I. OVERVIEW
Myoblast fusion is critical for the development, maintenance, and regeneration of skeletal muscles. Despite the identification of many fusion-related molecules in the past decades, the cellular mechanics of myoblast membrane fusion have just begun to be understood. Recent studies using the fruit fly Drosophila as a model system have revealed an asymmetric fusogenic synapse composed of an actin-enriched, invasive podosome-like structure (PLS) in one fusion partner and a thin sheath of actin underlying the apposing membrane in the other. The PLS consists of ring of cell adhesion molecules encircling the F-actin-enriched focus which propels finger-like protrusions from one cell to the other, and PLS invasion is critical for fusion pore formation. The discovery of the PLSs by both light and electron microscopy (EM) allows for the first time the identification of the sites of fusion at the ultrastructural level. These new findings pose challenges to a classic model on myoblast fusion, and provide the foundation for a new conceptual framework for understanding the cellular mechanisms of myoblast fusion in vivo.
II. INTRODUCTION
A fascinating feature of skeletal muscle development is the fusion of mononucleated muscle cells (myoblasts) to form multinucleated myotubes (Hauschka, 1994). Myoblast fusion is not only critical for myogenesis during embryonic development but also indispensible for skeletal muscle maintenance and repair during adult life (Bischoff, 1994). In addition, the fusogenic property of myoblasts has been exploited in cell-based gene therapy for the treatment of muscle degenerative disorders (Wagers & Conboy, 2005). Thus, understanding the mechanisms underlying myoblast fusion will provide significant insights into skeletal muscle biology and muscle regeneration.
Unlike epithelial cells that are tightly attached via adherence junctions, myoblasts are migratory mesodermal cells that do not have permanent adhesion partners. Accordingly, while fusion between epithelial cells “simply” involves targeting fusogenic proteins to the adherence junctions, fusion between muscle cells requires more sophisticated interactions between fusion partners in order to create a tight adhesive junction, followed by targeted transport of fusogenic materials to the sites of fusion, and ultimately cell membrane merger. Much of what we have learned about myoblast fusion to date concerns how muscle cells recognize their fusion partners and how they build a tight adhesive junction to promote membrane fusion.
III. MYOBLAST FUSION IN VERTEBRATES
Ever since the seminal finding that multinucleated skeletal muscle fibers are formed by fusion of individual myoblasts during myogenesis (Mintz & Baker, 1967), numerous studies have aimed to identify mechanisms underlying myoblast fusion. Early studies were mostly performed with cultured vertebrate myoblasts, including both primary cultures and transformed myoblast cell lines. Studies using inhibitory chemicals, antibodies and antisense oligos have provided valuable information on the types of macromolecules involved in myoblast fusion. These include cell adhesion molecules, phospholipases, protein kinases, metalloproteases and calcium binding proteins (for review see Knudsen, 1992; Wakelam, 1985). Recent studies with cultured mammalian myoblasts continued to reveal additional components involved in myoblast fusion (for review see Pavlath, 2010; Rochlin, Yu, Roy, & Baylies, 2010). Many of these molecules have been localized along the broad contact zone of muscle cells, consistent with their playing a role in myoblast fusion. However, it remains unclear how the fusion machinery is assembled by these molecules at the actual sites of membrane merger. In addition, the in vivo functions of many of these implicated molecules remain to be elucidated.
Besides studies identifying molecular components involved in myoblast fusion, electron microscopy (EM) has been employed to detect cellular changes during myoblast fusion at the ultrastructrual level. Classic EM studies revealed several intriguing structures along the muscle cell contact zone, including vesicles with or without electron-dense cores (Engel, Egar, & Przybylski, 1986; Kalderon & Gilula, 1979), gap junction-like structures (Rash & Fambrough, 1973), electron-dense plaques (Engel et al., 1986), and single or multiple openings along apposing membranes (Kalderon & Gilula, 1979; Lipton & Konigsberg, 1972; Shimada, 1971). The relevance of these intriguing structures to the actual fusion process remains uncertain, since the precise sites of fusion were not pinpointed by molecular or cellular markers in these studies. Thus, a comprehensive understanding of the fusion mechanism requires the identification of the sites of fusion at the ultrastructural level and the localization of implicated molecules relative to these sites.
IV. DROSOPHILA AS A MODEL SYSTEM TO STUDY MYOBLAST FUSION
Since the discovery of the first myoblast fusion mutant in Drosophila (Rushton, Drysdale, Abmayr, Michelson, & Bate, 1995), the fruit fly has been used as a model system to study this fusion process in vivo. In Drosophila, the larval body wall muscles start to develop during embryogenesis (Bate, 1990). Through the function of a set of transcription factors and a lateral inhibition process, two types of muscle cells are specified in the embryonic mesoderm, muscle founder cells and fusion competent myoblasts (FCMs) (for review see Baylies, Bate, & Ruiz Gomez, 1998). While muscle founder cells are divided into subgroups, each of which expresses a unique combination of transcription factors (muscle identity genes), all FCMs are specified by a single transcription factor, Lame duck (Lmd) (Duan, Skeath, & Nguyen, 2001). In the embryo, muscle founder cells reside in the outer mesodermal layer and act as sources of attraction for FCMs, which reside in deeper mesodermal layers. FCMs are attracted by founder cells, migrate toward, adhere and fuse with them to form multinucleated myotubes, each of which has its specific position, orientation, and size (Bate, 1990; Beckett & Baylies, 2007). As a consequence, the nucleus of a fused FCM adopts the transcription profile of the founder cell with which it has fused. The entire process of embryonic muscle development takes 5–6 h and each much fiber contains 3–25 nuclei (Bate, 1990), making this process a relatively simple system to dissect myoblast fusion in vivo.
At the early stage of Drosophila studies of myoblast fusion, it was not immediately apparent if mechanisms identified in flies would be readily applicable to vertebrates. This is because the first set of fusion-related genes identified in Drosophila did not correspond to the specific macromolecules revealed by myoblast culture studies. However, it rapidly became clear that most of the Drosophila fusion-related genes have vertebrate homologues, many of which have since then been demonstrated to play conserved roles in myoblast culture and/or zebrafish/mouse models (Bach et al., 2010; Kim et al., 2007; Laurin et al., 2008; Moore, Parkin, Bidet, & Ingham, 2007; Nowak, Nahirney, Hadjantonakis, & Baylies, 2009; Pajcini, Pomerantz, Alkan, Doyonnas, & Blau, 2008; Srinivas, Woo, Leong, & Roy, 2007; Vasyutina, Martarelli, Brakebusch, Wende, & Birchmeier, 2009). Thus, myoblast fusion in Drosophila shares highly conserved molecular components with vertebrates and the basic mechanisms revealed by fly studies appear to be readily applicable to higher organisms.
V. MYOBLAST FUSION IN DROSOPHILA
For any migratory cell to fuse with a partner, a key event is to establish a tight adhesive junction where the two cell membranes can be brought to intimate proximity. Studies of myoblast fusion in Drosophila to date suggest that formation of close membrane juxtaposition between muscle founder cells and migratory FCMs is a two-step process that involves the initial muscle cell attachment via cell adhesion molecules and a subsequent reinforcement of membrane apposition via the action of the actin cytoskeleton.
A. Initial Recognition/Attachment Between Muscle Cells
The initial recognition between founder cells and FCMs is mediated by cell type-specific cell adhesion molecules containing Ig domains (Fig. 1). In founder cells, a pair of paralogues, Dumbfounder (Duf)/Kin of IrreC (Kirre) and Rougest (Rst)/IrreC, have redundant functions during myoblast fusion (Ruiz-Gomez, Coutts, Price, Taylor, & Bate, 2000; Strunkelnberg et al., 2001). Thus, loss of either duf or rst does not lead to a fusion defect, and expression of either duf or rst can rescue the fusion defect in the duf rst double mutant embryos. Rst is expressed in both types of muscle cells, but is only demonstrated to function in founder cells (Shelton, Kocherlakota, Zhuang, & Abmayr, 2009; Strunkelnberg et al., 2001). FCMs specifically express a second pair of paralogues, Sticks and stones (Sns) (Bour, Chakravarti, West, & Abmayr, 2000) and Hibris (Hbs) (Artero, Castanon, & Baylies, 2001; Dworak, Charles, Pellerano, & Sink, 2001), which play partially redundant functions during myoblast fusion (Shelton et al., 2009). While the hbs mutant appears largely wild type (Artero et al., 2001; Dworak et al., 2001), the sns mutant exhibits a significant fusion defect (Bour et al., 2000), which can be partially rescued by Hbs overexpression (Shelton et al., 2009).
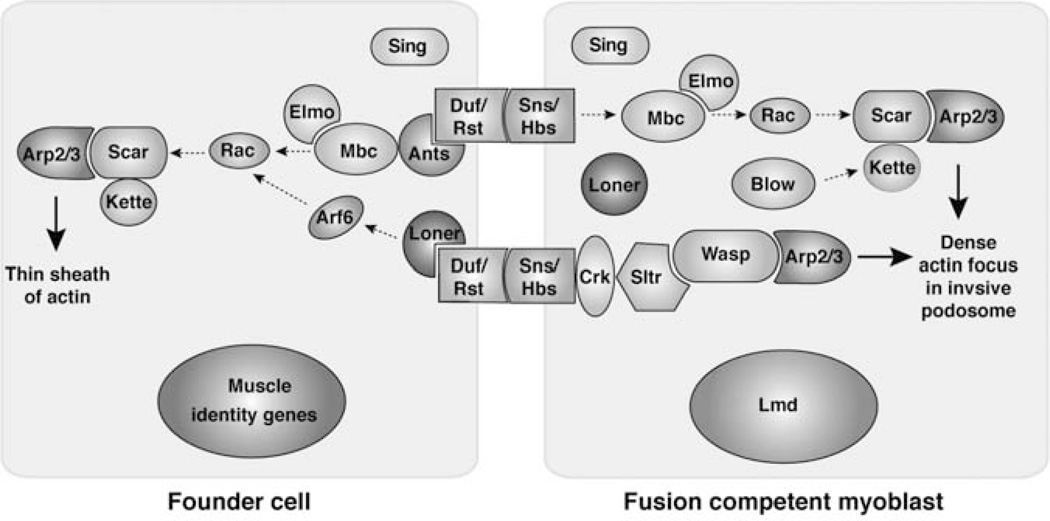
FIGURE 1.
Signaling pathways controlling myoblast fusion in Drosophila. A simplified diagram describing the signaling pathways in founder cell and fusion competent myoblasts. See text for details.
Among these cell adhesion molecules, the localization and function of Duf and Sns have been extensively characterized and both proteins have been localized to muscle cell contact sites. By confocal microscopy, Duf and Sns appear to colocalize at these contact sites despite the fact that they are expressed in different cell types (Galletta, Chakravarti, Banerjee, & Abmayr, 2004; Sens et al., 2010). This is likely due to the close membrane apposition at muscle cell contact sites and the limited resolution of the light microscopy (the thickness of cell membranes is ~10 nm and the resolution of light microscopy is 200 nm). Duf physically interacts with Sns, and this interaction has been proposed to mediate the adhesion between founder cells and FCMs (Galletta et al., 2004). Interestingly, ectopic expression of Duf (or Rst) in epithelial cells can attract FCMs to migrate toward these cells (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001), demonstrating that Duf (or Rst) expression is sufficient to attract the Sns-expressing FCMs and that founder cells are the organizing centers for myoblast fusion. It remains unclear, however, how Duf-expressing cells attract FCMs. One possibility is that both founder cells and FCMs extend filopodia-like membrane protrusions to probe their environment, and once the filopodia from a founder cell contacts those from an FCM, the interaction between Duf and Sns would lead to polarized migration of the FCM toward the founder cell. Alternatively, the Duf-expressing cells may send out a secreted signal that forms a concentration gradient to attract the FCMs. In this regard, Duf has been shown to be cleaved to release its ectodomain in cultured cells (Chen & Olson, 2001). Whether such a cleavage occurs in vivo and, if so, its physiological relevance requires future investigations.
B. Signal Transduction from the Cell Membrane to the Actin Cytoskeleton
Upon the engagement of transmembrane adhesion molecules, such as Duf and Sns, the cytoplasmic domains of these molecules are involved in transducing the signal for cell fusion to the actin cytoskeleton (Bulchand, Menon, George, & Chia, 2010; Galletta et al., 2004; Kocherlakota, Wu, McDermott, & Abmayr, 2008) (Fig. 1). In founder cells, Duf recruits Antisocial (Ants)/Rolling pebbles (Rols), an adaptor protein containing ankyrin repeat, TPR repeat, and coiled-coil domains (Chen & Olson, 2001; Menon & Chia, 2001; Rau et al., 2001), to sites of fusion via protein–protein interactions (Chen & Olson, 2001). In turn, Ants/Rols has been shown to be involved in recycling Duf to sites of fusion (Menon, Osman, Chenchill, & Chia, 2005). A potential link between Ants/Rols and the actin cytoskeleton was suggested by the biochemical interaction between Ants and Myoblast city (Mbc) (Chen & Olson, 2001), an actin cytoskeleton-associated protein required for fusion (Erickson et al., 1997; Rushton et al., 1995). Mbc is the Drosophila homologue of DOCK180, which, in complex with Elmo, functions as a bipartite guanine nucleotide exchange factor (GEF) for the small GTPase, Rac (Brugnera et al., 2002). Both Elmo and Rac have been shown to be required for myoblast fusion (Geisbrecht et al., 2008; Hakeda-Suzuki et al., 2002). Rac regulates a variety of cellular processes by promoting site-specific actin polymerization (for review see Etienne-Manneville & Hall, 2002). For example, Rac is thought to activate and/or localize Scar (also known as WAVE), a nucleation promoting factor (NPF) for the Arp2/3 complex, which is a seven-protein complex with actin nucleation activity (for review see Kurisu & Takenawa, 2009; Stradal & Scita, 2006; Takenawa & Suetsugu, 2007). Scar resides in a heteropentameric Scar complex with Kette (Nap1/Hem), Sra1 (PIR121/CYFIP), Abi, and HSPC300 (for review see Ibarra, Pollitt, & Insall, 2005). Both the Scar and Arp2/3 complexes are essential for myoblast fusion (Berger et al., 2008; Richardson, Beckett, Nowak, & Baylies, 2007; Schroter et al., 2004). In particular, the Scar complex has been shown to localize in a broad domain at muscle cell contact sites and is required in both founder cells and FCMs (Richardson et al., 2007; Sens et al., 2010). In addition to Mbc, a second GEF, Loner, has been shown to partially colocalize with Ants and is required for myoblast fusion (Bulchand et al., 2010; Chen, Pryce, Tzeng, Gonzalez, & Olson, 2003). Loner belongs to the Arf family of GEFs and has specific activity toward Arf6 in vitro (Chen et al., 2003). Although an Arf6 mutant did not exhibit a fusion defect (Dyer et al., 2007), overexpression of a dominant negative form of Arf6 partially blocked fusion (Chen et al., 2003), suggesting that there may be functional redundancy among Arf family members and that Loner may act through other Arfs in the absence of Arf6. A link between the Loner/Arf module and the actin cytoskeleton is shown by the ability of Loner to regulate the proper localization of Rac (Chen et al., 2003). Interestingly, Loner is also expressed in FCMs (Richardson et al., 2007), although its precise function in this cell population requires further investigations.
In FCMs, two Arp2/3 NPFs, Scar and WASP, function together to activate actin polymerization (Fig. 1). The recruitment of Scar to the site of fusion in FCMs may be mediated by Rac, since Scar’s enrichment at the leading edge of FCM disappears in rac mutant embryos (Gildor, Massarwa, Shilo, & Schejter, 2009). Notably, a member of the Scar complex, Kette, has been shown to genetically interact with a PH domain-containing protein, Blown fuse (Blow) (Doberstein, Fetter, Mehta, & Goodman, 1997), although the molecular function of Blow remains unknown (Schroter et al., 2004). While Scar is functionally required in both cell populations, WASP appears to function only in FCMs, since overexpression of a dominant-negative form of WASP (WASPDN) generated a fusion defect when WASPDN was expressed in all muscle cells, but not in founder cells alone (Schafer et al., 2007). Moreover, the WASP-interacting protein, Solitary (Sltr)/dWIP/Vrp1, is only expressed in FCMs (Kim et al., 2007), and is recruited to sites of fusion by the FCM-specific adhesion molecule Sns (Kim et al., 2007; Massarwa, Carmon, Shilo, & Schejter, 2007). The cytoplasmic domain of Sns binds Crk (Kim et al., 2007), a small SH2 and SH3 domain-containing adaptor protein (Galletta, Niu, Erickson, & Abmayr, 1999). Crk, in turn, interacts with Sltr, which forms a tight complex with WASP (Kim et al., 2007) and recruits WASP to sites of fusion (Massarwa et al., 2007; Schafer et al., 2007). The FCM-specific function of the WASP–Sltr complex raises an interesting possibility that FCMs may have a different pattern of actin polymerization than founder cells.
C. An Invasive Podosome-Like Structure at the Site of Fusion
1. F-Actin Foci are Cell-Type Specific
The recruitment of actin cytoskeleton-associated proteins to sites of fusion suggests that there may be active actin polymerization at these sites. Indeed, a dense, F-actin-enriched structure (referred to as an F-actin focus or plug) is observed between pairs of founder cells/myotubes and FCMs and is associated with the cell adhesion molecules Duf and Sns (Kesper et al., 2007; Kim et al., 2007; Richardson et al., 2007). Live imaging analyses revealed that these F-actin foci are transient structures that appear and dissolve at the site of fusion prior to each fusion event, with an average lifespan of ~11.9 min (ranging from 5.7 to 29.5 min) (Richardson et al., 2007). Initially, the F-actin foci were thought to be symmetrically localized across the adherent muscle cell membranes (Richardson et al., 2007). However, the extensive colocalization of the F-actin foci with the FCM-specific protein Sltr (Kim et al., 2007) suggests that the majority of the actin polymerization many occur in FCMs. Indeed, by expressing GFP-actin in specific muscle cell types and comparing the accumulation and localization of GFP-actin versus total F-actin (stained with phalloidin), it became clear that these actin-enriched structures were exclusively generated in FCMs (Sens et al., 2010). This conclusion was further supported by live imaging experiments in which dynamic GFP-actin foci were only observed when GFP-actin was specifically expressed in FCMs, but not in founder cells (Sens et al., 2010).
2. FCM-Specific Actin Foci are Invasive
A striking feature of the FCM-specific actin focus is its invasion into the territory of the apposing founder cell, which creates a V- or U-shaped dent on the founder cell membrane (Sens et al., 2010). Three-dimensional reconstruction of the F-actin foci showed that they reside in cup-shaped dents, with the wall of the cup lined with the cell adhesion molecules Duf and Sns (Fig. 2) (Sens et al., 2010). Thus, when viewed along the vertical/longitudinal axis of the cup, an actin focus appears to be encircled by the cell adhesion molecules, Duf and Sns inner (Fig. 2) (Kesper et al., 2007; Sens et al., 2010). Live imaging of these F-actin foci revealed extensive size and shape changes during their lifespan (Sens et al., 2010), suggesting a dynamic actin polymerization process. Taken together, these light microscopic studies suggest that the dynamic, FCM-specific actin foci can invade into the founder cell and deform the founder cell membrane.
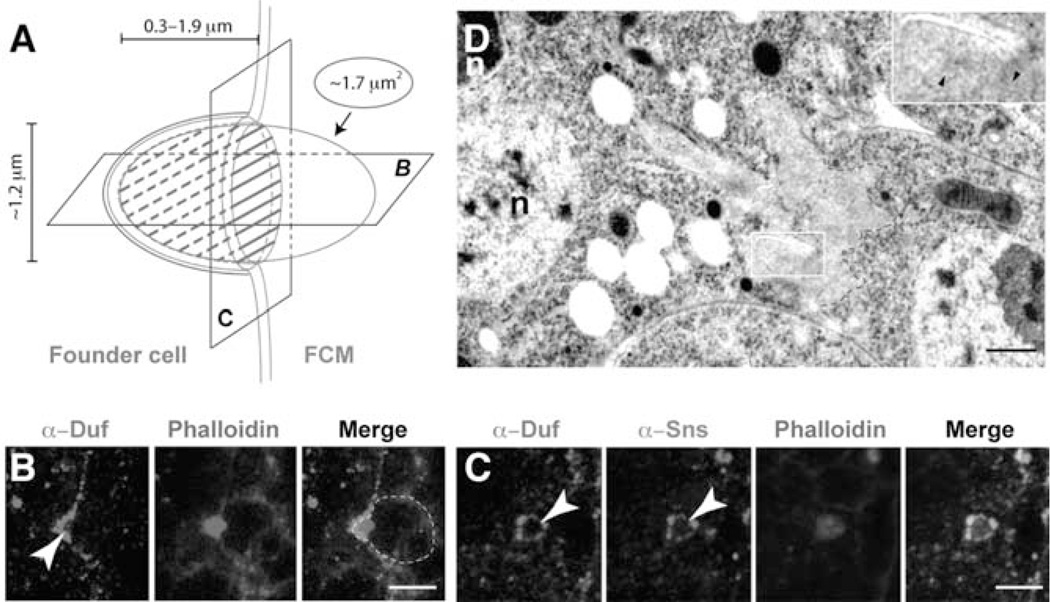
FIGURE 2.
The FCM-specific F-actin focus invades the apposing founder cell. (A) Schematic drawing of the asymmetric muscle cell adhesion junction. An F-actin focus (green oval) forms at the tip of the FCM(right) and invades the apposing founder cell (left) to create a cup-shaped dimple. The inner wall of the cup is lined with Sns (blue), and the outer wall with Duf (red). Numbers show average actin foci size (1.7 µm2), diameter of the adhesive rings (1.2 µm), and depth of invasion (0.3–1.9 µm). (B and C) Confocal images of wild-type embryos showing horizontal (B) and perpendicular (C) pairs of founder cell/myotube and FCM. Embryos labeled with α-Duf (red), phalloidin (green) and α-Sns (blue). (B) An F-actin focus in an FCM (outlined) invading a founder cell. Arrowhead indicates the inward curvature on the founder cell membrane. (C) An F-actin focus encircled by cell adhesion molecules (arrowheads). (D) Ultrastructural details of an invasive F-actin focus. An FCM (pseudo colored pink) projects multiple F-actin-enriched invasive fingers into a binucleated myotube in a wild-type (wt) embryo fixed by HPF/FS. The F-actin-enriched area within the FCMs (boundary marked by dashed green lines) is identified by the light gray coloration and lack of ribosomes and intracellular organelles. Magnified inset shows faint actin filaments (arrowheads) within an invasive finger. n: nuclei in myotube. Scale bars: (B and C) 5 µm; (D) 500 nm. (See Color Insert.)
Ultrastructural studies by EM also revealed an F-actin-enriched structure between founder cells/myotubes and FCMs (Sens et al., 2010). This structure is localized exclusively at the tip of FCMs, not in founder cells. The area of F-actin enrichment is similar to that of an actin focus under light microscopy. Most strikingly, the tip of the FCM-specific actin focus is composed of multiple finger-like protrusions invading into the apposing founder cell (Fig. 2) (Sens et al., 2010). The remarkable similarities in the sidedness, size, and invasiveness between the F-actin-enriched structures revealed by light microscopy and EM demonstrate that they correspond to the same cellular structure. Since the F-actin foci observed by light microcopy effectively mark the sites of fusion, the invasive F-actin-enriched structures identified by EM, therefore, provide the first unambiguous cellular marker for myoblast fusion sites at the ultrastructural level.
3. Invasive F-Actin Focus Resembles a Podosome
The actin-enriched foci that form at the junction between fusing muscle cells resemble podosomes, which have been identified in a variety of cultured cells, including monocytic, endothelial, smooth muscle, and skeletal muscle cells (for review see Gimona, Buccione, Courtneidge, & Linder, 2008; Linder, 2009). Podosomes, also called invadopodia in cancer cells, are bipartate structures consisting of an F-actin-enriched core encircled by cell adhesion molecules. They are associated with extracellular matrix (ECM) protease secretion and play important roles in cell adhesion, migration, and invasion (for review see Linder, 2009; Weaver, 2006). Tens or even hundreds of podosomes are often present in a belt-like pattern at the periphery of a single cultured cell. The FCM-specific actin focus encircled by Sns and Duf resembles a single podosome, judged by its size, dynamics, morphology, and protrusive behavior, and is therefore named a podosome- like structure (PLS) (Sens et al., 2010). The FCM-specific PLS represents the first PLS identified in an intact developing tissue. It will be interesting to determine whether this PLS is also associated with ECM protease secretion.
4. A Thin Sheath of Actin in Founder Cell
Founder cells do not form invasive PLSs during myoblast fusion. In fact, wild-type embryos expressing GFP-actin in founder cells do not show visible GFP-actin enrichment at sites of fusion (Sens et al., 2010). It is possible that actin polymerization at the sites of fusion in founder cells is transient and/or masked by the overexpressed GFP-actin. Consistent with this, in certain mutants where fusion is blocked, actin enrichment has been observed as a thin sheath that underlies the founder cell membrane apposing the FCM-specific actin focus (Sens et al., 2010). Thus, founder cells respond to the PLS invasion by making a transient thin sheath of actin at the sites of fusion (Fig. 2).
5. Molecular Components of the PLS
The formation of the F-actin foci with in the FCM-specific PLSs requires both the Scar and WASP complexes, based on the fact that the number and size of actin foci are greatly diminished in scar sltr, as well as in sltr kette, double mutant embryos (Sens et al., 2010; Sens & Chen, unpublished data). These results are in contrary to a previous report showing enlarged F-actin foci when both the Scar and WASP complexes were disrupted (Gildor et al., 2009). This discrepancy might be due to the differences in the way by which the Scar and WASP function were disrupted (mutant alleles versus overexpression of dominant negative forms of proteins) (Gildor et al., 2009; Sens et al., 2010). In support of the conclusion that both Scar and WASP play a role in F-actin foci formation, Sltr precisely colocalizes with the enlarged actin foci in scar or kette single mutant embryos (Sens et al., 2010), suggesting that the WASP–Sltr complex promotes actin polymerization in the absence of Scar or Kette. Conversely, recruitment of Scar to cell contact sites is greatly elevated in sltr mutant embryos (Sens et al., 2010), and is likely responsible for promoting actin foci formation in the absence of WASP–Sltr. Taken together, Scar and WASP are the major Arp2/3 NPFs activating actin polymerization at sites of fusion.
Although Scar and WASP play redundant roles in actin foci formation, overexpressing one of them in the absence of the other does not rescue the fusion defect in embryos (Sens et al., 2010), suggesting that Scar and WASP have distinct functions during the fusion process. Indeed, the WASP-Sltr complex, but not the Scar complex, promotes PLS invasion. Thus, while F-actin foci in scar and kette mutants are as invasive as wild type, those in wasp and sltr mutants are defective in their invasion (Sens et al., 2010). The latter has been confirmed by ultrastructural analysis, which revealed actin-enriched fingers from FCMs folding upon each other without protruding into the apposing founder cell (Sens et al., 2010). On the other hand, the Scar complex plays an independent role in founder cells to promote the formation of the transient thin sheath of actin along the F-actin focus (Sens et al., 2010).
D. Invasive PLSs Promote Fusion Pore Formation
Genetic and cell biological studies suggest that the invasive PLSs promote fusion pore formation. In sltr mutant embryos where PLS invasion is defective, cytoplasmic GFP failed to diffuse between myotubes and attached FCMs (Kim et al., 2007; Sens et al., 2010). Furthermore, EM analysis of sltr mutant embryos revealed the absence of pores on the cell membranes abutting the F-actin-enriched foci, which correspond to muscle cell contact sites, further supporting this conclusion (Sens et al., 2010). Two conflicting reports showing cytoplasmic GFP transfer from founder cells to FCMs in dWIP and wasp mutants led to the interpretation that the WASP complex was not required for fusion pore formation, but was required for fusion pore expansion (Gildor et al., 2009; Massarwa et al., 2007). However, these results can be explained by leaky GFP expression in FCMs by the founder cell driver (rP298-GAL4) (Sens et al., 2010). In addition to the invasive PLS, regulated actin polymerization by the Scar complex in founder cells also promotes fusion pore formation, since founder cell-expressed GFP does not diffuse into attached FCMs in kette mutant embryos (Gildor et al., 2009; Sens et al., 2010).
How does an invasive PLS promote fusion pore formation? Several potential mechanisms are considered here. First, the invasive PLS in the FCM and the thin sheath of actin in the apposing founder cell could mechanically push the two cell membranes against each other and bring them to very close proximity, thus priming them for fusion. Interestingly, podosome-like protrusions generated by leukocytes are thought to bring the inner leaflets of the apical and basal membranes of endothelial cells together for the formation of transcellular pores (Carman et al., 2007). Since PLSs with folded, noninvasive fingers in sltr mutant embryos fail to promote fusion pore formation, it is likely that the invasive tips of the protruding fingers in wild-type embryos are fusogenic. Second, the FCM-specific podosome could be associated with protease secretion, give that all podosomes studied so far are associated with protease activity (Linder, 2009). Secreted proteases may degrade ECM proteins and/or the ectodomains of the Ig domain-containing cell adhesion molecules, to remove any physical barriers between the two cell membranes, in order to achieve tight membrane juxtaposition. And third, rapid polymerization at the barbed ends of several actin filaments could generate a local membrane curvature at the tip of an invasive finger. In light of the proposed role of membrane curvature in fusion pore formation (Kozlov, McMahon, & Chernomordik, 2010), a sharply bent local curvature created by the corporative extension of several actin filaments may facilitate membrane fusion. These potential mechanisms are likely to be used in combination to regulate fusion pore formation.
E. A Single-Channel MACRO Fusion Pore Mediates Myoblast Fusion
Fusion pore formation is the hallmark of any membrane fusion event. A classic EM study of Drosophila myoblast fusion has led to the conclusion that fusion pores are multiple membrane discontinuities (MMDs) formed along the muscle cell contact zone (Doberstein et al., 1997). Each pore is 50–100 nm in width, and there are no cytoplasmic materials in the lumen (Doberstein et al., 1997). Several groups have since reproduced such fusion pore morphology (Berger et al., 2008; Kim et al., 2007; Massarwa et al., 2007), thus making it a widely accepted view. However, these EM studies were performed using conventional chemical fixation method at room temperature, which is known to generate artifacts including membrane discontinuities (McDonald & Auer, 2006; Zhang & Chen, 2008). Indeed, the same fixation method also resulted in the presence of MMDs between cells that do not normally fuse, as well as between muscle cells in fusion mutants (Sens et al., 2010). In contrast, wild-type and mutant embryos fixed by the high-pressure freezing and freeze substitution (HPF/FS) method, which provides better preservation of the cell membrane (McDonald & Auer, 2006; Zhang & Chen, 2008), do not show any MMDs (Sens et al., 2010). Additional EM studies using the HPF/FS method revealed single-channel macro fusion pores with a diameter between 300 nm and 1.5 µm (Sens et al., 2010). The lumens of these single-channel fusion pores are filled with evenly distributed cytoplasmic materials, indicating active cytoplasmic exchange between the two fusing cells. Interestingly, pores smaller than 300 nm have not been detected thus far (Sens et al., 2010), likely due to the rapid expansion of nascent fusion pores with a diameter smaller than 200 nm (Kaplan, Zimmerberg, Puri, Sarkar, & Blumenthal, 1991; Plonsky, Cho, Oomens, Blissard, & Zimmerberg, 1999; Plonsky & Zimmerberg, 1996).
It is unclear at this point how many nascent fusion pores form during each fusion event. Since each F-actin focus is composed of multiple (~ 4.3 on average) finger-like protrusions (Sens et al., 2010), multiple nascent fusion pores could potentially form during each fusion event. However, all protrusive fingers extend to different directions and exhibit different depth of invasion, making it unlikely that they would all promote fusion pore formation simultaneously. Alternatively, once a single nascent fusion pore has formed at one of the protrusive finger tips, downstream events, such as actin depolymerization and fusion pore expansion, may proceed and prevent the initiation of additional pores. If the latter is the case, why does each PLS generate multiple invasive fingers? It is conceivable that muscle cell membranes contain specific fusogenic domains of particular lipids and/or proteins required for fusion. Close contact between these fusogenic domains across the apposing cell membranes is likely to be a stochastic event mediated by the protrusive fingers. This possibility is consistent with the wide range of time (between 5.7 and 29.5 min) required for each PLS to promote fusion pore formation (Richardson et al., 2007). Thus, the presence of multiple exploratory fingers in each PLS ensure the successful engagement of fusogenic materials across the two cell membranes, and ultimately fusion pore initiation. Future studies are required to test this hypothesis.
What mediates the expansion of nascent fusion pores? Based on the observation of MMDs in wild-type and fusion mutant embryos (dWIP and wasp) prepared by chemical fixation, it was proposed that fusion pore expansion is mediated by membrane vesiculation and/or actin polymerization (Doberstein et al., 1997; Gildor et al., 2009; Massarwa et al., 2007). However, these proposals need to be reevaluated in light of the recent finding of invasive PLSs and single-channel macro fusion pores revealed by HPF/FS EM. The previously reported MMDs in mutant embryos of Arp2/3 NPFs (Gildor et al., 2009; Massarwa et al., 2007) were not associated with F-actin enrichment, which marks the muscle cell contact sites, consistent with the idea that these MMDs may have been caused by fixation artifacts on random segments of the cell membrane. Furthermore, single-channel macro fusion pores in wild-type embryos are not associated with vesicles/membrane sacs or F-actin (Sens et al., 2010), suggesting that neither membrane vesiculation nor actin polymerization plays a role in fusion pore expansion, at least for pores with a diameter of over 300 nm. These results are consistent with the findings by Chen et al. (2008) on virus (gp64)-induced fusion of Sf9 cells, which suggest that fusion pore expansion is not mediated by membrane vesiculation or actin polymerization.
F. The Elusive Fusogen
Although actin polymerization is essential for myoblast fusion and may play a role in bringing the adherent membranes into close proximity, it remains likely that additional proteins with transmembrane domains are involved in initiating the actual membrane merger. To date, none of the transmembrane proteins implicated in Drosophila myoblast fusion fulfill the definition of a “fusogen,” which should be both necessary and sufficient to induce membrane fusion. There are several plausible explanations for why a fusogen for myoblast fusion, if there is one, has not yet been identified by genetic approaches. First, genetic screens for fusion mutants have not been saturated. Second, the putative fusogenic proteins may be maternally contributed, which would be make it difficult to be identified by a zygotic screen in embryos. Third, there may be more than one fusogenic proteins that have redundant functions, thus a simple loss-function screen would not be sufficient to identity them. Finally, the putative fusogen may play an essential role during early embryonic development, such that mutants in the gene may not survive till the mid-embryogenesis stage to allow functional analyse in myoblast fusion.
If there is indeed a fusogen required for fusing muscle cell membranes, how is it delivered to the sites of fusion? Since all known fusogens are transmembrane proteins, the myoblast fusogen(s) is likely transported to the sites of fusion by vesicles trafficking. Early EM studies revealed clusters of paired vesicles with electron-dense rims aligned along the muscle cell contact sites (Doberstein et al., 1997). It was proposed that these sites correspond to sites of fusion and that exocytosis of these vesicles may release fusogenic materials to trigger fusion (Doberstein et al., 1997). However, two observations raised questions about the relevance of these vesicles to the sites of fusion in wild-type embryos. First, the frequency of the vesicle clusters shown in Doberstein et al. far exceeds that of the F-actin-enriched foci at any developmental stage in wild-type embryos (Kim et al., 2007; Zhang & Chen, 2008). Second, Doberstein et al. (1997) described that these vesicles are present during stage 13 and disappear by stage 14 in wild-type embryos, yet most of the fusion events occurs during stage 14 (Beckett & Baylies, 2007). The recent discovery of invasive PLSs that mark the sites of fusion at the ultrastructural level provides an opportunity to address these questions. Strikingly, the previously reported clusters of vesicles (Doberstein et al., 1997) are not associated with the F-actin-enriched invasive PLSs, and are therefore unlikely to be localized to the sites of fusion. Instead, these vesicles may have been mistargeted to other membrane adhesion sites, for example, between FCMs, as in sltr mutant embryos (Kim et al., 2007). Indeed, clusters of paired vesicles are not frequently observed in wild-type embryos (Estrada et al., 2007; Kim et al., 2007; Zhang & Chen, 2008), and the few vesicles seen in wild-type embryos are mostly localized in the cytoplasm but rarely at the PLS (Sens et al., 2010). Taken together, clusters of paired vesicles do not appear to mark the sites of fusion in wild-type embryos as previously suggested, and vesicle accumulation at ectopic sites only occurs in certain fusion mutants.
Although vesicles do not accumulate to the sites of fusion in wild-type embryos, several pieces of evidence suggest that vesicle trafficking plays a role in myoblast fusion. First, there is a gradual increase in the amount of the Ig domain-containing cell adhesion molecules at each site of fusion (Menon et al., 2005), indicating a continuous transport of these molecules to these sites. Second, a MARVEL domain protein, Singles Bar (Sing) is required for myoblast fusion (Estrada et al., 2007). Interestingly, MARVEL domain proteins in mammals have been implicated in vesicular trafficking and several members of the MARVEL protein family are components of transport vesicles (Sanchez-Pulido, Martin-Belmonte, Valencia, & Alonso, 2002). Although Sing does not appear to be required for transporting cell adhesion molecules to sites of fusion (Estrada et al., 2007), it could be a functional component of vesicles that trafficking other fusion-related components. And third, vesicles with electrondense rims have been observed at the vicinity of the Golgi and associated with microtubules that point toward the muscle cell periphery (Kim et al., 2007), suggesting that the Golgi-derived vesicles are likely transported by microtubules to the cell membrane in adherent muscle cells. If these vesicles are involved in myoblast fusion, why are they scarcely observed in muscle cells of wild-type embryos? The simplest explanation would be that they undergo rapid exocytosis once they reach the site of fusion, without accumulating in the cytoplasm or at the cell membrane. Additional studies will be required to clearly define the components and trafficking of these intriguing vesicles.
G. Challenge to an Old Model
For the past decade, a prevailing model has been proposed to describe the ultrastructural events associated with myoblast fusion in Drosophila (Doberstein et al., 1997). Upon the adhesion of a founder cell and an FCM, pairs of prefusion vesicles from the two muscle cells align across the apposed cell membranes. This prefusion complex resolves into dense membrane plaques between apposed cells. The adherent cells then establish cytoplasmic continuity by fusing the cell membranes at multiple positions along the cell contact zone, followed by membrane vesiculation and fusion pore expansion. However, in light of the unambiguous marking of the sites of fusion by the invasive F-actin foci at the ultrastructural level, much of this classic model needs to be revised.
First, despite the presence of prefusion vesicles in muscle cells, they do not appear to accumulate, singularly or as pairs, at the site of fusion in wild-type embryos. Extensive accumulation of these vesicles only occurs in certain mutant embryos and at membrane adhesion sites without enriched F-actin, presumably due to defects in vesicle targeting and/or exocytosis in these particular mutants. Second, the dense membrane plaques, if they exist in wild-type embryos, may result from the exocytosis of continuously trafficked prefusion vesicles, instead of a synchronized release from accumulated paired vesicles, given the scarcity of the latter. Third, fusion pores have a single-channel morphology, instead of containing multiple neighboring membrane openings. And finally, fusion pore expansion is not likely to be mediated by membrane vesiculation. Given the multiple inaccuracies of the classic model, a new conceptual framework is required to describe the fusogenic structure in Drosophila (see below).
H. The Asymmetric Fusogenic Synapse
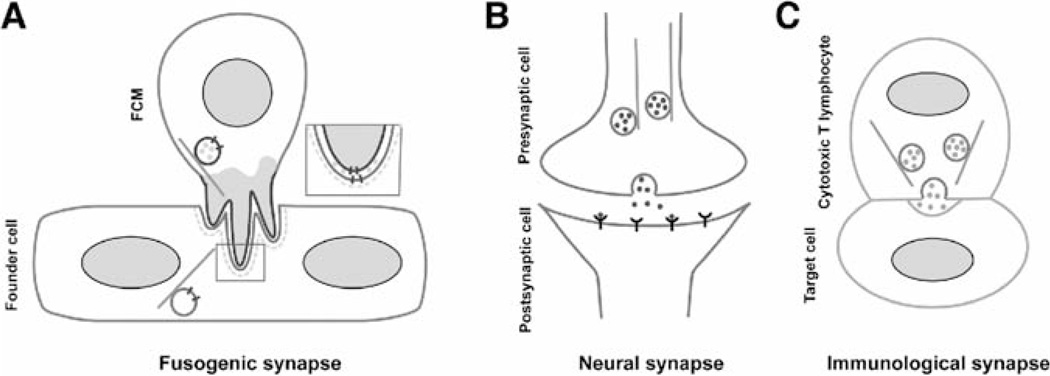
Based on the recent light and EM studies, the fusogenic structure mediating myoblast fusion can be described as follows (Fig. 3A). It is an asymmetric adhesive junction composed of an F-actin-enriched, invasive PLS from the FCM and a thin sheath of actin underlying the apposing founder cell membrane. Within the boundary of the adhesive rings (or “cups” viewed in 3D space), the PLS extends multiple protrusive fingers to palpitate the founder cell membrane. Meanwhile, prefusion vesicles that are trafficked from the Golgi to the cell membrane via microtubules continuously deposit cell adhesion molecules and perhaps fusogenic proteins and/or lipids to the fusion site. At the tip of one of the invasive fingers, the cell membranes are brought into intimate proximity, such that the putative fusogen can be engaged across the adherent cell membranes, leading to fusion pore formation. Once a nascent fusion pore is formed, rapid actin depolymerization occurs to allow fusion pore expansion. Ultimately, the FCM integrates into the founder cell/myotube and completes a round of fusion. The asymmetric fusogenic structure mediating myoblast fusion is analogous to two known synapses, neural synapse, and immunological synapse (Fig. 3) (Billadeau, Nolz, & Gomez, 2007; Dillon & Goda, 2005; Dustin, 2005; Salinas & Price, 2005; Stinchcombe & Griffiths, 2007). They are all relatively stable adhesive structures that are targets for polarized vesicle trafficking, despite the differences in cell type, adhesion molecule, and physiological function. Since the adhesive junction in myoblast fusion eventually leads to cell membrane fusion, it is accordingly referred to as a “fusogenic synapse” (Fig. 3).
FIGURE 3.
The fusogenic synapse shares common features with neural synapse and immunological synapse. Fusogenic synapse (A), neural synapse (B), and immunological synapse (C) are all relatively stable adhesive junctions that are targets for vesicle trafficking. In (A), Ig domain-containing cell adhesion molecules, Duf in the founder cell and Sns in the FCM, mediate the adhesion between the two muscle cells. Engagement of Duf and Sns leads to the formation of asymmetrically localized actin-based structures, an invasive PLS in the FCM (shaded area) and a thin sheath of actin (dashed line) in the founder cell. Adhesion molecules for neural and immunological synapses are not shown in (B and C). In (A–C), vesicles are trafficked along microtubules (light gray lines) to the adhesive junctions. (A) Vesicles in muscle cells may contain fusogenic protein(s) (short dark lines) and putative ECM proteases (gray dots) associated with the PLS. Boxed area is enlarged to show the putative fusogen on cell membranes and the ECM proteases in the intermembrane space. (B) Neural transmitters (gray dots) are transported to the synaptic cleft between the pre- and postsynaptic cells. (C) Pore-forming proteins and esterases (gray dots) are transported to the cleft between the cytotoxic T lymphocyte and the target cell.
Why is the fusogenic synapse asymmetric? In principle, following the adhesion between a founder cell and an FCM, the cell type-specific adhesion molecules could have organized symmetrical PLSs to push the membranes to closer proximity. However, a potential disadvantage of this configuration is that the two actively protruding structures may not be able to coordinate their activities to effectively respond to the invasion from the apposing cell. On the other hand, an invasive structure from only one of the fusing partners can create a “key and lock” configuration, if its fusion partner only needs to respond to its invasion. Thus, the tight membrane juxtaposition generated by an asymmetric invasion may be a more efficient way to initiate fusion pore formation.
I. Same Cellular Mechanism Underlying Each Fusion Event
An important yet unresolved question in myoblast fusion is whether each fusion event is mediated by the same molecular and cellular mechanisms. Previous studies have shown that myoblast fusion in Drosophila embryos proceeds in two temporal phases. In the first phase, bi- or tri-nucleated muscle precursor cells form from fusion between FCMs and mononucleated founder cells or bi-nucleated myotubes (Bate, 1990). These muscle precursor cells appear to pause for a period of time before starting the second phase of fusion, in which they fuse with additional FCMs to form multinucleated myotubes (Beckett & Baylies, 2007). Two different models have been proposed to describe the mechanisms underlying the two temporal phases of fusion based on genetic observations. One model states that the two phases of fusion utilize distinct molecular mechanisms, since some fusion mutants seem to block fusion at the stage of bi- or tri-nucleated myotubes (Rau et al., 2001). However, detailed analyses of these fusion mutants revealed that not all founder cells form bi- or tri-nucleated myotubes (some remain mononucleated) and the limited rounds of fusion are delayed in fusion mutants (Beckett, Rochlin, Duan, Nguyen, & Baylies, 2008). These observations led to a second model in which all fusion-related genes are equally required in the two temporal phases of fusion and that the formation of occasional bi- or tri-nucleated myotubes in fusion mutants is due to maternal contributions in the zygotic mutants (Beckett & Baylies, 2007).
The discovery of the asymmetric fusogenic synapse provides an opportunity to address this question at a cellular level and the new evidence so far lends support to the second model described above. EM studies with serial sections have shown that invasive PLSs are present not only between FCMs and multinucleated myotubes but also between FCMs and mononucleated founder cells (Luo and Chen, unpublished data; Sens et al., 2010). This is consistent with the live-imaging observation that each fusion event is mediated by an F-actin focus (Richardson et al., 2007). In addition, the formation of bi- or tri-nucleated myotubes in strictly zygotic fusion mutants is likely due to inefficient functional compensations by other components of the fusion machinery. For example, in sltr mutant, elevated level of the Scar complex promotes the formation of F-actin foci that only exhibit occasional invasive behavior toward founder cells with aberrant fingers (fewer, shorter, and wider) (Sens et al., 2010). These abnormal invasive fingers likely account for the delayed, inefficient, and somewhat random pattern of fusion (resulting in the formation of miniature myotubes with one to four nuclei) (Kim et al., 2007). Taken together, these findings support and extend the model that all fusion events in wild-type embryos (including those in both first and second temporal phases) use the same molecular and cellular machineries to induce myoblast fusion. Thus, FCMs do not need to reinvent themselves after a temporal delay (between the two phases of fusion) to create a molecularly and/or morphologically distinct fusogenic synapse to promote additional rounds of fusion.
VI. RELEVANCE TO VERTEBRATE MYOBLAST FUSION
The identification of PLSs at the sites of fusion in Drosophila myoblast fusion raises an interesting question of whether a similar structure is involved in promoting fusion pore formation during vertebrate skeletal muscle differentiation. Loss-function studies with RNAi in cultured mouse C2C12 myoblasts have demonstrated an essential role of the WASP and Scar complexes in myoblast fusion (Kim et al., 2007; Nowak et al., 2009). Given the highly conserved molecular function of the two complexes, it is conceivable that the fly and vertebrate myoblast fusion share a similar cellular mechanism underlying fusion pore initiation.
Studies using mammalian myoblast cultures have yet to pinpoint specific sites of fusion with molecular or cellular markers. Although many proteins, especially cell adhesion molecules, have been localized to the broad muscle cell contact zone along the long axes of the adherent, spindle-shaped muscle cells, none of them appears to be enriched at specific focal points within the contact zone (for review see Pavlath, 2010). A recent live imaging study with mouse C2C12 cells showed that fusion occurred in a region enriched with phosphotidylinositol 4,5-bisphosphate (PI(4,5)P2) (Nowak et al., 2009), although the region is still relatively large and no specific sites of fusion could be distinguished. With regard to the actin cytoskeleton, no significant changes were observed in cultured “wild-type” muscle cells, despite the presence of discrete F-actin-enriched foci in Kette/Nap1 RNAi cells that failed to fuse (Nowak et al., 2009). Taken together, these studies suggest that although the Arp2/3 NPFs are essential for myoblast fusion in cultured cells, actin cytoskeletal rearrangements may occur at a subtler scale in these cells compared with the formation of dense F-actin foci during myoblast fusion in intact embryos. Such a difference in actin cytoskeletal rearrangement could be due to the different cellular environment in cells residing in vitro versus in vivo. It is well known that cultured myoblasts adopt a spindle-like, elongated shape when they adhere to a 2D culture dish, whereas myoblasts in vivo are tear drop-shaped cells, as seen in the 3D environment of Drosophila embryos. As a consequence, cultured muscle cells adhere along a broad contact zone, whereas myoblasts in vivo attach to myotubes at a focal point. Different modes of cell adhesion, in turn, may lead to differential distributions of the actin cytoskeletal machinery and distinct cytoskeletal responses to fusion signals. Alternatively and/or in addition, the difference in actin cytoskeletal rearrangement could be due to differences in gene expression when muscle cells were transformed to become immortal. In this regard, it is known that transformed cultured myoblasts, which are widely used for myoblast fusion studies, are less efficient in fusion than the primary myoblasts (Wakelam, 1985), indicating a compromise in the fusogenic potential in the immortal cell lines. Thus, to gain a clear understanding of the cellular mechanism underlying myoblast fusion in a physiologically relevant condition, it would be important to examine the actin cytoskeletal changes during mouse embryogenesis and/or muscle repair in 3D tissues in vivo.
VII. CONCLUDING REMARKS
Our understanding of myoblast fusion has advanced significantly in recent years with the application of a versatile toolbox including genetics, immunohistochemistry, live imaging, EM, and biochemistry. The discovery of an invasive PLS allows, for the first time, a clear identification of the sites of fusion at the ultrastructural level and provides an unprecedented view of the asymmetric fusogenic synapse. This exciting new development also leads to many important questions for future investigations. For example, what is required for the formation of the invasive fingers and what controls the dynamics of the PLS invasion? Is there an ECM protease activity associated with myoblast fusion? How is vesicle trafficking regulated and what are the biochemical components of the vesicles? What controls fusion pore expansion? Are similar PLSs used in vertebrate myoblasts fusion in vivo? Answers to these questions will undoubtedly lead to significant new insights into the molecular and cellular mechanisms underlying myoblast fusion in flies and vertebrates.
Acknowledgments
I apologize to colleagues whose work could not be cited because of space limitations. I thank Drs. Eric Grote and Duojia Pan and members of the Chen lab for comments on the manuscript. Research in the Chen lab has been supported in part by grants from National Institute of Health, American Heart Association, Muscular Dystrophy Association, and the David and Lucile Packard Foundation.
References
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128(21):4251–4264. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Bach AS, Enjalbert S, Comunale F, Bodin S, Vitale N, Charrasse S, et al. ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Molecular Biology of the Cell. 2010;21(14):2412–2424. doi: 10.1091/mbc.E09-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110(3):791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: A view from Drosophila. Cell. 1998;93(6):921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Developmental Biology. 2007;309(1):113–125. doi: 10.1016/j.ydbio.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett K, Rochlin KM, Duan H, Nguyen HT, Baylies MK. Expression and functional analysis of a novel Fusion Competent Myoblast specific GAL4 driver. Gene Expression Patterns. 2008;8(2):87–91. doi: 10.1016/j.modgep.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Schafer G, Kesper DA, Holz A, Eriksson T, Palmer RH, et al. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. Journal of Cell Science. 2008;121(Pt 8):1303–1313. doi: 10.1242/jcs.022269. [DOI] [PubMed] [Google Scholar]
- Billadeau DD, Nolz JC, Gomez TS. Regulation of T-cell activation by the cytoskeleton. Nature Reviews Immunology. 2007;7(2):131–143. doi: 10.1038/nri2021. [DOI] [PubMed] [Google Scholar]
- Bischoff R. The satellite cell and muscle regeneration. In: Engel AG, Franszini-Armstrong C, editors. Myogenesis. New York: McGraw-Hill; 1994. pp. 97–118. Myogenesis. [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes& Development. 2000;14(12):1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature Cell Biology. 2002;4(8):574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. The intracellular domain of Dumbfounded affects myoblast fusion efficiency and interacts with Rolling pebbles and Loner. PLoS One. 2010;5(2):e9374. doi: 10.1371/journal.pone.0009374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman CV, Sage PT, Sciuto TE, de la Fuente MA, Geha RS, Ochs HD, et al. Transcellular diapedesis is initiated by invasive podosomes. Immunity. 2007;26(6):784–797. doi: 10.1016/j.immuni.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Leikina E, Melikov K, Podbilewicz B, Kozlov MM, Chernomordik LV. Fusion-pore expansion during syncytium formation is restricted by an actin network. Journal of Cell Science. 2008;121(Pt 21):3619–3628. doi: 10.1242/jcs.032169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Developmental Cell. 2001;1(5):705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114(6):751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- Dillon C, Goda Y. The actin cytoskeleton: Integrating form and function at the synapse. Annual Review of Neuroscience. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: Blown fuse is required for progression beyond the prefusion complex. Journal of Cell Science. 1997;136(6):1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128(22):4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- Dustin ML. A dynamic view of the immunological synapse. Seminars in Immunology. 2005;17(6):400–410. doi: 10.1016/j.smim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Dworak HA, Charles MA, Pellerano LB, Sink H. Characterization of Drosophila hibris, a gene related to human nephrin. Development. 2001;128(21):4265–4276. doi: 10.1242/dev.128.21.4265. [DOI] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Dominguez P, Elkhatib N, Chavrier P, Daviet L, et al. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134(24):4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Engel LC, Egar MW, Przybylski RJ. Morphological characterization of actively fusing L6 myoblasts. European Journal of Cell Biology. 1986;39(2):360–365. [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. Journal of Cell Biology. 1997;138(3):589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. The MARVEL domain protein, Singles Bar, is required for progression past the prefusion complex stage of myoblast fusion. Developmental Biology. 2007;307(2):328–339. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chakravarti M, Banerjee R, Abmayr SM. SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mechanisms of Development. 2004;121(12):1455–1468. doi: 10.1016/j.mod.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Niu XP, Erickson MR, Abmayr SM. Identification of a Drosophila homologue to vertebrate Crk by interaction with MBC. Gene. 1999;228(1–2):243–252. doi: 10.1016/s0378-1119(99)00010-4. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP, Abmayr SM. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Developmental Biology. 2008;314(1):137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Reports. 2009;10(9):1043–1050. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Current Opinion in Cell Biology. 2008;20(2):235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, et al. Rac function and regulation during Drosophila development. Nature. 2002;416(6879):438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hauschka SD. The embryonic origin of muscle. In: Engel AG, Franzini-Armstrong C, editors. Myology. New York: McGraw Hill Inc.; 1994. pp. 3–73. Myology. [Google Scholar]
- Ibarra N, Pollitt A, Insall RH. Regulation of actin assembly by SCAR/WAVE proteins. Biochemical Society Transactions. 2005;33(Pt 6):1243–1246. doi: 10.1042/BST0331243. [DOI] [PubMed] [Google Scholar]
- Kalderon N, Gilula NB. Membrane events involved in myoblast fusion. Journal of Cell Biology. 1979;81(2):411–425. doi: 10.1083/jcb.81.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D, Zimmerberg J, Puri A, Sarkar DP, Blumenthal R. Single cell fusion events induced by influenza hemagglutin: Studies with rapid-flow, quantitative fluorescence microscopy. Experimental Cell Research. 1991;195(1):137–144. doi: 10.1016/0014-4827(91)90509-s. [DOI] [PubMed] [Google Scholar]
- Kesper DA, Stute C, Buttgereit D, Kreiskother N, Vishnu S, Fischbach KF, et al. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS) Developmental Dynamics. 2007;236(2):404–415. doi: 10.1002/dvdy.21035. [DOI] [PubMed] [Google Scholar]
- Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, et al. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Developmental Cell. 2007;12(4):571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Knudsen KA. Fusion of myoblasts. In: Wilschut J, Hoekstra D, editors. Membrane fusion. New York, NY: Marcel Decker Inc; 1992. Membrane fusion. [Google Scholar]
- Kocherlakota KS, Wu JM, McDermott J, Abmayr SM. Analysis of the cell adhesion molecule sticks-and-stones reveals multiple redundant functional domains, protein-interaction motifs and phosphorylated tyrosines that direct myoblast fusion in Drosophila melanogaster. Genetics. 2008;178(3):1371–1383. doi: 10.1534/genetics.107.083808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends in Biochemical Sciences. 2010;35(12):699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu S, Takenawa T. The WASP and WAVE family proteins. Genome Biology. 2009;10(6):226. doi: 10.1186/gb-2009-10-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Cote JF. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S. Invadosomes at a glance. Journal of Cell Science. 2009;122(Pt 17):3009–3013. doi: 10.1242/jcs.032631. [DOI] [PubMed] [Google Scholar]
- Lipton BH, Konigsberg IR. A fine-structural analysis of the fusion of myogenic cells. Journal of Cell Science. 1972;53(2):348–364. doi: 10.1083/jcb.53.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Developmental Cell. 2007;12(4):557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- McDonald KL, Auer M. High-pressure freezing, cellular tomography, and structural cell biology. Biotechniques. 2006;41(2):137, 139, 141. doi: 10.2144/000112226. passim. [DOI] [PubMed] [Google Scholar]
- Menon SD, Chia W. Drosophila rolling pebbles: A multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Developmental Cell. 2001;1(5):691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- Menon SD, Osman Z, Chenchill K, Chia W. A positive feedback loop between Dumbfounded and Rolling pebbles leads to myotube enlargement in Drosophila. Journal of Cell Biology. 2005;169(6):909–920. doi: 10.1083/jcb.200501126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B, Baker WW. Normal mammalian muscle differentiation and gene control of isocitrate dehydrogenase synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1967;58(2):592–598. doi: 10.1073/pnas.58.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134(17):3145–3153. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. Journal of Cell Science. 2009;122(Pt 18):3282–3293. doi: 10.1242/jcs.047597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajcini KV, Pomerantz JH, Alkan O, Doyonnas R, Blau HM. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. Journal of Cell Biology. 2008;180(5):1005–1019. doi: 10.1083/jcb.200707191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlath GK. Spatial and functional restriction of regulatory molecules during mammalian myoblast fusion. Experimental Cell Research. 2010;316(18):3067–3072. doi: 10.1016/j.yexcr.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plonsky I, Cho MS, Oomens AG, Blissard G, Zimmerberg J. An analysis of the role of the target membrane on the Gp64-induced fusion pore. Virology. 1999;253(1):65–76. doi: 10.1006/viro.1998.9493. [DOI] [PubMed] [Google Scholar]
- Plonsky I, Zimmerberg J. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. Journal of Cell Biology. 1996;135(6 Pt 2):1831–1839. doi: 10.1083/jcb.135.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Fambrough D. Ultrastructural and electrophysiological correlates of cell coupling and cytoplasmic fusion during myogenesis in vitro. Developmental Biology. 1973;30(1):166–186. doi: 10.1016/0012-1606(73)90055-9. [DOI] [PubMed] [Google Scholar]
- Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, et al. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128(24):5061–5073. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134(24):4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: When it takes more to make one. Developmental Biology. 2010;341(1):66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: A myoblast attractant essential for fusion. Cell. 2000;102(2):189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121(7):1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Salinas PC, Price SR. Cadherins and catenins in synapse development. Current Opinion in Neurobiology. 2005;15(1):73–80. doi: 10.1016/j.conb.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Martin-Belmonte F, Valencia A, Alonso MA. MARVEL: A conserved domain involved in membrane apposition events. Trends in Biochemical Sciences. 2002;27(12):599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, et al. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Developmental Biology. 2007;304(2):664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schroter RH, Lier S, Holz A, Bogdan S, Klambt C, Beck L, et al. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131(18):4501–4509. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, et al. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. Journal of Cell Biology. 2010;191(5):1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C, Kocherlakota KS, Zhuang S, Abmayr SM. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 2009;136(7):1159–1168. doi: 10.1242/dev.026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y. Electron microscope observations on the fusion of chick myoblasts in vitro. Journal of Cell Biology. 1971;48(1):128–142. doi: 10.1083/jcb.48.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas BP, Woo J, Leong WY, Roy S. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nature Genetics. 2007;39(6):781–786. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Griffiths GM. Secretory mechanisms in cell-mediated cytotoxicity. Annual Review of Cell and Developmental Biology. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Current Opinion in Cell Biology. 2006;18(1):4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, et al. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128(21):4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nature Reviews Molecular Cell Biology. 2007;8(1):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(22):8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: Current concepts and controversies in adult myogenesis. Cell. 2005;122(5):659–667. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ. The fusion of myoblasts. Biochemical Journal. 1985;228(1):1–12. doi: 10.1042/bj2280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM. Invadopodia: Specialized cell structures for cancer invasion. Clinical & Experimental Metastasis. 2006;23(2):97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen EH. Ultrastructural analysis of myoblast fusion in Drosophila. In: Chen EH, editor. Cell fusion: Overviews and methods. New Jersey: Humana Press; 2008. pp. 275–297. [DOI] [PMC free article] [PubMed] [Google Scholar]