Abstract
The vascular plant Arabidopsis thaliana is a central genetic model and universal reference organism in plant and crop science. The successful integration of different fields of research in the study of A. thaliana has made a large contribution to our molecular understanding of key concepts in biology. The availability and active development of experimental tools and resources, in combination with the accessibility of a wealth of cumulatively acquired knowledge about this plant, support the most advanced systems biology approaches among all land plants. Research in molecular ecology and evolution has also brought the natural history of A. thaliana into the limelight. This article showcases our current knowledge of the natural history of A. thaliana from the perspective of the most closely related plant species, providing an evolutionary framework for interpreting novel findings and for developing new hypotheses based on our knowledge of this plant.
DOI: http://dx.doi.org/10.7554/eLife.06100.001
Research organism: Arabidopsis
Introduction
Arabidopsis thaliana is a small, annual or winter annual, rosette plant (Figure 1). It belongs to the taxonomic family of the Brassicaceae in the eudicotyledonous group of angiosperm vascular plants (see Box 1 for a glossary of specialist terms used in this article). This family also includes morphologically diverse but closely related oilseed crops, vegetables and spice plants, for example rapeseed, brussel sprouts, various cabbages, cauliflower, garden radish and mustard. First described by the physician Johannes Thal in the Harz Mountains of Northern Germany in 1577, A. thaliana featured in the Species Plantarum II published by Linnaeus in 1753, and it received its present name, Arabidopsis thaliana (L.) Heynh, from Gustav Heynhold in 1842 (Kück, 2005). Friedrich Laibach was the first researcher to publish an influential cytogenetic study on A. thaliana in 1907, to foster genetics in the 1940s, and to systematically collect accessions in the wild after emphasizing the extent of intraspecific phenotypic variation in A. thaliana in the 1950s (Somerville and Koornneef, 2002; Koornneef and Meinke, 2010).
Figure 1. Life cycle of Arabidopsis thaliana.
(A) A. thaliana of the accession Columbia (Col) at different stages of its life cycle, from seed (bottom left) to seedling (11 days), to vegetative growth (39 days), and to reproductive growth (45 days). Photographs of (B) a flower, (C) a pollen grain (scanning electron micrograph), and (D) mature siliques (seed pods; left: closed; right: open with a few remaining unshattered seeds) at higher magnification. Image credits: B and C, Maria Bernal and Peter Huijser; other photographs, Ines Kubigsteltig and Klaus Hagemann.
Box 1. Glossary.
Allopolyploidy: Following the hybridization of two different species, this term describes when two (or more) chromosome sets from each parent are maintained in the genome.
Autopolyploidy: Polyploidy arising from the multiplication of chromosome sets in a genome, often due to the formation of unreduced gametes in an aberrant meiosis.
Calcareous outcrops: Rocky areas with predominantly calcium carbonate-rich minerals, sparse vegetation, soils of high pH with poor biovailability of some nutrients and poor water-holding capacity.
Commensal: An organism that lives in close association with a host organism, from which it benefits—usually by receiving nutrients—without harming the host or being beneficial to it.
Effector-triggered immunity: A form of plant innate immunity mediated by plant NLR proteins (see below) of diverse recognition specificities, encoded by Resistance (R) genes organized in recombinogenic, but somatically stable, gene clusters. Each R protein detects a pathogen-derived effector molecule or its action on a host target protein.
Eudicotyledon: Angiosperm (flowering) plant species characterized by two cotyledons (embryonic leaves) and other morphological characteristics.
Glucosinolates: A group of secondary metabolites characteristic of the plant order Brassicales, including the Brassicaceae family.
Induced systemic resistance: Systemic resistance to infection by a broad spectrum of pathogens and to attack by herbivores. Its induction is triggered locally (usually in the root) by the presence of plant-associated beneficial microbes.
Metal hyperaccumulation: The accumulation in above-ground plant structures (usually leaves) of metals or metalloids at concentrations far higher than in non-accumulator plants at the same site. A rare trait found in ∼0.2% of angiosperms that acts as an elemental defense against biotic stress.
Molecular pattern-triggered immunity: A form of plant innate immunity mediated by transmembrane pattern recognition receptors that detect evolutionarily conserved molecules, called cell damage-associated or microbe-associated molecular patterns, usually on the plant cell surface.
NLR proteins: NLR (nucleotide-binding domain leucine-rich repeat) proteins function in plant effector triggered immunity. Also denotes a family of proteins found subsequently to act in animal innate immunity. All NLR proteins share a similar three-domain architecture.
Phenology: The timing, or the study, of periodic life cycle events in organisms in conjunction with seasonal cycles and/or inter-annual climatic variation.
Photosynthate: A product of photosynthesis, that is, an energy-rich carbon-containing molecule (most commonly a sugar).
Plant growth-promoting rhizobacteria: Beneficial bacteria that enhance plant growth and biomass production and live in association with plant roots in natural environments.
