Abstract
Autophagy is an evolutionarily conserved, selective degradation pathway of cellular components that is important for cell homeostasis under healthy and pathologic conditions. Here we demonstrate that an increase in the level of BAG3 results in stimulation of autophagy in glioblastoma cells. BAG3 is a member of a co-chaperone family of proteins that associate with Hsp70 through a conserved BAG domain positioned near the C-terminus of the protein. Expression of BAG3 is induced by a variety of environmental changes that cause stress to cells. Our results show that BAG3 overexpression induces autophagy in glioma cells. Interestingly, inhibition of the proteasome caused an increase in BAG3 levels and induced autophagy. Further analysis using specific siRNA against BAG3 suggests that autophagic activation due to proteosomal inhibition is mediated by BAG3. Analyses of BAG3 domain mutants suggest that the WW domain of BAG3 is crucial for the induction of autophagy. BAG3 overexpression also increased the interaction between Bcl2 and Beclin-1, instead of disrupting them, suggesting that BAG3 induced autophagy is Beclin-1 independent. These observations reveal a novel role for the WW domain of BAG3 in the regulation of autophagy.
Keywords: autophagy, BAG3, WW domain, glioblastoma
Introduction
Gliomas are the most common and lethal form of adult brain tumors with a median survival rate of 12 months. While gliomas are resistant to therapies that induce apoptosis, they seem to be less resistant to therapies associated with activating autophagy [1, 2]. Autophagy is an important cellular process that mainly mediates the basal turnover of long-lived proteins and removal of damaged and aged organelles by lysosomes [3, 4]. In general, autophagy is mediated through three pathways including macroautophagy, microautophagy, and chaperone mediated autophagy. Macroautophagy (hereafter called autophagy) involves the packaging of cargo into autophagosomes and its fusion with lysosomes. In microautophagy, the cargo enters lysosomes by invagination of the lysosomal membrane. Both processes result in degradation of the cargo content by lysosomal enzymes. In addition, there are many studies pointing to the importance of autophagy for the clearance of misfolded and aggregated proteins by chaperone-mediated autophagy that involves direct transport of the selected proteins across lysosomal membranes [4-7]. Protein quality control (PQC) is mainly achieved by the ubiquitin-proteosome system (UPS). While the UPS ensures the degradation of ubiquitinated misfolded or unfolded proteins through proteasomes, the aggresome-autophagy system initiates the degradation of aggresomes and protein aggregates through lysosomes. In both systems, chaperones and co-chaperones play important roles for the definition of the cargo content, which must be degraded to maintain cellular and physiological functions.
The very first autophagy gene to be discovered, Atg 1, was identified in 1993 by yeast genetic screening and cloned in 1997 [8,9]. Soon after, Beclin-1 was identified as a binding partner of Bcl2 by yeast two-hybrid screening [10]. Subsequent studies revealed that Beclin-1 is a functional ortholog of Atg6 and required for the induction of autophagy [8]. The initial discovery of Beclin-1 as a binding partner of Bcl2 suggested that the Beclin-1/Bcl2 complex may serve as a regulatory complex between autophagy and apoptosis. Indeed, later studies have demonstrated that the interaction of Bcl2 with Beclin-1 can inhibit autophagy [11-13]. Other studies revealed that under stress conditions, Bcl2 must be displaced from Beclin-1 to mediate the induction of autophagy, suggesting the possible involvement of other cellular proteins that physically and/or functionally communicate with Bcl2 in this event [14]. Recently, the Bcl2-associated athanogene 3 (BAG3), which is a member of the BAG family of co-chaperone proteins that interact with the ATPase domain of the heat shock protein 70 (Hsp70), has received special attention in the control of apoptosis and PQC [15, 16]). Similar to other members of the family, BAG3 is induced by a variety of stress stimuli and has been shown to reduce the chaperone activity of Hsp70 [17]. In addition to Hsp70, several binding partners of BAG3 have been identified, including PLC-γ, and results suggest that Bcl-2 that may serve as a survival signal for cells [18]. Recently, BAG3 stabilization of Bcl2 family proteins has been shown to protect cancer cells from apoptosis [19]. We also reported that downregulation of BAG3 sensitized primary microglial cells to caspase-3 activation following HIV-1 infection, suggesting a role for BAG3 in the balance of cell death versus survival during viral infection [20]. One of the important functions of BAG3 is related to its involvement in regulation of selective autophagy. Earlier studies have demonstrated that BAG3 forms a complex with HspB8 and mediates the degradation of Htt43Q, a pathogenic form of huntingtin, through an autophagic process that seems to be independent from its interaction with Bcl2 [5, 21]. Interestingly, BAG3 levels were shown to be increased during aging and BAG3 was required for the enhancement of age-dependent autophagic activity in I90 cells, a model culture system for aging [7].
BAG3 has a modular structure with a BAG domain at the C-terminus, a WW domain at the N-terminus, two IPV domains which are binding sites for HspB8 and HspB6, and a proline-rich domain (PXXP) in the central portion of the protein. While the BAG3 domain initiates the interaction with Hsp70, the PXXP domain of the protein is mainly involved in interactions with other cellular proteins [22, 23]. In addition to cellular proteins, BAG3 can also associate with viral transforming proteins including JC virus T-antigen, in a mouse model of medulloblastoma, and mediates its degradation through autophagy [24]. This observation suggested a novel role for BAG3 in controlling tumor growth by selective autophagic degradation of oncogenic proteins. Here we investigated the direct involvement of BAG3 in autophagy in human glioma cell lines. Our data suggests that expression of BAG3 in glioblastoma cell lines was sufficient to induce autophagy. Interestingly, our data showed that BAG3-mediated induction of autophagy was mediated through its WW domain. These observations provide a novel role of WW domain in the regulation of autophagy in glioma cells.
