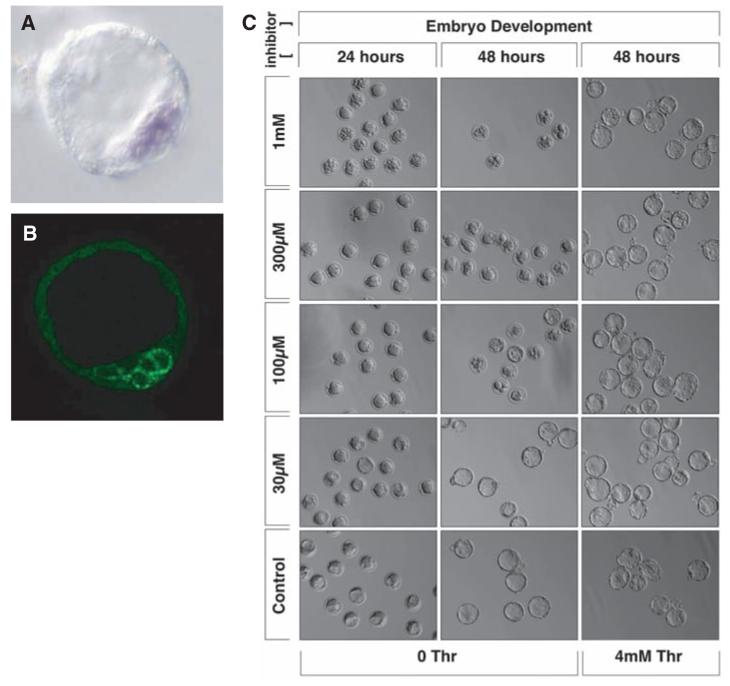
The availability of 3-HNV as a cell-permeable threonine analog facilitated experiments using living mouse embryos. Before testing the effects of 3-HNV on early mouse embryo development, we performed in situ hybridization and immunohistochemical staining assays to investigate the expression pattern of both TDH mRNA and TDH enzyme. Both assays revealed staining patterns within the inner cell mass (ICM) of blastocyst embryos (Fig. 5, A and B). Knowing that the ICM represents the in vivo source of ES cells (12), we hypothesized that the early development of mouse embryos might be inhibited by 3-HNV. When administered at doses of 1 mM and 300 μM, 3-HNV blocked the conversion of morula-staged embryos into cavitated blastocysts. Less inhibition was observed when embryos were incubated with 100 μM concentrations of the threonine analog, and no inhibition was observed at 30 μM. Complete rescue of normal development was effected, even at 1 mM 3-HNV, when embryos were incubated in culture medium supplemented by 4 mM threonine (Fig. 5C). These studies provide evidence that ICM cells of the early mouse embryo, like cultured mouse ES cells, are critically dependent on threonine catabolism.
Fig. 5.
3-Hydroxynorvaline arrests the early development of mouse embryos. Blastocyst-stage mouse embryos were assayed by in situ hybridization to localize TDH mRNA (A) and immunohistochemical staining with antibodies specific to the TDH enzyme (B). In situ and antibody staining signals localized the TDH mRNA (purple) and enzyme (green) to cells associated with the ICM. Precompacted morula-stage embryos were incubated in varying concentrations of the threonine analog 3-HNV. Embryos exposed to 1 mM and 300 μM 3-HNV were blocked from forming cavitated blastocysts (C). Incubation with 4 mM threonine resulted in complete rescue of embryonic development even at the highest concentration (1 mM) of 3-HNV.
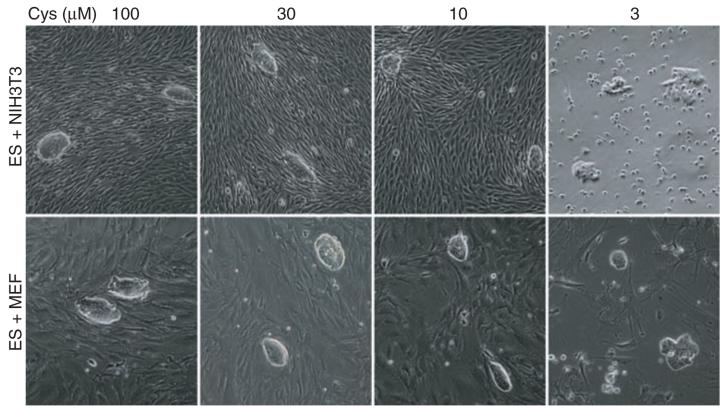
Neither MEF nor 3T3 cell growth was selectively sensitive to threonine deprivation. Threonine-low (Tl) culture medium could thus be anticipated to selectively favor the growth of embryonic fibroblasts over ES cells. Unlike the threonine dependence of ES cells, it was instead observed that both MEF and 3T3 cells failed to grow in cysteine-deprived medium (fig. S5, B and C). Thus, threonine-high:cysteine-low (ThCl) culture media could rationally be predicted to selectively support ES cell growth relative to that of differentiating or differentiated cells of the early mouse embryo. As an initial test of this hypothesis, small numbers of ES cells were mixed and cultured with a vast excess of either MEF or 3T3 cells and exposed to culture media supplemented with varying amounts of either threonine or cysteine. As expected, threonine deprivation severely limited the retrieval of ES clones from the mixed cultures under conditions in which both MEF and 3T3 cell growth was unimpeded (fig. S7). By contrast, deprivation of cysteine in the presence of 4 mM threonine favored the growth of ES cells relative to either MEF or 3T3 cells (Fig. 6).
Fig. 6.
Effects of cysteine deprivation on the growth of ES, MEF, and 3T3 cells. Cocultures of ES/MEF or ES/3T3 cells were subjected for 2 days to media containing varying amounts of supplemented cysteine. Cysteine deprivation severely impeded MEF cell growth at 10 and 3 μM and 3T3 growth at 3 μM (see also fig. S5). Although colony morphology was altered under the most severe conditions of cysteine deprivation, ES cell colonies were observed under all culture conditions tested.
Mouse ES cells grow well in culture medium supplemented with standard amounts of essential and nonessential amino acids, remaining continuously in a state of rapid, symmetric, self-renewing division in which all cells are maintained in the ES state. ES cells derived from other species, even rodents as closely related as rats, have a strong tendency toward differentiation. So profound is the difference between mouse ES cells and ES cells derived from other organisms that there has not been a single reported incidence of success in the use of ES cells from any other species for homologous recombination and subsequent generation of an animal bearing targeted changes in the genome. It is hoped that forced expression of the TDH gene, proper choice of ThCl culture medium, or both might favor the establishment of improved ES cell lines from species other than mice.
Humans are deficient in the threonine dehydrogenase enzyme
The human gene encoding TDH appears to be nonfunctional by virtue of three inactivating mutations (13). Although highly conserved in gene organization, as well as primary amino acid sequence of the predicted TDH open reading frame, the human TDH gene carries AG-to-GG splice acceptor mutations in exons 4 and 6, as well as a nonsense mutation within exon 6 wherein arginine codon 214 is replaced by a translational stop codon. Whereas polymorphic variation within the human population has been observed for the exon 4 splice acceptor mutation, with some individuals carrying the normal AG splice acceptor dinucleotide and others carrying the GG variant, all individuals genotyped to date carry both the splice acceptor and nonsense mutations in exon 6. Reverse transcriptase–PCR analysis of TDH transcripts expressed in human fetal liver tissue showed complete skipping of exon 4 and either complete skipping or aberrant splicing of exon 6 (fig. S8). Given that exons 4 and 6 encode segments of the enzyme critical to its function and that truncation via the nonsense codon at amino acid 214 would also be predicted to yield an inactive variant, it appears that the human gene is incapable of producing an active TDH enzyme. Remarkably, all metazoans whose genomes have been sequenced to date, including chimpanzees, appear to contain an intact TDH gene (14). Unless humans evolved adaptive capabilities sufficient to overcome three mutational lesions, it would appear they are TDH deficient.
Human ES cells grow at a far slower rate than mouse ES cells, with a doubling time of 35 hours (15). Whether the slower growth rate of human ES cells reflects the absence of a functional TDH enzyme can perhaps be tested by introducing, into human ES cells, either a repaired human TDH gene or the intact TDH gene of a closely related mammal. That this strategy might work is supported by the expression in human cells of a functional form of the 2-amino-3-ketobutyrate-CoA ligase enzyme that converts the short-lived product of TDH-mediated catabolism of threonine into acetyl-CoA and glycine (Fig. 1B). It is possible that the culture conditions used to grow human ES cells do not match the ICM environment of the human embryo, in which the cell division cycle is more rapid than the 35-hour doubling time of cultured human ES cells (16). If human ES cells do not use the TDH enzyme to acquire an advantageous metabolic state for rapid growth, and if conditions can be adapted to facilitate the rapid proliferation of human ES cells in culture, the tools and approaches that we describe here may prove useful. As often happens in science, findings made in one experimental system can open avenues of investigation useful for other matters of inquiry. Finally, it is important to consider whether humans benefit from some form of selective advantage as a consequence of mutational inactivation of the TDH gene.
Supplementary Material
Acknowledgments
We thank B. Tu for the help with LC-MS/MS analysis; J. De Brabander and J. Ready for guidance on the chemical properties of threonine analogs; E. Olson for providing the AOK-5P line of iPS cells; D. Chong for technical assistance with in situ hybridization assays; and J. Goldstein, M. Rosen, and W. Neaves for helpful scientific input. This work was funded by an NIH Directors Pioneer Award, and unrestricted funds provided to S.L.M. by an anonymous donor.
Footnotes
References and Notes
- 1.Alberts B, et al. Molecular Biology of the Cell. ed. 5 Garland, New York: 2007. p. 1. [Google Scholar]
- 2.Snow MHL. J. Embryol. Exp. Morphol. 1977;42:293. [Google Scholar]
- 3.Smith AG. Annu. Rev. Cell Dev. Biol. 2001;17:435. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Tu BP, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16886. [Google Scholar]
- 5.Warren L, Buchanan JM. J. Biol. Chem. 1957;229:613. [PubMed] [Google Scholar]
- 6.Weissbach H, Peterkofsky A, Redfield BG, Dickerman H. J. Biol. Chem. 1963;238:3318. [PubMed] [Google Scholar]
- 7.Phear EA, Greenberg DM. J. Am. Chem. Soc. 1957;79:3737. [Google Scholar]
- 8.Dale RA. Biochim. Biophys. Acta. 1978;544:496. doi: 10.1016/0304-4165(78)90324-0. [DOI] [PubMed] [Google Scholar]
- 9.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Nature. 2004;427:839. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 10.Hartman JL. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11,700. [Google Scholar]
- 11.Kazuoka T, et al. J. Bacteriol. 2003;185:4483. doi: 10.1128/JB.185.15.4483-4489.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brook FA, Gardner RL. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5709. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar AJ. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2007;35:D61. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amit M, et al. Dev. Biol. 2000;227:271. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 16.Hardy K, Handyside AH, Winston RMT. Development. 1989;107:597. doi: 10.1242/dev.107.3.597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.