Abstract
Protein kinases transfer a phosphoryl group from ATP onto target proteins and play a critical role in signal transduction and other cellular processes. Here, we review the kinase kinetic and chemical mechanisms and their application in understanding kinase structure and function. Aberrant kinase activity has been implicated in many human diseases, in particular cancer. We highlight applications of technologies and concepts derived from kinase mechanistic studies that have helped illuminate how kinases are regulated and contribute to pathophysiology.
1. INTRODUCTION
The discovery of protein kinases in the 1950s led to a massive influence on clarifying biological pathways and disease mechanisms and developing therapies over the subsequent six decades (Hunter, 2000; Krebs & Beavo, 1979). Eukaryotic protein kinases are enzymes that catalyze phosphoryl transfer from MgATP to Ser/Thr and Tyr side chains in proteins. Their importance is in part evidenced by their frequency in eukaryotic genomes, typically representing 2–3% of the genes, including in human where 518 protein kinases have been annotated (Manning, Whyte, Martinez, Hunter, & Sudarsanam, 2002). While each specific kinase is thought to have a specialized function, there are many conserved features among kinases regarding their structures and catalytic mechanisms (Hanks, Quinn, & Hunter, 1988). This protein kinase chapter is written from an enzymology perspective and will cover the kinetic and chemical mechanisms of kinases and how an understanding of these features has been used to explore the structure, function, and regulation of these important catalysts.
2. KINETIC MECHANISM
Protein kinases operate on two substrates, proteins, and MgATP and produce phosphoproteins and MgADP (Adams, 2001; Taylor & Kornev, 2011). While it is sometimes the case that free ATP rather than Mg-bound ATP is thought of as the phosphoryl-donor substrate, the affinity of Mg for ATP is high enough that there is only a low concentration of non-Mg-bound ATP in cells. Thus with one apparent exception (Mukherjee et al., 2008), protein kinases require at least one divalent ion, Mg or Mn, for catalysis. Two substrate group transfer enzymes like kinases can be classified into two general types, those that follow ternary complex mechanisms and those that follow ping-pong mechanisms (Segel, 1993). Ternary complex mechanisms typically involve direct reaction between the two substrates to afford the two products, whereas ping-pong mechanisms proceed through a covalent enzyme intermediate, which in the case of kinases would be a phosphoenzyme species.
Classical two substrate steady-state kinetics experiments revealing an intersecting line pattern in double reciprocal plots (Segel, 1993) as well as more technically sophisticated stereochemical studies showing inversion at the phosphoryl group (Knowles, 1980) helped define protein kinase A (PKA) as following a ternary complex mechanism. Subsequently, two substrate kinetic studies on a variety of Ser/Thr and Tyr kinases and many X-ray structures of these enzymes in complex with substrate analogs have confirmed this to be a general feature of the kinase superfamily (Zheng et al., 1993). However, recently, an X-ray crystal structure of an atypical kinase showed the surprising finding that an active site aspartate was phosphorylated (Ferreira-Cerca et al., 2012). This phosphoAsp was proposed to correspond to a phosphoenzyme intermediate that could deliver the phosphoryl group to a protein substrate, though further experiments will be needed to establish this mechanism. Of note, nucleoside diphosphokinase does proceed through a phosphohistidine intermediate so there is enzymatic precedence for a small-molecule kinase using a related mechanism (Admiraal et al., 1999).
For the vast majority of protein kinases that involve direct phosphoryl transfer through a ternary complex, other kinetic mechanism issues that have been addressed are whether there is a preference for MgATP or protein substrate to bind first and what step(s) is rate-limiting for catalysis? These features have been analyzed for a variety of protein kinases and the results are somewhat enzyme and reaction condition dependent. For example, PKA displays a clear preference for MgATP binding prior to peptide substrate whereas Csk kinase shows no apparent-binding preference between nucleotide or peptide substrates (Cole, Burn, Takacs, & Walsh, 1994; Qamar, Yoon, & Cook, 1992; Zheng et al., 1993). Interestingly, experiments on p38 MAP kinase have led to contradictory models. Models in which protein substrate binds first, MgATP binds first, or random order binding have all been proposed for p38 MAP kinase (LoGrasso et al., 1997; Szafranska & Dalby, 2005). While these different models could be traced to the distinct methods used for measurement, they also highlight the limitation of steady-state kinetic approaches to provide unambiguous mechanistic portraits. The most comprehensive studies on p38 MAP kinase that include complementary methods including calorimetry and structural considerations point to a random order of substrate binding for this enzyme (Szafranska & Dalby, 2005). One potential practical application that emanates from such models relates to the development of specific kinase inhibitors that target the ATP pocket (Noble, Endicott, & Johnson, 2004). If the protein substrate binds to the kinase in the absence of MgATP, there may be an influence on drug affinity.
Regarding rate-limiting steps, a combination of viscosity effects, presteady-state kinetic techniques, and alternate substrates have been employed with various kinases to define the microscopic rate constants. To some extent, the kinetic models not only depend on the conditions of the kinase assay conditions (salt concentration, divalent ion (Mg vs. Mn), peptide, or protein substrate), but they also show differences among the kinases themselves. With PKA, MgADP release is fully rate determining (Adams & Taylor, 1992; Qamar & Cook, 1993), whereas for Csk phosphoryl transfer is partially or fully rate limiting depending on whether MgATP or MnATP is used as the substrate (Grace, Walsh, & Cole, 1997). When Mg is used with Csk, product release is fast and chemistry is rate determining, whereas when Mn is employed, product release slows down, presumably because of metal–enzyme interactions. Increasing the ionic strength of the buffer can also speed product release, possibly by weakening the interactions between nucleotide and enzyme. Some kinases such as Ser–Arg protein kinase or Src protein tyrosine kinase can show processive phosphorylation of its protein substrate, effectively indicating that protein substrate/product release is the slow step in turnover (Aubol et al., 2003; Pellicena & Miller, 2001). Furthermore, many protein kinases like the insulin receptor tyrosine kinase (IRK) are regulated by accessory domains, phosphorylation, or allosteric ligands which can dramatically impact the nature of the rate-limiting steps (Ablooglu, Frankel, Rusinova, Ross, & Kohanski, 2001; Hubbard & Miller, 2007).
