The authors compared survival prediction using machine-learning data-mining techniques versus traditional histopathologic analysis in a group of 74 tumors. They looked at 55 imaging variables, extracting the most important ones and comparing these with standard histology. The most important variables were: extent of resection, mass effects, volume of enhancing tumor, maximum T2 intensity, and mean trace intensity in non-enhancing or edematous surrounding areas. The accuracy of the data-mining method was 85% and based on Kaplan-Meier curves this predicted survival better than histology.
Abstract
BACKGROUND AND PURPOSE:
The prediction of prognosis in HGGs is poor in the majority of patients. Our aim was to test whether multivariate prediction models constructed by machine-learning methods provide a more accurate predictor of prognosis in HGGs than histopathologic classification. The prediction of survival was based on DTI and rCBV measurements as an adjunct to conventional imaging.
MATERIALS AND METHODS:
The relationship of survival to 55 variables, including clinical parameters (age, sex), categoric or continuous tumor descriptors (eg, tumor location, extent of resection, multifocality, edema), and imaging characteristics in ROIs, was analyzed in a multivariate fashion by using data-mining techniques. A variable selection method was applied to identify the overall most important variables. The analysis was performed on 74 HGGs (18 anaplastic gliomas WHO grades III/IV and 56 GBMs or gliosarcomas WHO grades IV/IV).
RESULTS:
Five variables were identified as the most significant, including the extent of resection, mass effect, volume of enhancing tumor, maximum B0 intensity, and mean trace intensity in the nonenhancing/edematous region. These variables were used to construct a prediction model based on a J48 classification tree. The average classification accuracy, assessed by cross-validation, was 85.1%. Kaplan-Meier survival curves showed that the constructed prediction model classified malignant gliomas in a manner that better correlates with clinical outcome than standard histopathology.
CONCLUSIONS:
Prediction models based on data-mining algorithms can provide a more accurate predictor of prognosis in malignant gliomas than histopathologic classification alone.
Brain neoplasms are considered one of the most lethal and difficult-to-treat forms of cancer. The prognosis of brain cancer varies on the basis of the type of neoplasm and is poor for the most malignant types, with a median survival of approximately 3 years for anaplastic astrocytoma and 1 year for glioblastoma.1 A better prognosis based on routine studies, including mostly MR imaging, may help treatment planning. Tumor grade, imaging properties, and clinical data have been studied by many groups in relationship to survival.2,3 A study including 34 patients with gliomas showed that rCBV values in astrocytomas are predictive for recurrence and 1-year survival and may be more accurate than histopathologic grading.2 This was not observed for tumors with oligodendroglial components.2
A larger study including 189 HGGs and LGGs showed that patients with a high rCBV have a significantly more rapid time to progression than patients who have gliomas with a low rCBV.4 HGGs have been studied by Mills et al,5 who examined the direct relationship between the contrast transfer coefficient and length of survival in 27 adult patients. CBV related directly to histologic grade but provided no independent prognostic information over and above that provided by grade.5 A different small study6 showed that CBV is a stronger predictor of survival than enhancement. Also it has been shown in 28 patients with GBM that the volume after surgery, but before pre-external beam radiation therapy of the metabolic abnormality, and the nADC value within the T2 hyperintense region may be valuable in predicting outcome.7 On the other hand, a previous study monitoring evolution of 41 GBMs showed that perfusion and diffusion techniques cannot be used to anticipate tumor progression and that classic T1 and T2 imaging remain the criterion standard.8
Besides advanced MR imaging characteristics, clinical characteristics and tumor descriptors were correlated with survival time. A multivariate analysis of 416 patients with GBM3 identified 5 independent predictors of survival: age, Karnofsky Performance Scale score, extent of resection, and the degree of necrosis and enhancement on preoperative MR imaging studies.
In this work, we investigate whether patient characteristics, as well as DTI and rCBV measurements in HGGs, can serve as an adjunct to conventional imaging characteristics in predicting overall survival. Specifically, we combined clinical findings, tumor pathology descriptors, and imaging characteristics from 2 ROIs and explored data-mining techniques9 for assessing total survival time after first-time treatment (resection). Data mining primarily assists in the analysis of large quantities of data, aiming to extract interesting patterns. This has been shown to be helpful in tissue segmentation,10 variable selection,11 and tumor classification.11 In this study, a segmentation method based on multiparametric MR imaging intensities was used for ROI segmentation, requiring less user interaction than most reported studies based on ROI analysis. The combined set of variables was reduced by a variable-selection method, and a classification tree was constructed by using the reduced set of significant variables.