Results
Colocalization of BAG3 with LC3 in autophagosomes
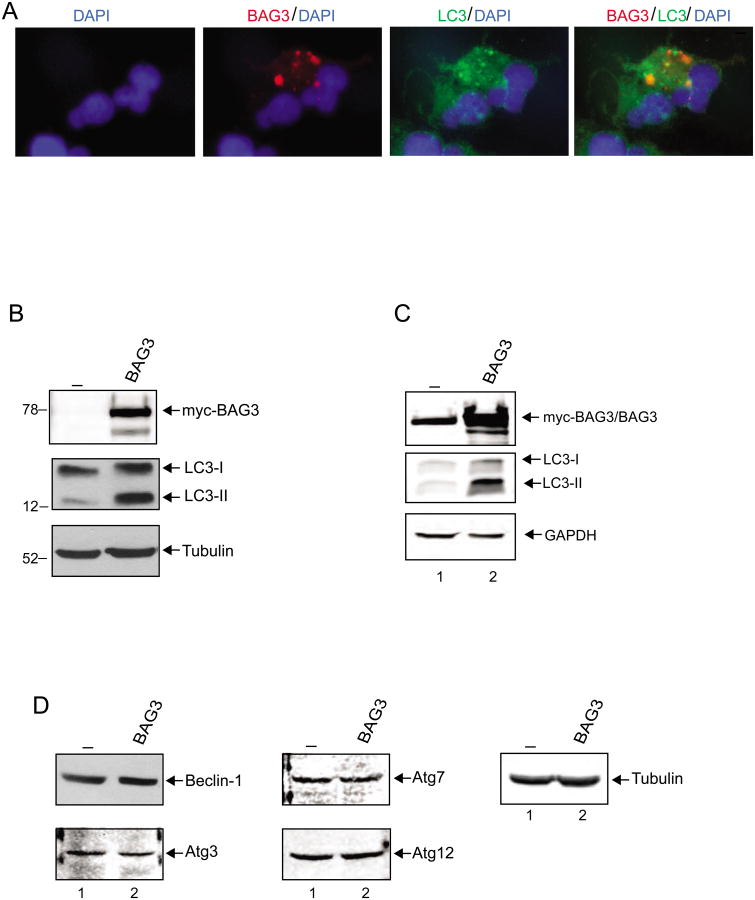
To investigate the impact of BAG3 on autophagy in glioma cells, we utilized T98G cells, a cell line derived from a human glioblastoma multiforme tumor. Transfection of T98G cells with a myc-tagged BAG3 expression plasmid induced the appearance of typical LC3 puncta formation in the cytoplasm, a reliable marker of autophagosomes (Figure 1A). Interestingly, BAG3 co-localized with autophagosomes labeled with LC3 suggesting a possible role for BAG3 in the formation of autophagosomes in these cells. Western blot analysis further confirmed the induction of autophagy by BAG3 as evident by the characteristic enhancement of LC3-II levels (Figure 1B). In parallel, we investigated the impact of BAG3 overexpression on the state of primary human glioblastoma cells. We found that, similar to T98G cells, overexpression of BAG3 in a primary culture of grade IV human glioblastoma also lead to the induction of the autophagic marker, LC3-II (Figure 1C), suggesting that BAG3-mediated stimulation of autophagy was not restricted to the T98G cell line and may have a broader impact on tumors of glial origin. Expression of other cellular proteins including GAPDH remained unchanged. Examination of Beclin-1 levels, a protein that has a central role in autophagy, showed no noticeable changes in cells with elevated levels of BAG3 in comparison to that in control cells, suggesting that BAG3 induces autophagy through a pathway in which the level of endogenous Beclin-1 remains unchanged (Figure 1D). Similarly, no noticeable changes in the levels of various other proteins associated with autophagy such as Atg3, Atg7 and Atg12 were detected (Figure 1D).
Figure 1. Colocalization of BAG3 with LC3 in autophagosomes.
A. Co-localization of LC3-II and BAG3. T98G cells were transfected with myc-BAG3, and fixed at 48h post-transfection. Cells were co-stained with polyclonal LC3 antibody (green) and monoclonal myc-tag antibody (red). Nuclei were also visualized by DAPI staining (blue). B. T98G cells were transfected with either expression plasmid encoding myc-BAG3 (lane 2) or vector alone (lane1) and subsequently processed for whole-protein extraction at 48h post-transfection for Western blot analysis of autophagy marker, LC3. C. GLI87 primary human glioblastoma cells (grade IV glioblastoma) were either transfected with expression plasmid encoding myc-BAG3 (lane 1) or vector alone (lane1). Cells were harvested at 48h post-transfection and total protein extracts were subjected for western blot analysis of autophagy marker. D. Expression of autophagy-related genes (ATGs) was analyzed by western blot using the same protein extracts that were used in Panel B.
High levels of BAG3 induces autophagolysosome formation in glioma cells
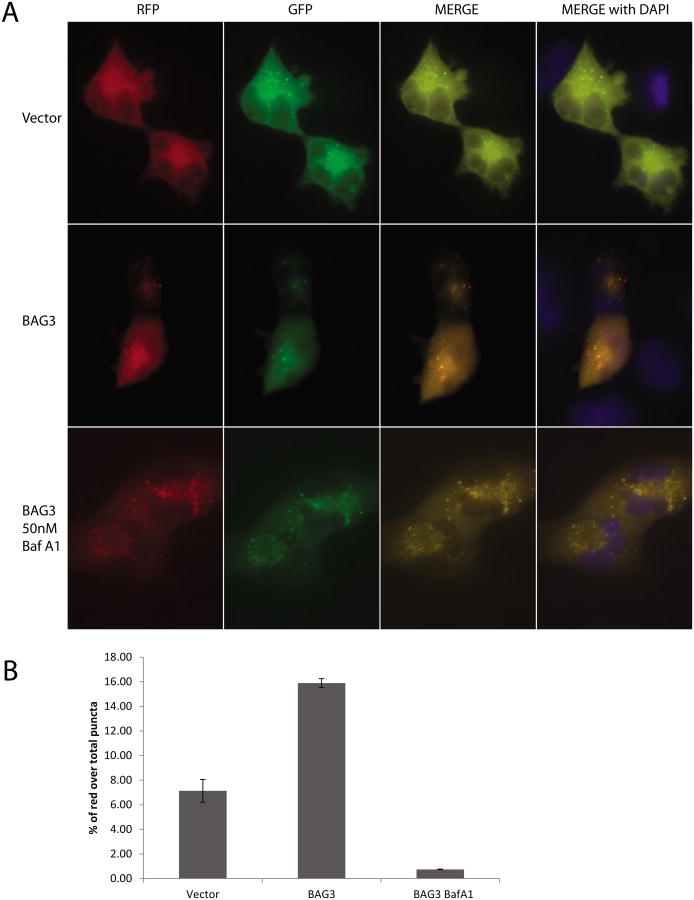
In order to gain more insight into autophagy induced by BAG3, autophagic activity was monitored by ptfLC3 plasmid encoding LC3 fused to mRFP and EGFP. The ptfLC3 construct provides the advantage of distinguishing autophagosomes from autophagolysosomes by a change in fluorescence emission based on pH which results in a loss of GFP-LC3 signal integrity within the lysosomal environment, whereas the RFP-LC3 signal persists [25]. T98G cells were co-transfected with ptfLC3 and an expression plasmid encoding BAG3. At 24 h post-transfection, cells were fixed and analyzed for the mRFP and EGFP signals. As shown in Figure 2A, cells transfected with control vector showed only GFP-LC3 puncta in the cytoplasm of the cells with limited expression of RFP-LC3 in the autophagolysosomes (upper panels) underlying the baseline of autophagic activity in these cells. On the other hand, cells overexpressing BAG3 showed a significant increase in the number of puncta expressing RFP-LC3 and GFP-LC3 (Figure 2A, middle panels). Although the GFP signals were colocalized with RFP signal in the cytoplasm (merge, orange puncta), a significant subpopulation of puncta with RFP expression was not colocalized with GFP signal, suggesting the formation of autophagolysosomes. Cells were then incubated with Bafilomycin A1 (Baf A1) which blocks the fusion of autophagosomes with lysosomes. As expected, BAG3 overexpression induced the formation of GFP-LC3 as well as RFP-LC3 in the cytoplasm of the cells in the presence of Baf A1 (Figure 2A, bottom panels). Merged images of RFP and GFP signals revealed that majority of the GFP signals were colocalized with RFP signal in the cytoplasm of the cells, suggesting that BAG3-induced autophagolysosome formation was suppressed by Baf A1.