3. CHEMICAL MECHANISM OF KINASE PHOSPHORYL TRANSFER
Despite the apparent simplicity of the reaction chemistry, there has been significant effort to understand the details of how the phosphoryl group moves from ATP to the protein substrate hydroxy group in the kinase active site. This interest stems from several considerations. One is the fundamental challenge in defining the catalytic mechanism of an important family of enzymes. A second factor relates to our fascination with how kinase enzymes interconvert between more active and less active forms. Kinase regulation by ligands, phosphorylation, as well as mutation can alter the alignment of active site residues, which ultimately translates to effects on the chemistry of phosphoryl transfer. A third reason for interest in the chemical mechanism is to aid in the design of synthetic compounds which can artificially switch kinase activity on or off. Such mechanism-inspired chemical biology approaches can and have shed light on the biological functions of kinases in cellular signaling.
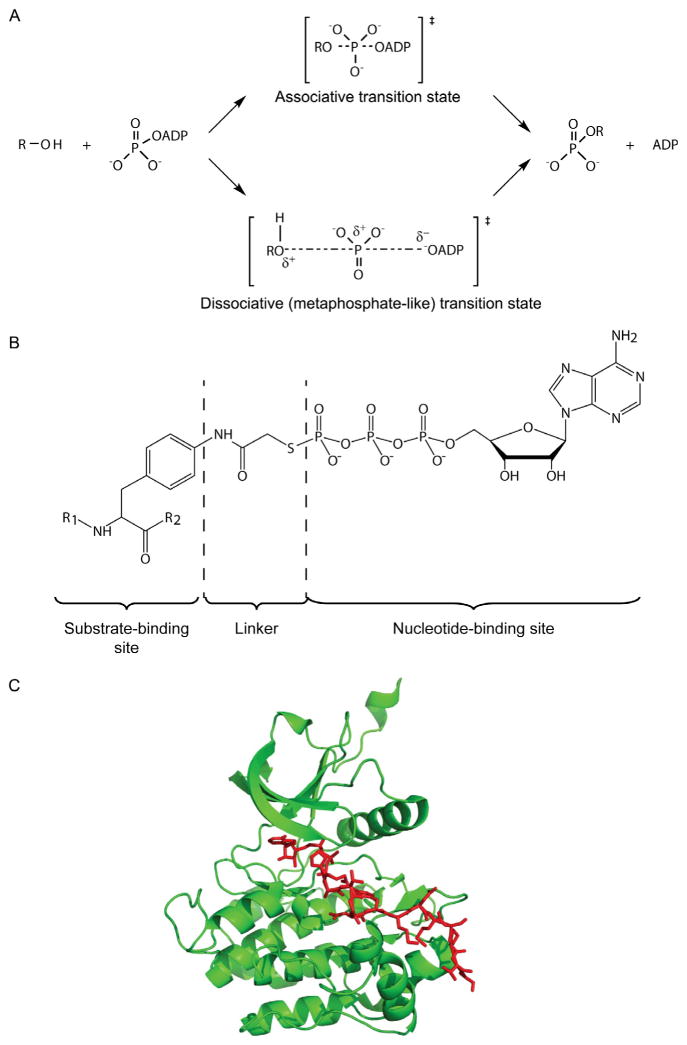
A central issue in defining the kinase mechanism is clarifying the nature of the phosphoryl transfer transition state. In the study of nonenzymatic phosphoryl transfer mechanisms, research dating back to the 1960s showed that phosphate monoesters like phenol phosphates display “dissociative” transition states (Kirby & Jencks, 1965). A dissociative transition state is one in which the bond between the phosphorus atom and the leaving group is largely broken prior to significant bond formation between the incoming nucleophile and the phosphorus, involving a metaphosphate-like intermediate (Admiraal & Herschlag, 1995; Mildvan, 1997; Fig. 1.1A). In contrast, nonenzymatic phosphate triester reactions typically show an associative transition state where the nucleophile forms a substantial bond with the phosphorus prior to leaving group departure. This dissociative character of the monoester is rationalized based on its negatively charged nonbridging oxygens repelling nucleophiles and stabilizing the partial positive charge on the metaphosphate-like intermediate.
Figure 1.1.
Protein kinase mechanism and bisubstrate analog inhibitors. (A) Mechanistic scheme of associative and dissociative transition states of phosphoryl transfer. A dissociative state model is proposed for the transfer of the γ-phosphoryl group of ATP to hydroxy group in a kinase–substrate. (B) Designed bisubstrate analog inhibitor for a protein tyrosine kinase. R1 and R2 amino acid sequences are derived from efficient substrate motifs. The linker is predicted to result in a 5–5.7 Å spacer between the substrate-binding site and the nucleotide-binding site, compatible with a dissociative transition state. ATPγS was used as the ATP-mimic analog. (C) The crystal structure of IRK (green ribbon) in complex with the above peptide–ATP conjugate (red stick), determined at 2.7 Å resolution (PDB code: 1GAG; Parang et al., 2001).
Relative to their nonenzymatic counterparts, transition states for enzyme reactions are much harder to analyze because of the complexity of the large protein catalyst and the more limited repertoire of methods available. Based on the early X-ray crystal structures of PKA, a chemical phosphoryl transfer mechanism with associative character was implied (Knighton et al., 1991). However, analysis of a static view of an enzyme complexed with substrate analogs can be an imprecise way for probing the dynamics of rapid chemical processes. Linear-free energy studies designed to measure the Bronsted nucleophile coefficient (βnuc) gave a different interpretation of the transition state. By analyzing the tyrosine kinase activity dependence with a series of tyrosine analogs substituted with fluorine atoms on the phenol ring, it was established that phosphoryl transfer rate is nearly independent of the nucleophile pKa, that is a near zero value for βnuc (Kim & Cole, 1997, 1998). In addition, the unexpected observation that the neutral phenol, rather than the phenoxide anion, was the obligate functional substrate form based on pH-rate studies further underscored the minimal role, the nucleophile likely plays in the transition state (Kim & Cole, 1998).
Other experiments have corroborated that tyrosine kinase transition states are likely dissociative, analogous to their nonenzymatic counterparts (Ablooglu et al., 2000; Parang et al., 2001; Sondhi, Xu, Songyang, Eck, & Cole, 1998; Williams & Cole, 2002). Interestingly, rate reductions of kinases with MgATPγS were initially considered as evidence against kinase-dissociative transition states (Cole, Grace, Phillips, Burn, & Walsh, 1995), since nonbridging sulfur substitution of phosphate monoesters can accelerate nonenzymatic phosphoryl transfer through stabilization of a metaphosphate-like species. However, follow-up measurements with various divalent ions showed that thiophilic metals like Ni and Co could complement the assays with ATPγS (Grace et al., 1997). Of note, it should be mentioned that kinase-mediated thiophosphate installation has been elegantly used for protein substrate identification in chemoselective labeling (Allen, Lazerwith, & Shokat, 2005).
The totality of the solution-phase evidence strongly supports a dissociative transition state for the protein kinase reactions that have been studied. Perhaps this result was to be expected since it would be energetically costly to force the phosphoryl transfer down a mechanistic pathway that traverses a steeper energetic barrier. Furthermore, based on the conserved features of all ~500 human kinases, it is reasonable to assert that dissociative character is a general property of these reactions.
This model does raise the somewhat untidy question of how a protein kinase would catalyze a dissociative transition state based on the architecture and electrostatics of the kinase active site. Since the enzyme is organizing a ternary complex, the templating of the ATP and protein substrate in position for reaction could be thought most compatible with a more associative reaction, as the crystallographic interpretations appeared to suggest. Perhaps an overly simple explanation for what kinases need to do to facilitate dissociative reactions is that they must focus positive charge on the departing ADP-anionic oxygens to sever the gamma-phosphoryl bond. Indeed, positive charges including a Lys side chain and Mg ion are pointed toward this β-phosphate and are observed in many kinase crystal structures in an arrangement that could stabilize the buildup of negative charge in the leaving group. It is also true that many kinases show substantial ATP-hydrolytic activity, which is consistent with generation of a metaphosphate-like species that can be intercepted by water in the absence of precise positioning of a protein hydroxyl (Rominger et al., 2007).
A number of computational studies have been performed to understand the kinase phosphoryl transfer mechanism (Cheng, Zhang, & McCammon, 2005; Valiev, Yang, Adams, Taylor, & Weare, 2007). Many of these simulations have analyzed the contributions of specific active site interactions, as well as geometry and charge states as a function of phosphoryl transfer progress. Such calculations generally are consistent with a dissociative transition-state mechanism. While these studies are generally not readily testable with precise experiments, site-directed mutagenesis and substrate analog experiments generally support their plausibility. Most fully active states of kinases are defined crystallographically by macroscopic and microscopic features that seem to be crucial for phosphoryl transfer. There is a particular alignment of the N-terminal and C-terminal lobes, generating a hydrophobic spine, that correspond to active kinase conformations (Kornev, Haste, Taylor, & Eyck, 2006). The formation of a salt bridge between conserved catalytic site Glu and Lys correlates well with active enzymes (Huse & Kuriyan, 2002). Availability of the catalytic base (Asp residue) for hydrogen bonding to the substrate hydroxyl also seems to be an important feature from structural studies.
Small molecule tools have also been used to decipher the nature of kinase catalytic mechanisms. The phosphate mimic aluminum fluoride has been crystallized with PKA in complex with ADP and a peptide substrate analog (Madhusudan, Akamine, Xuong, & Taylor, 2002). Despite the weak affinity of AlF3 for ADP in the absence of enzyme, the fact that the crystal structure captures an arrangement expected for catalysis suggests that the enzyme has selected a transition-state conformation. The distance between the substrate Ser nucleophile and the aluminum ion in this structure is sufficiently long to suggest a dissociative transition state in the phosphoryl transfer reaction (Mildvan, 1997).
Bisubstrate analogs for kinases have been designed based on a dissociative transition state. For enzymes that obey ternary complex mechanisms, covalent linkage of the two substrates can generate high-affinity inhibitors, if the linkers are installed to approximate the geometry of the reaction coordinate. In favorable cases, bisubstrate analog-binding energies to enzymes can equal or exceed the sum of the energies of the individual substrates, since the entropic penalty of assembling the three components of the reaction is reduced. Early work on PKA bisubstrate analog inhibitors involved synthetic compounds in which the peptide Ser oxygen was covalently linked to ADP, ATP, and adenosine-tetraphosphate via a phosphodiester with the terminal phosphate (Medzihradszky, Chen, Kenyon, & Gibson, 1994). While none of these compounds was a stronger PKA inhibitor than ADP alone, the tetraphosphate-containing bisubstrate analog was more potent than the shorter phosphate strings. This suggests that an extended spacer between the two substrates better approximates the reaction coordinate distance of kinase reaction. A theoretical analysis by Mildvan of the preferred reaction coordinate distance for a dissociative transition state, which is the optimal distance between the entering oxygen and the attacked phosphorus, should be 4.5–6 Å(Mildvan, 1997). The tetraphosphate compound is in this range.
Another bisubstrate analog approach that has been applied to several kinases employs a thioacetyl-bridge and replaces the entering oxygen with a nitrogen atom (Cheng et al., 2006; Levinson et al., 2006; Parang et al., 2001; Zhang, Gureasko, Shen, Cole, & Kuriyan, 2006). This spacer places the nitrogen and ATP gamma phosphorus about 5 A ° apart (Fig. 1.1B). This design has produced some very potent kinase inhibitors, some with Kis in the low nanomolar range. Thus, it appears that bisubstrate analogs with geometries matching the Mildvan reaction coordinate model for a dissociative mechanism are most effective.