The method was applied in a population of 67 patients with HGGs (histologically diagnosed according to the WHO system) from June 2006 to December 2007. Seven patients had multiple2 tumors, which were considered independent masses, resulting in 74 samples in total (18 anaplastic gliomas grade III, 55 GBMs grade IV, and 1 gliosarcoma grade IV). The robustness and accuracy of the proposed classification scheme were assessed by cross-validation of the data.
Materials and Methods
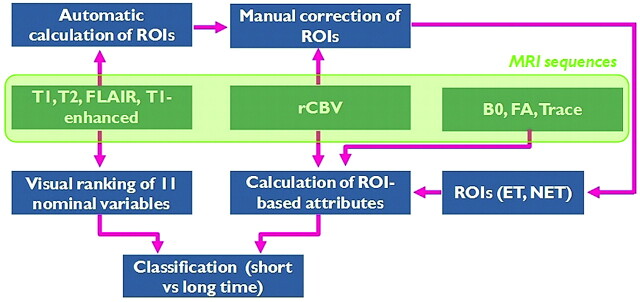
A computer-assisted classification scheme combining conventional MR imaging, perfusion MR imaging, and DTI was developed and used for survival analysis. Data analysis was performed in several stages, as shown in Fig 1. First, the data were preprocessed, and ROIs were semiautomatically segmented. Then, ROI-based analysis was performed to extract imaging characteristics from rCBV and DTI. The imaging characteristics were combined with clinical findings to retrieve a complete set of diagnostic variables describing the pathology. Subsequently, due to the substantial number of variables, data-mining procedures were used to discern relevant patterns of relationships within the dataset and eliminate irrelevant variables, as will be described in “Variable Selection and Classification.” Last, once a final set of variables was found, the datasets were classified by using a decision tree algorithm. The calculation of the imaging characteristics was performed by using Matlab (MathWorks, Natick, Massachusetts), whereas variable selection and classification were implemented in the WEKA platform.12
Fig 1.
Pipeline of the computer-based methodology for prediction of survival (long/short = more/less than 18 months). Conventional MR imaging was used to visually rank 11 variables characterizing the tumor, whereas DTI and rCBV were used to extract ROI-based imaging attributes.
Data
We examined patients with HGGs who underwent preoperative conventional and advanced MR imaging (perfusion, DTI). LGGs were not analyzed due to the small number of cases. The patient demographics are shown in Table 1. Exclusion criteria were any surgery, radiation therapy, or chemotherapy of a brain tumor before inclusion in the study as well as lack of histopathologic diagnoses, missing imaging data (pre- or postoperational), or presence of artifacts. Treatment was not used to exclude patients. All patients were treated under the same protocol. After resection, they received chemotherapy/radiation, including bevacizumab (Temodar). All patients underwent biopsy or surgical resection of the tumor with histopathologic diagnosis based on WHO criteria. Postoperative scans were analyzed to determine the extent of resection. The study was approved by the institutional review board and was compliant with the Health Insurance Portability and Accountability Act.
Table 1:
Patient demographics
| Demographics | Data |
|---|---|
| No. of patients | 67 |
| Mean age (yr) | 56 (25–86) |
| No. of tumors | 74 |
| Pathologic finding | 18 anaplastic gliomas (III/III), 55 GBM (IV/IV), 1 gliosarcoma (IV/IV) |
The patients were imaged by using a 3T MR imaging scanner system (Magnetom Trio Tim; Siemens, Erlangen, Germany) with a 12-channel phased-array coil. The imaging acquisition protocol was the same for all patients and included the following sequences: axial 3D T1-weighted (magnetization prepared rapid acquisition of gradient echo; TR/TE/TI, 1760/3.1/950 ms; matrix size, 192 × 256; pixel spacing, 0.9766 × 0.9766 mm; section thickness, 1 mm); sagittal 3D T2-weighted (matrix size, 256 × 320; pixel spacing, 0.8969 × 0.8969 mm; section thickness, 0.9 mm); FLAIR (TR/TE/TI, 9420/141/2500 ms; matrix size, 192 × 256; pixel spacing, 0.9375 × 0.9375 mm; section thickness, 3 mm); and DTI. Axial 3D contrast-enhanced T1-weighted images were obtained after administration of a standard dose (0.1 mmol/kg) of gadodiamide with a power injector (Medrad, Indianola, Pennsylvania).