Synanthropy: Occurrence of an organism in association with human settlement, where human activities directly or indirectly generate favorable environmental conditions for the organism.
Ultramafic soils: Infertile, nutrient poor soils derived from so-called serpentine minerals, which are common in zones of tectonic activity.
The popularity of A. thaliana as a model organism took off in the 1980s, and then rapidly gained momentum when researchers successfully began to combine genetics with powerful molecular biology methods. The Arabidopsis reference genome sequence was published as the first nuclear genome of a flowering plant in 2000 (http://www.arabidopsis.org/) (Arabidopsis Genome Initiative, 2000; Somerville and Koornneef, 2002; Koornneef and Meinke, 2010). The ease and speed with which experiments can be conducted on A. thaliana has allowed enormous fundamental progress in our knowledge of the molecular principles of plant development, cell biology, metabolism, physiology, genetics and epigenetics (http://www.arabidopsisbook.org/). The many uses of Arabidopsis as the universal reference plant continue to expand, particularly in the field of systems biology (Brady et al., 2007; Endo et al., 2014), and more recently provide a means of placing molecular functions into an ecological and evolutionary context in natural environments (Tian et al., 2003; Mitchell-Olds and Schmitt, 2006; Alonso-Blanco et al., 2009; Atwell et al., 2010; Bergelson and Roux, 2010; Fournier-Level et al., 2011; Hancock et al., 2011; Züst et al., 2012).
Many of the traits that render A. thaliana an attractive model plant also constitute adaptations that reflect the ecological niche occupied by this species, such as the short generation time and the ability to self-fertilize (selfing). Indeed, the natural history of A. thaliana differs notably from that of the species in its sister group of the most closely related existing plant species, which includes Arabidopsis lyrata, Arabidopsis arenosa and Arabidopsis croatica, and the three species Arabidopsis halleri, Arabidopsis cebennensis and Arabidopsis pedemontana (Koch and Matschinger, 2007; Hohmann et al., 2014) (Figure 2). Within these six evolutionary lineages, a number of phylogenetically (as yet) unresolved taxa have been described, as have auto- and allopolyploidization events (see Glossary). The divergence time of the common ancestor of these species and the A. thaliana lineage was estimated at around 5 million years ago (Mya) (Koch et al., 2000), with upper and lower bounds of 17.9 and 3.1 Mya (Koch et al., 2000; Beilstein et al., 2010; Ossowski et al., 2010). Thus, divergence occurred immediately before or during the transition from a warm period to progressively colder climates, followed by rapid glacial cycles from 3 Mya until the latest glacial maximum about 18,000 years ago. There has even been a natural allopolyploidization event between A. arenosa and A. thaliana 10,000–300,000 years ago, giving rise to Arabidopsis suecica (Sall et al., 2003; Jakobsson et al., 2006).
Figure 2. A. thaliana and a subset of species from its sister clade.
From left to right: A. thaliana (Col), A. halleri (ssp. halleri; individual Lan5, Langelsheim, Harz, Germany), A. lyrata (ssp. lyrata; selfing accession Great Lakes, North America), and A. croatica (Baške Oštarje/Ljubičko Brdo, Croatia). A. thaliana was grown from seed to early reproductive stage, and the other species were propagated vegetatively and grown for 3–6 months. The individuals shown here do not reflect the large within-species morphological diversity, particularly in leaf shape, among different accessions of A. halleri and A. lyrata. Image credit: Ute Krämer and Klaus Hagemann.
Work in recent years has increasingly emphasized the relevance of conducting experiments on A. thaliana in natural or semi-natural settings (Wilczek et al., 2009; Brachi et al., 2010; Chiang et al., 2013; Karasov et al., 2014). This important development will benefit from a more detailed knowledge of the unique and shared features of its natural and evolutionary history, as well as of the natural environments that host populations of A. thaliana.