4. APPLICATIONS OF MECHANISTIC STUDIES IN UNDERSTANDING KINASE FUNCTION AND REGULATION
4.1. Bisubstrate analogs
As discussed, peptide–ATP conjugates inspired by a dissociative transition state have been developed for several protein kinases. A principal motivation for developing these analogs is for their use in structural biology studies to clarify the basis of protein substrate recognition by kinases. The field is still lacking a comprehensive understanding of how particular protein substrates are selected by kinases. Various bioinformatic and peptide library approaches have been applied to deduce specificity but they have enjoyed a limited impact. Despite the thousands of X-ray crystal structures of protein kinases, there are only a small number with kinases complexed with protein or peptide substrates. As a result, for the vast majority of kinases, we have little insight into the molecular basis of substrate recognition. The difficulty in obtaining kinase–substrate crystal complexes is thought to emanate from the relatively low affinity of most kinase–substrate binding interactions. From an efficiency perspective, it is logical that enzyme–substrate complexes not be too tight since this could limit catalytic turnover.
Bisubstrate analogs facilitate the anchoring of the peptide moiety to the kinase surface involved in substrate recognition, often allowing for enhanced cocrystallization (Fig. 1.1C). To date, this strategy has been used to capture four kinase–substrate crystal structures: insulin receptor, Abl, EGFR, and cyclin-dependent kinase (Bose, Holbert, Pickin, & Cole, 2006; Levinson et al., 2006; Parang et al., 2001; Zhang et al., 2007). These structures have revealed several of the contacts that give rise to substrate selectivity for these enzymes.
In the case of the Abl and EGFR tyrosine kinases, these cocrystal structures provided helpful clues to activity regulation. One of the pharmacologically important features of Abl is its propensity to populate a conformation which can bind tightly to the drug gleevec. This conformation, known as DFG (Asp–Phe–Gly) out, leads to inactivation of the kinase. A crystal structure of the Abl-kinase domain in complex with the peptide–ATP bisubstrate analog captured Abl in a novel conformation, a catalytically inactive state reminiscent of the downregulated Src tyrosine kinase (Levinson et al., 2006). Based on this unexpected structure, a new framework was proposed for how kinases shuttle between various activation states.
The cocrystal structure of EGFR kinase domain complexed with a peptide–ATP bisubstrate analog showed EGFR kinase in its likely active conformational state (Zhang et al., 2006). This structure also showed high-quality electron density for an apparent asymmetric dimer between two neighboring EGFR kinase molecules. The dimeric interactions are somewhat akin to how a cyclin binds to a cyclin-dependent kinase where it allosterically activates the enzyme. Using a combination of mutagenesis and vesicle-binding studies, it was revealed that the asymmetric EGFR kinase dimer is critical for the mechanisms of EGF ligand-induced activation of EGFR.
4.2. Oncogenic kinase mutants
Enzyme mechanism studies on kinases have also been pursued to understand how mutations can stimulate kinase activity and altered drug sensitivity. A battery of structural, chemical, and kinetic experiments has been channeled to address the functional effects of mutation. Two protein kinases that have received considerable attention in this regard are EGFR tyrosine kinase and B-RAF Ser/Thr kinase, and the experimental progress in these areas is highlighted later.
EGFR mutation has been shown to drive oncogenesis, with L858R point mutation and Δ(746–750) deletion the most prevalent EGFR mutations in nonsmall cell lung cancer (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004). Patients with tumors carrying these mutations are particularly responsive to small-molecule tyrosine kinase inhibitors erlotinib and gefitinib, which bind the ATP pocket. The ATP site-targeting drug lapatinib inhibits WT EGFR and Her2/Neu and is approved for the treatment of breast cancer (Cameron & Stein, 2008; Wood et al., 2004). In vitro studies on isolated EGFR kinase domains showed that the L858R EGFR kinase domain is about ~50-fold more active relative to the wild-type EGFR kinase domain (Yun et al., 2007; Zhang et al., 2006). In addition, L858R EGFR kinase domain has a dramatically increased ATP Km for ATP and a lower gefitinib IC50 relative to the corresponding parameters for wild type. More recent experiments on near full-length EGFRs (tEGFRs), which lack part of the C-terminal tail, investigated the mechanistic basis for oncogenic activation by mutation (Qiu et al., 2009; Wang et al., 2011). These tEGFR studies revealed that L858R and Δ (746–750) are about as active as wild type in the presence of EGF, but retained their full activity when EGF was replaced by the competitive antibody antagonist, cetuximab, which blocks EGF from binding to the EGFR and prevents ectodomain dimerization. In contrast, wild-type tEGFR in complex with cetuximab showed <1% activity relative to its EGF-bound form.