Also, after an initial loading dose of 3-mL gadodiamide and a 5-minute delay, T2*-weighted dynamic susceptibility perfusion MR imaging was performed by using a gradient-echo EPI acquisition during bolus injection of 12-mL gadodiamide. Twenty sections were acquired with the following parameters: TR/TE, 2000/45 ms; matrix size, 128 × 128; pixel spacing, 1.7 × 1.7 mm; section thickness, 3.0 mm; flip angle, 90°. DTI data were acquired by using a single-shot spin-echo echo-planar sequence with parallel imaging by using generalized autocalibrating partially parallel acquisition and an acceleration factor of 2. Sequence parameters were the following: TR/TE, 5000/86 ms; NEX, 3; FOV, 22 × 22 cm2; section thickness, 3 mm; matrix size, 128 × 128; 40 sections covering the whole brain with an acquisition time of 8 minutes. The diffusion-weighting gradients were applied in 30 isotropically distributed directions by using a b-value of 1000 s/mm2.
Preprocessing of DTI and Perfusion
DTI provides a powerful imaging tool for macroscopically studying the neuronal organization, by capturing microscopic water diffusion characteristics along different orientations. DTI models these local diffusion characteristics through a 3 × 3 positive definite symmetric matrix, known as a diffusion tensor. A diffusion tensor may equivalently be represented through an orthonormal basis formed by its eigenvectors and the corresponding eigenvalues. Several quantities can be extracted from the eigenvalues and eigenvectors for characterization of tissue structure. In this study, we used the B0 and also calculated, with our own software, the voxelwise FA map and the trace of the diffusion tensor. FA represents the amount of deviation from isotropic diffusion. It assumes values in the range of 0–1, with values close to zero denoting isotropic diffusion and values close to 1 representing highly anisotropic diffusion. Trace is the sum of the eigenvalues and is a more suitable feature when one is interested in the magnitude of water diffusivity at a voxel.
Furthermore, rCBV maps were generated off-line from perfusion MR imaging by calculating the change in relaxation rate by using the equation ΔR2*(t)=−ln(S(t)/S0) / TE, where S(t) is the signal intensity at time t and S0 is the baseline signal intensity, and then by integrating under the ΔR2*(t) curve over time points corresponding to the first pass of the contrast bolus. The B0, FA, trace, and rCBV maps were rigidly registered to the T1 postcontrast image to be in the space of the ROIs.
ROI Segmentation
The DTI and rCBV imaging characteristics were extracted from only 2 ROIs, including the enhancing neoplastic tissue and all hypointense tissue (neoplastic, edematous, and necrotic). The ROI extraction was performed by an automatic segmentation method combining all conventional MR images into a machine-learning method.10 This method segments the brain into 6 tissue types (enhancing neoplastic, nonenhancing neoplastic, edema, white matter, gray matter, and CSF) by using a previously trained segmentation model. The segmentation model was trained by means of support vector machines by using samples drawn by an expert for the tumorous tissue types and automatically extracted by FAST13 (a K-means segmentation algorithm provided by the Functional MRI of the Brain Software Library, http://www.fmrib.ox.ac.uk/fsl/) for the healthy tissue types. Postprocessing was applied to refine the segmentation of the pathologic tissue, while the normal tissue was not used further in this study. Specifically, small isolated tumor or edema clusters, located far from the main tumor bulk, were removed. Also, regions in falx, which are often wrongly segmented as enhancing tumor due to the vivid enhancement they show on the T1 contrast-enhanced images, were corrected. This was performed by warping an atlas with presegmented falx to the subject image. Any region segmented as enhancing tumor overlapping with the warped falx was replaced by CSF. Due to the uncertainty in differentiating nonenhancing neoplastic tissue from edematous tissue, these 2 ROIs were merged into 1.
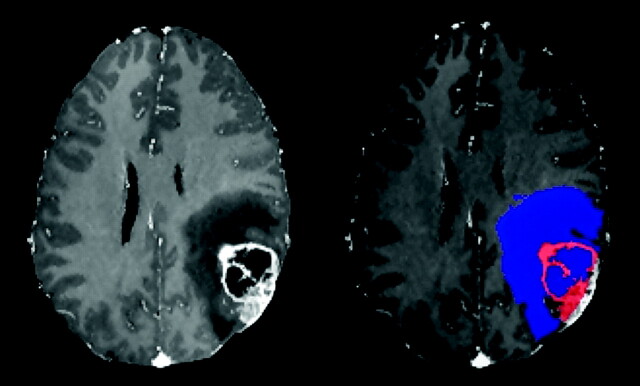
Finally, an expert visually inspected the 2 extracted ROIs, denoted as ET and NET, and manually corrected these wherever necessary by examining the rCBV and DTI maps. The manual correction is rarely needed and does not require a lot of expertise due to the definition of a single ROI for all NET. A segmentation example is shown in Fig 2.
Fig 2.
T1 contrast-enhanced image of a patient with GBM and corresponding ROIs. The enhancing neoplastic tissue (in red) and the nonenhancing/edematous/necrotic tissue (in blue) are segmented as shown on the right.