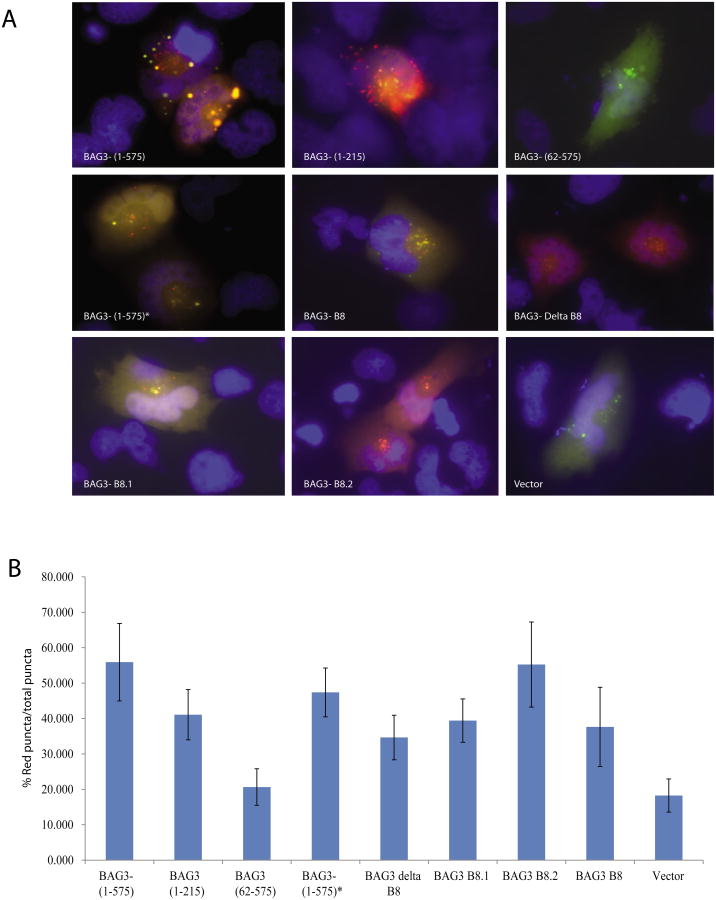
Figure 2. BAG3 overexpression induces autophagolysosome formation in T98G cells.
A. T98G cells were cotransfected with full length BAG3 or control vector and ptfLC3 plasmid encoding LC3 fused to mRFP and EGFP, which results in a loss of GFP-LC3 signal integrity within the low pH of the lysosomal environment, whereas the RFP-LC3 signal persists. Cells were either treated with 50 nM Bafilomycin A1 (Baf A1) or left untreated. B. % of red puncta signal over total puncta (red+orange) per cell in merged images was calculated and shown as a bar graph. Data are shown as mean ± SD and are representative of at least 10 different cells from 10 different fields.
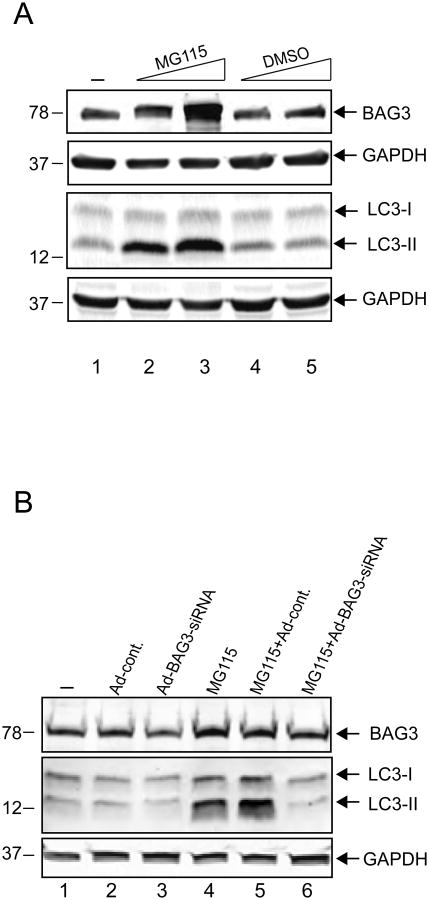
In the next series of experiments, we treated cells with MG115, a proteasome inhibitor, which lead to enhanced levels of BAG3 and examined its impact on autophagy. As seen in Figure 3A, induction in the level of endogenous BAG3 upon treatment of cells with MG115, but not DMSO vehicle, was associated with an increase in the level of LC3-II while no changes in the level of GAPDH were detected. To further establish the involvement of BAG3 with autophagy, T98G cells were treated with Adenovirus expressing BAG3-specific siRNA and as seen in Figure 3B, a substantial decrease in the level of LC3-II induced by MG115 treatment was observed when BAG3 expression was inhibited by siRNA, but not the Adenovirus control, suggesting that BAG3 is required for LC3-II induction by MG115.
Figure 3. Induction of LC3-II by the proteasome inhibitor, MG115, is BAG3-dependent.
A. T98G cells were treated with MG-115 or DMSO at the indicated doses for 16 hours and whole-cell protein extracts were subjected to SDS-PAGE and western blot analysis for BAG3, LC3, and GAPDH as a loading control. B. Western blot analysis of protein extracts from T98G cells treated either with a BAG3-specific small interfering (si) RNA (Ad-BAG3siRNA) at 50 MOI or with a control Adenovirus (Ad-cont.). Cells were either treated with MG115 for 16 hours or left untreated as indicated. Whole-cell protein extracts were subjected to SDS-PAGE and western blot analysis for BAG3, LC3, and GAPDH as a loading control.
Identification of the domain(s) within BAG3 that induces autophagy in glioblastoma cells
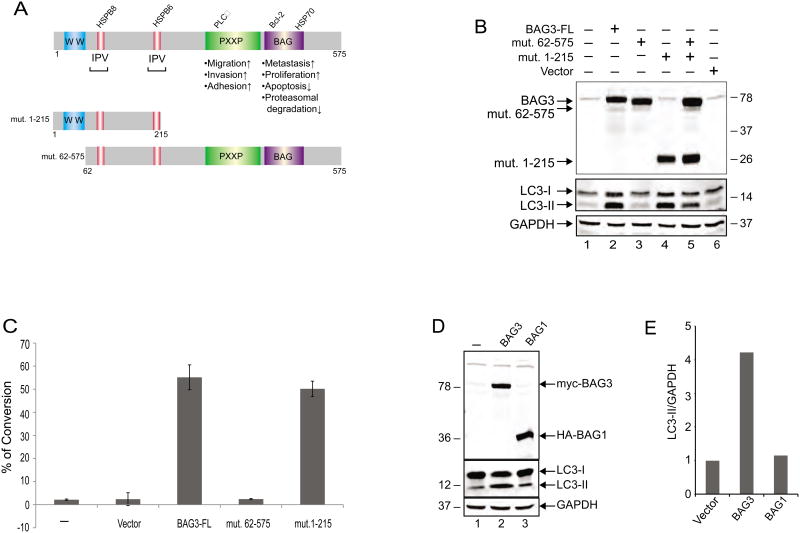
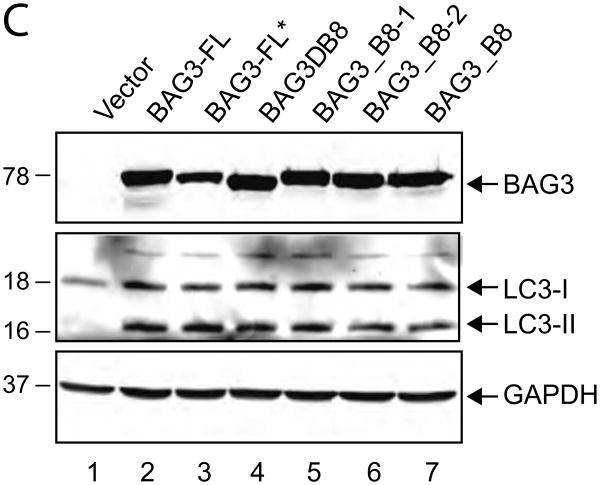
As illustrated in Figure 4A, BAG3 has multiple domains that are associated with several biological functions. The region spanning amino acids 21 to 55 of the N-terminal domain of the protein encompasses the WW domain with no well-defined function described thus far, although through its proline-rich domain may interact with the SH3 motif. In addition, two IPV domains exist that are recognized by HspB8 and HspB6. Initially, we became interested in the N-terminal region of BAG3 in part due to the presence of the two IPV domains that have been previously implicated in protein quality control and upregulation of autophagy [26, 27]. As a first step, we created several mutant constructs spanning various lengths of the N-terminus of the protein, i.e. residues 1-60 and 1-101 to assess their ability to induce autophagy. Our repeated efforts showed that neither of these short mutant proteins could be stably produced in transfected cells, hence we changed our strategy and created mutant proteins encompassing residues 1 to 215 and 62 to 575, both of which could be stably produced in cells. As shown in Figure 4B, only mutant 1 to 215, which contains the WW domain along with the two IPV domains, but not mutant 62 to 575 which encompasses the IPV motifs and the remainder of BAG3, was able to induce LC3-II production (Figure 4B). We also investigated the impact of these BAG3 mutants on autophagy. In this respect, the full length BAG3 or mutants (1-215) and (62-575) were introduced into cells either alone or in combination. Co-transfection of mutant (62-575) along with mutant (1-215) modestly reduced the autophagy levels as evidenced by the intensity of the band corresponding to LC3-II (Figure 4B, lane 5). These observations point to the importance of the extreme N-terminal domain of BAG3 that includes the WW domain in stimulating autophagy in the cells.