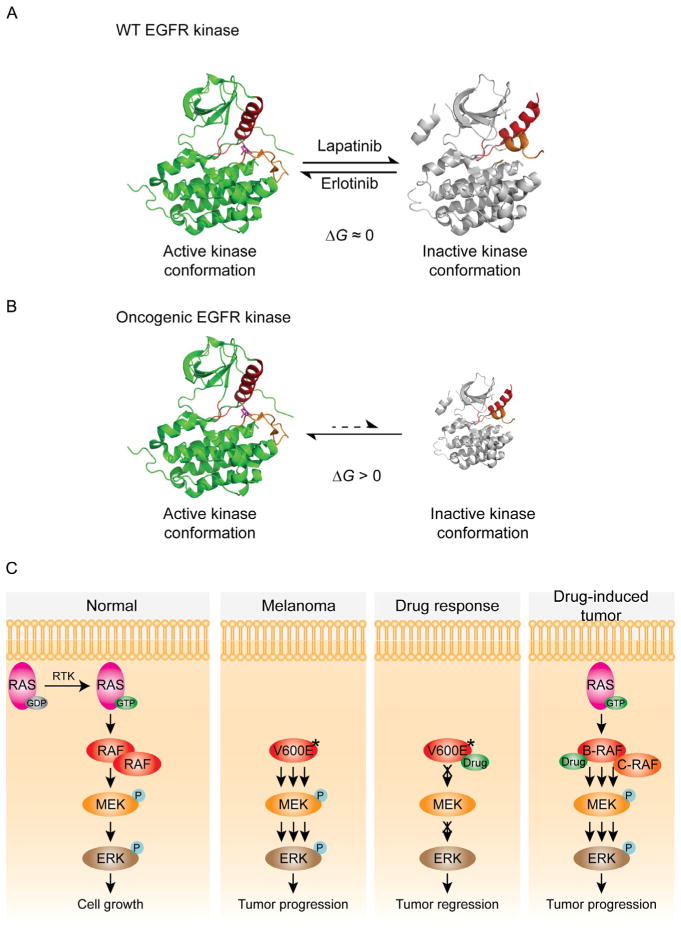
Interestingly, in the presence of EGF, wild-type and oncogenic mutant tEGFRs were equally susceptible to strong erlotinib inhibition but the oncogenic mutant tEGFRs were quite resistant to lapatinib (Wang et al., 2011). Wild-type tEGFR was about as potently inhibited by lapatinib whether in complex with EGF or cetuximab (Wang et al., 2011). Since lapatinib is thought to have a strong binding preference for the inactive conformation of EGFR (Wood et al., 2004), while erlotinib selectively binds the active conformation of EGFR (Stamos, Sliwkowski, & Eigenbrot, 2002), this was puzzling initially. However, a model to explain this data hypothesizes that the active conformation of wild-type EGF/tEGFR complex is roughly isoenergetic with the inactive kinase conformation (Fig. 1.2A; Wang et al., 2011). In contrast, the oncogenic tEGFR mutants greatly favor their active over inactive kinase conformations, regardless of the engagement with EGF (Fig. 1.2B). Separate studies involving negative staining electron microscopy and computational analysis are consistent with this model (Mi, Lu, Nishida, Walz, & Springer, 2011; Shan et al., 2012). tEGFR enzyme kinetic experiments also helped to illuminate why both L858R and Δ (746–750) have very high ATP Kms, even though these mutations are somewhat remote from the adenine pocket. It appears that the reduced binding affinity of the oncogenic tEGFRs for the nucleotide per se is only a minor part of the answer. The greater impact appears to be related to a faster chemical (phosphoryl transfer) step with these oncogenic mutants. By increasing the chemical step rate, the Km will be elevated, independent of the dissociation constant of EGFR for ATP.
Figure 1.2.
Oncogenic kinase mutant mechanisms. (A) The crystal structures of active and inactive EGFR kinase conformations are shown in ribbon representation. Two key structural elements within the kinase domain, the activation loop and helix, are colored orange and red. For WT EGF-bound EGFR, the active kinase conformation (green ribbon, PDB code: 1M17) (Stamos et al., 2002) is nearly isoenergetic to the inactive kinase conformation (gray ribbon, PDB code: 1XKK). Erlotinib selectively binds and stabilizes the active conformation; therefore, the active conformation is dominant in the presence of erlotinib. Similarly, the inactive conformation is dominant in the presence of lapatinib (Wood et al., 2004). Erlotinib and lapatinib are both potent inhibitors against WT EGFR. (B) Oncogenic mutations alter the energy landscape to favor the active conformation. As a result, lapatinib has little access to the majority of oncogenic EGFR mutants, which are locked into the active conformation. The size of each structure has been schematized to depict the relative population of active and inactive conformations. (C) In normal cells, activated RAS recruits B-RAF from the cytosol to the plasma membrane for dimerization and activation. Once activated, B-RAF phosphorylates MEK, which in turn phosphorylates and activates ERK. Oncogenic forms of B-RAF (marked with V600E*) highly activate the ERK pathway in a RAS-independent manner. In B-RAF mutant cancer cells, such as melanoma, B-RAF inhibitor efficiently blocks ERK activation. In RAS mutant cancer cells, the same ATP-competitive inhibitor unexpectedly activates the ERK pathway by promoting RAF dimerization.
In principle, oncogenic mutation could drive activation in a dimer-dependent or dimer-independent fashion. The tEGFR oncogenic mutants were resistant to the protein inhibitor Mig6, which blocks dimerization by coating the EGFR kinase domain C-lobe surface (Zhang et al., 2007). These findings suggested that oncogenic kinase domain dimerization could be dispensable for activation. Alternatively, resistance to Mig6 could suggest that oncogenic kinase domain asymmetric dimerization could be very tight. The fact that Mig6 is a potent inhibitor of the isolated L858R EGFR kinase domain, whereas tEGFR L858R is resistant to Mig6 suggests that the isolated kinase domain could be misleading as a model system compared with the full-length EGFR (Wang et al., 2011). In fact, N-lobe and C-lobe mutations that disrupt the asymmetric kinase dimer interface caused steep reductions in kinase activity for both L858R and Δ(746–750) tEGFRs (Wang et al., 2011). These experiments establish that oncogenic mutants drive activation through the asymmetric kinase dimer, and that the dimer is likely too tight to be effectively blocked by Mig6.