Variables
We calculated and analyzed the relationship of 55 study variables, which included clinical findings and tumor pathology descriptors obtained by visual inspection of conventional MR imaging and also imaging characteristics calculated from DTI and rCBV maps. Variables such as molecular profile, performance status (eg, Karnofsky Performance Scale score), and therapy-associated factors such as radiation dose or the use of adjuvant therapy, were not considered. Typical variables that have been used by others include localization, mass effect, T1 contrast enhancement, T2, diffusion, and perfusion signal intensity. We have combined such nominal variables ranked by an expert with attributes extracted automatically from ROIs, to achieve a more complete representation. The scoring pattern of the variables selected in this study is shown in Table 2. Specifically, the scoring of edema was visually performed as described next. In case of enhancing tumors, the tumor lesion was extracted from the contrast-enhanced T1-weighted image and subtracted from the abnormal tissue traced on FLAIR (which includes tumor and edema). In case of nonenhancing tumors, edema was traced in T1 by detecting signal-intensity differences between the tumor lesion and edema. The extent of resection was rated on the basis of the postoperative scans. Undefined variables (ie, when some ROIs [ET or NET] did not exist) were marked as missing, whereas the volume of those ROIs was marked as zero. The complete set of variables was used to predict survival.
Table 2:
Scoring pattern of variables
| Patterns | |
|---|---|
| Clinical findings | |
| Age | |
| Sex | |
| Nominal variables ranked by visual inspection of conventional MR imaging | |
| Side | Left, right |
| Site | Superficial (only cortical, not in basal ganglia and outside the insula), deep (involves the insula, thalamus, basal ganglia, or posterior fossa) |
| Multi-focality | 0 (no), |
| 1 (yes) | |
| Functional location | Noneloquent, near-eloquent, eloquent brain |
| Mass effect | 0 (not apparent), |
| 1 (deformity of ventricle, no midline shift), | |
| 2 (moderate, <5 mm), | |
| 3 (significant, >5 mm) | |
| Edema | 0 (not apparent), |
| 1 (<tumor volume), | |
| 2 (≥tumor volume) | |
| Cyst | 0 (no), |
| 1 (yes) | |
| Necrosis | 0 (none), |
| 1 (<25% of tumor volume), | |
| 2 (25%–50% of tumor volume), | |
| 3 (>50% of tumor volume) | |
| Enhancement | 0 (none), |
| 1 (low-to-medium signal intensity), | |
| 2 (medium-to-high signal intensity) | |
| Enhancing margin | 0 (well-defined), |
| 1 (none or poorly defined) | |
| Extent of resection | Based on postoperative scans: |
| 0 (total resection), | |
| 1 (90%–99%), | |
| 2 (20%–89%), | |
| 3 (<20%) | |
| Volumes of ROIs | |
| ET (neoplastic) | |
| NET, including neoplastic, edematous, necrotic, or cystic | |
| Intensity characteristics of DTI (B0, FA, trace) and rCBV over the 2 ROIs (ET, NET) | |
| Mean | |
| Minimum | |
| Maximum | |
| Peak of histogram | |
| Variance |
Definition of Survival Time
For the purpose of this study, the preoperative images acquired at the time of the first operation were considered as the baseline and were used to monitor the evolution of the disease. Overall survival was evaluated from the baseline to death or, for cases that were not followed until death (eg, living patients), from the baseline to the time of last available follow-up. A time threshold of 18 months was defined to differentiate the patients into 2 groups, those with short- or long-term survival. Living patients with last follow-up earlier than the examined period (18 months) were excluded from the study because their vital status at the end of the period was unknown. The study of survival was performed as a binary classification problem rather than a regression problem to include living patients in the dataset, for whom the actual survival time was unknown at the time of the analysis. The threshold of 18 months represents patients with survival that is longer than the median survival time of HGGs and was chosen to divide the patient population into an approximately equal number of cases in both groups (37 cases in each group). Thus, the definition of long-term survival here is different from that in other studies,14–18 where the long-term survivors are those who survive ≥3 years after the initial diagnosis.
Variable Selection and Classification
Variable or feature selection is an important processing step, during which a subset of the most informative variables is chosen and then used for the final analysis. By discarding the redundant variables, one can construct more cost-effective predictors with improved performance.19 We tested several variable selection algorithms and selected the one with overall best classification performance. The selected method searched over the variables following the “scatter search” algorithm20 and defined the predictive value of each subset of variables by using a “wrapper approach.”21 Scatter search operates on a group of subsets of variables, which constitute good solutions, in respect to special criteria such as diversity. The subsets are linearly combined, and a local search procedure is applied to update the initial group and incorporate good solutions. On each subset of variables, the wrapper builds a classifier by applying an induction learning algorithm. The variables subset with the highest classification accuracy, estimated by cross-validation, is selected. These steps are repeated until a stopping condition is met.