Figure 4. Mapping the domain of BAG3 associated with autophagic induction.
A. Schematic diagram of full-length BAG# and two deletion mutants used in the study. B. T98G cells were transfected with expression vectors encoding full-length BAG3 and its truncated forms as indicated. Whole-cell protein extracts were prepared at 48h post-transfection, and analyzed by western blot using anti-myc and anti-LC3 antibodies. Western blot analysis of the same membranes with anti-GAPDH antibody was used as loading control. C. T98G cells were transfected with expression plasmids encoding full length BAG3 (BAG3-FL and its deletion mutants as indicated. At 48 hours post-transfection, cells were incubated with acridine orange and processed for FACS analysis as described in Materials and Methods. Conversion of acridine orange to bright red color, which is indicative of the formation of acidic vesicular organelles was quantified and is presented as a bar graph. Data are shown as mean ± SD and are representative of at least three independent experiments. D. Western blot analysis of myc-BAG3, HA-BAG1, and LC3 expression upon transfection of T98G cells with the plasmid constructs. E. BAG1 overproduction does not induce autophagy as illustrated by the level of LC3 and its quantitative evaluation using GAPDH as a loading control.
In an alternative approach, we stained T98G cells with acridine orange to determine the formation of acidic vesicular organelles, indicative of autophagy, upon the expression of full-length BAG3 and the two deletion mutants. The protonated acridine orange, which accumulates and forms aggregates in the acidic compartment of the cells fluoresces bright red and is detected by flow cytometry. As seen in Figure 4C, more than 50% of the cells expressing full-length BAG3 or mutant (1-215) showed green to red color conversion, indicative of the degree of acidity and/or the volume of the cellular acidic compartment. The expression of mutant (62-575) showed no effect on the formation of acidic vesicular organelles and the acidity of the cells. These observations point to the initiation of the process that leads to autophagy by the N-terminal region of BAG3.
In a different set of experiments, we investigated the ability of BAG1, a member of the BAG family with similar structural features of BAG3, yet lacking a WW domain, to induce autophagy in T98G cells. Our results show that unlike BAG3, overexpression of HA tagged BAG1 in these cells has no impact on the production of LC3-II (Figure 4, panels D and E).
To examine the potential role of the IPV domains in the induction of LC3-II, we utilized deletion and point mutation mutants of BAG3 in which both IPV motifs were deleted (BAG3 delta B8), both IPV motifs were mutated to abrogate HspB8 and HspB6 interaction with BAG3 (BAG3-B8), or either the first or second IPV motifs were mutated (BAG3-B8.1 and BAG3-B8.2, respectively) and assessed their ability to induce puncta formation [26]. The ptfLC3 plasmid encoding LC3 in fusion with EGFP and mRFP and the BAG3 IPV mutants described above were introduced into T98G cells by co-transfection. As seen in Figure 5, all mutant BAG3 constructs except for the mutant BAG3 lacking the WW domain [BAG3 (62-575)] were able to induce the formation of autophagolysosomes. Interestingly overexpression of the various mutants of BAG3 IPV motifs induced autophagy as evidenced by the RFP signal exclusively in the autophagolysosomes. We also tested BAG3 IPV mutants for autophagy by immunoblotting. All the IPV mutants induced autophagy as evidenced by LC3-II formation similar to full length BAG3 construct (BAG3-FL) (Figure 5C). These results suggested that BAG3-induced autophagy was not associated with the IPV motifs.
Figure 5. BAG3 IPV motifs are not responsible for autophagolysosome formation in glioma cells.
A. T98G cells were co-transfected with the ptfLC3 plasmid encoding mRFP-GFP-LC3 and either wild-type BAG3 (BAG3 1-575) (cloned in our laboratory) or its mutant forms as indicated. BAG3-(1-575)* donates the BAG3 plasmid encoding full-length BAG3 provided by Jacques Landry, [27]. At 24hr hours post-transfection, cells were fixed and analyzed as described in the Materials and Methods. B. Percentage of red puncta over total puncta signal from cells represented in Panel A was calculated and presented as a bar graph. Data are shown as mean ± SD and are representative of at least 10 different cells from 10 different fields. C. T98G cells were transfected with empty vector or expression vectors encoding full-length BAG3 and BAG3 IPV mutants as indicated. Whole-cell protein extracts were prepared at 48h post-transfection, and analyzed by western blot using anti-myc and anti-LC3B antibodies. Western blot analysis of the same membranes with anti-GAPDH antibody was used as a loading control.
Interaction of Bcl2 and Beclin-1 in the presence of BAG3
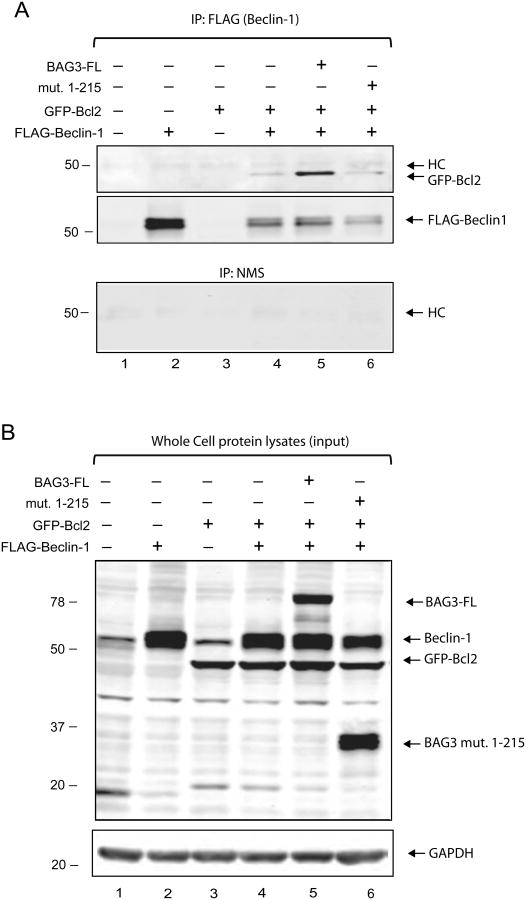
The anti-apoptotic protein, Bcl2, interacts with Beclin-1 and this interaction is crucial to determine the fate of a cell with respect to apoptosis and autophagy which are tightly regulated processes [28]. Since Bcl2 also interacts with BAG3, we decided to examine the effect of BAG3 overexpression on the interaction between Bcl2 and Beclin-1. Upon transfection of Beclin-1 or Bcl2 expression vectors in HEK293T cells alone or in combination, co-transfection of Bcl2 along with Beclin-1 led to downregulation of exogenous Beclin-1 expression (data not shown). This is similar to previously reported results suggesting that 3 times more Beclin-1 DNA was needed to reach equal amount of Bcl-xl when Beclin-1 and Bcl-xl overexpressed in HeLa cells [29]. To study BAG3, Bcl2, and Beclin-1 interactions by co-immunoprecipitation, we chose HEK293T cells since this cell line can express larger quantities of the aforementioned proteins with minimal induction of apoptotic effects. A ratio of DNA concentrations of (6:1) for Beclin-1 and Bcl2 were used to produce equal levels of these proteins in HEK293T cells (Figure 6B, Lane 4). Since Beclin-1 was tagged with a FLAG epitope, anti-FLAG antibody was used to immunoprecipitate Beclin-1 in complex with any other associated proteins. Our results indicated that when co-IP with anti-FLAG antibody using cell lysates co-expressing full length wt BAG3, GFP-Bcl2 and Beclin-1, more Bcl2 is immunoprecipitated (Figure 6A Lane 5, upper panel) indicating that interaction between Bcl2 and Beclin-1 became stabilized in the presence of full length BAG3. However the BAG3 mutant (1-215) did not produce the same results (Figure 6A, Lanes 6). The BAG3 (1-215) mutant showed no effect on Beclin-1 and Bcl2 interaction, which was expected since this mutant lacks the BAG domain and it is not able to interact with Bcl2. The unrelated protein, GAPDH, used as a loading control was unaffected (Figure 6B). We did not observe any BAG3 in the immunoprecipitated Beclin-1-Bcl2 complex indicating that BAG3 does not interact with Beclin-1 directly. Co-IP with control normal mouse serum (NMS) showed no non-specific interactions with Bcl2 or Beclin-1 (Figure 6A, bottom panel). Although BAG3 overexpression with Bcl2 stabilized the Beclin-1-Bcl2 interaction, a ternary complex containing BAG3, Beclin-1, and Bcl2 was not detectable under the conditions we used. Likewise, co-immunoprecipitation of Beclin-1 with BAG3 showed that Beclin-1 and BAG3 do not interact with each other directly (data not shown).
Figure 6. Overexpression of full length wt BAG3 stabilizes the interaction of Bcl2 and Beclin-1.
A. Co-immunoprecipitation (co-IP) of Beclin-1 and Bcl2 in HEK293T cells. Lysates from cells transfected with Bcl2, Beclin-1 and BAG3 expression constructs (200 μg total protein) were used in co-IP experiments using antibodies to the FLAG tag on Beclin-1 or normal mouse serum (NMS) instead of FLAG antibody as a control for specificity of the complexes. Precipitated complexes were subjected to western blot analysis using antibodies to detected GPF (for Bcl2), Beclin-1 or myc (for BAG3). Note that FLAG-Beclin-1 migrates slightly above the endogenous Beclin-1 exhibiting a double protein band. B. Western Blot for Bcl2, Beclin-1 and BAG3 expression in HEK293T cells. Aliquots of HEK293T cell lysates (40 μg total protein) for each co-IP experiment were analyzed by western blot in parallel, resolved on the same SDS-PAGE, and probed against the same antibodies. GAPDH antibody was used on the same membrane as a loading control (Figure 6B, bottom panel).
Co-localization of BAG3 with Bcl2 and Beclin-1
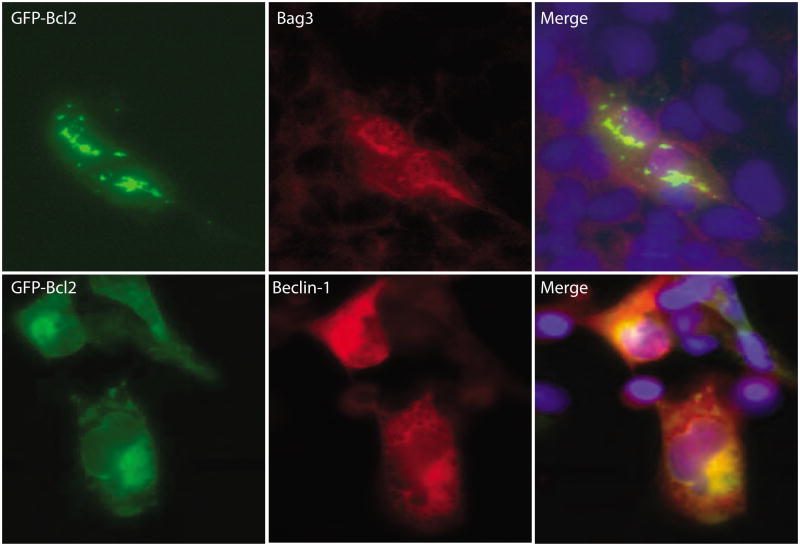
The subcellular localization of BAG3, Bcl2, and Beclin-1 were carried out using T98G cells transfected with expression constructs by immunocytochemistry following fixation in ice-cold acetone. BAG3 with Bcl2 were partially co-localized and exhibited an aggregated pattern as seen in Figure 7, upper panels. This pattern was predominant where GFP-Bcl2 and BAG3 were highly accumulated indicating a strong interaction between Bcl2 and BAG3. Both proteins were in the cytoplasm and appeared to localize around the nucleus. Bcl2 and Beclin-1 also show co-localization in the cytoplasm near the nucleus, the pattern appeared more diffuse rather than aggregated (bottom panels).
Figure 7. Co-localizations of BAG3, Bcl2 and Beclin-1.
Top Panel: Co-localization of BAG3 with GFP-Bcl2 in T98G cells. GFP-Bcl2 was detected with mouse anti-Bcl2 antibodies; BAG3 was detected with rabbit anti-BAG3 antibodies. Bottom Panel: Co-localization of GFP-Bcl2 with Beclin-1 in T98G cells. Similar to the top panel, Bcl2 was detected with mouse anti-Bcl2 antibodies; Beclin-1 was detected with rabbit anti-Beclin-1 antibodies. Secondary antibodies conjugated either with FITC or rhodamine were used each case depending upon the primary antibody as described in the Materials and Methods. Cells were mounted in medium containing DAPI which stains nuclei in blue. Co-localization of two expressed proteins is represented by yellow in the merged images.
Discussion
Autophagy is an essential cellular pathway which plays a major role in maintaining cellular homeostasis under various stress conditions. Many studies have pointed to a possible role for BAG3 in regulation of the autophagic pathway [5, 7, 21-24]. However, the actual mechanism that may be affiliated with this regulatory event in glioblastoma has not been investigated. Here we provide evidence that the expression of BAG3 is sufficient to induce autophagy in glioblastoma cells as measured by autophagolysosome formation and LC3-II induction upon BAG3 overproduction and its suppression by autophagic inhibitors. All of the BAG family of proteins contains at least one copy of an evolutionarily conserved BAG domain that allows them to interact with and regulate proteins in the Hsp70 family of molecular chaperones. Among these family members, BAG3 is the only one that contains a WW domain and it is present at the N-terminus of the protein [30]. Recent studies have suggested that BAG3 interacts with synaptopodin-2 (SYNPO2) through its WW domain and this interaction is required for autophagosome formation during chaperone-assisted selective autophagy (CASA) [31]. Consistent with these observations, our data reveal that the WW domain of BAG3 is required for the selective induction of autophagy in glial cells suggesting a novel role for this domain in BAG3-mediated regulation of autophagy.