The most frequently mutated protein Ser/Thr kinase in human cancer, B-RAF, has been studied intensively over the past decade (Dhomen & Marais, 2007; Greenman et al., 2007; Wellbrock, Karasarides, & Marais, 2004). B-RAF and its homologs, A-RAF and C-RAF (also called Raf-1), as well as a related kinase KSR1 are involved in the RAS/RAF/MEK/ERK signaling cascade which is activated by growth factors, hormones, and cytokines (Matallanas et al., 2011; Mercer & Pritchard, 2003; Roskoski, 2010; Stephens et al., 1992; Fig. 1.2C). B-RAF mutations have been found in 7% of human cancers and in 66% of malignant melanomas, among which the hyperactivating mutation V600E is the predominant form (Davies et al., 2002). Oncogenic B-RAF mutations are within the catalytic domain, and most enhance B-RAF’s ability to phosphorylate MEK, the only well-established substrate of B-RAF (Wan et al., 2004). Cell-based assays showed that the presence of V600E B-RAF results in constitutive ERK activation (Davies et al., 2002). Whereas wild-type B-RAF seems to require dimerization of the kinase domain for activation, a series of elegant cellular experiments suggest that the “gain-of-function” of V600E B-RAF is independent of RAS and dimerization (Davies et al., 2002; Freeman, Ritt, & Morrison, 2013; Poulikakos et al., 2011). The crystal structures of the catalytic domains of WT and V600E B-RAFs verified that replacing the hydrophobic Val600 with a charged residue destabilizes the interactions that maintain the DFG motif in an inactive conformation and favors conversion of B-RAF into an active, closed conformation (Wan et al., 2004). This “gain-of-function” mutation presumably mimics activation loop phosphorylation.
In addition to mutation affording enhanced kinase activity, several studies also suggest that protein–protein interaction functions of B-RAF may confer cell transformation. Some oncogenic B-RAF mutants have decreased or undetectable kinase activity but retain the ability to activate the ERK pathway (Dhomen & Marais, 2007). These mutations have been suggested to stimulate C-RAF activity through the formation of B-RAF/C-RAF heterodimers (Garnett, Rana, Paterson, Barford, & Marais, 2005; Moretti et al., 2009; Rushworth, Hindley, O’Neill, & Kolch, 2006). Various RAF inhibitors have been developed and are in different stages of preclinical and clinical developments. Treatment with vemurafenib (PLX-4032), a small-molecule B-RAF inhibitor, was shown to be beneficial in melanoma patients harboring the V600E B-RAF mutant and recently was approved for the treatment of metastatic melanoma (Bollag et al., 2010; Flaherty et al., 2010; Joseph et al., 2010; Yang et al., 2010). Unexpectedly, B-RAF inhibitors caused adverse effects in tumor cells carrying wild-type B-RAF. Follow-up studies demonstrated that inhibitor binding to RAF promotes formation of dimeric RAF complexes and activation of the ERK pathway (Hatzivassiliou et al., 2010; Heidorn et al., 2010; Poulikakos, Zhang, Bollag, Shokat, & Rosen, 2010). It remains unresolved precisely how various drug resistance mutants evade kinase inhibition and whether RAF dimerization is thermodynamically coupled to activation loop phosphorylation.
4.3. Chemical rescue of tyrosine kinases
A crystal structure of the IRK domain in its active conformation in complex with a peptide substrate and ATP analog revealed a well-defined triangle of hydrogen bonds with vertices involving the side chains of the catalytic base Asp, the catalytic loop Arg, and the substrate phenol (Parang et al., 2001). Site-directed mutagenesis on this Arg in several tyrosine kinases showed that it is critical for activity, with 100-fold or greater rate reductions associated with its mutation to Ala (Muratore et al., 2009; Qiao, Molina, Pandey, Zhang, & Cole, 2006; Williams, Wang, & Cole, 2000). It is also interesting that this Arg, while present in the sequences in all canonical tyrosine kinases, is flexibly located in two spots in the catalytic loop, either +2 or +4 upstream of the catalytic base Asp (Manning, Plowman, Hunter, & Sudarsanam, 2002). It has been shown previously with various mutant enzymes that loss of activity can sometimes be rescued by the addition of small molecules that complement the missing residue. To assess the specific role of the Arg side chain in the tyrosine kinase reaction, a series of nitrogen-containing small molecules were studied to see if they could complement the Arg to Ala mutant. In fact, several diamino and triamino compounds could at least partially rescue activity. Interestingly, the small-molecule imidazole, the functional group of the side chain of histidine, is the most efficient rescue agent for the tyrosine kinases Csk, Src, and Abl (Muratore et al., 2009; Qiao et al., 2006; Williams et al., 2000). A combination of pH-rate measurements and structure–activity relationship studies indicated that it is the imidazolium (protonated imidazole) state rather than the neutral imidazole form that is necessary for chemical rescue. The positively charged imidazolium would be predicted to be the stronger hydrogen bond donor and acceptor and a better mimic of the Arg guanidinium, which has a higher pKa than imidazole.
It is noteworthy that replacement of the conserved catalytic Arg by His fails to confer enzymatic activity, despite the fact that imidazole is more efficacious in chemical rescue than guanidinium, the functional group of the Arg side chain (Muratore et al., 2009). It is hypothesized that the histidine side chain is simply too short to extend into hydrogen-bonding position when located at the Arg position. Nevertheless, X-ray crystallographic studies of the Src Arg/Ala mutant reveal that a distantly located histidine side chain from an adjacent molecule in the crystal lattice can occupy the cavity associated with Arg removal (Muratore et al., 2009).
Interestingly, where investigated, the chemical rescue of Arg/Ala tyro-sine kinases preserves the natural substrate specificity and regulation of the enzymatic activity. Because imidazole is relatively nontoxic (at less than 30 mM) and is cell permeable (Iguchi, Usui, Ishida, & Hirano, 2002), it was tested in cells stably expressing tyrosine kinases bearing the Arg/Ala replacements in place of the natural wild-type enzymes. The most complete studies have been carried out analyzing Arg/Ala cellular Src (c-Src) and v-Src in mouse embryonic fibroblasts lacking the three major Src tyrosine kinases, Src, Yes, and Fyn (Ferrando et al., 2012; Qiao et al., 2006). c-Src is the proto-oncoprotein that, after passage in Rous sarcoma virus, can become v-Src and thereby induce sarcoma in chickens. v-Src can no longer be downregulated by C-terminal phosphorylation and is a hyperactive tyrosine kinase. Chemical rescue studies of Arg/Ala c-Src and v-Src with imidazole treatment showed that both c-Src and v-Src Arg/Ala mutants could have their kinase activity robustly restored with chemical rescue (Ferrando et al., 2012; Qiao et al., 2006). The findings with c-Src complementation were somewhat unexpected because, in the absence of cell stimulation by growth factors, the basal activity of c-Src was believed to be very low. It must be remembered that cellular protein tyrosine phosphatase activity is robust and so at baseline, probably achieves an equilibrium with protein tyrosine kinase activity. Chemical rescue of the mutant kinase unmasks acutely this substantial enzymatic protein phosphorylation.
