Abstract
Lysophosphatidic acid (LPA) is a naturally occurring phospholipid that initiates a broad array of biological processes, including those involved in cell proliferation, survival and migration via activation of specific G protein-coupled receptors located on the cell surface. To date, at least five receptor subtypes (LPA1-5) have been identified. The LPA1-3 receptors are members of the endothelial cell differentiation gene (Edg) family. LPA4, a member of the purinergic receptor family, and the recently identified LPA5 are structurally distant from the canonical Edg LPA1-3 receptors. LPA4 and LPA5 are linked to Gq, G12/13 and Gs but not Gi, while LPA1-3 all couple to Gi in addition to Gq and G12/13. There is also evidence that LPA4 and LPA5 are functionally different from the Edg LPA receptors. Computational modeling has provided useful information on the structure-activity relationship (SAR) of the Edg LPA receptors. In this work, we focus on the initial analysis of the structural and ligand-binding properties of LPA4, a prototype non-Edg LPA receptor. Three homology models of the LPA4 receptor were developed based on the x-ray crystal structures of the ground state and photoactivated bovine rhodopsin and the recently determined human β2-adrenergic receptor. Docking studies of LPA in the homology models were then conducted, and plausible LPA binding loci were explored. Based on these analyses, LPA is predicted to bind to LPA4 in an orientation similar to that reported for LPA1-3, but through a different network of hydrogen bonds. In LPA1-3, the ligand polar head group is reported to interact with residues at positions 3.28, 3.29 and 7.36, whereas three non-conserved amino acid residues, S114(3.28), T187(EL2) and Y265(6.51), are predicted to interact with the polar head group in the LPA4 receptor models.
Keywords: Lysophosphatidic acid (LPA), G Protein-coupled receptors (GPCRs), LPA4/p2y9/GPR23, Homology modeling, Automated docking
1. Introduction
Lysophosphatidic acid (LPA, 1, Figure 1) is an important signaling molecule and a member of the phospholipid growth factor family, exerting its effects via Class A rhodopsin-like G protein-coupled receptors (GPCRs). Through activation of its receptors, LPA elicits a multitude of biological actions, including cell proliferation, cell survival, cell migration/invasion and regulation of immune responses [1]. LPA is thus involved in diverse physiological effects including neurogenesis, myelination, angiogenesis, spermatogenesis, embryo implantation, platelet aggregation, brain development, smooth muscle contraction, and apoptosis, and a number of biological processes including reproduction, hair growth, neuropathic pain, cardiovascular diseases and cancer have also been linked to the physiological action of LPA and its associated receptors [2]. So far, at least five GPCRs have been cloned and characterized as high affinity LPA receptors. Three of them, LPA1, LPA2 and LPA3, belong to the endothelial cell differentiation gene (Edg) receptor family [3]. More recently, two orphan GPCRs were found to be additional LPA receptors (LPA4/p2y9/GPR23 and LPA5/GPR92) [4,5]. However, LPA4 and LPA5 are more closely related to the purinergic P2Y receptor family than to the Edg family of LPA receptors. In addition to LPA4 and LPA5, other orphan receptors in the purinergic receptor family such as GPR87 and P2Y5 may also be high-affinity receptors for LPA [6,7]. However, independent studies are necessary to confirm their identity as bona fide LPA receptors.
Figure 1.

The structure of 1-oleoyl-lysophosphatidic acid (LPA(1:18)), commonly known as LPA. The ‘stereospecific numbering’ (sn) identifiers are shown for the glycerol carbon atoms.
The LPA4 and LPA5 receptors share less than 20% amino acid sequence homology with the LPA1-3 receptors. There is also a difference in G protein coupling selectivity between the Edg LPA receptors and the novel LPA4 or LPA5 receptors. LPA4 and LPA5 are primarily linked to Gq, G12/13 and Gs but not Gi while LPA1-3 couple to Gi as well as Gq and G12/13. The expression patterns of LPA4 and LPA5 are not coincidental with any of the LPA1-3 receptors. Consistent with these differences in structures, G protein coupling, and tissue distribution, these novel LPA receptors may have different functionalities. For example, LPA4 inhibits LPA-dependent cell migration and invasion in contrast to the motility-stimulating LPA1-3 receptors [8,9]. Therefore, it is important to explore structural and signaling characteristics of the novel LPA receptors.
The LPA receptors are members of a larger rhodopsin-like GPCR subfamily whose members can efficiently bind various functionalized fatty acid derivatives. As noted above, these molecules often act as potent signaling molecules in addition to serving as integral components in lipid membranes [10]. Phylogenetic analysis of related sequences (Figure 2) shows that the LPA receptors fall into one of two main classes of phospholipidergic receptor. The first group includes the Edg receptors (LPA1-3 and sphingosine-1-phosphate S1P1-5). The second group, which includes LPA4-5, consists of nucleotide-binding P2Y and cysteinyl-leukotriene (CysLT) receptors. The second group can be further subdivided into those that signal primarily through Gq (P2Y1,2,4,6,11) and through Gi (P2Y12-14) [11,12]. The ligand selectivity among the receptors depends largely on the degree of similarities in the region(s) comprising the ligand binding site. As will be discussed later, there are both regions of similarities and differences among these closely related receptors.
Figure 2.
Tree diagram showing the phylogenetic interrelationships among members of the lysophosphatidic acid (LPA), sphingosine-1-phosphate (S1P), cannabinoid (CNR), purinergic nucleotide (P2Y) and Cys-leukotriene (CysLT) receptor families. Also included are the sequences of bovine rhodopsin (OPSD_BOVIN), the β2-adrenoceptor (ADRB2) and two recently de-orphanized receptors (CPR87 and GPR91). The diagram was generated using the Geneious software package (see Experimental Methods), and the corresponding aligned sequences are presented as Supplemental Figure S2. The scale bar corresponds to a maximum likelihood branch of 0.1 inferred substitutions per site.
In order to visualize which amino acids likely participate in ligand binding, homology models of the LPA4 receptor would be helpful in inferring the possible interaction between LPA and its receptor and in designing LPA receptor subtype selective ligands. Since an experimentally determined structure of an LPA4 receptor is not yet available, we may rely on comparative modeling techniques combined with other experimental results to arrive at a plausible solution structure. It is believed that with all the lessons learned from previous experience, GPCR homology modeling based on the bovine rhodopsin and/or beta-adrenergic X-ray crystal structures will help in structure-based drug design and virtual screening for therapeutic applications and in fact such molecular modeling studies have been successfully applied to other GPCRs [13]. Furthermore, molecular modeling combined with site-directed mutagenesis study has aided significantly in our understanding of the binding mode of GPCR ligands to their receptors [14]. One of the challenges in homology modeling of GPCRs is the less predictable conformation of the extracellular loop regions [15]. In this work, we have identified loci on EL2 and the nearby regions on TM3 and TM6 potentially responsible for ligand binding.