Classification of the datasets into short- and long-term survivors is performed with a J48 classification tree (http://www.opentox.org/dev/documentation/components/j48).22 J48 produces decision trees from a set of labeled training data by examining the normalized information gain (difference in entropy) that results from choosing a variable for splitting the data into smaller subsets. To make the decision, the variable with the highest normalized information gain is used.
Results
Univariate Statistical Analysis of the Image-Intensity Characteristics
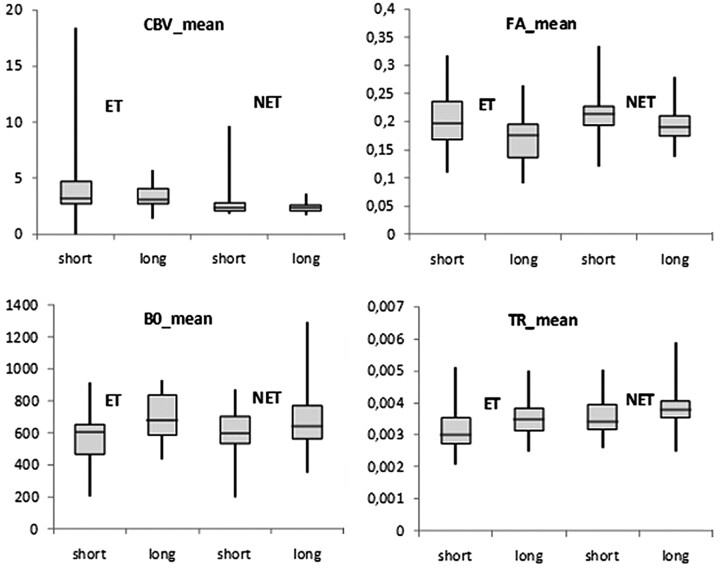
A 2-tailed t test was performed on each image-intensity characteristic in the segmented ROIs between short- and long-term survivors by assuming that the data came from normal distributions with unknown but equal variances. There was no significant difference (at significance level α = .05) in all variables (mean, minimum, maximum, peak of histogram, variance) extracted from the rCBV maps from either of the segmented ROIs (ET and NET). In contrast, the mean B0 and FA intensity values showed significant differences in both regions (P < .01 for ET and P < .04 for NET), whereas the mean trace value was significantly different only in NET. Similarly, the peak of histogram of FA and trace showed significant differences (P < .022 in both regions in FA, and P = .01 in NET in trace), whereas the minimum-intensity value showed marginally significant differences (P = .047) only in NET in FA. The variance and the maximum value did not demonstrate significant differences in any of the images and regions. The region-wise comparison of the mean intensities is shown in Fig 3.
Fig 3.
Boxplots of rCBV, B0, FA, and trace (TR) mean values in HGGs with short and long survival. Boxes represent the 25th and 75th percentiles with the medians indicated by the middle lines in the boxes. Vertical bars (whiskers) indicate the range of data. ET is indicated in the first and second box in each figure. NET includes neoplastic, edematous, necrotic, and cystic tissue (third and fourth box in each figure).
Group separation was not feasible by analyzing the variables independently. In the next section, classification results of the proposed scheme implementing machine-learning tools for variable selection and classification are presented.
Variable Selection and Classification
Multivariate analysis with variable selection was applied to classify the 2 groups of patients. The method was evaluated with leave-3-out cross-validation on the decision tree (ie, 3 samples were left out each time for testing and the rest were used for training). The results for every test set were finally averaged. The average classification accuracy (percentage of correctly classified samples) was 85.1%, and the AUC of the ROC was 0.84. Because variable selection was internal in the cross-validation process, the selected variables and the decision tree varied for each fold. To summarize the results in a single decision tree, we also applied the method on all samples together. In this case (without cross-validation), the variables selected as the most discriminative were the extent of resection, mass effect, volume (in cubic millimeters) of ET, maximum B0 intensity in NET, and mean trace intensity in NET. The classification accuracy on the training set was 90.5%, and the AUC was 0.96. The histograms of these variables for short- and long-term survival did largely overlap; thus, class separation would not have been possible on the basis of univariate analysis. Also, these variables (selected by the scatter search algorithm) differed from the variables that demonstrated the most significant group differences by t test (shown in “Univariate Statistical Analysis of the Image-Intensity Characteristics”).