Chemical rescue in combination with mass spectrometry has been used to characterize previously unrecognized functions and targets of Src-mediated phosphorylation. Using stable isotope labeling in cell culture and mass spectrometry, chemical rescue can identify phosphorylation events kinetically linked to a specific nonreceptor kinase, bypassing the need for growth factor stimulation. In particular, chemical rescue of Arg/Ala c-Src has been used to suggest a role for Src-mediated tyrosine phosphorylation of the Rap1 guanine nucleotide exchange factor, C3G, in cellular adhesion regulation (Ferrando et al., 2012). In this fashion, chemical rescue is highly complementary to other powerful chemical genetic approaches that allow for specific inhibition of wild type or mutant kinases.
5. SUMMARY AND OUTLOOK
Advances in our understanding of the enzymology of protein kinases have led to new insights into their catalytic mechanisms in terms of kinetic steps, transition-state stabilization, substrate specificity, and basis of regulation. Yet, much of the effort in the analysis of kinases has centered, understandably, on a small proportion of the kinome that have been identified as important in biology and diseases. Hundreds of kinases have not yet been analyzed in enzymologic depth. In addition, there has been a limited work in exploring kinase mechanisms in the larger protein complexes in which they are often found or bearing the rich tapestry of posttranslational modifications (PTMs) that adorn many of them. Regarding the influence of PTMs’, emerging methods to introduce these PTMs site specifically into proteins using unnatural amino acid mutagenesis (Park et al., 2011; Wang, Xie, & Schultz, 2006) or expressed protein ligation (Muir, Sondhi, & Cole, 1998) offers the potential to dissect their effects in greater detail. Thus, despite the major progress that has been made in the kinase field, there is far more work to do before a comprehensive understanding of these fascinating signaling enzymes is achieved.
References
- Ablooglu AJ, Frankel M, Rusinova E, Ross JB, Kohanski RA. Multiple activation loop conformations and their regulatory properties in the insulin receptor’s kinase domain. Journal of Biological Chemistry. 2001;276:46933–46940. doi: 10.1074/jbc.M107236200. [DOI] [PubMed] [Google Scholar]
- Ablooglu AJ, Till JH, Kim K, Parang K, Cole PA, Hubbard SR, et al. Probing the catalytic mechanism of the insulin receptor kinase with a tetrafluorotyrosine-containing peptide substrate. Journal of Biological Chemistry. 2000;275:30394–30398. doi: 10.1074/jbc.M003524200. [DOI] [PubMed] [Google Scholar]
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chemical Reviews. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- Adams JA, Taylor SS. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry. 1992;31:8516–8522. doi: 10.1021/bi00151a019. [DOI] [PubMed] [Google Scholar]
- Admiraal SJ, Herschlag D. Mapping the transition state for ATP hydrolysis: Implications for enzymatic catalysis. Chemistry & Biology. 1995;2:729–739. doi: 10.1016/1074-5521(95)90101-9. [DOI] [PubMed] [Google Scholar]
- Admiraal SJ, Schneider B, Meyer P, Janin J, Veron M, Deville-Bonne D, et al. Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase. Biochemistry. 1999;38:4701–4711. doi: 10.1021/bi9827565. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal affinity purification of direct kinase substrates. Journal of the American Chemical Society. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, et al. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R, Holbert MA, Pickin KA, Cole PA. Protein tyrosine kinase–substrate interactions. Current Opinion in Structural Biology. 2006;16:668–675. doi: 10.1016/j.sbi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Cameron DA, Stein S. Drug Insight: Intracellular inhibitors of HER2-clinical development of lapatinib in breast cancer. Nature Clinical Practice Oncology. 2008;5:512–520. doi: 10.1038/ncponc1156. [DOI] [PubMed] [Google Scholar]
- Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, et al. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. Journal of Biological Chemistry. 2006;281:23167–23179. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhang Y, McCammon JA. How does the cAMP-dependent protein kinase catalyze the phosphorylation reaction: An ab initio QM/MM study. Journal of the American Chemical Society. 2005;127:1553–1562. doi: 10.1021/ja0464084. [DOI] [PubMed] [Google Scholar]
- Cole PA, Burn P, Takacs B, Walsh CT. Evaluation of the catalytic mechanism of recombinant human Csk (C-terminal Src kinase) using nucleotide analogs and viscosity effects. Journal of Biological Chemistry. 1994;269:30880–30887. [PubMed] [Google Scholar]
- Cole PA, Grace MR, Phillips RS, Burn P, Walsh CT. The role of the catalytic base in the protein tyrosine kinase Csk. Journal of Biological Chemistry. 1995;270:22105–22108. doi: 10.1074/jbc.270.38.22105. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Current Opinion in Genetics & Development. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Ferrando IM, Chaerkady R, Zhong J, Molina H, Jacob HK, Herbst-Robinson K, et al. Identification of targets of c-Src tyrosine kinase by chemical complementation and phosphoproteomics. Molecular and Cellular Proteomics. 2012;11:355–369. doi: 10.1074/mcp.M111.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Sagar V, Schafer T, Diop M, Wesseling AM, Lu H, et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nature Structural and Molecular Biology. 2012;19:1316–1323. doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New England Journal of Medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Molecular Cell. 2013;49:751–758. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Molecular Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grace MR, Walsh CT, Cole PA. Divalent ion effects and insights into the catalytic mechanism of protein tyrosine kinase Csk. Biochemistry. 1997;36:1874–1881. doi: 10.1021/bi962138t. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: Mechanisms of activation and signaling. Current Opinion in Cell Biology. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Iguchi K, Usui S, Ishida R, Hirano K. Imidazole-induced cell death, associated with intracellular acidification, caspase-3 activation, DFF-45 cleavage, but not oligonucleosomal DNA fragmentation. Apoptosis. 2002;7:519–525. doi: 10.1023/a:1020691026578. [DOI] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Cole PA. Measurement of a Brønsted nucleophile coefficient and insights into the transition state for a protein tyrosine kinase. Journal of the American Chemical Society. 1997;119:11096–11097. [Google Scholar]
- Kim K, Cole PA. Kinetic analysis of a protein tyrosine kinase reaction transition state in the forward and reverse directions. Journal of the American Chemical Society. 1998;120:6851–6858. [Google Scholar]
- Kirby AJJ, Jencks WP. Reactivity of nucleophilic reagents toward p-nitrophenyl phosphate dianion. Journal of the American Chemical Society. 1965;87:3209. [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- Knowles JR. Enzyme-catalyzed phosphoryl transfer reactions. Annual Review of Biochemistry. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs EG, Beavo JA. Phosphorylation–dephosphorylation of enzymes. Annual Review of Biochemistry. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biology. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGrasso PV, Frantz B, Rolando AM, O’Keefe SJ, Hermes JD, O’Neill EA. Kinetic mechanism for p38 MAP kinase. Biochemistry. 1997;36:10422–10427. doi: 10.1021/bi9706778. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. New England Journal of Medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Madhusudan S, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nature Structural Biology. 2002;9:273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends in Biochemical Sciences. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, et al. Raf family kinases: Old dogs have learned new tricks. Genes & Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky D, Chen SL, Kenyon GL, Gibson BW. Solid-phase synthesis of adenosine phosphopeptides as potential bisubstrate inhibitors of protein kinases. Journal of the American Chemical Society. 1994;116:9413–9419. [Google Scholar]
- Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochimica et Biophysica Acta. 2003;1653:25–40. doi: 10.1016/s0304-419x(03)00016-7. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Lu C, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nature Structural & Molecular Biology. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan AS. Mechanisms of signaling and related enzymes. Proteins. 1997;29:401–416. [PubMed] [Google Scholar]
- Moretti S, De Falco V, Tamburrino A, Barbi F, Tavano M, Avenia N, et al. Insights into the molecular function of the inactivating mutations of B-Raf involving the DFG motif. Biochimica et Biophysica Acta. 2009;1793:1634–1645. doi: 10.1016/j.bbamcr.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Sudhof TC, et al. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore KE, Seeliger MA, Wang Z, Fomina D, Neiswinger J, Havranek JJ, et al. Comparative analysis of mutant tyrosine kinase chemical rescue. Biochemistry. 2009;48:3378–3386. doi: 10.1021/bi900057g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: Insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Mechanism-based design of a protein kinase inhibitor. Nature Structural Biology. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicena P, Miller WT. Processive phosphorylation of p130Cas by Src depends on SH3-polyproline interactions. Journal of Biological Chemistry. 2001;276:28190–28196. doi: 10.1074/jbc.M100055200. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF (V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar R, Cook PF. pH dependence of the kinetic mechanism of the adenosine 3′,5′-monophosphate dependent protein kinase catalytic subunit in the direction of magnesium adenosine 5′-diphosphate phosphorylation. Biochemistry. 1993;32:6802–6806. doi: 10.1021/bi00077a035. [DOI] [PubMed] [Google Scholar]
- Qamar R, Yoon MY, Cook PF. Kinetic mechanism of the adenosine 3′,5′-monophosphate dependent protein kinase catalytic subunit in the direction of magnesium adenosine 5′-diphosphate phosphorylation. Biochemistry. 1992;31:9986–9992. doi: 10.1021/bi00156a018. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Boronina T, Longo PA, Kavran JM, Cole RN, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rominger CM, Schaber MD, Yang J, Gontarek RR, Weaver KL, Broderick T, et al. An intrinsic ATPase activity of phospho-MEK-1 uncoupled from downstream ERK phosphorylation. Archives of Biochemistry and Biophysics. 2007;464:130–137. doi: 10.1016/j.abb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr RAF protein-serine/threonine kinases: Structure and regulation. Biochemical and Biophysical Research Communications. 2010;399:313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Molecular and Cellular Biology. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel IH. Enzyme kinetics. New York: Wiley; 1993. [Google Scholar]
- Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Peptide and protein phosphorylation by protein tyrosine kinase Csk: Insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. Journal of Biological Chemistry. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Stephens RM, Sithanandam G, Copeland TD, Kaplan DR, Rapp UR, Morrison DK. 95-Kilodalton B-Raf serine/threonine kinase: Identification of the protein and its major autophosphorylation site. Molecular and Cellular Biology. 1992;12:3733–3742. doi: 10.1128/mcb.12.9.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska AE, Dalby KN. Kinetic mechanism for p38 MAP kinase alpha. A partial rapid-equilibrium random-order ternary-complex mechanism for the phos-phorylation of a protein substrate. FEBS Journal. 2005;272:4631–4645. doi: 10.1111/j.1742-4658.2005.04827.x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends in Biochemical Sciences. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. Journal of Physical Chemistry B. 2007;111:13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF–ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nature Structural and Molecular Biology. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Annual Review of Biophysics and Biomolecular Structure. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nature Reviews Molecular Cell Biology. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Williams DM, Cole PA. Proton demand inversion in a mutant protein tyrosine kinase reaction. Journal of the American Chemical Society. 2002;124:5956–5957. doi: 10.1021/ja025993a. [DOI] [PubMed] [Google Scholar]
- Williams DM, Wang D, Cole PA. Chemical rescue of a mutant protein-tyrosine kinase. Journal of Biological Chemistry. 2000;275:38127–38130. doi: 10.1074/jbc.C000606200. [DOI] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Research. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Yang H, Higgins B, Kolinsky K, Packman K, Go Z, Iyer R, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in pre-clinical melanoma models. Cancer Research. 2010;70:5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, et al. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]