There are now several crystal structures of GPCRs that may be used as templates for homology modeling, and the appropriateness of a given template for a particular GPCR model target has begun to be addressed in the literature [16]. The highest-resolution (2.2 Å) crystal structure of bovine rhodopsin in its dark state was reported in 2004 [17], and a lower-resolution (4.2 Å) photoactivated structure of rhodopsin was reported in 2006 [18]. In this context, we propose the first homology model of the LPA4 receptor based on the photoactivated rhodopsin crystal structure to simulate an active state of the LPA4 receptor. In order to compare with the photoactivated model, we further develop a second model of the LPA4 receptor in an inactive state using the high-resolution (2.2 Å) dark state rhodopsin as a homology template [17]. Two β2-adrenergic receptor (β2AR) crystal structures were reported in 2007 [19–21]. Both β2AR structures are bound with the inverse agonist carazolol, and as such these structures are presumably more representative of an inactive state of β2AR. However, the reported crystal structures feature broken “ionic locks” [22], and the increased affinity of the β2AR-T4 fusion protein for agonists compared to the wild type β2AR suggest that these β2AR crystal structures may be more closely related to an active state of the β2AR receptor, or a state between active and inactive, consistent with the binding of a functionally weak inverse agonist. This suggests that a homology model based on β2AR structure could also be useful as a template to model the LPA4 receptor.
Therefore, in order to determine which crystal structure would provide the most appropriate template for the modeling of LPA4 and to gain insight into its interaction with its endogenous ligand LPA, we constructed three separate homology models using a) the high-resolution “inactive” or dark-state rhodopsin (PDB code = 1U19), b) the “active” rhodopsin (PDB code = 2I37), and c) the β2-adrenoceptor-T4 lysozyme fusion protein (PDB code = 2RH1) [19]. Other GPCR crystal structures, including the recently-determined β1- [23] and β2-adrenergic [24] receptors and squid rhodopsin [25], are structurally similar to one of these three, and were not considered in this study. The unliganded opsin [26,27] and the A2A adenosine [28] receptors were also not considered as we are more interested in an agonist-occupied receptor model of LPA4. Automated docking studies of LPA in the three previously mentioned models were then conducted using the GOLD program [29], and the possible binding modes and critical interactions between LPA and the LPA4 receptor were explored.
2. Materials and methods
2.1. Modeling platform
All computations were performed on Hewlett-Packard dual-CPU, dual-core AMD Opteron 2216-based xw9400 workstations running Red Hat Enterprise Linux Workstation 4 (RHEL-WS4).
2.2 Sequence alignment
All primary amino acid sequences were retrieved from the SwissProt/TrEMBL database at www.expasy.org (bRh, P02699; hB2AR, P07550; LPA1, Q92633; LPA2, Q9HBW0; LPA3, Q9UBY5; LPA4, Q99677; LPA5, Q9H1C0). The alignment was performed using ClustalX 2.0.5 [30] using default parameters, except that the gap opening penalty was increased to 15 (for both pairwise and multiple alignments) and an alternate weight matrix was employed (BLOSUM series for multiple alignments and BLOSUM 30 for pairwise alignments). The LPA1-5 sequences were aligned to a profile of pre-aligned diverse rhodopsin-like GPCR sequences [31]. Manual adjustments were made as necessary in the EL2 loop regions to properly align the cysteine residues that comprise the disulfide linkage that tethers the EL2 loop to the extracellular portion of TM3. Additionally, a second non-profile multiple sequence alignment was performed with bRh, hβ2AR and LPA1-5 using the parameters described above. This alignment was identical to the previous profile alignment in the TM helical regions, whose boundaries are defined in this work as in Bissantz, et al. [31]. The results of this alignment are shown in Figure 3. Pairwise alignments are shown in Supplemental Figure S1. The tree diagram of Figure 2 was created using Geneious 3.6.2 (BioMatters Ltd., Auckland, NZ) with ClustalW [30] 2.0.5 (BLOSUM series similarity matrices, gap opening penalty = 5). The sequences used in this alignment were also retrieved from the SwissProt/TrEMBL database. Supplemental Figure S2 shows the results of this alignment.
Figure 3.
Sequence alignment of bRh, hβ2AR and LPA receptor subtypes LPA1-5. Transmembrane and loop regions are labeled. Red blocks denote sequence identity among the seven sequences, orange blocks denote strong similarity and yellow blocks denote weak similarity. Ballesteros-Weinstein [34] reference positions for each TM region are indicated. Amino acid numbering corresponds to the LPA4 sequence. Green blocks indicate residues that are critical for LPA binding to LPA1-3; yellow blocks indicate residues proposed to be critical for LPA binding to LPA4. The figure was prepared using ALSCRIPT 2.07a [68].
2.3 Homology modeling
The homology modeling template structures were retrieved from the PDB Data Bank at http://www.rcsb.org (PDB codes = 1U19, 2.2 Å, “dark-state” bRh; 2I37, 4.2 Å, “activated” bRh; 2RH1, 2.4 Å, hβ2AR). The homology models were constructed using SYBYL v7.3 (Tripos, St. Louis, MO, USA). Because there was no appropriate structure template available, the 35 amino acid residues preceding F36(1.30) in the N-terminal region and 42 amino acid residues following Ile328 in C-terminal region were not included in the LPA4 homology models. Residues in the TM helices, Helix 8 and the first intracellular loop (IL1) were directly mutated from the template receptor to the cognate residues in LPA4. The remaining loops were modeled using the loop search function within SYBYL. The EL2 loop required two separate searches, one for the sequence preceding and another for the sequence following the TM3-EL2 disulfide linkage. For the β2AR-based model, the original EL2 loop was replaced with the EL2 loop from rhodopsin (1U19) by first aligning the TM helical regions of the β2AR-based model and bRh, then merging the EL2 loop from bRh onto the β2AR-based model. A subsequent loop search was performed to address the difference in sequence lengths of the bRh and LPA4 in the TM4-EL2 region. The resulting models were renumbered and the side chain conformations assigned using the program SCWRL 3.0 [32]. The structures were then energy-minimized using the Tripos Force Field (TFF) in SYBYL v7.3. Gasteiger-Hückel charges were assigned and a distance-dependent dielectric constant = 4.0 was employed, with termination occurring when the energy gradient fell below 0.05 kcal/(mol×Å) or the number of iterations exceeded 100,000. PROCHECK [33] were used to assess the stereochemical quality of the resulting receptor models. PROCHECK Ramachandran plots are included as Supplemental Figure S3. In addition, the stereochemical quality of these models were checked using the WHAT IF software and the full WHAT IF summaries were included in Supplementary Information. The results indicated that the overall quality of each model was comparable to the parent crystal templates from which the model was derived.