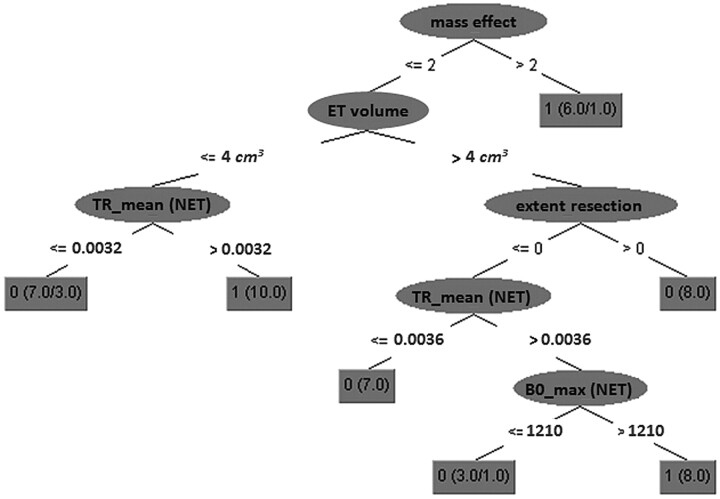
The decision tree constructed by all training samples by using the 5 variables selected by the variable-selection method is visualized in Fig 4. Mass effect is on top of the tree, associating significant displacement with long-term survival (mass effect likely being partially a surrogate for lower tumor diffusivity and infiltration). The rest of the cases exhibit long-term survival if the mean trace intensity in the nonenhancing region is high and either the enhancing part is small and/or it is large preoperationally but is then completely resected and also the maximum B0 intensity in the nonenhancing region is high. Similarly, short survival is associated with the rest of the cases having small or moderate mass effect, low mean trace intensity in the nonenhancing region, and enhancing volume <4 cm3 or enhancing volume >4 cm3 and not completely resected, or completely resected but with low maximum B0 intensity in the nonenhancing region.
Fig 4.
A 5-node decision tree calculated from all training data (no cross-validation). The numbers in each gray box indicate the predicted class label (0, survival time <18 months; 1, survival time >18 months), the total number of samples from both classes satisfying the conditions of each tree path, and the number of misclassified samples (shown only if nonzero). TR indicates trace intensity; BO_max indicates maximum BO intensity.
Kaplan-Meier Curves
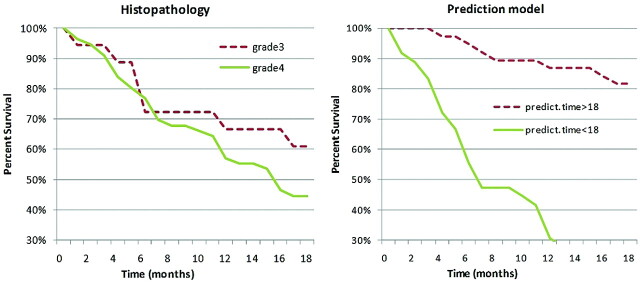
It is well accepted that prognosis depends on histopathology or tumor grade (though recent studies on HGGs suggest that the association might not be significant23). We compared the predictive ability of tumor grade with the power of the model (which is not based on histopathology). The prediction model was applied in a leave-1-out cross-validation scheme by using the 5 variables previously described. The survival curves for each case are shown in Fig 5.
Fig 5.
Kaplan-Meier survival curves of patients with 56 gliomas of grade 4 and 18 anaplastic gliomas of grade 3. Survival curves were plotted according to classifications based on either traditional histopathology (dashed line, grade 3; solid line, grade 4) or the class prediction model (dashed line, predicted survival time >18 months; solid line, predicted survival time <18 months).
When tumors were classified according to histopathology (grade 3 versus grade 4), the survival of patients was not significantly different (P = .17), whereas class distinctions according to the prediction model were significantly associated with survival outcome (P < 0.00001).
Discussion
In this study, we examined whether prediction models based on machine-learning algorithms provide a more accurate predictor of prognosis in malignant gliomas than does histopathologic classification alone. Furthermore, we investigated whether DTI and rCBV measurements in HGGs may serve as an adjunct to conventional imaging in predicting overall survival. The explored variables included clinical data, descriptive tumor characteristics as they appear on conventional MR imaging, and imaging properties in selective regions of advanced MR imaging. The computer-based method consisted of 2 main steps: First, the most informative variables were identified; then, a decision tree algorithm was applied to differentiate short- and long-term survival.
Two ROIs were selected that included the enhancing neoplastic tissue and all hypointense tissue (neoplastic, edematous, and necrotic). The possibly heterogeneous hypointense tissue was examined together into a single ROI to simplify the data-preparation steps and also avoid segmentation errors because differentiation between edema and tumor infiltration is often ambiguous. The assumption is that the tissue heterogeneity will be captured by the extracted statistical characteristics (mean, minimum, maximum, histogram peak, and variance) of the imaging profile (eg, a low minimum value indicates the presence of necrosis).
Five variables were selected as the most informative, providing the highest classification accuracy, namely the volume of enhancing tissue, mass effect, extent of resection, and B0 and trace values in the nonenhancing/edematous ROIs. Thus, the results support our hypothesis that DTI contains information on HGG prognosis similar to its potential in evaluating cellularity and grading of gliomas.24 This information seems to be mostly contained in the nonenhancing or edematous area. The volume of enhancing tissue, as segmented from the T1 contrast-enhanced image, was also an important predictor, as observed in similar studies investigating MR imaging characteristics of brain tumors.3,11,25 The extent of resection was also selected as an important variable and was characterized by a binary decision function, incomplete or complete resection. Similarly in the study of Lacroix et al,3 univariate and multivariate analyses showed that resection of ≥98% of the tumor volume was associated with a significant survival advantage. On the contrary, the extent of resection was not a statistically meaningful predictor of survival in the study of Pope et al.26 The mass effect was placed on top of the decision tree in this study and related to survival in an opposite way than might be expected. Specifically, the data show that most the cases with significant mass effect (>5 mm) were associated with long survival time. A potential reason could be that aggressive gliomas, which usually have the worst prognosis, tend to be more diffuse and infiltrative, thereby causing relatively lower mass effect. This hypothesis, however, remains to be confirmed in future prospective studies. In previous studies,3,23 mass effect did not show any association with survival when multivariate analysis was applied, whereas univariate Cox regression demonstrated statistical significance for a large mass effect (>10 mm).23
ROI-based characteristics in rCBV were not among the top-ranked variables, whereas rCBV statistics have been associated with brain tumor type in previous work11 and with histologic changes such as angiogenesis.27 Direct comparison with such studies, however, should not be performed because tumor classification11 and tumor growth27 are different targets from survival prediction, which was the aim in this work.
Conclusions
Kaplan-Meier survival curves demonstrated that the constructed prediction model provides a more accurate predictor of prognosis in high-grade gliomas than pathologic classification. Further investigation with a larger patient population and histologic validation will be necessary to determine the robustness of the selected variables in predicting survival and to increase understanding of the morphologic and functional characteristics of brain tumors. In the future we would also like to include in the analysis therapy-associated factors, such as radiation dose or adjuvant chemotherapy, as well as genotypic information because it has been shown to be an important factor for clinical outcome in certain brain tumor types.28–30
ABBREVIATIONS:
- AUC
area under the curve
- B0
baseline (T2-weighted) image
- ET
enhancing tissue
- FA
fractional anisotropy
- GBM
glioblastoma multiforme
- HGG
high-grade glioma
- LGG
low-grade glioma
- NET
nonenhancing tissue
- rCBV
relative cerebral blood volume
- ROC
receiver operating characteristic analysis
- WHO
World Health Organization
Footnotes
The work was supported in part by R01NS042645.
Paper previously presented in part at: 33rd International Conference of the Institute of Electrical and Electronics Engineers Engineering in Medicine and Biology Society, August 30–September 3, 2011; Boston, Massachusetts.
Disclosures: Donald O'Rourke—UNRELATED: Board Membership: American Association for Cancer Research (Editorial Board), American Journal of Neuroradiology (former Editorial Board member), Consultancy: Celldex Therapeutics, Comments: EGFRvIII vaccine company focused on treatment of malignant gliomas in phase II/III clinical trials, Grants/Grants Pending: National Institutes of Health,* Patents: US patent on combination ErbB inhibition and radiation,* Comments: US patent issued EPO patent, pending appeal; No direct relationship to study relevant to cancer in general, Royalties: Ception Pharmaceuticals* (due to above patent), Comments: Monies to University of Pennsylvania in licensing, Stock/Stock Options: Celldex Therapeutics, Comments: see above concerning consultancy. *Money paid to the institution.
References
- 1. DeAngelis LM. Brain tumors. N Engl J Med 2001;344:114–23 [DOI] [PubMed] [Google Scholar]
- 2. Bisdas S, Kirkpatrick M, Giglio P, et al. Cerebral blood volume measurements by perfusion-weighted MR imaging in gliomas: ready for prime time in predicting short-term outcome and recurrent disease? AJNR Am J Neuroradiol 2009;30:681–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg 2001;95:190–98 [DOI] [PubMed] [Google Scholar]
- 4. Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247:490–98. Epub 2008 Mar 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mills SJ, Patankar TA, Haroon HA, et al. Do cerebral blood volume and contrast transfer coefficient predict prognosis in human glioma? AJNR Am J Neuroradiol 2006;27:853–58 [PMC free article] [PubMed] [Google Scholar]
- 6. Lev MH, Ozsunar Y, Henson JW, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas. AJNR Am J Neuroradiol. 2004;25:214–21 [PMC free article] [PubMed] [Google Scholar]
- 7. Oh J, Henry RG, Pirzkall A, et al. Survival analysis in patients with glioblastoma multiforme: predictive value of choline-to-N-acetylaspartate index, apparent diffusion coefficient, and relative cerebral blood volume. J Magn Reson Imaging 2004;19:546–54 [DOI] [PubMed] [Google Scholar]
- 8. Leimgruber A, Ostermann S, Yeon EJ, et al. Perfusion and diffusion MRI of glioblastoma progression in a four-year prospective temozolomide clinical trial. Int J Radiat Oncol Biol Phys 2006;64:869–75. Epub 2005 Oct 13 [DOI] [PubMed] [Google Scholar]
- 9. Kantardzic M. Data Mining: Concepts, Models, Methods, and Algorithms. John Wiley & Sons; 2003. [Google Scholar]
- 10. Verma R, Zacharaki EI, Ou Y, et al. Multi-parametric tissue characterization of brain neoplasms and their recurrence using pattern classification of MR images. Acad Radiol 2008;15:966–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zacharaki EI, Wang S, Chawla S, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009;62:1609–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11:10–18. [Google Scholar]
- 13. Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans Med Imaging 2001;20:45–57 [DOI] [PubMed] [Google Scholar]
- 14. Scott JN, Rewcastle NB, Brasher PM, et al. Which glioblastoma multiforme patient will become a long-term survivor? A population-based study. Ann Neurol 1999;46:183–88 [PubMed] [Google Scholar]
- 15. Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004;64:6892–99 [DOI] [PubMed] [Google Scholar]
- 16. Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol 2008;10:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sonoda Y, Kumabe T, Watanabe M, et al. Long-term survivors of glioblastoma: clinical features and molecular analysis. Acta Neurochir (Wien) 2009;151:1349–58 [DOI] [PubMed] [Google Scholar]
- 18. Glas M, Happold C, Rieger J, et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. J Clin Oncol 2009;27:1257–61 [DOI] [PubMed] [Google Scholar]
- 19. Guyon I, Elisseeff A. An introduction to variable and feature selection. Journal of Machine Learning Research 2003;3:1157–82 [Google Scholar]
- 20. Laguna M, Mart R. Scatter Search: Methodology and Implementations in C. Norwell, Massachusetts: Kluwer Academic Press; 2003 [Google Scholar]
- 21. Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence 1997;97:273–324 [Google Scholar]
- 22. Gandhi GM, Srivatsa SK. Adaptive machine learning algorithm (AMLA) using J48 classifier for an NIDS environment. Advances in Computational Sciences and Technology 2010;3:291–304 [Google Scholar]
- 23. Ramnarayan R, Dodd S, Das K, et al. Overall survival in patients with malignant glioma may be significantly longer with tumors located in deep grey matter. J Neurol Sci 2007;260:49–56. Epub 2007 May 1 [DOI] [PubMed] [Google Scholar]
- 24. Sugahara T, Korogi Y, Kochi M, et al. Usefulness of diffusion weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging 1999;9:53–60 [DOI] [PubMed] [Google Scholar]
- 25. Al-Okaili RN, Krejza J, Woo JH, et al. Intra-axial brain masses: MR imaging-based diagnostic strategy—initial experience. Radiology 2007;243:539–50 [DOI] [PubMed] [Google Scholar]
- 26. Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol 2005;26:2466–74 [PMC free article] [PubMed] [Google Scholar]
- 27. Principi M, Italiani M, Guiducci A, et al. Perfusion MRI in the evaluation of the relationship between tumour growth, necrosis, and angiogenesis in glioblastomas and grade 1 meningiomas. Neuroradiology 2003;45:205–11. Epub 2003 Mar 5 [DOI] [PubMed] [Google Scholar]
- 28. Sundgren PC, Junck L, Meyer CR, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol 2010;28:2293–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanner AA, Staugaitis SM, Castilla EA, et al. The impact of genotype on outcome in oligodendroglioma: validation of the loss of chromosome arm 1p as an important factor in clinical decision making. J Neurosurg 2006;104:542–50 [DOI] [PubMed] [Google Scholar]
- 30. Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998;90:1473–79 [DOI] [PubMed] [Google Scholar]