The natural history of A. thaliana in an evolutionary and ecological context
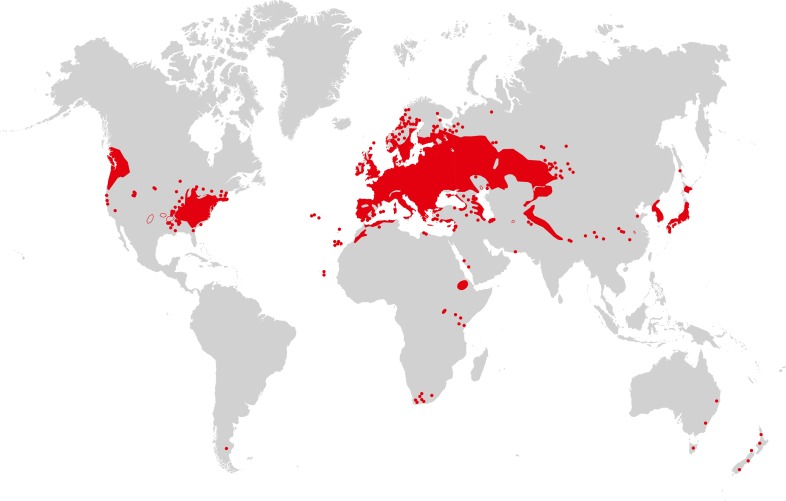
Arabidopsis thaliana is recognized as native to Western Eurasia (Figure 3). The species is a colonizer and pioneer plant of disturbed, poor, stony or shallow soils, and it can also be found in nutrient-poor, often sandy, meadow and forest habitats (Mitchell-Olds and Schmitt, 2006). Similarly, the species closely related to A. thaliana are able to colonize bare rocky habitats. Sequence polymorphism data provide evidence for a historical expansion of the A. thaliana species population (Nordborg et al., 2005; Schmid et al., 2005), thus differing from the known demographic histories of the non-hybrid species in its sister group in the Arabidopsis genus (Wright et al., 2003; Heidel et al., 2010). It has been suggested that A. thaliana re-colonized Central Europe from glacial refugia in the Caucasus or Balkans between ∼10,000 and 5000 years ago, possibly accompanying the spread of human farming from the Near East (Francois et al., 2008). There is also clear evidence for additional glacial refugia for A. thaliana in the Mediterranean region, including the North African Atlas Mountains (Brennan et al., 2014), and for additional migration (for example, from the Iberian peninsula in a northeastern direction [Sharbel et al., 2000; Pico et al., 2008]), as well as for population structure reflecting isolation by distance in Eurasia (Nordborg et al., 2005). This reflects an evolutionary history of vast climatic and environmental fluctuations, likely including a series of alternating phases of population size contraction and expansion with migration and admixture following glacial cycles (Beck et al., 2008). In the past few 100 years, A. thaliana has further expanded its geographic and climatic range in synanthropy (see Glossary), migrating to and across North America, as well as southward in Africa, and more recently to East Asia (see Figure 3). The present climatic and geographic range of A. thaliana is larger than that of any of its close relatives, which have either not expanded into warmer climates or are local endemics (Koch and Matschinger, 2007; Hohmann et al., 2014).
Figure 3. Map of A. thaliana worldwide distribution.
Areas colored in red correspond to the continuous distribution of A. thaliana; red circles mark additional sites. This map is based on a partial map kindly provided by Matthias Hoffmann (personal communication, November 2014), with manual additions to the southern hemisphere (Bresinsky et al., 2008). Image credit: Ute Krämer and Klaus Hagemann.
In addition to—and likely permitting—the rapid expansion of its range, A. thaliana has undergone transitions to reproduction by self-fertilization (selfing). Estimates place these transitions as having occurred, based on different approaches, at around 0.4 Mya or around ≥1 Mya (Bechsgaard et al., 2006; Tang et al., 2007). By contrast, most species closely related to A. thaliana are either predominantly or strictly outcrossing (the latter if species are strongly self-incompatible). In addition to selfing, A. thaliana is an annual plant with a rapid life cycle that can be as short as 6–8 weeks in some accessions, whereas most of its close relatives are biannuals or herbaceous perennials (Al-Shehbaz and O'Kane, 2002) (see Figures 1, 2). Vegetative A. thaliana plants are smaller in size than most of its close relatives, and their survival and reproduction are comparably robust in a variety of environments. The leaves of A. thaliana are positioned to form a more compact rosette, and inflorescences are more erect and elongated, than in its close relative A. halleri. These morphological differences are likely to constitute adaptations to differing environments. By comparison to its sister group of species, the flowering of A. thaliana is highly synchronized, with the formation of a larger number of siliques (seed pods) per plant, each containing a higher, or far higher, number of seeds that are smaller in size compared to those of its relatives. Seeds of A. thaliana are long-lived and highly viable by comparison to those of A. halleri and A. lyrata (Ute Krämer, personal observation). These features of A. thaliana render it ideal for mutagenesis, genetics, and for the identification, selection and propagation of mutant or transgenic lines, as well as for stock maintenance.
The complexity of organ architecture appears to be reduced in A. thaliana. Notably, its roots mostly contain only a single layer of each specialized cell type, a feature that is optimal for in vivo imaging (Figure 4) (De Lucas and Brady, 2013). This organization differs from the roots of its close relative A. halleri, which contain two additional cell layers (Hanikenne et al., 2008), and is in stark contrast to root tissues composed of multiple cell layers, as found in many other plants. The metabolic cost of building an A. thaliana root is evidently minimal, and this is likely to have ecological implications which have yet to be determined.
Figure 4. The simple anatomy of A. thaliana roots.
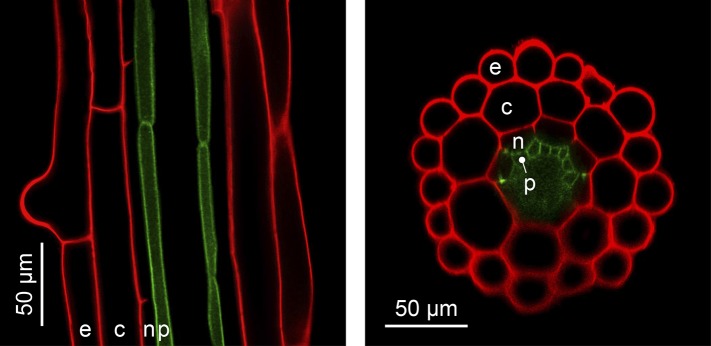
Longitudinal (left) and transverse (right) confocal sections of A. thaliana roots. Green fluorescence highlights the plasma membrane of the pericycle cells of an A. thaliana hma2hma4 double mutant line (Sinclair et al., 2007) (accession Wassilewskija). Red fluorescence of propidium iodide (PI) as a stain is overlaid to visualize cell walls. The root cell layers (consecutive outward to inward) are: epidermis (e), cortex (c), endodermis (n) and pericycle (p). Note that the Casparian Strip surrounding the endodermis cells forms an apoplastic diffusion barrier (Roppolo et al., 2011) that blocks the movement of PI further inward. Image credit: Ute Krämer and Scott A. Sinclair.
The ancestral number of chromosomes in the Brassicaceae (n = 8) is conserved in A. thaliana's closest relatives. By comparison, the genome of the diploid A. thaliana is rearranged, with two sets of n = 5 chromosomes. This is associated with a pronounced reduction in genome size in A. thaliana, down to 157 Mbp (Bennett et al., 2003) (reference genome assembly of 135 Mbp)—one of the smallest known genomes among flowering plants. For comparison, nuclear genome sizes of its closest relatives are larger, mostly due to a higher abundance of transposable elements in euchromatic regions, with 207 Mbp for the reference assembly of the nuclear genome of A. lyrata (Hu et al., 2011), and about 260 Mbp estimated for A. halleri (Johnston et al., 2005; Peer et al., 2006; Hohmann et al., 2014). These properties of A. thaliana have greatly facilitated the sequencing, assembly and annotation of its genome, as well as molecular genetics and genomics approaches.
All of these characteristic features of A. thaliana are likely to constitute adaptations that jointly define the ecological niche of the species and reflect an ecological strategy that has apparently been highly successful in recent evolutionary history.
Plant development and its environmental control
Classical molecular genetics approaches have been used in A. thaliana to dissect the patterning and development of flowers (Meyerowitz et al., 1991), embryos (Mayer et al., 1991), leaves and roots (Di Laurenzio et al., 1996), and of the maintenance of stem cell niches at the plant's growing tips. These stem cells fuel the ongoing (indeterminate) growth that is characteristic of plants (Clark et al., 1997; Lenhard et al., 2001). The biology of A. thaliana is rich in temperature- and light-dependent regulation, some of which might be triggered by photosynthates (see Glossary) rather than by light itself (Haydon et al., 2013). Important work has investigated the molecular networks that mediate environmentally controlled developmental switches in A. thaliana, as well as natural intraspecific variation in phenology (see Glossary). Examples include the light-regulated development of seedlings (photomorphogenesis) (Deng et al., 1992; Nemhauser, 2008), the transition from vegetative to reproductive development, also termed flowering time control (Simpson and Dean, 2002; Valverde et al., 2004; Andres and Coupland, 2012), and seed dormancy and germination control (Bentsink et al., 2006; Graeber et al., 2012), as well as natural variation in these traits (Brachi et al., 2010; Chiang et al., 2013). The need for developmental transitions to be timed accurately and robustly is reflected in the emerging complex, multi-layered epigenetic regulation of some key loci, for example the flowering control gene Flowering Locus C (FLC) (Simpson and Dean, 2002; Csorba et al., 2014). Such complex regulation in A. thaliana may have evolved in a step-wise fashion alongside recurrent climatic fluctuations encountered by A. thaliana during its evolutionary history. Nevertheless, contemporary global warming appears to proceed faster than the local adaptation of A. thaliana populations (Wilczek et al., 2014), based on the reproductive fitness of plants grown at locations across the European distribution range from stored seeds, which were collected in different geographic regions between 4 and 70 years earlier.
Phenotypic plasticity and utilizing limited local resources in bare habitats for rapid and ample reproduction appear to be characteristics of the life history of A. thaliana. The plant also seemingly evades periodic or episodic environmental adversities, including between-plant competition, via a rapid life cycle. Consequently, the adjustment of developmental timing to environmental conditions is likely to be of major importance in the phenotypic plasticity of A. thaliana (Martinez et al., 2004; Riboni et al., 2014), and an apparent target of selection (Horton et al., 2012). This might also contribute further to the observed regulatory complexity of flowering time control (see preceding paragraph).
Other interactions with the environment
In contrast to the vast majority of flowering plants, the roots of the Brassicaceae generally do not undergo mycorrhizal or rhizobial symbiosis. This is likely to make A. thaliana comparably independent of specific soil microbes or microbiomes, which may be advantageous for a colonizer of recently disturbed and exposed soils. Additionally, it allows the plant to commit all energy expenditure to its own growth and reproduction, but heightens the demands on the endogenous machinery for nutrient acquisition and efficiency, and for protecting roots from environmental stress. Some of the central molecular components of these pathways have been identified in A. thaliana before their discovery in other kingdoms of life, for example, the biosynthesis of low-molecular-weight chelators for micronutrient metal cations through nicotianamine synthase (Trampczynska et al., 2006; Schuler et al., 2012), and for both micronutrient and non-essential toxic metal cations through phytochelatin synthase (Clemens, 2006; Tennstedt et al., 2009). Moreover, the effective and balanced acquisition of adequate quantities of inorganic nutrients, such as phosphate, nitrogen and iron, and their efficient utilization by A. thaliana plants have been observed and characterized at the molecular level (Abel, 2011; Hindt and Guerinot, 2012; Araya et al., 2014). When natural accessions of A. thaliana are cultivated under uniform controlled conditions, genetically controlled within-species variation is observed in leaf nutrient concentrations (Atwell et al., 2010), which appears to be adaptive at least in some cases (Poormohammad Kiani et al., 2012).
Key traits of A. thaliana are the timing of germination to coincide with favorable environmental conditions, a short life cycle, and the ability to tolerate unfavorably hot or dry conditions in the form of de-hydrated seeds. Consequently, we would expect stress tolerance to be less elaborate in A. thaliana than in some other plants. Indeed, despite a very wide geographic distribution range, there are no reports of A. thaliana accessions that exhibit extreme physiological traits, different from its sister group of plant species. As one out of a total of ∼500 known metal hyperaccumulator plant species (see Glossary), A. halleri accumulates extraordinarily high concentrations of zinc and cadmium in leaves where these metals act as a so-called elemental defense against biotic stress (Boyd, 2010; Krämer, 2010). In addition, several species in the sister clade of A. thaliana are well-known members of plant communities found in extreme habitats. Calamine soils rich in zinc, cadmium and lead—commonly as a result of anthropogenic pollution—are readily colonized by A. halleri (Ernst, 1974; Krämer, 2010), and occasionally by A. arenosa (Przedpełska and Wierzbicka, 2007). In addition, there are some natural populations of A. arenosa and A. lyrata on nutrient-poor ultramafic soils (Holubover serpentinite, 2002; Turner et al., 2010), which contain high levels of nickel and magnesium originating from the local bedrock (see Glossary). Finally, populations of A. lyrata (Al-Shehbaz and O'Kane, 2002) and A. croatica (Plants, 2014) characteristically inhabit calcareous outcrops (see Glossary).
Above-ground (Vorholt, 2012; Horton et al., 2014) and below-ground organs of A. thaliana, and their immediate vicinity, provide habitats for microbial communities, which appear to be more strongly influenced by soil type than by plant genotype (Bulgarelli et al., 2012; Lundberg et al., 2012; Schlaeppi et al., 2014). These microbial communities can include commensals (see Glossary), as well as various naturally occurring viral, bacterial, fungal and oomycete pathogens. Arabidopsis possesses a wealth of general molecular pattern-triggered innate immunity responses (Schwessinger and Ronald, 2012) and more-specific, effector-triggered and Resistance (R) gene-dependent, defenses against microbes (Cui et al., 2014) (see Glossary). Their elucidation has extensively contributed to our understanding of plant defenses against pathogens. By comparison, we understand far less about how pathogens shape genetic diversity in A. thaliana natural populations (Karasov et al., 2014) and about the establishment and maintenance of beneficial interactions of A. thaliana with microbes, such as plant growth-promoting rhizobacteria and microbes that trigger induced systemic resistance (Lugtenberg and Kamilova, 2009; Pieterse et al., 2014) (see Glossary). Once A. thaliana has initiated reproductive development, reproduction is favored over defense (Winter et al., 2011). This is consistent with the general ecological strategy of A. thaliana towards an escape from stress through resource allocation that prioritizes reproduction, followed by a rapid conclusion of the life cycle, which differs from its longer-lived relatives. Similarities have been noted between plant defenses and animal innate immunity. These relate primarily to the domain architectures of NLR (nucleotide-binding domain leucine-rich repeat) protein receptors (see Glossary) that function in the detection of biotic attack, and the programmed cell death that occurs as a host defense response (Maekawa et al., 2011).
Arabidopsis possesses effective defenses against herbivory, for example by insects and snails, with an important role for secondary metabolites. Most prominently, a class of sulfur-rich compounds, called glucosinolates (see Glossary), are converted to pungent and toxic breakdown products in damaged tissues (Halkier and Gershenzon, 2006). These compounds are agronomically important determinants of taste and are receiving much attention as health-protective, anti-cancer components of ‘functional foods’ (Sonderby et al., 2010). Upon leaf wounding, the systemic activation of defenses in distant leaves involves the long-distance movement of electrical surface potential changes at velocities between 43 and 150 µm s−1 involving ionotropic glutamate receptor (iGluR)-related membrane proteins (Mousavi et al., 2013). Strikingly, this bears some similarity to the role of vertebrate iGluRs in fast excitatory synaptic transmission in the central nervous system. The fast systemic movement of electrical signals turns out to be of broader importance in plant responses to the environment. For example, upon exposure of A. thaliana to salt stress, a systemic signal moves from root to shoot in the form of a Ca2+ wave at a velocity of 400 µm s−1, requiring the vacuolar ion channel TPC1 (Two Pore Ca2+ Channel protein 1) (Choi et al., 2014). These findings underpin the central importance of A. thaliana as a model for obtaining molecular insights into fundamentally novel aspects of plant biology.
Metabolism in the context of natural history
Metabolic properties are noteworthy components of natural history because they are likely to facilitate, constrain or even prevent specific adaptations in a given lineage sharing them. With respect to nutrient ions, plants of the Brassicaceae family were classified as calciotrophic, based on their ability to store comparably large quantities of dissolved calcium (Ca2+) (Kinzel, 1982). This is physiologically challenging because cytosolic free Ca2+ levels have to be maintained at very low (<0.1 µM) resting levels continuously in order to allow for Ca2+-mediated signaling to occur (Choi et al., 2014). By contrast, calciophobic plants contain Ca in the form of insoluble precipitates, mostly oxalates. In calciotrophic plants, leaf Ca concentrations are generally higher than leaf potassium concentrations, as is common in the Brassicaceae, Papaveraceae and Resedaceae families, but rare in other plant families. Calcium cations are stored inside cell vacuoles alongside other solutes, including the organic acid anions malate or citrate, which act as both chelators and charge balance for Ca2+. The large capacity that Brassicaceae family members have for storing divalent cations in their vacuoles might explain why this family comprises ∼50% of all known metal hyperaccumulator plant taxa (Krämer, 2010). Moreover, the absence of adaptation to extreme osmotic conditions in A. thaliana could result from its metabolism, which appears to generally avoid high internal osmotic potentials by storing osmotically inactive starch instead of osmotically active photosynthetic sugars (Streb and Zeeman, 2012). Finally, in association with the high sulfur content of glucosinolates, the Brassicaceae exhibit an active sulfur metabolism and a comparably elevated requirement for sulfate as a nutrient in the soil (Marschner, 1995). More data are required for future comparisons between A. thaliana and its sister clade.
Conclusions
Arabidopsis thaliana, a well-established model in molecular genetics and plant cell biology, is progressively being adopted by ecologists and evolutionary biologists, who require far more information on the sites of origin of natural accessions than is contained in the written records of the earlier collections. Insights into the molecular functions of a multitude of individual genes, as well as the elucidation of biosynthetic pathways and regulatory networks in A. thaliana, have proven invaluable for identifying the genetic basis of agronomically important traits in crops, such as plant height and flowering time (Doebley et al., 2006; Olsen and Wendel, 2013). The successful transfer of more complex molecular knowledge from A. thaliana to crops requires us to account for between-species differences in natural history. Future advances in several disciplines will thus involve, for example, an improved understanding of the diversity of abiotic environments where A. thaliana grows naturally, and plant-associated microbial communities, disease and herbivory at these sites (see Box 2). Recently published work highlights the ecological relevance and evolutionary consequences of the relationships between local temperature profiles, plant immunity and hybrid fitness in the field (Chae et al., 2014). In contrast to some endemic and highly specialized plant species among its closest relatives, A. thaliana can physiologically acclimate and evolutionarily adapt to a broad range of environments, and the genetic basis underlying this difference is of major interest for future research.
Box 2. Outstanding questions about the natural history of A. thaliana.
- To understand the genetic diversity between and within populations of A. thaliana in the context of their natural environments of origin, with respect to, for example: abiotic factors (including soil composition), plant-associated microbial communities, disease, and herbivory.
- To investigate the natural history of the species closely related to A. thaliana for subsequent comparative studies in order to identify the genetic basis of traits that cannot be studied in A. thaliana.
- To explore the vast between- and within-species phenotypic variation in the sister clade of A. thaliana for the genetic dissection of these traits, and to analyze their ecological roles and the evolutionary history of the underlying polymorphisms. This will additionally provide key insights into the natural history of A. thaliana.
However, the natural history of A. thaliana is likely to impose some fundamental limitations on its validity as a model for investigating agronomically relevant traits related to the maintenance of growth and survival under stress. Comparative approaches studying A. thaliana alongside closely related species will enable us to unravel the molecular basis of traits that are absent in A. thaliana, as well as the implications of between-species differences in natural history. Yet our knowledge of the natural history of the species in the sister clade of A. thaliana is sketchy at best (see Box 2). Finally, in some cases, it may become relevant to apply the concept of natural history not exclusively at the species level, but also, for example, at the broader genus scale or at the smaller population scale.
Acknowledgements
I am grateful to the following colleagues for their contributions to the figures: Matthias Hoffmann (Martin Luther University Halle-Wittenberg; Figure 3); Peter Huijser (Max Planck Institute for Plant Breeding Research, Cologne) and Maria Bernal (Figure 1B,C); Klaus Hagemann (all Figures); Ines Kubigsteltig (Figure 1A,D); and Scott A. Sinclair (Figure 4) (MB, KH, IK, SAS: Ruhr University Bochum). I also thank Miltos Tsiantis (Max Planck Institute for Plant Breeding Research) for kindly providing A. lyrata seeds, and Marcus Koch (University of Heidelberg) for an A. croatica plant (Heidelberg Botanical Garden, code HEID006009), both used to generate Figure 2. I apologize to all colleagues whose important work was not cited here because of restrictions in space.
Funding Statement
The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Competing interests:
The author declares that no competing interests exist.
Author contributions
UK, Drafting or revising the article.
Funding Information
This paper was supported by the following grants:
Deutsche Forschungsgemeinschaft (DFG) SPP1529 ADAPTOMICS, Kr1967/10-2 to Ute Krämer.
Deutsche Forschungsgemeinschaft (DFG) SPP1529 ADAPTOMICS, Kr1967/12-1 to Ute Krämer.
References
- Abel S. Phosphate sensing in root development. Current Opinion in Plant Biology. 2011;14:303–309. doi: 10.1016/j.pbi.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? The Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA, O'Kane SL. The Arabidopsis Book. The American Society of Plant Biologists; 2002. Taxonomy and phylogeny of Arabidopsis (Brassicaceae) pp. 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences of USA. 2014;111:2029–2034. doi: 10.1073/pnas.1319953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JD, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz JO, Bergelson J, Nordborg M. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Molecular Biology and Evolution. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- Beck JB, Schmuths H, Schaal BA. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Molecular Ecology. 2008;17:902–915. doi: 10.1111/j.1365-294X.2007.03615.x. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences of USA. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Price HJ, Johnston JS. Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb. Annals of Botany. 2003;91:547–557. doi: 10.1093/aob/mcg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences of USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nature Reviews Genetics. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- Boyd R. Elemental defenses of plants by metals. Nature Education Knowledge. 2010;3:57. [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLOS Genetics. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brennan AC, Mendez-Vigo B, Haddioui A, Martinez-Zapater JM, Pico FX, Alonso-Blanco C. The genetic structure of Arabidopsis thaliana in the south-western Mediterranean range reveals a shared history between North Africa and southern Europe. BMC Plant Biology. 2014;14:17. doi: 10.1186/1471-2229-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresinsky A, Neuhaus G, Körner C, Sonnewald U, Kadereit JW. Strasburger Lehrbuch der Botanik. 36th edition. Heidelberg, Germany: Spektrum Akademischer Verlag Heidelberg; 2008. [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, Lechner S, Demar M, Habring-Müller A, Lanz C, Rätsch G, Weigel D. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159:1341–1351. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Dittmar E, Kramer EM, de Casas RR, Donohue K. Pleiotropy in the wild: the dormancy gene DOG1 exerts cascading control on life cycles. Evolution. 2013;67:883–893. doi: 10.1111/j.1558-5646.2012.01828.x. [DOI] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proceedings of the National Academy of Sciences of USA. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Clemens S. Evolution and function of phytochelatin synthases. Journal of Plant Physiology. 2006;163:319–332. doi: 10.1016/j.jplph.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proceedings of the National Academy of Sciences of USA. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology. 2014 doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- De Lucas M, Brady SM. Gene regulatory networks in the Arabidopsis root. Current Opinion in Plant Biology. 2013;16:50–55. doi: 10.1016/j.pbi.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-Q. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/S0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515:419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst WHO. Schwermetallvegetationen der Erde. Stuttgart, Germany: Gustav Fischer Verlag; 1974. [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Francois O, Blum MG, Jakobsson M, Rosenberg NA. Demographic history of european populations of Arabidopsis thaliana. PLOS Genetics. 2008;4:e1000075. doi: 10.1371/journal.pgen.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. Molecular mechanisms of seed dormancy. Plant, Cell & Environment. 2012;35:1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–692. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Ramos-Onsins SE, Wang WK, Chiang TY, Mitchell-Olds T. Population history in Arabidopsis halleri using multilocus analysis. Molecular Ecology. 2010;19:3364–3379. doi: 10.1111/j.1365-294X.2010.04761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt MN, Guerinot ML. Getting a sense for signals: regulation of the plant iron deficiency response. Biochimica et Biophysica Acta. 2012;1823:1521–1530. doi: 10.1016/j.bbamcr.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann N, Schmickl R, Chiang TY, Lu Anova M, Kola F, Marhold K, Koch MA. Taming the wild: resolving the gene pools of non-model Arabidopsis lineages. BMC Evolutionary Biology. 2014;14:224. doi: 10.1186/s12862-014-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubover serpentinite Sdružení Oficiálního informačního systému Český Krumlov. 2002 http://www.ckrumlov.cz/de/region/soucas/t_holhad.htm. [Google Scholar]
- Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, Bergelson J. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nature Communications. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, Nordborg M, Borevitz JO, Bergelson J. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nature Genetics. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KF, Van de Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Hagenblad J, Tavare S, Sall T, Hallden C, Lind-Hallden C, Nordborg M. A unique recent origin of the allotetraploid species Arabidopsis suecica: evidence from nuclear DNA markers. Molecular Biology and Evolution. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ. Evolution of genome size in Brassicaceae. Annals of Botany. 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov TL, Kniskern JM, Gao L, DeYoung BJ, Ding J, Dubiella U, Lastra RO, Nallu S, Roux F, Innes RW, Barrett LG, Hudson RR, Bergelson J. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature. 2014;512:436–440. doi: 10.1038/nature13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel H. Pflanzenökologie und Mineralstoffwechsel. Stuttgart: Verlag Eugen Ulmer; 1982. [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Molecular Biology and Evolution. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Koch MA, Matschinger M. Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of USA. 2007;104:6272–6277. doi: 10.1073/pnas.0701338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Meinke D. The development of Arabidopsis as a model plant. The Plant Journal. 2010;61:909–921. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- Krämer U. Metal hyperaccumulation in plants. Annual Review of Plant Biology. 2010;61:517–534. doi: 10.1146/annurev-arplant-042809-112156. [DOI] [PubMed] [Google Scholar]
- Kück U. Praktikum der Molekulargenetik. Heidelberg: Springer-Verlag Berlin Heidelberg; 2005. [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/S0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annual Review of Microbiology. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nature Immunology. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. 2nd edition. London: Academic Press Ltd; 1995. [Google Scholar]
- Martinez C, Pons E, Prats G, Leon J. Salicylic acid regulates flowering time and links defence responses and reproductive development. The Plant Journal. 2004;37:209–217. doi: 10.1046/j.1365-313X.2003.01954.x. [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. doi: 10.1038/353402a0. [DOI] [Google Scholar]
- Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D. A genetic and molecular model for flower development in Arabidopsis thaliana. Development. 1991;1:157–167. [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature. 2013;500:422–426. doi: 10.1038/nature12478. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL. Dawning of a new era: photomorphogenesis as an integrated molecular network. Current Opinion in Plant Biology. 2008;11:4–8. doi: 10.1016/j.pbi.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, Jakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao K, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J. The pattern of polymorphism in Arabidopsis thaliana. PLOS Biology. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF. A bountiful harvest: genomic insights into crop domestication phenotypes. Annual Review of Plant Biology. 2013;64:47–70. doi: 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Mahmoudian M, Freeman JL, Lahner B, Richards EL, Reeves RD, Murphy AS, Salt DE. Assessment of plants from the Brassicaceae family as genetic models for the study of nickel and zinc hyperaccumulation. The New Phytologist. 2006;172:248–260. doi: 10.1111/j.1469-8137.2006.01820.x. [DOI] [PubMed] [Google Scholar]
- Pico FX, Mendez-Vigo B, Martinez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180:1009–1021. doi: 10.1534/genetics.108.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- Plants Northern Velebit National Park Public Institution. 2014 http://www.np-sjeverni-velebit.hr/park/zivapriroda/biljke/?lang=en. [Google Scholar]
- Poormohammad Kiani S, Trontin C, Andreatta M, Simon M, Robert T, Salt DE, Loudet O. Allelic heterogeneity and trade-off shape natural variation for response to soil micronutrient. PLOS Genetics. 2012;8:e1002814. doi: 10.1371/journal.pgen.1002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przedpełska E, Wierzbicka M. Arabidopsis arenosa (Brassicaceae) from a lead–zinc waste heap in southern Poland – a plant with high tolerance to heavy metals. Plant Soil. 2007;299:43–53. doi: 10.1007/s11104-007-9359-5. [DOI] [Google Scholar]
- Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L. Environmental stress and flowering time: the photoperiodic connection. Plant Signaling & Behavior. 2014;9 doi: 10.4161/psb.29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Denervaud Tendon V, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- