Summary
Genetically encoded effectors are important tools for probing cellular function in living animals, but improved methods for directing their expression to specific cell types are required. Here we introduce a simple, versatile method for achieving cell type-specific expression of transgenes that leverages the untapped potential of “coding introns” (i.e. introns between coding exons). Our method couples the expression of a transgene to that of a native gene expressed in the cells of interest using intronically inserted “plug-and-play” cassettes (called “Trojan exons”) that carry a splice acceptor site followed by the coding sequences of T2A peptide and an effector transgene. We demonstrate the efficacy of this approach in Drosophila using lines containing suitable MiMIC transposons and a palette of Trojan exons capable of expressing a range of commonly used transcription factors. We also introduce an exchangeable, MiMIC-like Trojan exon construct that can be targeted to coding introns using the Crispr/Cas system.
Introduction
Genetically-based tools for perturbing cellular function are increasingly used to study the contributions of different cell types to development, physiology, and behavior. The utility of these tools depends critically on the cell-type specificity of their expression, and there is considerable demand for targeting methods with greater selectivity than is currently available. In general, selectivity of expression is achieved by using DNA regulatory elements of one or more genes normally expressed by a cell type of interest to drive the expression of a primary transgene, such as Gal4 or Cre; the primary transgene can then activate the expression of secondary transgenes that mediate functional perturbations (Branda and Dymecki, 2004; Venken et al., 2011b; Yizhar et al., 2011).
Co-opting a gene’s full complement of regulatory elements to faithfully target all the cells that express it has been best achieved by inserting a transgene coding sequence into one of its translated exons (Demir and Dickson, 2005; Diao and White, 2012; Taniguchi et al., 2011). This approach, however, is both labor-intensive and lacks the convenient modularity of less-precise targeting systems, such as transposon-based systems, in which recombinase-mediated cassette exchange (RMCE) can be used to swap primary transgenes (Gohl et al., 2011).
A targeting method that combines the simplicity of RMCE with the precision of directly coupled transgene and native gene expression was recently described in Drosophila, for use with fly lines that carry the engineered transposable element MiMIC (i.e. Minos-Mediated Integration Cassette) (Venken et al., 2011a). When a MiMIC insertion is in the 5′ untranslated region (UTR) of the gene of interest, RMCE can be used to replace the MiMIC cassette with an artificial exon encoding a primary transgene, preceded by a universal splice acceptor. The splice acceptor insures inclusion of the transgene coding sequence in the mature message of the native gene, and the transgene’s start methionine, rather than the native gene’s, directs its translation. Although a similar strategy can be used to introduce artificial exons into MiMIC insertions within coding introns, which are two-and-a-half times more numerous than 5′ UTR intron insertions (Venken et al., 2011a), co-translation of these artificial exons produces fusion proteins that will not predictably retain the function of a primary transgene’s product. Therefore the MiMIC method’s utility for gaining genetic access to cell types of interest is currently limited to genes with MiMIC insertions in 5′ UTR introns, and most Drosophila genes lack any MiMIC insertion.
To overcome these limitations, we have created an integrated toolkit of artificial exons that capitalize on the ability of the viral T2A peptide to promote the translation of a second protein product from a single transcript (Diao and White, 2012; Tang et al., 2009). Incorporation of the T2A sequence permits transcriptional effectors encoded by our artificial exons to be expressed from sites within coding introns and thus extends the “plug-and-play” flexibility of the MiMIC system to this important class of intron. We have further extended the capabilities of our toolkit by creating an artificial exon that can be targeted to a coding intron in any gene of interest by using the recently developed Crispr/Cas technology (Gratz et al., 2013). This allows most genes in the fly genome to serve as gateways to genetically access the cells that express them.
By analogy to the Trojan horse, we call our artificial exons “Trojan exons” because they gain access to cells through their insertion into coding introns and “release” transcriptional effectors that render the cells susceptible to exploitation by other transgenes. The palette of Trojan exons presented here includes most of the transcriptional effectors commonly used in the fly and enables the fast and easy creation of driver lines with high-fidelity expression in the pattern of an endogenously expressed gene. Although developed for use in Drosophila, the Trojan exon approach should be readily adaptable for use in other genetic model organisms.
Results
Tools for Creating Gal4 Drivers from MiMIC Lines with Inserts into Coding Introns
We have previously shown that the T2A peptide can effectively promote the expression of a Gal4 transgene in the pattern of a native Drosophila gene of interest when the T2A and Gal4 coding sequences are fused in-frame to the coding sequence of the native gene by homologous recombination (Diao and White, 2012). To simplify this procedure for Drosophila genes that contain MiMIC transposons in coding introns, we have adapted the protein-trap vectors previously developed by Venken et al. (2011a) to permit insertion of the T2A-Gal4 sequences directly into the mature message of the native gene by mRNA splicing (Fig. 1A; Table 1). Embryonic injection of such a vector into germline cells expressing ΦC31 integrase leads to exchange of the T2A-Gal4-containing cassette, thus producing a Gal4 driver line that has an expression pattern corresponding to that of a native gene.
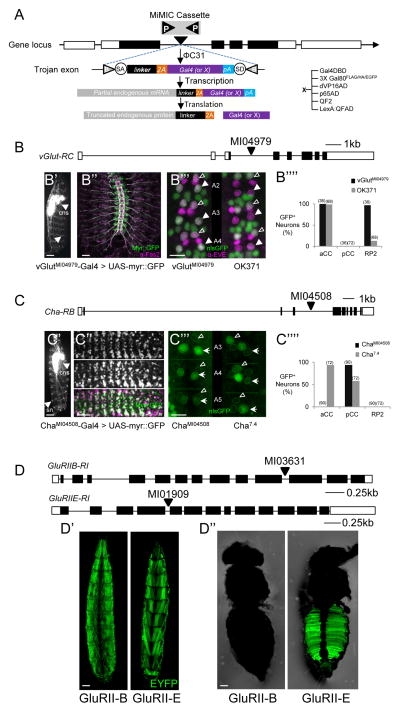
Fig. 1. Creating Trojan Gal4 driver lines using MiMIC insertions into coding introns.
A) Trojan exon structure, integration, and expression. The flanking inverted attP sites (P) of a MiMIC cassette (gray) within a coding intron of a gene (coding exons, black) permit ΦC31 integrase-mediated exchanges for a Trojan exon cassette, via its flanking attB (B) sites. The splice acceptor site (SA) insures incorporation of the T2A-transgene sequence into the gene’s mRNA; a short linker with one of three possible lengths maintains the reading frame of the native message; the T2A sequence causes truncation of the native gene product and promotes the translation of Gal4 (or other transgene, X). pA, Hsp70 polyadenylation signal. See also Fig. S1A and S6.
B) Schematic of the vGlut gene (RC isoform) and site of MiMICMI04979 insertion in a coding intron common to all splice isoforms. B′) vGlutMI04979-Gal4 drives expression of a UAS- myr::GFP reporter in many neurons of the larval CNS (central nervous system, arrowhead) including motor neurons, which project axons (arrow) to bodywall muscles. Scale bar, 200 μm. (See also Fig. S2) B″) Motor nerves (labeled as in B′) in an embryonic fillet showing peripheral nerves double-labeled with anti-Fas2 antibody (magenta). Scale bar: 25 μm. B′″) Comparison of the vGlutMI04979-Gal4 (left) and OK371-Gal4 (right) expression patterns (green, UAS-nlsGFP) in three hemisegments of the larval VNC, labeled with an anti-Eve antibody (magenta). Two of the three Eve-immunopositive neurons located dorsally at the ventral midline in each hemisegment, aCC (open arrowhead) and RP2 (filled arrowhead), are glutamatergic, but OK371-Gal4 consistently fails to label RP2. Scale bar, 10 μm. B″″) Bar graph summarizing the results of labeling experiments of the type shown in B′″ from vGlutMI04979-Gal4 (n=36) and OK371-Gal4 (n=72) hemisegments.
C) Schematic of the Cha gene (RB isoform) and site of the MiMIC MI04508 insertion in a coding intron common to all splice isoforms. C′) ChaMI04508 -Gal4 drives expression of a UAS-myr::GFP reporter in most neurons of the larval CNS (arrowhead), and sensory neurons of the peripheral nervous system (sn, arrow). Scale bar, 200 μm: C″) Co-localization of vChaT mRNA, visualized by embryonic fluorescence in situ hybridization (top) and ChaMI04508 -Gal4 driven UAS-myr::GFP (middle). Bottom, merged images: green, UAS-myr::GFP; magenta, vChaT mRNA. Scale bar: 25 μm. C′″) Comparison of the ChaMI04508 -Gal4 (left) and Cha7.4-Gal4 (right) expression patterns (green, UAS-nlsGFP) in three hemisegments of the larval VNC labeled with anti-Eve antibody. Of the three Eve+ neurons (outlined), Cha7.4-Gal4 consistently labels the glutamatergic aCC neurons (open arrowheads) but not the putative cholinergic pCC neurons. Scale bar: 10 μm. C″″) Bar graph summarizing the results of labeling experiments of the type shown in C′″ from ChaMI04508-Gal4 (n=90) and Cha7.4-Gal4 (n=72) hemisegments.
D) Schematics of the GluRIIB and GluRIIE genes and the insertion sites of MiMICMI03631 and MiMICMI01909. D′) Expression of GluRIIBMI03631-Gal4 (left panel; dorsal view) and GluRIIEMI01909-Gal4 (right panel; ventral view) in larval bodywall muscles. D″) GluRIIEMI01909-Gal4 (right panel), but not GluRIIBMI03631-Gal4 (left panel) drives expression in adult ventral abdominal muscles. Scale bar in D′, D″: 150 μm. Green, UAS-EYFP.
Table 1.
Trojan exons, fly lines, and MiMIC insertions used or generated
| Transgene | Trojan Exon Plasmid | Genes/MiMIC Inserts for Fly Lines Made |
Developmentally Lethal2 |
Poor Viability3 |
|---|---|---|---|---|
| Gal4 | pBS-KS-attB2-SA(0)-T2A-Gal4-Hsp70 | RdlMI02620, ChaMI04508 | CCAP MI01341 (no recombinants) | |
| pBS-KS-attB2-SA(1)-T2A-Gal4-Hsp70 | GluRIIEMI01909, ShawMI01735, CCAPRMI05804, HR46MI04877, jebMI03124 | HR46MI00362, E75MI04895, abMI01579 | jebMI03124 | |
| pBS-KS-attB2-SA(2)-T2A-Gal4-Hsp70 | GluRIIBMI03631, dVGlutMI004979, Gad1MI09277 | Gad1MI09277 | ||
| pBS-KS-attB2-SA(0)-T2A-Gal41 | ||||
| pBS-KS-attB2-SA(1)-T2A-Gal41 | HR46MI04877, CCAPRMI05804 | E75MI04895, crcMI02300 | ||
| pBS-KS-attB2-SA(2)-T2A-Gal41 | dVGlutMI004979 | |||
| pC-(loxP2-attB2-SA(0)-T2A-Gal4-Hsp70) (i.e. “Singlet Donor”) | ||||
| pC-(loxP2-attB2-SA(1)-T2A-Gal4-Hsp70) (i.e. “Singlet Donor”) | ||||
| pC(loxP2-attB2-SA(2)-T2A-Gal4-Hsp70) (i.e. “Singlet Donor”) | ||||
| pC-(lox2-attB2-SA-T2A-Gal4-Hsp70)3 (i.e. “Triplet Donor”) | amonMI00899, RdlMI02957, Gycβ100B/STOPSMI01568 | CCAP MI01341 (no recombinants) | amonMI00899 | |
| pT-GEM(attP2-SA(0)-T2A-Gal4-Hsp70-lox2-3XP3-RFP) | pburs-Intron 1 | |||
| pT-GEM(attP2-SA(1)-T2A-Gal4-Hsp70-lox2-3XP3-RFP) | ||||
| pT-GEM(attP2-SA(2)-T2A-Gal4-Hsp70-lox2-3XP3-RFP) | ||||
| 3XGal80 | pBS-KS-attB2-SA(0)-T2A-3XGAL80-HSP70 | |||
| pBS-KS-attB2-SA(1)-T2A-3XGAL80-HSP70 | ||||
| pBS-KS-attB2-SA(2)-T2A-3XGAL80-HSP70 | dvGlutMI004979 | |||
| Gal4DBD | pBS-KS-attB2-SA(0)-T2A-GAL4DBD-Hsp70 | cacMI02836, ChaMI04508 | ||
| pBS-KS-attB2-SA(1)-T2A-GAL4DBD-Hsp70 | ||||
| pBS-KS-attB2-SA(2)-T2A-GAL4DBD-Hsp70 | dVGlutMI004979 | |||
| dVP16AD | pBS-KS-attB2-SA(0)-T2A-dVP16AD-Hsp70 | |||
| pBS-KS-attB2-SA(1)-T2A-dVP16AD-Hsp70 | ShawMI01735 | |||
| pBS-KS-attB2-SA(2)-T2A-dVP16AD-Hsp70 | ||||
| p65AD | pBS-KS-attB2-SA(0)-T2A-P65AD-Hsp70 | amonMI00899 | ||
| pBS-KS-attB2-SA(1)-T2A-P65AD-Hsp70 | ShawMI01735, RdlMI02957 | |||
| pBS-KS-attB2-SA(2)-T2A-P65AD-Hsp70 | Gad1MI09277 | |||
| QF2 | pBS-KS-attB2-SA(0)-T2A-QF2-Hsp70 | ChaMI04508 | amonMI00899 | |
| pBS-KS-attB2-SA(1)-T2A-QF2-Hsp70 | ||||
| pBS-KS-attB2-SA(2)-T2A-QF2-Hsp70 | dvGlutMI004979, Gad1MI09277 | |||
| LexA:QFAD | pBS-KS-attB2-SA(0)-T2A-LexA:QFAD-Hsp70 | ChaMI04508 | ||
| pBS-KS-attB2-SA(1)-T2A-LexA:QFAD-Hsp70 | ||||
| pBS-KS-attB2-SA(2)-T2A-LexA:QFAD-Hsp70 | dvGlutMI004979, Gad1MI09277 |
These Trojan Gal4 vectors lack the Hsp70 pA and are designed to retain a gene’s native transcription termination signals (e.g. when miRNA binding sites are present in the 3′ UTR). For two genes analyzed, we found no detectable differences in the expression patterns obtained with these constructs compared with those obtained with constructs containing the Hsp70 pA. We recommend use of the latter constructs to insure robust transcription termination unless mRNA downregulation is a known or suspected problem.
Lines for which recombinants died developmentally (or, as for CCAPMI01341, no recombinants were recovered)
Lines in which animals had impaired viability
Because approximately 10% of Drosophila genes currently have MiMIC insertions into coding introns, a wide selection of genes is amenable to this technique (Nagarkar et al., 2015; see also http://flypush.imgen.bcm.tmc.edu/pscreen/mimic.html). To test its efficacy, we focused on genes of neurobiological interest because of the nervous system’s large array of cell types and also because cell-type specific targeting has found its greatest use in mapping neuronal circuits. Two genes of considerable neurobiological interest for which high-fidelity Gal4 lines have been lacking are those encoding the vesicular glutamate transporter (i.e. vGlut; Fig. 1B), and Cholineacetyl transferase (i.e. Cha; Fig. 1C), which are expressed in neurons that use glutamate and acetylcholine as neurotransmitters, respectively. Both genes contain intronic MiMIC inserts (vGlutMI04979, phase 2; ChaMI04508, phase 0) that are common to all splice isoforms of the respective genes, and for both, there are Gal4 driver lines made by other methods (Mahr and Aberle, 2006; Salvaterra and Kitamoto, 2001). These lines thus permit the direct comparison of commonly used drivers with those generated by the Trojan-mediated conversion of MiMIC (Trojan-MiMIC) technique described here.
Targeting glutamatergic and cholinergic neurons using a Trojan Gal4 exon
Trojan-MiMIC lines were created as described in the Experimental Procedures, with recombinant flies identified directly by Gal4-driven expression of a fluorescent reporter in the G1 generation (Fig. S1A). Cassette replacement was also later verified by PCR. The vGlut and Cha genes are broadly expressed in the nervous system, so reporter-mediated fluorescence was readily detectable in live larvae and driver lines could be established within two generations. Both Trojan-MiMIC drivers exhibited expression patterns broadly consistent with the expression of their respective genes (Daniels et al., 2008; Yasuyama et al., 1995): vGlutMI04979-Gal4 targets GFP reporter expression to larval motor neurons (Fig. 1B′, B″), while ChaMI04508-Gal4 targets most central neurons and also peripheral sensory neurons (Fig. 1C′). Comparison of these patterns with those obtained for the Cha gene by in situ hybridization at the embryonic stage (Fig. 1C″) or for the vGlut protein by immunostaining at the larval stage (data not shown) confirmed their general fidelity, as did the substantial rescue from embryonic lethality of expression of a UAS-Cha transgene under the control of ChaMI04979- Gal4 (Table 2). The one noted exception to fidelity of expression was a set of neurons in the posterior optic lobe that is clearly labeled by the vGlutMI04979-Gal4 driver, despite not being readily detected by anti-vGlut antiserum (Fig. S2). These neuronal somata, which may simply stain below the detection threshold of the anti-vGlut antibody, are likewise included in the expression pattern of an enhancer-trap line widely-used to target glutamatergic neurons, OK371-Gal4 (Mahr and Aberle, 2006).
Table 2.
UAS-Cha rescues the hatching deficits of ChaMI04508-Gal4/ChaL13 mutants.
| Cross1 | Expected % Cha MI04508-Gal4/Cha L13 | Mutant Phenotype2 | ||
|---|---|---|---|---|
| Control |
|
25% | 29% (n=120/416) | |
| Rescue |
|
25% | 1% (n=4/368) |
Embryos from the indicated crosses were collected for 1 hour and incubated for 22.5 hours at 25°C. Unhatched embryos were dechorionated and individually prodded to test for mutant-like phenotypes (i.e. no response or incomplete peristalsis).
Only progeny with ChaMI04508-Gal4/ChaL13 genotypes, which should occur at a frequency of 25% assuming Mendelian segregation ratios, are expected to have mutant phenotypes. The almost complete absence of mutant phenotypes when UAS-Cha is included in the cross indicate strong rescue of the embryonic lethality associated with Cha null mutations.
For a more detailed assessment of the fidelity of the vGlutMI04979-Gal4 and ChaMI04508-Gal4 lines, we examined their expression in three pairs of medial, segmentally repeated neurons in the larval ventral nerve cord (VNC) identifiable by their expression of the Even-skipped protein, Eve (Fujioka et al., 2003). Two pairs of Eve+ neurons, aCC and RP2, are known glutamatergic motor neurons (Vanvactor et al., 1993), while the third, pCC, is a putative cholinergic interneuron. Consistent with their neurotransmitter phenotypes, both aCC and RP2 motor neurons are within the expression pattern of the vGlutMI04979-Gal4 line (Fig. 1B‴, left panel; 1B″″), while only pCC is included in the pattern of the ChaMI04508-Gal4 line (Fig. 1C‴, left panel), strongly suggesting that pCC is cholinergic. An existing cholinergic driver, Cha7.4-Gal4, which was made using a 7.4 kb genomic fragment located upstream of the Cha coding sequence, also labels the pCC pair, but inconsistently, and often labels the glutamatergic aCC motor neurons (Fig. 1C‴, right panel, 1C″″). Similarly, OK371-Gal4 fails to express consistently in the glutamatergic motor neurons, often not labeling RP2 (Fig. 1B‴, right panel; 1B″″). The two driver lines made using the Trojan-MiMIC method thus display a high degree of fidelity when compared with similar drivers made by other methods. The consistent labeling of pCC interneurons by ChaMI04508-Gal4 also suggests that Trojan-MiMIC drivers can serve as highly sensitive detectors of gene expression to identify neurotransmitter and other cellular phenotypes.
Targeting larval muscle using a Trojan Gal4 exon
In addition to targeting neurons based on their neurotransmitter phenotypes, the Trojan-MiMIC technique can similarly be used to target cells that receive input from such neurons by exploiting MiMIC insertions into genes that encode subunits of specific neurotransmitter receptors. The larval bodywall muscles innervated by glutamatergic motor neurons express five different glutamate receptor genes (DiAntonio, 2006), two of which—GluRIIB and GluRIIE—have MiMIC insertions in coding introns (Fig. 1D). We created the GluRIIBMI03631- and GluRIIEMI01909-Gal4 driver lines, which, as expected, expressed Gal4 in all larval bodywall muscles (Fig. 1D′), though the patterns generated by GluRIIEMI01909-Gal4 were weaker than those generated by GluRIIBMI03631-Gal4 and exhibited some variegation.
We also examined the expression of the GluRIIB and GluRIIE genes in adult animals, where the subunit composition of glutamate receptors at neuromuscular junctions is largely unknown. While GluRIIBMI03631-Gal4 does not drive observable reporter expression in any muscle in one-week old adults, GluRIIEMI01909-Gal4 produced a distinct pattern of expression in ventral muscles of the abdomen (Fig. 1D″). We again observed variegation, sometimes quite pronounced, which has been previously documented for other Gal4 lines (Skora and Spradling, 2010). However, such variation is the exception, and most Trojan-MiMIC Gal4 insertions yield robust and stable expression patterns.
Efficiency of Generating Trojan-MiMIC Gal4 Lines
We have successfully generated two or more Trojan-MiMIC lines for each of the Trojan Gal4 exons in the three possible reading frames (Table 1). The efficiency of cassette exchange using the microinjection protocol ranged from 3–33% (i.e. percentage of fertile adults from Trojan-Gal4-injected embryos that yielded Gal4-expressing progeny). Overall, recombinant progeny were identified for 15 of 16 independent MiMIC insertions tested, although for several genes we could not establish stocks and in some cases recombinant animals were poorly viable. While truncation of the endogenous gene product by the introduction of T2A-Gal4 may in some cases have created a dominant-negative version of the endogenous protein with deleterious effects, such effects were not consistently observed for all Trojan exons introduced into the same locus. For example, heterozygous Gad1MI09277–Gal4 animals were severely impaired, but heterozygous Gad1MI09277–p65AD animals were viable and healthy (Table 1) suggesting that the Trojan exons encoding the different transcriptional effectors may be spliced with different efficacies. In any case, our findings suggest that the majority of MiMIC sites in coding introns can be converted into viable Trojan-MiMIC Gal4 lines by microinjection techniques.
In Vivo Generation of Trojan-MiMIC Gal4 Driver Lines
Because microinjection is both expensive and laborious, we also designed a cheaper system for generating Trojan-MiMIC Gal4 flies that relies on simple genetic crosses and that can produce large numbers of recombinants. We created fly lines carrying either single or “triplet” Trojan donor constructs (Table 1), which generate attB-flanked integration cassettes when excised from the genome. The more universally useful “triplet donor” has three cassettes (Fig. 2A), each containing a Trojan Gal4 exon in one of the three reading frames. Each Trojan exon is flanked by attB sites and a pair of uniquely compatible lox sites, which are substrates of the Cre recombinase (Livet et al., 2007). By crossing flies from this triplet donor line to flies with germline expression of both Cre recombinase and ΦC31 integrase, one can induce Cre-mediated excision and circularization of the three cassettes, which can then be integrated into the attP sites of a MiMIC insertion by ΦC31 integrase (Bischof et al., 2007).
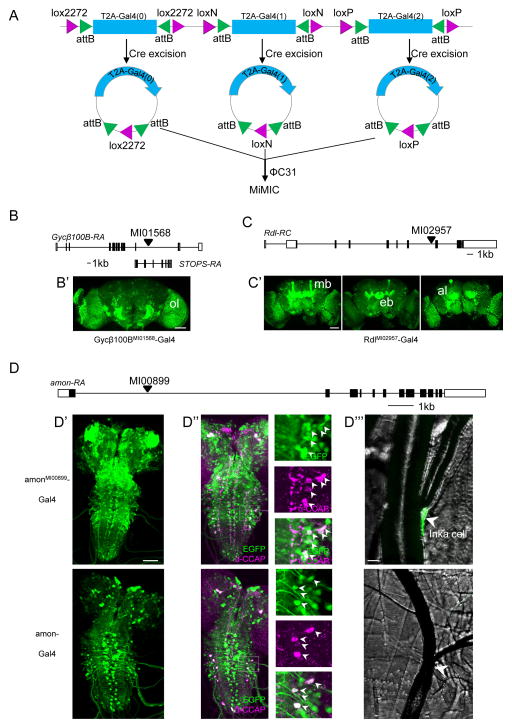
Fig. 2. Creating Gal4 drivers using the in vivo system for Trojan exon exchange.
A) Schematic of the triplet donor construct consisting of three tandem Trojan Gal4 cassettes, one in each reading frame, each flanked by attB sites nested within uniquely compatible pairs of lox sites, recognized by Cre recombinase. See Figure S1B for details of genetic crosses.
B) Schematic of the Gycβ100B gene locus and the site of the MiMICMI01568 insertion, which also lies within a coding intron of the STOPS gene on the opposite strand. (See also Fig. S3.) B′) Expression of a UAS-EYFP reporter driven by Gycβ100BMI01568-Gal4 in the optic lobe (ol) of the adult brain.
C) Schematic of the Rdl gene and the MiMICMI 02957 insertion site. C′) Confocal images from an adult brain show prominent RdlMI02957-Gal4 driven expression of a UAS-EYFP reporter in the mushroom bodies (mb), ellipsoid body (eb), and antennal lobes (al). Scale bars: 50 μm. (See also Fig. S4.)
D) Schematic of the amon gene and the MiMIC MI00899 insertion site. D′–D″′) Comparison of larval expression patterns of amonMI00899-Gal4 (top panels) and amon-Gal4 (bottom panels). D′) Expression patterns in CNS whole mounts (green, UAS-EGFP). D″) Expression within the subset of CCAP-expressing neurons (anti-CCAP staining, magenta). Insets show higher magnification images of the boxed regions. D″′) Expression of amonMI00899-Gal4, but not amon-Gal4, in the endocrine Inka cells located at the base of the dorsal tracheal trunks (arrowheads, see also Fig. S5). Scale bars: D′, D″: 50 μm; D′″: 10 μm.
To test this “in vivo” Trojan-MiMIC system, we selected MiMIC insertions in three genes, each of which required a T2A-Gal4 construct in a different reading frame: 1) Guanylyl Cyclase Beta-100B (Gycβ100BMI01568; ph 2; Fig. 2B), a subunit of the NO-sensitive soluble guanylyl cyclase (Shah and Hyde, 1995); 2) Resistant to Dieldrin (RdlMI02957, ph 1; Fig. 2C) which encodes a well-characterized Drosophila GABA-A receptor α-subunit (Buckingham et al., 2005); and 3) amontillado (amonMI00899, ph 0; Fig. 2D), which encodes the Drosophila ortholog of Prohormone Convertase 2, an enzyme critical for the processing of neuropeptides (Wegener et al., 2011). Gal4 lines were successfully generated for these three genes (Fig. S1B), confirming that all three Trojan Gal4 constructs of the triplet donor are functional. In addition, all three appeared to be excised from the donor locus and re-integrated into the MiMIC site with similar efficiency, as determined by PCR analysis of all 61 independent exchange events produced in the generation of the Gycβ100BMI01568-Gal4 line (Fig. S3). In total, eight exchange events resulted in phase 2 insertions with the robust Gycβ100BMI01568-Gal4 CNS expression pattern seen in Fig. 2B′. Interestingly, nine phase 1 insertions in the opposite direction gave expression patterns that were largely non-neural (data not shown) due to the fortuitous intronic localization of MI01568 in a second gene (i.e. STOPS) encoded on the opposite strand.
The expression patterns of both the RdlMI02957- and amonMI00899-Gal4 lines conform closely to those reported for the Rdl and amon genes, respectively (Harrison et al., 1996; Wegener et al., 2011). RdlMI02957-Gal4 drives reporter expression broadly in the adult brain, most notably in structures previously identified by antibody staining to express Rdl protein, including the Mushroom Body lobes, the Ellipsoid Body, and the antennal lobes (Fig. 2C′). In addition, expression of a UAS-Rdl-RA transgene (Sanchez-Soriano and Prokop, 2005) eliminated larval lethality in animals mutant for Rdl (data not shown). Despite some lethality later in development, surviving adults bearing the rescue transgene were fertile and could be propagated as a stable stock, indicating that the RdlMI02957-Gal4 expression pattern includes all essential Rdl-expressing neurons. Notably, the expression pattern of the RdlMI02957-Gal4 line contrasts to the more restricted patterns produced by two previously published promoter-fusion Rdl-Gal4 lines (Hekmat-Scafe et al., 2010; Yuan et al., 2014), neither of which exhibited expression in all previously characterized sites of Rdl expression (Fig. S4). The Rdl-Gal42-2 line, which has a second chromosome transgene insertion and could therefore be readily assayed for its ability to rescue the larval lethality of Rdl mutations, failed to do so (data not shown).
As expected, the amonMI00899-Gal4 line expression pattern included many neurons with the large somata characteristic of neurosecretory cells (Fig. 2D′, upper panel), including a well-known group that express Crustacean Cardioactive Peptide (i.e. CCAP; Fig. 2D″, upper panels). This group is likewise present in the expression pattern of a Gal4 line made by P-element transgenesis using a putative amon enhancer element (Rhea et al., 2010) (amon-Gal4; Fig. 2D′, D″, lower panels). However, the latter line has both weaker and sparser expression than amonMI00899-Gal4 in the nervous system, and, unlike amonMI00899-Gal4, fails to label the peripheral endocrine cells (i.e. Inka cells) that make the peptide Ecdysis Triggering Hormone (ETH; Fig. 3D″′). Evidence for amon gene expression in Inka cells has been ambiguous: amon mRNA is found in embryonic Inka cells by in situ hybridization (Siekhaus and Fuller, 1999), but larval Inka cells are reported to lack Amon immunoreactivity (Park et al., 2004; Rayburn et al., 2009). To determine whether Inka cells express the amon gene, we selectively knocked down its expression using an Inka-cell specific driver (ETH-Gal4) and UAS-amon-RNAi. We found substantial developmental deficits in ecdysis at all stages of development as is expected for loss of ETH function (Fig. S5). This result strongly suggests that Amon protein is present at levels below the threshold of antibody detection, and that the amonMI00899-Gal4, but not the amon-Gal4, line faithfully reports its expression.
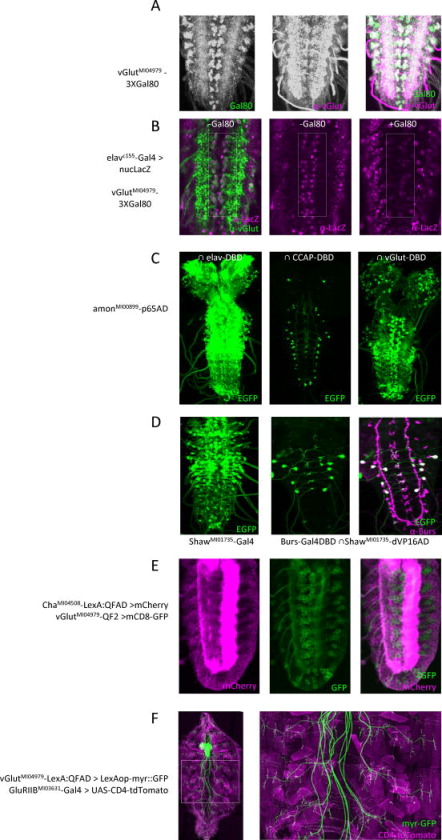
Fig. 3. Combinatorial gene targeting using Gal80, Split Gal4, QF2, and LexA::QFAD Trojan exons.
A–B) Suppression of Gal4 activity in glutamatergic neurons using 3XGal80FLAG/HA/EGFP. A) vGlutMI04979- 3XGal80FLAG/HA/EGFP expresses Gal80 (left; anti-GFP immunostaining) in glutamatergic motor neurons of the larval VNC labeled with anti-vGlut antibody (middle); merged images (right). Scale bar (panels A–E): 25 μm. B) elav-Gal4 drives UAS-LacZ reporter in the glutamatergic motor neurons (rectangle: anti-LacZ immunoreactivity, magenta; anti-vGlut, green), but Gal4 activity is absent when the vGlutMI04979-3XGal80 transgene is present (right).
C) Identification of neuronal subsets within the amonMI00899-p65AD expression pattern. Left: The entire complement of amon-expressing neurons in the larval CNS, visualized using the pan-neuronal elav-VP16AD hemidriver and a UAS-EGFP reporter. Middle: The CCAP-expressing complement, visualized using a CCAP-Gal4DBD hemidriver. Right: The vGlut-expressing complement, visualized using a Trojan-MiMIC vGlutMI04979-Gal4DBD hemidriver.
D) Split Gal4 isolation of the subset of neurons that express both the Shaw K+ channel and the hormone Bursicon. Left: neurons expressing the RA, RC, and RE splice isoforms of the Shaw gene, visualized using a Trojan-MiMICShawMI01735-Gal4 driver line and a UAS-EGFP reporter. Middle: The Shaw-expressing neurons that also express Burs, isolated using a Trojan-MiMIC ShawMI01735-dVP16AD in combination with a Burs-Gal4DBD hemidriver; green, UAS-EGFP. Right: anti-Burs immunostaining (magenta) of the section shown in middle panel (double-labeled neurons are white).
E) Double-labeling of the embryonic VNC to identify subsets of cholinergic (LexAop-mCherry, magenta) and glutamatergic (QUAS-mCD8-GFP, green) neurons using orthogonal Trojan-MiMIC drivers. Left: Expression pattern of a ChaMI04508-LexA::QFAD driver in a confocal section through the ventral midline. Middle: Same section showing expression of a vGlutMI04979-QF2 driver. Right: Merged images; note the two expression patterns do not overlap.
F) Double-labeling of glutamatergic neurons in the larval CNS and body wall muscles using the orthogonal Trojan-MIMC drivers vGlutMI04979-T2A-LexA::QFAD (green, LexAop2-myr::GFP) and GluRIIBMI03631-T2A-Gal4 (magenta, UAS-CD4-tdTomato). Right panel: magnified image of the boxed region at left showing neuromuscular synapses. Scale bar: 100 μm.
Creating Trojan-MiMIC Lines for Combinatorial Expression
It is often necessary to genetically target specific cells based not on their expression of a single gene of interest, but on their expression of two such genes (Luan and White, 2007; Pfeiffer et al., 2010; Potter et al., 2010). To permit such combinatorial targeting, we created Trojan exons for several transgenes other than Gal4 (Fig. 1A; Table 1), including the Gal4 inhibitor, Gal80, which effects a Boolean NOT operation when used in conjunction with Gal4 drivers, and components of the Split Gal4 system (Gal4DBD, dVP16AD, and p65AD), which can be used in combination to perform a Boolean AND operation and identify neurons common to the patterns of expression of two genes.
The 3XGal80 Trojan exon (see Experimental Procedures) is designed to express three copies of Gal80, one of which is tagged with epitopes for HA and FLAG, and another of which is tagged with EGFP. A vGlutMI04979-3XGal80 line generated with this construct has the same fidelity of expression as vGlutMI04979-Gal4, as demonstrated by double-labeling with anti-vGlut and anti-GFP antibodies (Fig. 3A), and effectively subtracts glutamatergic neurons from the expression pattern of the pan-neural elav-Gal4 driver line (Fig. 3B). Similarly, an amonMI00899-p65AD line has a neuronal expression pattern that matches that of the previously described amonMI00899-Gal4 line (Fig. 3C, left) as revealed by using it in conjunction with an elav-Gal4DBD hemidriver, and can be used to target the subpopulation of CCAP-expressing neurons (Fig. 3C, middle), when used in conjunction with a CCAP-Gal4DBD hemidriver. Importantly, restriction of expression can be used to identify and manipulate previously inaccessible subsets of neuropeptidergic neurons. For example, amonMI00899-p65AD together with vGlutMI04979–Gal4DBD unveils the class of peptidergic neurons that also use the neurotransmitter glutamate. This class can be seen to include putative motor neurons along the ventral nerve cord that express the neuropeptide proctolin (Anderson et al., 1988), and numerous other central neurons (Fig. 3C, right). In the same way, a ShawMI01735-dVP16AD line in conjunction with a Burs-Gal4DBD line isolates a subpopulation of neurons that expresses both the Shaw K+ channel and the Burs subunit of the hormone bursicon (Fig. 3D).
Overlap of two expression patterns and a variety of dual manipulations can also be accomplished using orthogonal binary expression systems (Lai and Lee, 2006; Potter et al., 2010), and to facilitate such applications we created Trojan exons for the transcriptional activators QF2 and LexA::QFAD (Riabinina, 2015), which drive the expression of transgenes placed downstream of QUAS and LexAop sequences, respectively (Table 1). We applied these reagents to the question of whether acetylcholine and glutamate are used as neurotransmitters by distinct populations of neurons. While this is thought to be the case, a comprehensive comparison has not been possible with existing tools. Using ChaMI04508-LexA::QFAD and vGlutMI04979-QF2 driver lines together, we surveyed the neurons of early first instar larvae and found no segmentally reiterated overlap in expression (Fig. 3E), suggesting that specification of the main transmitter systems in the Drosophila nervous system is mutually exclusive. Simultaneous targeting of different cell types can also be used to label or manipulate both upstream and downstream partners within an interacting pair. To demonstrate this application, we made a vGlutMI04979-LexA::QFAD line to drive expression of LexAop-transgenes in motor neurons and used it together with the previously described GluRIIBMI03631-Gal4 line to simultaneously label both pre- and post-synaptic elements at the larval neuromuscular junction (Fig. 3F).
T-GEM: a MiMIC-independent Application of the Trojan Exon Approach
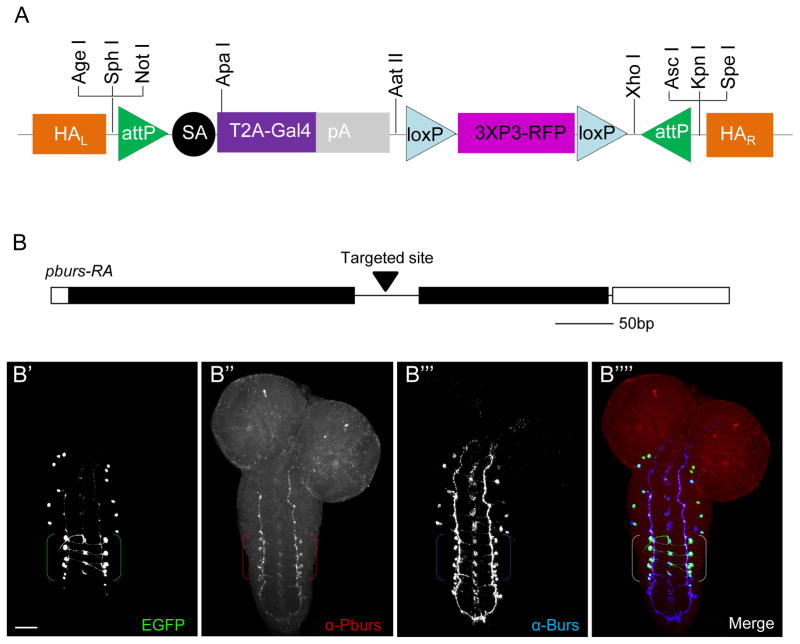
While the growing collection of MiMIC insertions in Drosophila is an invaluable resource for the implementation of the Trojan-MiMIC technique, MiMIC insertions into coding introns are still not available for all fly genes. To make the “plug-and-play” versatility of Trojan exons more generally serviceable—both in the fly and potentially in other model organisms—we created a MiMIC-like Trojan Gal4 construct that can be targeted to genomic loci by homologous recombination using the Crispr/Cas technology (Gratz et al., 2013). Targeted insertion of this Trojan-Gal4 Expression Module (T-GEM; Fig. 4A) into an intronic locus generates a Gal4 driver line that can be easily converted by cassette exchange into any other type for which Trojan exons have been created. To demonstrate the use of T-GEM, we targeted its insertion to an intron in the pburs gene, which encodes a subunit of the heterodimeric hormone, bursicon (Honegger et al., 2008) (Fig. 4B). The expression pattern of the resulting PbursTGEM-Gal4 line includes four pairs of segmentally-repeated neurons in the larval ventral nerve cord previously shown to exhibit anti-Pburs immunoreactivity (Luo et al., 2005) (Fig. 4B, B′, brackets). In addition, the pattern includes several other pairs that are not immunoreactive for Pburs, but which are immunoreactive for Burs, the heterodimeric partner of Pburs (Fig. 4B″′, B″″). The observation that PbursTGEM-Gal4 driven reporter expression coincides with Burs expression in these neurons is consistent with reports that only Burs/Pburs heterodimers have biological activity (Luo et al., 2005), and indicates, as in other cases, that the Trojan exon driver is a more sensitive indicator of gene expression than a corresponding antibody.
Fig. 4. Targeting coding introns using the pT-GEM vector and the Crispr/Cas system.
A) Schematic of the Trojan-Gal4 Expression Module (T-GEM), which includes a Red Fluorescent Protein (RFP) selection marker expressed in the adult eye that can be used to monitor genomic insertion. The attP sites permit cassette exchange with any other Trojan exon, and unique restriction sites at the 5′ and 3′ ends of the construct permit the insertion of suitable homologous arms (HAR and HAL) for targeting T-GEM to the desired intronic locus.
B) Schematic of the pburs gene, showing the intronic site targeted by Crispr/Cas for T-GEM insertion. B′-B″″) Confocal micrographs of a larval CNS whole mount triple-labeled for pbursTGEM-Gal4 (B′), Pburs (B″), and Burs, which heterodimerizes with Pburs to form the hormone Bursicon (B″′); merged image of all three labels (B″″). Brackets: Pburs-immunopositive cell bodies. Green, UAS-EGFP; red, anti-Pburs immunoreactivity; blue, anti-Burs immunoreactivity. Scale bar: 25 μm.
Discussion
The Trojan exon tools introduced here permit the rapid production of transgenic fly lines for cell-type specific manipulation. The advantages of the Trojan exon system include: 1) fully modular design using off-the-shelf, exchangeable Trojan constructs; 2) the ease of ΦC31-mediated cassette exchange after the initial creation of a suitable target site; 3) the availability in Drosophila of thousands of ready-made target sites in the form of MiMIC lines with insertions into coding introns; 4) the ability to create novel intronic target sites using the T-GEM vector, which simultaneously creates a Gal4 driver; and 5) high fidelity co-expression of transgene and targeted native gene. While genes lacking introns, or genes for which the elimination of a single allele is lethal are not amenable to the Trojan exon technique, such genes are not numerous in Drosophila, and special cases, such as essential genes on the X chromosome, can be easily accommodated by the Trojan exon strategy (Fig. S6). In cases where the mutagenic nature of Trojan insertions adversely affects viability, we find that different Trojan exons vary in their degree of lethality, and the range of such exons introduced here gives experimenters considerable latitude in making gene- and cell-type specific reagents.
For the majority of Drosophila genes tested, we found that Trojan exon-generated driver lines duplicated the expression pattern of the native gene with higher fidelity, and detected its expression with greater sensitivity, than existing reagents (see also, Gnerer et al. 2015). As such, Trojan lines should serve as a new “gold standard” in determining the expression patterns of genes of interest. In addition, we have shown that Trojan lines can be readily generated for intersectional analysis to isolate previously inaccessible cell groups for characterization. Overall, the Trojan exon technology introduced here facilitates a range of cellular and genetic manipulations and allows them to be performed with unprecedented speed and accuracy. It does so by leveraging the largely unexploited capacity of coding introns to provide genetic access to specific cell types in vivo. Its implementation in other organisms using Crispr/Cas-mediated genome editing (Seruggia and Montoliu, 2014) should be possible using strategies similar to the T-GEM technology introduced here.
Experimental Procedures
Molecular Biology
Trojan Exon Constructs
All Trojan exons except the pT-GEM construct were made by incorporating primary transgene coding sequences into the protein trap vectors introduced by Venken et al. (2011a) for all three reading frames (Drosophila Genomics Resource Center, clones # 1297, 1305 and 1313; Indiana University). Transgene coding sequences were amplified by PCR from the following sources: pC-attB-bursα-mCD8-EGFP-T2A-Gal4 (Addgene #39463) (Diao and White, 2012) for Gal4; pBPZpGAL4DBDUw (Addgene #26233) (Pfeiffer et al, 2011) and pBPp65ADZpUw (Addgene # 26234) (Pfeiffer et al, 2011) for Gal4DBD and p65AD, respectively; pCAST-CCAP-dVP16ADZip—used to create pP{y+, dVP16AD}(Gao et al., 2008; Luan et al., 2006)—for dVP16AD; pattb-nsyb-QF2-Hsp70 (Addgene #46115) for QF2 (Riabinina, 2015); and pattb-nsyb-Lex::QF-Hsp70 (Addgene #46123) for LexA::QFAD. The T2A-Gal80FLAG/HA-P2A-Gal80EGFP-T2A-Gal80-Hsp70 sequence was synthesized by Epoch Life Science, Inc., using the P2A and T2A sequences described in Diao and White (2012) and the Drosophilized Gal80 sequence of Pfeiffer et al. (2010).
The pT-GEM vectors were assembled in pBlueScript from T2A-Gal4-Hsp70 sequences made as described above and a 3XP3-RFP selection marker (Bischof et al., 2007) with flanking inverted loxP sites synthesized by Epoch Life Science, Inc. To insert the pT-GEM construct into the pburs locus, homology arms of approximately 1 kb were amplified by PCR from genomic DNA of the {nos-Cas9} attP2 transgenic fly line used for embryo microinjection (Ren et al., 2013). The guide RNA expression construct (sgRNA) was made by inserting a DNA fragment, produced by annealing the oligonucleotides ttcgAGAGTTCGCTAACTCTATAG and aaacCTATAGAGTTAGCGAACTCT, into the BbsI (New England Biolabs)-digested U6b-sgRNA-short plasmid of Ren et al. (2013).
Triplet and singlet donor constructs
The triplet donor construct was assembled from several synthesized gene fragments. The attB2 and SA sequences were from Venken et al. (2011a); the sequences of the loxP, loxN, and lox2272 sites were identical to those in Livet et al. (2007); the unique 100 bp spacer sequences separating each pair of lox and attB sites were derived from a 600 bp sequence in the promoter of nerfin1, which lacks DNA regulatory elements (Kuzin et al., 2009); the T2A-Gal4 sequence was the same as used for the Trojan Gal4 constructs. After synthesis, the fragments were assembled and cloned into pCAST (Luan et al., 2006) to make the final construct, pC-(lox2-attB2-SA-T2A-Gal4-Hsp70)3.
Singlet donor constructs were made similarly to the triplet donor constructs. First, backbone fragments consisting of loxP-flanked attB2-SA sequences were synthesized in all three phases and subcloned into the pCAST vector (Luan et al., 2006) to make the pC-(loxP2-attB2-SA). A T2A-Gal4-Hsp70 fragment derived from the Trojan Gal4 construct by BamH I digestion was then cloned into these vectors to make the pC-(loxP2-attB2-SA-T2A-Gal4-Hsp70) singlet donor constructs.
TubP-dVP16AD, TubP-Gal4DBD, and ETH-Gal4 constructs
Constructs in which the dVP16ADZip and ZipGal4DBDflUw sequences described above were expressed under the tubulin-1α promoter (TubP) were made by replacing the Gal80 sequence in the pCaSpeR4-tubP-Gal80 vector (gift of Tzumin Lee). The ETH-Gal4 construct was made by cloning a 397-bp promoter fragment lying just upstream of the ETH start methionine into the pPTGAL vector (Sharma et al., 2002).
Genbank Accession Codes
KP686436: pC-(loxP2-attB2-SA(0)), KP686437: pC-(loxP2-attB2-SA(1)), KP686438: pC-(loxP2-attB2-SA(2)), KP686439: pC-(lox2-attB2-SA-Hsp70)3, KP686440: pT-GEM(0), KP686441: pT-GEM(1), KP686442: pT-GEM(2)
Fly Lines
MiMIC lines were obtained from the Bloomington Stock Center at Indiana University or the Bellen Lab of Baylor College of Medicine. The TubP-dVP16AD, TubP-Gal4DBD, and ETH-Gal4 lines were made by P-element transformation, and the insertions of TubP-dVP16AD and TubP-Gal4DBD on chromosomes X, II, and III were isolated and characterized. An ETH-Gal4 insertion on chromosome II was used in the experiments described here. The following lines were described previously: w;+;Burs-Gal4DBD (Luan et al., 2012) and w;Sp/CyO;CCAP-Gal4DBD and yw;elav-Gal4DBD;Dr/TM3,Sb (Luan et al., 2006). w;+;Rdl2-1-Gal4 and w;Rdl2-2-Gal4/CyO;+ were gifts from Dr. Julie Simpson at JFRC, HMHI. All other stocks were obtained (or derived from stocks) from the Bloomington Drosophila Stock Center at Indiana University. All plasmid injections to generate Trojan-MiMIC lines were performed by Rainbow Transgenic Flies, Inc. A complete list of transgenic fly lines developed for use in implementing the Trojan system is shown in Table S1.
Lines made by ΦC31-mediated conversion of MiMIC lines by plasmid injection
Midiprep plasmid DNA containing the desired Trojan exon in the correct reading frame was injected together with ΦC31 plasmid DNA into the embryos of flies bearing MiMIC insertions in genes of interest. Recombinant transformants were isolated as described in Figure S1, using UAS-2XEYFP, UAS-2XEGFP, 13XLexAop2-myr-tdTomato, and QUAS-mCD8-GFP as fluorescent expression markers. In some cases, developmental lethality was observed that appeared to derive from high levels of fluorescent protein expression. In these cases, G0 flies were crossed with flies from the yw;Sp/CyO;Dr/TM3,Sb double balancer line, and y− recombinant progeny were isolated based on the loss of the y+ selection marker associated with the MiMIC insertion. The orientation of the Trojan exon insert was then determined by PCR amplification of the insertion. Since in general, the endogenous fluorescence of the EGFP-tagged Gal80 moiety was too weak to detect in live animals, this strategy was also used to screen for MiMIC-3XGal80FLAG/HA/EGFP-Hsp70 flies.
Lines made in vivo using the triplet donor line
Flies bearing inserts of the triplet donor transgene were made by P-element mediated transformation using pC-(lox2-attB2- SA-T2A-Gal4-Hsp70)3. Lines were established with insertions of the triplet donor on chromosomes X, II, and III. A transgenic line with the 2nd chromosome insert was used as the Trojan Gal4 donor to make the Trojan-MiMIC Gal4 lines as described in Figure S1.
PbursTGEM-Gal4 line made using pT-GEM
The PbursTGEM-Gal4 transgenic flies were made by microinjection into embryos of {nos-Cas9} attP2 flies (Ren et al., 2013), which were co-injected with sgRNA and pT-GEM plasmid DNA. The adults were screened by fluorescence for eye-specific expression of RFP.
Immunostaining, Fluorescent In Situ Hybridization, and Confocal Microscopy
Fixation, staining, and confocal imaging of whole-mount CNS preparations from third-instar larvae or adults were carried out as described (Luan et al., 2012). Primary antibodies were used as described in the Supplemental Experimental Procedures and were the kind gifts of the following individuals: rabbit anti-vGlut (Aaron DiAntonio); guinea pig anti-Eve (Kosman et al., 1998) (Dr. John Reinitz); mouse anti-Pburs (Aaron Hsueh). The rabbit anti-Bursα and anti-CCAP antibodies were the same in Luan et al. (2006), and mouse anti-LacZ and mouse anti-EGFP were from Promega and Invitrogen, respectively. AlexaFluor 488, 568, and 647 secondary antibodies were from Invitrogen. Embryonic fluorescent in situ hybridization (FISH) to confirm the fidelity of expression in the ChaMI04508-Gal4 line used a riboprobe synthesized from the coding sequence of the vChaT gene, which is co-expressed from the same genomic locus as Cha (Kitamoto et al., 1998). T7 RNA polymerase (New England Biolabs) was used to generate the riboprobe, and vChaT mRNA was visualized using a sheep anti-DIG-POD antibody (Roche). See the Supplemental Experimental Procedures for more details. Fluorescent imaging of larval and adult CNS preparations was performed with a Nikon C-1 confocal microscope using a 20X objective, and the images shown are composites of separately acquired volume-rendered images. A Leica TCS SP5 confocal microscope with a 63X oil-immersion objective was used for imaging late embryonic and early larval ventral nerve cords.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Mental Health (B.H.W.) and by grants from the Whitehall Foundation (C.J.P), NIH (R01DC013070, C.J.P.), the Welcome Trust (H.I. and M.L.), the Sir Isaac Newton Trust, Cambridge (M.L.). J.E. was supported by FONDECYT #1141278 and the CINV, which is supported by the Millennium Scientific Initiative of the Ministerio de Economía, Fomento y Turismo. We thank the Bellen laboratory and the Drosophila Gene Disruption Project at Baylor College of Medicine, the Bloomington Stock Center (NIH P40OD018537), and Julie Simpson, for fly lines. Thanks also to Aaron DiAntonio, Aaron Hsueh, and John Reinitz for antibodies and to the NINDS Sequencing Core Facility for DNA sequencing. Finally, thanks to Sarah Naylor for technical help and to Grace Gray, Herman Dierick, Koen Venken, and Hugo Bellen for comments on the manuscript and productive discussions.
Footnotes
Author Contributions: Fengqiu Diao and BHW conceived the technique, planned experiments, and drafted the manuscript; Fengqiu Diao, Feici Diao, HI, HL, and EM made and characterized constructs and fly lines; Fengqiu Diao, HI, ML, CJP, and WS planned and conducted experiments; JE contributed unpublished reagents; and CJP, ML, HI, and JE made intellectual contributions and contributed to the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson MS, Halpern ME, Keshishian H. Identificiation of the Neuropeptide Transmitter Proctolin in Drosophila Larvae - Characterization of Muscle Fiber-specific Neuromuscular Endings. J Neurosci. 1988;8:242–255. doi: 10.1523/JNEUROSCI.08-01-00242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phi C31 integrases. P Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Buckingham SD, Biggin PC, Sattelle BM, Brown LA, Sattelle DB. Insect GABA receptors: Splicing, editing, and targeting by antiparasitics and insecticides. Mol Pharmacol. 2005;68:942–951. doi: 10.1124/mol.105.015313. [DOI] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, Diantonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- DiAntonio A. Glutamate receptors at the Drosophila neuromuscular junction. In: Budnik V, RuizCanada C, editors. Fly Neuromuscular Junction: Structure and Function. 2. San Diego: Elsevier Academic Press Inc; 2006. pp. 165–179. [DOI] [PubMed] [Google Scholar]
- Diao FQ, White BH. A Novel Approach for Directing Transgene Expression in Drosophila: T2A-Gal4 In-Frame Fusion. Genetics. 2012;190:1139–U1356. doi: 10.1534/genetics.111.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics. 2013;194:1029-+. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JB, Chen HH, Sattelle E, Barker PJ, Huskisson NS, Rauh JJ, Bai D, Sattelle DB. Immunocytochemical mapping of a C-terminus anti-peptide antibody to the GABA receptor subunit, RDL in the nervous system of Drosophila melanogaster. Cell Tissue Res. 1996;284:269–278. doi: 10.1007/s004410050587. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Mercado A, Fajilan AA, Lee AW, Hsu R, Mount DB, Tanouye MA. Seizure Sensitivity Is Ameliorated by Targeted Expression of K+-Cl- Cotransporter Function in the Mushroom Body of the Drosophila Brain. Genetics. 2010;184:171–183. doi: 10.1534/genetics.109.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:989–1005. doi: 10.1007/s00359-008-0386-3. [DOI] [PubMed] [Google Scholar]
- Gnerer Joshua P, Venken Koen J T, Dierick Herman A. Gene-specific cell labeling using MiMIC transposons. Nucl Acids Res. doi: 10.1093/nar/gkv113. first published online February 20, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Wang W, Salvaterra PM. Structure and organization of the Drosophila cholinergic locus. J Biol Chem. 1998;273:2706–2713. doi: 10.1074/jbc.273.5.2706. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Development genes and evolution. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Kuzin A, Kundu M, Ekatomatis A, Brody T, Odenwald WF. Conserved sequence block clustering and flanking inter-cluster flexibility delineate enhancers that regulate nerfin-1 expression during Drosophila CNS development. Gene Expr Patterns. 2009;9:65–72. doi: 10.1016/j.gep.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang HN, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56-+. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Luan H, White BH. Combinatorial methods for refined neuronal gene targeting. Curr Opin Neurobiol. 2007;17:572–580. doi: 10.1016/j.conb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Luan HJ, Diao FQ, Peabody NC, White BH. Command and Compensation in a Neuromodulatory Decision Network. J Neurosci. 2012;32:880–889. doi: 10.1523/JNEUROSCI.3707-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan HJ, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, Hsueh AJ. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci U S A. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: A marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S, Lee P-T, Campbell ME, Chen K, Anguiano-Zarate S, et al. A library of MiMICs allows tagging of genes and reversible spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4:e05338. doi: 10.7554/eLife.05338. doi: http://dx.doi.org/10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DK, Han M, Kim YC, Han KA, Taghert PH. Ap-let neurons - a peptidergic circuit potentially controlling ecdysial behavior in Drosophila. Dev Biol. 2004;269:95–108. doi: 10.1016/j.ydbio.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo LQ. The Q System: A Repressible Binary System for Transgene Expression, Lineage Tracing, and Mosaic Analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn LYM, Rhea J, Jocoy SR, Bender M. The proprotein convertase amontillado (amon) is required during Drosophila pupal development. Dev Biol. 2009;333:48–56. doi: 10.1016/j.ydbio.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhea JM, Wegener C, Bender M. The Proprotein Convertase Encoded by amontillado (amon) Is Required in Drosophila Corpora Cardiaca Endocrine Cells Producing the Glucose Regulatory Hormone AKH. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 2015 doi: 10.1038/nmeth.3250. http://dx.doi.org/10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed]
- Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Sanchez-Soriano N, Prokop A. The influence of pioneer neurons on a growing motor nerve in Drosophila requires the neural cell adhesion molecule homolog FasciclinII. J Neurosci. 2005;25:78–87. doi: 10.1523/JNEUROSCI.2377-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seruggia D, Montoliu L. The new CRISPR-Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals. Transgenic Res. 2014;23:707–716. doi: 10.1007/s11248-014-9823-y. [DOI] [PubMed] [Google Scholar]
- Shah S, Hyde DR. 2 Drosophila Genes That Encode the Alpha-Subunit and Beta-Subunit of the Brain Soluble Guanylyl Cyclase. J Biol Chem. 1995;270:15368–15376. doi: 10.1074/jbc.270.25.15368. [DOI] [PubMed] [Google Scholar]
- Sharma Y, Cheung U, Larsen EW, Eberl DF. PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in drosophila. Genesis. 2002;34:115–118. doi: 10.1002/gene.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekhaus DE, Fuller RS. A role for amontillado, the Drosophila homolog of the neuropeptide precursor processing protease PC2, in triggering hatching behavior. J Neurosci. 1999;19:6942–6954. doi: 10.1523/JNEUROSCI.19-16-06942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora AD, Spradling AC. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. P Natl Acad Sci USA. 2010;107:7389–7394. doi: 10.1073/pnas.1003180107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ehrlich I, Wolff SBE, Michalski AM, Wolfl S, Hasan MT, Luthi A, Sprengel R. Faithful Expression of Multiple Proteins via 2A-Peptide Self-Processing: A Versatile and Reliable Method for Manipulating Brain Circuits. J Neurosci. 2009;29:8621–8629. doi: 10.1523/JNEUROSCI.0359-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu JT, Lin Y, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanvactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that Control Neuromuscular Specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011a;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011b;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener C, Herbert H, Kahnt J, Bender M, Rhea JM. Deficiency of prohormone convertase dPC2 (AMONTILLADO) results in impaired production of bioactive neuropeptide hormones in Drosophila. J Neurochem. 2011;118:581–595. doi: 10.1111/j.1471-4159.2010.07130.x. [DOI] [PubMed] [Google Scholar]
- Yasuyama K, Kitamoto T, Salvaterra PM. Immunocytochemical Study of Choline-Acetyltransferase in Drosophila melanogaster- An Analysis of Cis-regulatory Regions Controlling Expression in the Brain of cDNA-transformed Flies. J Comp Neurol. 1995;361:25–37. doi: 10.1002/cne.903610103. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Song Y, Yang CH, Jan LY, Jan YN. Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat Neurosci. 2014;17:81–88. doi: 10.1038/nn.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.