Beclin-1 is an important cellular protein which plays a direct role in the formation of autophagosomes [8, 10-14, 32-34]. Bcl2 has been shown to be a negative regulator of autophagy and this function has been attributed, at least in part, to its association with Beclin-1 [8, 11-14]. Under stress conditions, several mechanisms mediate the disruption of Beclin-1/Bcl2 interaction leading to autophagy. Proposed mechanisms include DAPK-mediated phosphorylation of Beclin-1, JNK-mediated phosphorylation of the non-structured loop of Bcl2, competition with Bad and Bax for Bcl2/Bcl-xL binding, and binding of the DAMP molecule HMGB-1 to Beclin-1 [35]. On the other hand, BAG3 is a known binding partner of Bcl2 and it is believed that the interaction of Bcl2 with BAG3 exhibits anti-apoptotic activity [36-38].
Even though we did not observe any interaction between BAG3 and Beclin-1, our data indicate that in the presence of exogenous BAG3, more Bcl2 was found to be bound to Beclin-1 suggesting that overexpression of BAG3 can stabilize the interaction between Bcl2 and Beclin-1. Furthermore, we found that overexpressed BAG3 had no effect on endogenous levels of Beclin-1. The stabilizing effect of BAG3 on the Bcl2/Beclin-1 complex may not be directly related to its autophagy inducing function. In fact, while the BAG3 (1-215) mutant induced autophagy, it failed to stabilize the Bcl2:Beclin-1 complex. Obviously, the BAG domain in BAG3 is responsible for Bcl2 binding, but it is surprising that the interaction between BAG3 and Bcl2 can render the Beclin-1:Bcl2 complex more stable. The stabilizing effect of BAG3 has recently been described in protecting colon cancer cells from apoptosis by stabilizing anti-apoptotic Bcl-2 proteins [20]. Taken together, these findings lead us believe that BAG3 induced autophagy is Beclin-1 independent. There are many examples in the literature describing Beclin-1 independent autophagy pathways. It is possible that BAG3 interaction with Bcl2 can lead to autophagy independently from Beclin-1. This notion is supported by data demonstrating that a compound which targets Bcl2 proteins induced autophagy in a Beclin-1-independent manner [39]. However, our data obtained with the BAG3 mutant (1-215) did not corroborate this claim. Our finding indicates that the WW domain is responsible for autophagy induction and the mutant carrying this domain but lacking the BAG domain cannot stabilize Beclin-1:Bcl2 interaction, which are similar to the recent finding that BAG3 induces autophagy in HEPG2 cells by proteasome inhibition independent of Beclin-1 [40]. We also observed that autophagy was induced using a proteasomal inhibitor which up-regulated BAG3 expression leading to autophagy. Although we have started to see an emerging picture how BAG3 regulates autophagy, the details of this pathways remain to be elucidated.
Material and Methods
Ethics statement
Human brain tumor tissue was obtained from surgical resections or brain biopsy under the approval of the Temple University Institutional Review Board (IRB). Written informed consent for all samples was obtained and any clinical samples presented in this study have been coded and de-identified.
Cell lines
The human glioblastoma cell line, T98G (Cat.# CRL-1690™) and the human embryonic kidney cells HEK293T, were from ATCC (Cat# CRL-11268) were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) and 100 U/mL penicillin and 100 μg/mL streptomycin, or 25 μg/mL gentamicin and 10 μg/ml ciproflaxin HCl. GLI87 cells were prepared as described previously [25]. Briefly, fresh glioblastoma tissue was separated by mechanical disruption using sterile scalpels. After tissue dissociation, cells were subjected to further separation using 70 μM nylon mesh and by passing cells through fire-polished Pasteur pipettes. Cells were cultured in 1:1 DMEM: F12 (Gibco) supplemented with 10% FBS, 2 mM L-glutamine, insulin, 50 μg/ml gentamicin and 50 μg/ml amphotericin-B. All cell lines used were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS and supplemented with antibiotics either with penicillin/streptomycin or gentamicin/ciproflaxin HCl.
Plasmids and reagents
pcDNA6 myc-His-BAG3WT-(1–575) and pcDNA6 myc-His-BAG3-(62–575), expression plasmids were described previously [42, 43]. pcDNA6 myc-His-BAG3-(1–215) was cloned into pCDNA6 myc-His expression vector using the following primers: BAG3-Forward (BamH1) -(1-575); 5′- ACCTATCAGGATCCAT GAGCGCCGCCACCCACTCG- 3, and BAG3-Reverse (XhoI)-(1-215) 5′- ATCTATCACTC GAGGTTCTGCTCGTGTATCACCGG-3′. ptfLC3 (Addgene plasmid 21074) was characterized by Kimura et al. [25]. pcDNA3/HA–BAG1 (wild type) was kindly provided by Dr. R. Morimoto [44]. The plasmids encoding myc-tagged human BAG3 (30, 31) and the BAG3 IPV motif mutants BAG3-B8, BAG3-deltaB8, BAG3-B8.1, and BAG3-B8.2 were previously described [Fuchs et al, Biochem J. 2009]. pCDNA3-Beclin-1 (Plasmid 21150, provided by Junying Yuan) and GFP-Bcl2 (Plasmid 17999, provided by Clark Distelhorst) were all purchased from Addgene. IRDye® 800CW Goat Anti-Mouse and IRDye® 680RD Goat Anti-Rabbit secondary antibodies were purchased from LI-COR, Inc. and used for detection by Odyssey® CLx Imaging System (LI-COR, Inc., Lincoln, NE). Bradford Reagent was from Bio-Rad. Fugene 6 was purchased from Promega (Madison, WI). Polyclonal anti-BAG3 antibody was from Proteintech Group (Chicago, IL). Polyclonal Living Color® full length GFP antibodies were from Clontech, Monoclonal Anti Flag® antibodies (M2) were from Sigma. Monoclonal antibodies; anti-myc Tag (9B11), anti-GAPDH, anti-alpha-tubulin (B-7) and polyclonal antibodies; anti-Beclin-1, anti-Atg3, anti-Atg7, anti-Atg12, Cleaved Caspase-3 (Asp175) were from Cell Signaling (Danvers, MA). Polyclonal anti-LC3 antibody was from Sigma-Aldrich (Saint Louis, MO). Polyclonal anti-BAG-1 Antibody (C-16) and monoclonal anti-Bcl-2 antibody (C-2) were from Santa–Cruz (Dallas, TX). Goat anti-(mouse IgG)-peroxidase conjugate and goat anti-(rabbit IgG)-peroxidase conjugate were from Pierce (Rockford, IL). Acridine orange was purchased from Polysciences, Inc. (Warrington, PA); MG115 was from Calbiochem (Darmstadt, Germany). 3-methyl adenine was purchased from Sigma-Aldrich (St. Louis, MO).
Adenoviral vectors
Adeno-BAG3 siRNA construct was described previously [20]. Briefly, Adeno-BAG3 siRNA construct was made using the BD Adeno-X Expression Systems 2 PT3674-1 (Pr36024) and BD knockout RNAi Systems PT3739 (PR42756) (BD Biosciences-Clontech, Palo Alto, CA). The BAG3 siRNA (5′-AAG GUU CAG ACC AUC UUG GAA-3) was inserted in RNAi-ready pSIREN-DNR vector, which was then used to transfer the shRNA expression cassette to the Adenoviral Acceptor Vector pLP-Adeno-X-PRLS viral DNA (BD Adeno-X Expression Systems 2), containing ΔE1/ΔE3 Ad5 genome, by Cre-loxP mediated recombination. An AdNull empty adenoviral vector was used as control. T98G cells were transduced by thawing the titrated virus stocks at 37°C, mixing the appropriate volume of virus (50 PFU for single cell) in serum free medium Opti-MEM® (Invitrogen, Grand Island, NY) and adding the mixture to the cells. After incubation for 1h, mixture was replaced by regular growth medium.
Western blots
For the preparation of whole-cell extract, cells were lysed with TNN lysis buffer (40 mM Tris-HCL pH 7.4, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, 1% NP40 and 1% protease inhibitors cocktail). Protein extracts were eluted with Laemmli sample buffer, heated at 95°C for 10 min, resolved by SDS–PAGE and transferred to reinforced supported nitrocellulose membranes (Whatman™, Germany) for 2 h at 4°C in a transfer buffer containing 25 mM Tris (pH 7.4), 193 mM glycine, and 20% methanol. Membranes were blocked for 1 hour at room temperature with 10% nonfat dry milk in 1 × phosphate-buffered saline with 0.1% Tween-20 (PBST), washed and incubated with primary antibodies for 2 h in 5% nonfat dry milk. The blots were subsequently washed three times and incubated with IRDye® 800CW goat anti-mouse and IRDye® 680RD goat anti-rabbit secondary antibodies and visualized with an Odyssey® CLx Imaging System (LI-COR, Inc., Lincoln, NE). Some of the blots were incubated with secondary anti-mouse or anti-rabbit antibody conjugated to horseradish peroxidase. Proteins were detected with ECL enhanced chemiluminescence detection kit (GE Healthcare Life Sciences, Piscataway, NJ) according to the manufacturer's recommendations.
Co-immunoprecipitation
HEK293T cells were grown in DMEM + 10% FBS supplemented with antibiotics in 100 mm dishes pre-treated with poly-D-Lysine at 37°C under 7% CO2. Transfections using Lipofectamine 2000 were performed when the cells reached 90-95 % confluency where transfection reagent: DNA ratio was 2:1. Before the transfections, DNA concentration of the each plasmid was checked carefully and quality of the DNA was also confirmed by agarose gel electrophoresis. The concentration of Bcl2 (GFP-Bcl2) plasmid DNA is critical to achieve efficient co-expression with Beclin-1 (pCDNA3-Beclin-1). The high amounts of Bcl2 expression lead to apoptosis and down regulation of Beclin-1 expression. Therefore plasmid DNA concentrations were adjusted to obtain equal amounts of Bcl2 and Beclin-1 expression; GFP-Bcl2: 0.5 μg /100 mm plate; pCDNA3-Beclin-1: 3 μg/100 mm plate (Bcl2: Beclin-1 DNA ratio was 1:6). For BAG3 expression BAG3 wt (pCDNA 6.1-myc BAG3 (WT) and its mutants [pCDNA 6.1-BAG3 (1-215) and pCDNA 6.1-BAG3 (62-575)] were used in 2 μg per 100 mm dish. BAG3 (62-575) mutant was unstable when co-expressed with Bcl2 and Beclin-1. Twenty hours after transfection, the cells were trypsinized, resuspended in DMEM + 10 % FBS and washed with 1× PBS, before being lysed in 1 ml TNN + 1 % NP40 with protease inhibitors. The clear lysates were obtained after centrifugation at max speed in an Eppendorf centrifuge. The cell lysates containing Bcl2, Beclin-1 and BAG3 proteins were first separated on a 10% SDS-PAGE and western blot experiments were performed as described above to ensure the equal Bcl2 and Beclin-1 overexpression. Once the equal amount of protein expression obtained, the co-immunoprecipitation was performed using monoclonal anti-FLAG antibodies which recognize FLAG tagged Beclin-1. First, 1.5 μg monoclonal antibody was incubated with Protein G Sepharose resin (20 mL) in TNN + 1% NP40 for an hour, and then the clear lysates were added to final 200 μg total protein concentration. As a control co-IP with normal mouse serum was done parallel to co-IP with anti-FLAG antibody. All immuno-precipitation experiments were carried out by gentle mixing in a 360° rotator at 4°C for 20 hours. After 20 hours, the resins containing immune-precipitates were extensively washed three times with TNN + 1 % NP40 buffer and then denatured at 95°C for 4 minutes with 20 ml 2 × SDS loading dye. The denatured samples were resolved on a 10 % SDS –PAGE followed by a transfer onto a 0.4 μ Odissey® nitrocellulose membrane at 250 mA for 2 hours at 4°C. The membranes were then blocked with 10 % milk in TBST for an hour and incubated with 1/1000 diluted polyclonal Living Color ® antibodies which recognize the GFP-Bcl2. The same membrane was also probed with polyclonal anti-Beclin-1 and monoclonal anti-myc for BAG3 antibodies (1/1000 dilutions). As a loading control, GAPDH primary antibodies were used (1/1000 dilution). All primary antibody incubations were carried out overnight at 4°C. Excess antibodies were washed with 1x TBST buffer three times and the blots were incubated with secondary antibodies [IRDye® 800 CW goat anti-rabbit IgG, or IRDye® 680 RD goat anti-mouse IgGs (1/10,000 dilutions)] for 1 hour in dark at RT. After washing 3 times with TBST and once with 1 × PBS, the wet membranes were scanned and analyzed using Odyssey® CLx Imaging System (LI-COR, Inc., Lincoln, NE).
Immunocytochemistry
T98G cells were plated in 2-well chamber slides and transiently transfected with ptfLC3 and pcDNA6 myc-His-BAG3-WT and its various mutations as described in the text. At 24hr post-transfections, cells were fixed with 4% (w/v) paraformaldehyde and mounted with Vectashield mounting medium containing DAPI. RFP-LC3 and GFP-LC3 signals were determined by fluorescence microscopy. For the experiments involving BAG3 and LC3 co-localization studies, T98G cells were plated on glass chamber slides. The next day cells were transfected with pcDNA6 Myc-His-BAG3-WT using Fugene 6 reagent. At 48 hours post-transfection, cells were washed with 1× PBS three times and fixed with 4% (w/v) paraformaldehyde and permeabilized by incubation with 0.1% Triton X-100 in PBS for 5 min. Cells were then blocked with 10% BSA in PBS for 2 h and incubated with mouse monoclonal anti-Myc Tag (9B11) antibody diluted 1:400 in 5% BSA/1× PBS and polyclonal anti-LC3 antibody diluted 1:200 in 5% BSA/1× PBS for 2h at room temperature. Control cells were processed without primary antibody. For BAG3 and Bcl2 co-localization experiments, T98G cells were plated on 2 chamber slides as described earlier and co-transfected with pcDNA6 Myc-His-BAG3-WT and GFP-Bcl2 plasmids; for Bcl2 and Beclin-1 co-localization experiments GFP-Bcl2 and pcDNA3-Beclin-1 plasmids were used (Lipofectamine 2000: DNA, 2:1). Twenty four hour after transfections, slides were washed with 1× PBS and fixed with ice cold acetone for 3 min. The fixed cells were then washes 3 times with 1× PBS and incubated in 1 × TBST buffer containing 1 % BSA for 1 hour. Polyclonal antibodies against Beclin-1 (α-Beclin-1), polyclonal antibodies against BAG3, monoclonal antibodies against myc tag (for BAG3) and monoclonal antibodies against Bcl2 (α-Bcl2) were used as needed in 1/200 dilutions in TBST buffer containing 1 % BSA. The slides were incubated overnight at RT followed by 3 times wash with TBST buffer. For secondary antibody treatment, cells were washed five times with 1× PBS and incubated with rhodamine-conjugated anti-mouse secondary antibody (Pierce, Rockford, IL) diluted 1:500 in 5% BSA/1× PBS and FITC-conjugated anti-rabbit secondary antibody (Pierce) diluted 1:200 in 5% BSA/1× PBS for 1h at RT in the dark. Slides were washed three times with PBS, mounted with Vectashield mounting medium for fluorescence containing DAPI (Vector Laboratories Burlingame, CA), and examined by fluorescence microscopy.
Fluorescence-activated cell sorting (FACS) analysis using acridine orange
The cellular acidic compartments, as a marker of autophagy, were assessed by supravital cell staining with acridine orange (AO) (Polysciences, Inc. Warrington PA). AO moves freely across biological membranes and stains the acidic compartments in the cell bright red, whereas the nucleus and cytoplasm are stained to bright green and dim red fluorescence, respectively. Therefore, the intensity of the red staining is proportional to the degree of acidity and can be quantified. T98G cells were plated on 12-well plate 0.1×106 cells /well in duplicate. Cells were transfected either with pcDNA6 Myc-His-BAG3-WT or the plasmids encoding each of BAG3 deletion mutants (pcDNA6-Myc-His-BAG3-(1–215), pcDNA6-Myc-His-BAG3-(62–575), and pcDNA6- Myc-His vector alone. Cells were incubated with growth medium containing AO at a final concentration of 1 μg/ml for 15 min, harvested with TrypLeExpress (Invitrogen) and quantified based on percent of cells displaying red (>650nm) fluorescence using Guava Express Software.
Acknowledgments
The authors wish to thank past and present members of the Department of Neuroscience/Center for Neurovirology for sharing of reagents and ideas. We also thank Dr. Satish Deshmane for the Adenoviral constructs for BAG3 siRNA, Dr. Martyn K White for critical reading of this manuscript, and C. Papaleo for editorial assistance. This study utilized services offered by core facilities of the Comprehensive NeuroAIDS Center (CNAC NIMH Grant Number P30MH092177) at Temple University School of Medicine. This work was made possible by grants awarded by NIH to KK.
Footnotes
Author Contributions: Conceived and designed the experiments: IKS, (A.S.S) and KK. Performed the experiments: TK, A.S.S., NM. Analyzed the data: IKS, A.S.S., J.G., and KK. Contributed reagents/materials/analysis tools: MW, JL, MCT. Wrote the paper: IKS, A.S.S., J.G. and KK.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Jiang H, White EJ, Conrad C, Gomez-Manzano C, Fueyo J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009;453:273–286. doi: 10.1016/S0076-6879(08)04013-5. [DOI] [PubMed] [Google Scholar]
- 3.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 4.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008a;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 6.Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 7.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 9.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 10.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 12.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, et al. Functional and physical interaction between BclX(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 14.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Doong H, Vrailas A, Kohn EC. What's in the ‘BAG’? – a functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188:25–32. doi: 10.1016/s0304-3835(02)00456-1. [DOI] [PubMed] [Google Scholar]
- 16.Rosati A, Ammirante M, Gentilella A, Basile A, Festa M, et al. Apoptosis inhibition in cancer cells: a novel molecular pathway that involves BAG3 protein. Int J Biochem Cell Biol. 2007a;39:1337–1342. doi: 10.1016/j.biocel.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 18.Lee JH, Takahashi T, Yasuhara N, Inazawa J, Kamada S, et al. Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene. 1999;18:6183–6190. doi: 10.1038/sj.onc.1203043. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem. 2009 Apr 3;284(14):9176–83. doi: 10.1074/jbc.M808656200. Epub 2009 Jan 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosati A, Khalili K, Deshmane SL, Radhakrishnan S, Pascale M, et al. BAG3 protein regulates caspase-3 activation in HIV-1-infected human primary microglial cells. J Cell Physiol. 2009;218:264–267. doi: 10.1002/jcp.21604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carra S, Seguin SJ, Landry J. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008b;4:237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 22.Beere HM. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J Clin Invest. 2005;115:2633–2639. doi: 10.1172/JCI26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doong H, Rizzo K, Fang S, Kulpa V, Weissman AM, et al. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly ubiquitinated Hsp90 client proteins. J Biol Chem. 2003;278:8490–28500. doi: 10.1074/jbc.M209682200. [DOI] [PubMed] [Google Scholar]
- 24.Sariyer IK, Merabova N, Patel PK, Knezevic T, Rosati A, et al. Bag3-induced autophagy is associated with degradation of JCV oncoprotein, T-Ag. PLoS One. 2012;7:e45000. doi: 10.1371/journal.pone.0045000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2010;430:559. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2009;425:245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 28.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis Cell Death and Differentiation. 2011;18:571–580. doi: 10.1038/cdd.2010.191. published online 11 February 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo S. Rubinsztein DC Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010 Feb;17(2):268–77. doi: 10.1038/cdd.2009.121. Epub 2009 Aug 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 31.Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 32.Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclinphosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001a;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001b;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3(1):17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antoku K, Maser RS, Scully WJ, Jr, Delach SM, Johnson DE. Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein. Biochem Biophys Res Commun. 2001;286:1003–1010. doi: 10.1006/bbrc.2001.5512. [DOI] [PubMed] [Google Scholar]
- 37.Wang HQ, Meng X, Gao YY, Liu BQ, Niu XF, et al. Characterization of BAG3 cleavage during apoptosis of pancreatic cancer cells. J Cell Physiol. 2010;224:94–100. doi: 10.1002/jcp.22097. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvationinduced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian S, Lin J, Zhou J, Wang X, Li Y, Ren X, Yu W, Zhong W, Xiao J, Sheng F, et al. Beclin 1-independent autophagy induced by a Bcl-XL/Bcl-2 targeting compound, Z18. Autophagy. 2010;6:1032–1041. doi: 10.4161/auto.6.8.13336. http://dx.doi.org/10.4161/auto.6.8.1333642. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Du Z, Zong Z, Li C, Li N, Zhang Q, Kong D, Wang H. BAG3-dependent noncanonical autophagy induced by proteasome inhibition in HepG2 cells. Autophagy. 2013;9:905–916. doi: 10.4161/auto.24292. http://dx.doi.org/10.4161/auto.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noch E, Bookland M, Khalili K. Astrocyte-elevated gene-1 (AEG-1) induction by hypoxia and glucose deprivation in glioblastoma. Cancer Biol Ther. 2011;11:32–39. doi: 10.4161/cbt.11.1.13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gentilella A, Khalili K. Autoregulation of co-chaperone BAG3 gene transcription. J Cell Biochem. 2009;108:1117–11124. doi: 10.1002/jcb.22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentilella A, Khalili K. BAG3 expression in glioblastoma cells promotes accumulation of ubiquitinated clients in an Hsp70-dependent manner. J Biol Chem. 2011;286:9205–9215. doi: 10.1074/jbc.M110.175836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song J, Takeda M, Morimoto RI. Bag1-Hsp70 mediates a physiological stress signaling pathway that regulates Raf-1/ERK and cell growth. Nat Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]