The Ballesteros-Weinstein [34] GPCR receptor residue numbering system is used in this work to identify corresponding residues in different GPCRs. This index is shown in parentheses following the traditional one-letter amino acid identifier (i.e. R136(3.50)).
2.4 Docking of LPA
The structure of LPA (18-1) was sketched, Gasteiger-Hückel charges assigned and its conformation energy-minimized with the TFF in SYBYL. The program GOLD 3.1 [29] was used to perform the docking studies with standard default settings. In all three receptor models, the binding site was defined to include all atoms within 18 Å of the carbon atom of D113(3.32). Based on the fitness scores and the binding orientation of LPA within the binding cavity, the best GOLD-docked solution was selected and merged into the LPA4 receptor. The conformation of LPA was adjusted and the combined receptor-ligand structure was energy-minimized using the parameters described above in order to remove clashes and minimize strain energy, optimizing the interaction of LPA within the LPA4 receptor binding pocket.
3. Results and discussion
3.1. Sequence analysis
In order to determine which of the two template receptor sequences is more similar to LPA4 (bRh or β2AR), we compared the sequence identities and similarities between LPA4 and either bRh or β2AR, and the results are presented in Table 1 and Supplemental Figure S1. Overall, the percent identity and similarity is roughly equal for both receptor types (24% and 53% for bRh; 24% and 50% for β2AR). TM1 and TM5 in LPA4 are more closely related to bRh, and TM3 is closer to that of β2AR. The increased similarity to bRh in TM1 is due largely to the presence of strong similarity at its extracellular end. This is significant because the extracellular region of TM1 is different in bRh and β2AR, the latter having a more severe tilt angle that directs the top of TM1 away from the helical bundle. The orientation of TM1 could affect the packing of the receptor into cholesterol-mediated dimeric units, as has been experimentally suggested for the β2AR [24]. The greater similarity of LPA4-TM5 to bRh-TM5 is most noticeable in the residues immediately flanking the conserved proline at position 5.50. TM5 is a region where differences in amino acid composition frequently affect ligand selectivity among GPCR subtypes, as its extracellular segment often constitutes part of the orthosteric binding site [35]. The greater sequence identity of LPA4 and β2AR in TM3 is attributable to a higher number of conserved residues in the intracellular “arginine cage” [36] region surrounding R(3.50). Considering their overall sequence similarity to LPA4, it seems plausible that both bRh and β2AR might provide suitable templates with which to construct useful homology models of the LPA4 receptor.
Table 1.
Sequence comparison of LPA4 with bovine rhodopsin and the human β2-adrenoceptor. Bold values indicate the higher identity (or similarity) when comparing the bRh alignment with the corresponding β2AR alignment.
| TM | Lengtha | bovine rhodopsin | β2-adrenoceptor | ||
|---|---|---|---|---|---|
| Identity (%)b | Similarity(%)c | Identity (%)b | Similarity (%)c | ||
| 1 | 30 | 20(6) | 60(18) | 13(4) | 40(12) |
| 2 | 30 | 27(8) | 50(15) | 27(8) | 50(15) |
| 3 | 33 | 15(5) | 48(16) | 27(9) | 58(19) |
| 4 | 23 | 13(3) | 43(10) | 13(3) | 39(9) |
| 5 | 26 | 38(10) | 54(14) | 27(7) | 46(12) |
| 6 | 26 | 31(8) | 69(18) | 31(8) | 62(16) |
| 7 | 21 | 29(6) | 48(10) | 29(6) | 57(12) |
| Total | 189 | 24(46) | 53(101) | 24(45) | 50(95) |
Number of amino acid positions in the transmembrane helix.
Percent identity. Values in parenthesis indicate the number transmembrane helix positions where the amino acids are identical.
Percent similarity, as determined by ClustalX 2.0.5. Two amino acids are considered to be similar if they are identical or if they are both members of the same strongly conserved group. Values in parenthesis indicate the number transmembrane helix positions where the amino acids are similar.
The LPA1, LPA2, and LPA3 receptors share about 50% sequence identity [37] amongst themselves, but only about 17% sequence identity to the LPA4 and LPA5 receptors [4]. On the other hand, the LPA4 and LPA5 receptors share about 35% sequence identity [5]. A profile sequence alignment was performed that included LPA1-5, bRh and hβ2AR (Figure 3). From the resulting sequence alignment, it is seen that the LPA4 receptor possesses all of the conserved residues (among rhodopsin-like GPCRs) that are used as reference positions in the Ballesteros-Weinstein numbering system [34]. There are three apparent differences between the LPA4 receptor and LPA1-3 receptors. First, the normally-conserved proline residue at position 5.50 (including LPA4) is replaced with methionine in LPA1-3, indicating that TM5 in these subtypes possibly does not possess the kink [38] and resulting “π-bulge” or “aneurism” [39] that their proline-containing counterparts do. However, it should be noted that the proline residue is not absolutely necessary for the kink to be retained [40]. Second, the conserved C(3.25) in LPA4 is replaced with tryptophan in LPA1-3. This is the residue that is normally part of a disulfide linkage that tethers the top of TM3 to EL2. On the other hand, LPA4 has only one conserved cysteine residue in EL2 (xl2.50 in Figure 3) while LPA1-3 have three, which may lead to a disulfide linkage proposed to exist between EL2 and EL3. Third, P(7.50) is replaced with alanine in the ‘N(7.49)PxxY(7.53)’ region in LPA2, indicating that there may be significant structural differences in this region compared to other GPCRs. Parrill et al. [37] reported for the LPA1-3 receptors that the conserved residues R(3.28), Q(3.29), K(7.36) in LPA1-2, and K(7.35) in LPA3 form a polar binding pocket that specifically binds with the polar head group of LPA through multiple hydrogen bonding interactions. However, in the LPA4-5 receptors, these mostly basic residues are not conserved. Instead, the cognate residues in LPA4 are S114(3.28), G115(3.29), Y293(7.35), and P294(7.36), indicating that LPA might bind to the LPA4 receptor in a different mode than in LPA1-3.
3.2. Homology modeling of the LPA4 receptor
Using the sequence alignment of Figure 3, three separate models of LPA4 were developed using homology modeling techniques (See Experimental Methods). These are designated bRhI-LPA4 for the model based on “dark state” rhodopsin, bRhA-LPA4 for the model based on “activated” rhodopsin, and β2AR-LPA4 for the β2AR-based model.
The most significant difference between the crystal structures of rhodopsin and β2AR is the presence of a short α-helical segment in the EL2 loop region, compared to the beta-hairpin structure of bRh. For many if not most GPCRs, the EL2 loop is very important for ligand recognition [15]. We thus made an attempt to rationally determine whether a β2AR-like α-helical or a bRh-like β-hairpin EL2 structure would be more appropriate for the LPA4 receptor. The possibility of α-helix formation within the EL2 loop of LPA4 receptor was predicted using a freely-available online package, PROF (Profile network prediction HeiDelberg; part of the PredictProtein Server) [41]. The results are presented in Figure 4, and show that the bRh EL2 loop was predicted to contain loop structures, while the β2AR EL2 loop was predicted (correctly) to contain a short alpha-helical segment. Short lengths of extended beta-strand were predicted for the LPA4 EL2, consistent with a rhodopsin-like beta-hairpin loop. Therefore in our β2AR-LPA4 model, the EL2 loop was replaced by the EL2 loop structure of rhodopsin template (See Experimental Methods).
Figure 4.
Summary of PROF secondary structure prediction results for the EL2 loops of bRh, β2AR and LPA4. Cysteine residues involved in disulfide linkages are colored blue (EL2-TM3) and orange (EL2-EL2).
The differences among the models were then compared (Figure 5). Reflecting its parent β2AR conformation, TM1 in β2AR-LPA4 retains a more nearly straight alpha-helical backbone conformation than in bRhI-LPA4 or bRhA-LPA4, effectively projecting the extracellular end of TM1 away from the helical bundle. TM3, TM5, TM6 and TM7 of the bRhA-LPA4 model are somewhat more loosely packed, consistent with the conformation that was inherited from its parent bRh crystal structure (2I37) [18]. This resulted in a larger cavity between TM4, TM5, TM6 and TM7 in bRhA-LPA4 when compared to the inactive bRhI-LPA4 model. A similar approach was previously described to generate an experimentally-guided active rhodopsin-based model of an agonist-bound κ-opioid receptor [42] in which the increase in intrahelical volume was shown to have physical meaning for the activated receptor state. In addition to the above differences and similarities attributable to their mother templates, somewhat different amino acid sidechain orientations were observed in the three models’ TM helices, which resulted in binding pockets with somewhat different distributions of hydrophilic and hydrophobic surface area (vide infra).
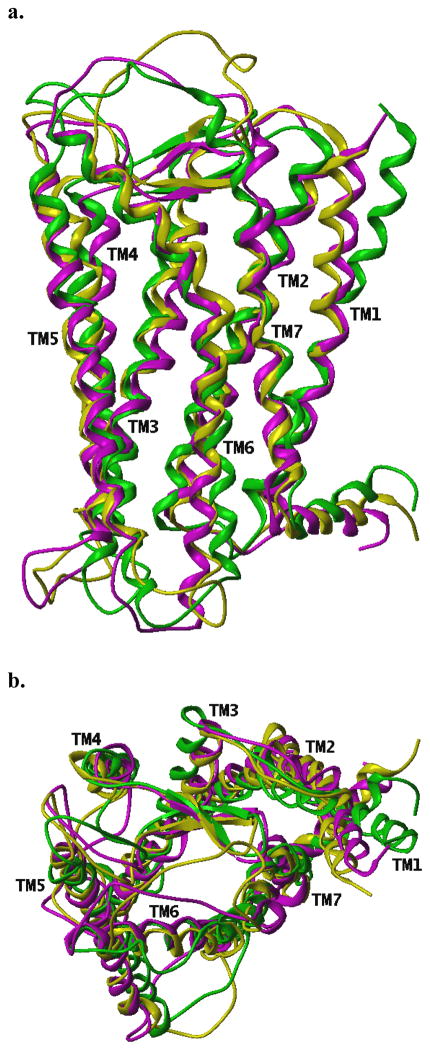
Figure 5.
Superimposition of backbone atoms of the three LPA4 receptor homology models using standard ribbon representations (magenta = bRhI-LPA4; yellow = bRhA-LPA4; green = β2AR-LPA4). RMSD values for equivalent backbone atoms in the transmembrane helical regions (as defined in [31]) among the LPA4 models: bRhI-bRhA, 1.41 Å; bRhI-β2AR, 2.06 Å; bRhA-β2AR, 2.20 Å. RMSD values comparing the original crystal structures with the corresponding final LPA4 models: bRhI, 1.10 Å; bRhA, 1.24 Å; β2AR, 1.02 Å.
The activation of rhodopsin-like GPCRs by ligands exhibiting agonist affinity is thought to occur through the stabilization of receptor conformations that can bind to G proteins, and the Gα subunit in particular. The ability of a ligand to stabilize (or possibly induce) an active conformation has been proposed [43] to be directly related to the rotameric state of the residues in the conserved ‘aromatic cluster’ [44] of TM6 that modulate its associated proline-induced kink. The aromatic residue at position 6.48 appears to be critical; the Cα-Cβ torsion angle χ1 = gauche+ has been associated with the inactive state(s), whereas the χ1 = trans form is associated with the active state(s) [45]. LPA is an endogenous agonist at LPA4 receptors, and might be expected to stabilize the χ1 = trans form of 6.48. In fact, mutagenesis studies in both LPA- [46] and P2Y-type [11,47] receptors indicate that the residue at position 6.48 does act as a molecular toggle switch. However, to date none of the crystallized GPCR structures (including the ‘active’ 2I37 [18] and 3DQB [27]) feature a χ1 = trans 6.48. Nonetheless, the gauche+ conformation was retained in all three models.
The kink and ‘π-bulge’ induced by the proline residue in TM2 may also be modified, often by nearby serine and threonine residues, thereby changing the shape and/or electrostatic character of the binding site [48]. The presence of T91(2.61) in LPA4 forms a ‘TxP’ motif, which would be expected to increase the bend angle of the helix by 7 to 10° compared to a standard proline-induced kink [49]. However, the TxP motif is not found in either the rhodopsin or β2AR TM2 helices, so the extracellular region of TM2 in LPA4 might be differently oriented than in the template structures.
The TM2 π-bulge also effectively rotates the extracellular portion of the helix compared to a regular α-helix [50]. This is important because it in part determines the position of K95(2.60) in LPA4 (and R in LPA5), whose basic-cationic sidechain may interact with the anionic phosphate or other polar groups of LPA. Such ligand-receptor ionic interactions are routinely proposed for phospholipid receptors (though the cognate residue at the LPA1-3 receptors is leucine, and as such cannot ion-pair with the ligand at position 2.60). For example, lysine is also found at position 2.60 in P2Y12-14. At least one very recent model (of GPR17, a receptor closely related to both P2Y receptors and LPA4) has suggested that the residue at position 2.60 (arginine) is part of the ligand binding site [51]. Another model of a closely-related and recently de-orphaned receptor (GPR87) includes position 3.26 (K112 in LPA4) prominently in its putative 1U19-derived binding site model [7]. However, polar and charged sidechains are also commonly found at the ends of transmembrane helices, where they stabilize the orientation of the helix within the membrane by interacting with the polar head groups of the lipids that make up the membrane bilayer [52,53]. Such is the case for K95(2.60) and K112(3.26) of LPA4 in our models; as presented here, these sidechains do not directly interact with the LPA ligand.
Incorporation of experimentally determined information about the conformation of a particular receptor and its interaction with high-affinity ligands, such as mutagenesis or substituted cysteine accessibility method (SCAM) studies, can greatly improve the accuracy of a model. Unfortunately, at the present time we are not aware of any such information directly relating to the LPA4 receptor. However, LPA4 is closely related to several other lipidergic receptor subtypes (Figure 2) that have been more fully studied, and it is possible to infer useful information from these studies and apply it to the modeling of the LPA4 receptor, and/or to use this information in selecting the most appropriate model from the three models. This was the strategy employed in this work.
3.3 Docking studies of LPA at the LPA4 receptor and putative binding pocket analysis
GOLD, a genetic algorithm-based automated docking routine, was employed to dock (find plausible binding modes for) the LPA molecule into the three receptor models. The results of the automated GOLD docking experiments are reported in Table 2 and shown in Figure 6. The scores in Table 2 correspond to the top-ranked chemically reasonable docked solution (pose). A more complete list of the top ten scores for each model are presented in Supplemental Table S1. Although LPA scored reasonably well in all three models, the highest score was obtained for the active bRhA-LPA4. Not only did this receptor-ligand complex produce the highest overall GoldScore (98.27), it also achieved the highest scores for each of the individual GoldScore components: receptor-ligand H-bonding (S(hb-ext), 13.79), receptor-ligand van der Waals contacts (S(vdw-ext), 69.52), and the lowest internal energy term, corresponding mainly to ligand intramolecular atom clashes and strained torsion angles (S(int), −11.11). The superior performance of the bRhA-LPA4 model is most likely a result of the receptor’s ability to effectively bury the large hydrophobic tail portion in an extended lipophilic binding pocket (Figure 6c) as well as coordinate the polar head group through strong H-bonds (Figure 6d). In the bRhA-LPA4 model, H-bonds are formed between terminal phosphate oxygen atoms and both S114(3.28) and T187(EL2), between the sn-2 hydroxyl group of LPA and Y265(6.51) and between the LPA carbonyl oxygen atom and the backbone NH group of E190(EL2).
Table 2.
GoldScores and components of LPA (18-1) binding to LPA4 receptor models.
| Model | GoldScore | S(hb-ext)a | S(vdw-ext)a | S(hb-int)a | S(int)a |
|---|---|---|---|---|---|
| bRhI-LPA4 | 70.78 | 10.80 | 57.96 | 0.00 | −19.71 |
| bRhA-LPA4 | 98.27 | 13.79 | 69.52 | 0.00 | −11.11 |
| β2AR-LPA4 | 83.00 | 3.05 | 68.19 | 0.00 | −13.81 |
GoldScore components: S(hb-ext): protein-ligand hydrogen bond energy (external H-bond); S(vdw-ext): protein-ligand van der Waals energy (external vdw); S(hb-int): ligand intramolecular hydrogen bond energy (internal H-bond); S(int): ligand internal energy term.
Figure 6.
The position and orientation of LPA (ball-and-stick) within the three LPA receptor models. a–b. bRhI-LPA4 (magenta); c.–d. bRhA-LPA4 (yellow); e.–f. β2AR-LPA4 (green). Important putative head group-interacting residues (capped sticks) and the F262(6.48) ‘toggle switch’ (space-filled) are depicted.
The other two models did not score as well. In the inactive rhodopsin-based bRhI-LPA4 model, H-bonding (Figure 6b) takes place between the terminal phosphate oxygen atoms and both T187 and N182 on EL2, between the sn-1 hydroxyl oxygen atom and Y265(6.51), and between the sn-2 hydroxyl group and the backbone carbonyl oxygen atom of G115(3.29), and the H-bonding score is relatively good (10.80). On the other hand, significantly higher internal strain energy contributes to the lower score in the bRhI-LPA4 model (−19.71), and the tail portion of LPA extends through TM4 and TM5, the tip of the tail protruding out into the space normally occupied by the transmembrane lipid bilayer (Figure 6a). The most likely cause of this is that the TM helices are too close together to accept the entire fatty LPA sidechain. As a result, the external vdw component of the GoldScore is reduced (57.96). However, this situation may not be totally unrealistic. The recently determined structure of opsin [26] has shown that GPCRs may (perhaps intentionally) form openings between TM helices to facilitate the movement of ligands in and out of the binding site. In this scenario, the lipophilic tail of LPA would be buried in the lipids of the membrane bilayer in an energetically favorable fashion.
In the β2AR-LPA4 model, the fatty LPA tail is contained within the TM helical bundle (Figure 6e), but the receptor’s interaction with the polar head group is much weaker (3.05) than in the rhodopsin-based models. Putative H-bonds are formed between phosphate oxygens and T187 and N182 in EL2 and Y293(7.35). The sn-2 hydroxyl group is associated with S114(3.28) (Figure 6f).
3.4 Binding site of the polar head group in other receptors
The binding mode of LPA in the LPA1-3 receptors has been reported by Parrill and Tigyi et al. [37]. The amino acid residues R(3.28), Q(3.29), K(7.35) and/or K/R(7.36) were found to be responsible for interaction with the polar head group of the LPA molecule. This places the head group in the same relative location as is proposed for the LPA4 models presented here, since most of the interacting residues are identical (notably 3.28, 3.29 and 7.35) to those proposed by Parrill and Tigyi et al. In contrast, the aforementioned authors do not propose a direct interaction of LPA with any residues on EL2. Several subtype-selective agonists and antagonists of LPA1-3 receptors have been designed and synthesized so far [54–57]. A pharmacophore including many of the same or nearby residues (including 3.28, 3.29 and 7.34) has also been proposed [46,58–60] for the binding of the structurally-related sphingosine-1-phosphate (S1P) at S1P receptors, which, like LPA1-3, are also members of the Edg gene family. In a similar fashion to LPA1-3, the putative tail-binding locus is the interhelical space between TM3, TM5 and TM6.
LPA4 is even more closely related phylogenetically to the purinergic nucleotide-binding P2Y receptor family than to the Edg family (Figure 2). As mentioned earlier, the P2Y receptors can be classified into two main subgroups (P2Y1-like and P2Y12-like) based on sequence analysis [61] and their signaling properties. Jacobson, et al. [62] have studied the P2Y receptor family extensively, and evidence suggests that the manner in which these receptors recognize their nucleotide ligands also differs based on this classification [11]. A common nucleotide phosphate-binding motif in the P2Y receptors, not found in LPA4 or the Edg subfamily, is H(6.52)xxR/K(6.55). Beyond this motif the proposed binding sites of the two subfamilies are thought to be different. The P2Y1 subfamily interacts preferentially at R(3.29) and R(7.39) [47,63,64]. In the P2Y12 subfamily, these residues are glycine and leucine, respectively, and instead ligands are proposed to interact with a lysine residue on EL2 and K(7.35) [7,12,51,65–67]. The lysine residue in the P2Y12 subfamily corresponds to T187(EL2) in LPA4, and K(7.35) in P2Y12 corresponds to Y293(7.35) in LPA4. The binding sites of the P2Y receptors are therefore quite similar to that proposed for LPA4. The degree of similarity may be greater for the P2Y12 group since the putative ligand binding site also includes EL2.
3.5 Binding site of the LPA lipophilic tail in other receptors
In their work to date, Parrill and Tigyi et al. (vide supra) propose that for LPA1-3, the lipophilic tail occupies a narrow lipophilic binding site described by TM3, TM5 and TM6, very similar to the tail-binding pocket of our LPA4 models presented here. Figure 7 shows the residues comprising the LPA tail binding site of the bRhA-LPA4 receptor model. Although reported to a lesser extent, ligands with long lipophilic tails have also been docked into P2Y family receptors. For example, the proposed binding site for leukotriene E4 (LTE4) in the human P2Y12 receptor is also circumscribed by TMs 3, 5 and 6 [12], as is the putative binding site of LPA in the orphan receptor GPR87 [7]. There is thus a consensus about where the tail portion binds within the helical bundle. The location seems quite reasonable, since a) LPA has agonist activity at LPA receptors, b) the putative binding site is dominated by hydrophobic sidechains and c) it is thought that upon activation of a typical rhodopsin-like GPCR, TM5 and TM6 move away from the other helices (including TM3), thus exposing the G protein (C-terminus of the α subunit) binding site [27] and initiating the signaling cascade.
Figure 7.
Residues of the extended binding pocket that are proposed to interact with the acyl tail portion of LPA (bRhA-LPA4 model).
4. Conclusions
Three homology models of the LPA4 receptor were built based on the crystal structures of the inactive and active bovine rhodopsin and human β2AR. In the highest-ranked docked LPA-LPA4 receptor complex based on the light form of rhodopsin, we observed that three non-conserved amino acid residues in LPA4, S114(3.28), T187(EL2) and Y265(6.51), engage in hydrogen bonding interactions with the polar head group of the LPA molecule, a result also observed in homology models derived from the crystal structures of dark-state rhodopsin and β2-adrenoceptor. These hydrogen bonding patterns were found to contribute significantly to the recognition of LPA within the LPA4 receptor. The location of the LPA binding site in LPA4 is consistent with the LPA binding sites that have been proposed for other LPA-activated LPA1-3 receptor models as well as for other phylogenetically-related lipidergic and purinergic GPCRs, although there is variation with respect to the specific ligand-receptor sidechain interactions among the various subtypes. Interestingly, it was observed that no basic residues directly interact with the polar head group in any of the three models, suggesting the possibility that multiple receptor conformational states may recognize LPA. In addition, our docking results suggest that the hydrophobic tail may protrude through an opening between TM5 and TM6, a result that is consistent with the recent crystal structures of opsin.
Supplementary Material
Acknowledgments
This work was partially supported by Pilot Project Concept Award (XF and YZ), Massey Cancer Center, VCU.
Appendix A. Supplementary data
GoldScores for the top ten ligand-receptor complexes, pairwise sequence alignments of LPA4 with either bRh or β2AR, the aligned family of lipidergic receptor sequences and PROCHECK Ramachandran plots for each receptor model.
References
- 1.Fang X, Yu S, LaPushin R, Lu Y, Furui T, Penn LZ, Stokoe D, Erickson JR, Bast RC, Jr, Mills GB. Lysophosphatidic Acid Prevents Apoptosis in Fibroblasts via Gi-protein-mediated Activation of Mitogen-activated Protein Kinase. Biochem J. 2000;352:135–143. [PMC free article] [PubMed] [Google Scholar]
- 2.Noguchi K, Herr D, Mutoh T, Chun J. Lysophosphatidic Acid (LPA) and its Receptors. Curr Opin Pharmacol. 2009;9:15–23. doi: 10.1016/j.coph.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Kano K, Arima N, Ohgami M, Aoki J. LPA and its Analogs — Attractive Tools for Elucidation of LPA Biology and Drug Development. Curr Med Chem. 2008;15:2122–2131. doi: 10.2174/092986708785747562. [DOI] [PubMed] [Google Scholar]
- 4.Noguchi K, Ishii S, Shimizu T. Identification of p2y9/GPR23 as a Novel G Protein-coupled Receptor for Lysophosphatidic Acid, Structurally Distant from the Edg Family. J Biol Chem. 2003;278:25600–25606. doi: 10.1074/jbc.M302648200. [DOI] [PubMed] [Google Scholar]
- 5.Lee CW, Rivera R, Gardell S, Dubin AE, Chun J. GPR92 as a New G12/13- and Gq-coupled Lysophosphatidic Acid Receptor That Increases cAMP, LPA5. J Biol Chem. 2006;281:23589–23597. doi: 10.1074/jbc.M603670200. [DOI] [PubMed] [Google Scholar]
- 6.Pasternack SM, von Kügelgen I, Al Aboud K, Lee Y-A, Rüschendorf F, Voss K, Hillmer AM, Molderings GJ, Franz T, Ramirez A, Nürnberg P, Nöthen MM, Betz R. G Protein-Coupled Receptor P2Y5 and Its Ligand LPA Are Involved in Maintenance of Human Hair Growth. Nat Genetics. 2008;40:329–334. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 7.Tabata KI, Baba K, Shiraishi A, Ito M, Fujita N. The Orphan GPCR GPR87 Was Deorphanized and Shown To Be a Lysophosphatidic Acid Receptor. Biochem Biophys Res Comm. 2007;363:861–866. doi: 10.1016/j.bbrc.2007.09.063. [DOI] [PubMed] [Google Scholar]
- 8.Lee Z, Cheng CT, Zhang H, Subler MA, Wu J, Mukherjee A, Windle JJ, Chen CK, Fang X. Role of LPA4/p2y9/GPR23 in Negative Regulation of Cell Motility. Mol Biol Cell. 2008;19:5435–5445. doi: 10.1091/mbc.E08-03-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagida K, Ishii S, Hamano F, Noguchi K, Shimizu T. LPA4/p2y9/GPR23 Mediates Rho-dependent Morphological Changes in a Rat Neuronal Cell Line. J Biol Chem. 2007;282:5814–5824. doi: 10.1074/jbc.M610767200. [DOI] [PubMed] [Google Scholar]
- 10.Piomelli D, Astarita G, Rapaka R. A Neuroscientist’s Guide to Lipidomics. Nat Rev Neurosci. 2007;8:743–754. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- 11.Costanzi S, Mamedova L, Gao ZG, Jacobson KA. Architecture of P2Y Nucleotide Receptors: Structural Comparison Based on Sequence Analysis, Mutagenesis, and Homology Modeling. J Med Chem. 2004;47:5393–5404. doi: 10.1021/jm049914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nonaka Y, Hiramoto T, Fujita N. Identification of Endogenous Surrogate Ligands for Human P2Y12 Receptors by In Silico and In Vitro Methods. Biochem Biophys Res Comm. 2005;337:281–288. doi: 10.1016/j.bbrc.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 13.Patny A, Desai PV, Avery MA. Homology Modeling of G-Protein-Coupled Receptors and Implications in Drug Design. Curr Med Chem. 2006;13:1667–1691. doi: 10.2174/092986706777442002. [DOI] [PubMed] [Google Scholar]
- 14.Shi L, Javitch JA. The Binding Site of Aminergic G Protein-coupled Receptors: the Transmembrane Segments and Second Extracellular Loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 15.de Graaf C, Foata N, Engkvist O, Rognan D. Molecular Modeling of the Second Extracellular Loop of G-protein Coupled Receptors and Its Implication on Structure-based Virtual Screening. Proteins. 2008;71:599–620. doi: 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- 16.Yuzlenko O, Kieć-Kononowicz K. Molecular Modeling of A1 and A2A Adenosine Receptors: Comparison of Rhodopsin- and β2-Adrenergid-Based Homology Models Through Docking Studies. J Comput Chem. 2008;30:14–32. doi: 10.1002/jcc.21001. [DOI] [PubMed] [Google Scholar]
- 17.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The Retinal Conformation and Its Environment in Rhodopsin in Light of a New 2.2 Å Crystal Structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 18.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal Structure of a Photoactivated Deprotonated Intermediate of Rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-Resolution Crystal Structure of an Engineered Human β2-Adrenergic G Protein-Coupled Receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal Structure of the Human β2 Adrenergic G-protein-coupled Receptor. Nature. 2007;450:383–388. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR Engineering Yields High-Resolution Structural Insights into β2-Adrenergic Receptor Function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 22.Audet M, Bouvier M. Insights Into Signaling from the β2-Adrenergic Receptor Structure. Nat Chem Biol. 2008;4:397–403. doi: 10.1038/nchembio.97. [DOI] [PubMed] [Google Scholar]
- 23.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GF. Structure of a β1-Adrenergic G-Protein-Coupled Receptor. Nature. 2008;454:486–492. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A Specific Cholesterol Binding Site is Established by the 2.8Å Structure of the Human β2-Adrenergic Receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami M, Kouyama T. Crystal Structure of Squid Rhodopsin. Nature. 2008;253:363–368. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 26.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal Structure of the Ligand-Free G-Protein-Coupled Receptor Opsin. Nature. 2008;454:183–188. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 27.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauß N, Choe HW, Hofmann KP, Ernst OP. Crystal Structure of Opsin in its G-Protein-Interacting Conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 28.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Izjerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and Validation of a Genetic Algorithm for Flexible Docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-Based Virtual Screening of Chemical Databases. II. Are Homology Models of G-Protein Coupled Receptors Suitable Targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 32.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A Graph-theory Algorithm for Rapid Protein Side-chain Prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Ballesteros JA, Weinstein H. In: Methods in Neurosciences. Sealfon SC, editor. Academic Press; San Diego: 1995. pp. 366–428. [Google Scholar]
- 35.Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A Chemogenomic Analysis of the Transmembrane Binding Cavity of Human G-Protein-Coupled Receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- 36.Ballesteros JA, Kitanovic S, Guarnier F, Davies P, Fromme BJ, Konvicka K, Chi L, Millar RP, Davidson JS, Weinstein H, Sealfon SC. Functional Microdomains in G-Protein-Coupled Receptors: The Conserved Arginine-Cage Motif in the Gonadotropin-Releasing Hormone Receptor. J Biol Chem. 1998;273:10445–10453. doi: 10.1074/jbc.273.17.10445. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara Y, Sardar VM, Tokumura A, Baker D, Murakami-Murofushi K, Parrill AL, Tigyi G. Identification of Residues Responsible for Ligand Recognition and Regioisomeric Selectivity of Lysophosphatidic Acid Receptors Expressed in Mammalian Cells. J Biol Chem. 2005;280:35038–35050. doi: 10.1074/jbc.M504351200. [DOI] [PubMed] [Google Scholar]
- 38.Sansom MSP, Weinstein H. Hinges, Swivels and Switches: The Role of Prolines in Signalling via Transmembrane α-Helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 39.Fowler CB, Pogozheva ID, Levine H, III, Mosberg HI. Refinement of a Homology Model of the μ-Opioid Receptor Using Distance Constraints from Intrinsic and Engineered Zinc-Binding Sites. Biochemistry. 2004;43:8700–8710. doi: 10.1021/bi036067r. [DOI] [PubMed] [Google Scholar]
- 40.Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The Evolution of Transmembrane Helix Kinks and the Structural Diversity of G Protein-Coupled Receptors. Proc Natl Acad Sci USA. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost B, Yachdav G, Liu J. The PredictProtein Server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F, Mosier PD, Westkaemper RB, Roth BL. Gα-Subunits Differentially Alter the Conformation and Agonist Affinity of κ-Opioid Receptors. Biochemistry. 2008;47:1567–1578. doi: 10.1021/bi701476b. [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharya S, Hall SE, Vaidehi N. Agonist Induced Conformational Changes in Bovine Rhodopsin: Insight Into Activation of G-Protein Coupled Receptors. J Mol Biol. 2008;382:539–555. doi: 10.1016/j.jmb.2008.06.084. [DOI] [PubMed] [Google Scholar]
- 44.Javitch JA, Ballesteros JA, Weinstein H, Chen J. A Cluster of Aromatic Residues in the Sixth Membrane-Spanning Segment of the Dopamine D2 Receptor is Accessible in the Binding-Site Crevice. Biochemistry. 1998;37:998–1006. doi: 10.1021/bi972241y. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. β2 Adrenergic Receptor Activation: Modulation of the Proline Kink in Transmembrane 6 by a Rotamer Toggle Switch. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 46.Fujiwara Y, Osborne DA, Walker MD, Wang D, Bautista DL, Liliom K, van Brocklyn JR, Parrill AL, Tigyi G. Identification of the Hydrophobic Ligand Binding Pocket of the S1P1 Receptor. J Biol Chem. 2007;282:2374–2385. doi: 10.1074/jbc.M609648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zylberg J, Ecke D, Fischer B, Reiser G. Structure and Ligand-binding Site Characteristics of the Human P2Y11 Nucleotide Receptor Deduced from Computational Modelling and Mutational Analysis. Biochem J. 2007;405:277–286. doi: 10.1042/BJ20061728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deupi X, Dlker N, Lpez-Rodrguez ML, Campillo M, Ballesteros JA, Pardo L. Structural Models of Class A G Protein-Coupled Receptors as a Tool for Drug Design: Insights on Transmembrane Bundle Plasticity. Curr Top Med Chem. 2007;7:991–998. doi: 10.2174/156802607780906799. [DOI] [PubMed] [Google Scholar]
- 49.Deupi X, Olivella M, Govaerts C, Ballesteros JA, Campillo M, Pardo L. Ser and Thr Residues Modulate the Conformation of Pro-Kinked Transmembrane α-Helices. Biophys J. 2004;86:105–115. doi: 10.1016/S0006-3495(04)74088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riek RP, Finch AA, Begg GE, Graham RM. Wide Turn Diversity in Protein Transmembrane Helices Implications for G-Protein-Coupled Receptor and Other Polytopic Membrane Protein Structure and Function. Mol Pharmacol. 2008;73:1092–1104. doi: 10.1124/mol.107.043042. [DOI] [PubMed] [Google Scholar]
- 51.Parravicini C, Ranghino G, Abbracchio MP, Fantucci P. GPR17: Molecular Modeling and Dynamics Studies of the 3-D Structure and Purinergic Ligand Binding Features in Comparison with P2Y Receptors. BMC Bioinformatics. 2008;9:263. doi: 10.1186/1471-2105-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Killian JA, von Heijne G. How Proteins Adapt to a Membrane-Water Interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 53.Monné M, Nilsson I, Johansson M, Elmhed N, von Heijne G. Positively and Negatively Charged Residues Have Different Effects on the Position in the Membrane of a Model Transmembrane Helix. J Mol Biol. 1998;284:1177–1183. doi: 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- 54.Durgam G, Tsukahara R, Makarova N, Walker MD, Fujiwara Y, Pigg KR, Baker DL, Sardar VM, Parrill AL. Synthesis and Pharmacological Evaluation of Second-generation Phosphatidic Acid Derivatives as Lysophosphatidic Acid Receptor Ligands. Bioorg Med Chem Lett. 2006;16:633–640. doi: 10.1016/j.bmcl.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Jiang G, Xu Y, Fujiwara Y, Tsukahara T, Tsukahara R, Gajewiak J, Tigyi G, Prestwich GD. α-Substituted Phosphonate Analogues of Lysophosphatidic Acid (LPA) Selectively Inhibit Production and Action of LPA. ChemMedChem. 2007;2:679–690. doi: 10.1002/cmdc.200600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamaruya Y, Suzuki M, Kamura G, Kanai M, Hama K, Shimizu K, Aoki J, Arai H, Shibasaki M. Identifying Specific Conformations by Using a Carbohydrate Scaffold: Discovery of Subtype-Selective LPA-Receptor Agonists and an Antagonist. Angew Chem Int Ed. 2004;43:2834–2837. doi: 10.1002/anie.200454065. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Jiang G, Tsukahara R, Fujiwara Y, Tigyi G, Prestwich GD. Phosphonothioate Fluoromethylene Phosphonate Analogues of Cyclic Phosphatidic Acid: Novel Antagonists of Lysophosphatidic Acid Receptors. J Med Chem. 2006;49:5309–5315. doi: 10.1021/jm060351+. [DOI] [PubMed] [Google Scholar]
- 58.Holdsworth G, Osborne DA, Pham TCT, Fells JI, Hutchinson G, Milligan G, Parrill AL. A Single Amino Acid Determines Preference Between Phospholipids and Reveals Length Restriction for Activation of the S1P4 Receptor. BMC Biochemistry. 2004;5:12. doi: 10.1186/1471-2091-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naor MM, Walker MD, van Brocklyn JR, Tigyi G, Parrill AL. Sphingosine 1-Phosphate pKa and Binding Constants: Intramolecular and Intermolecular Influences. J Mol Graph Model. 2007;26:519–528. doi: 10.1016/j.jmgm.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pham T-CT, Fells JI, Osborne DA, North EJ, Sr, Naor MM, Parrill AL. Molecular Recognition in the Sphingosine 1-Phosphate Receptor Family. J Mol Graph Model. 2007 doi: 10.1016/j.jmgm.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schöneberg T, Hermsdorf T, Engemaier E, Engel K, Liebscher I, Thor D, Zierau K, Römpler H, Schulz A. Structural and Functional Evolution of the P2Y12-Like Receptor Group. Purinergic Signal. 2007;3:255–268. doi: 10.1007/s11302-007-9064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanov AA, Costanzi S, Jacobson KA. Defining the Nucleotide Binding Sites of P2Y Receptors Using Rhodopsin-Based Homology Modeling. J Comput-Aided Mol Des. 2006;20:417–426. doi: 10.1007/s10822-006-9054-2. [DOI] [PubMed] [Google Scholar]
- 63.Ivanov AA, Ko H, Cosyn L, Maddileti S, Besada P, Fricks I, Costanzi S, Harden K, van Calenbergh S, Jacobson KA. Molecular Modeling of the Human P2Y2 Receptor and Design of a Selective Agonist, 2′-Amino-2′-deoxy-2-thiouridine 5′-Triphosphate. J Med Chem. 2007;50:1166–1176. doi: 10.1021/jm060903o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Major DT, Fischer B. Molecular Recognition in Purinergic Receptors. 1. A Comprehensive Computational Study of the h-P2Y1-Receptor. J Med Chem. 2004;47:4391–4404. doi: 10.1021/jm049772m. [DOI] [PubMed] [Google Scholar]
- 65.Ivanov AA, Fricks I, Harden K, Jacobson KA. Molecular Dynamics Simulation of the P2Y14 Receptor. Ligand Docking and Identification of a Putative Binding Site of the Distal Hexose Moiety. Bioorg Med Chem Lett. 2007;17:761–766. doi: 10.1016/j.bmcl.2006.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ko H, Fricks I, Ivanov AA, Harden K, Jacobson KA. Structure-Activity Relationship of Uridine 5′-Diphoshoglucosde Analogues as Agonists of the Human P2Y14 Receptor. J Med Chem. 2007;50:2030–2039. doi: 10.1021/jm061222w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhan C, Yang J, Dong XC, Wang YL. Molecular Modeling of Purinergic Receptor P2Y12 and Interaction With Its Antagonists. J Mol Graph Model. 2007;26:20–31. doi: 10.1016/j.jmgm.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Barton GJ. ALSCRIPT: A Tool to Format Multiple Sequence Alignments. Protein Engineering. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.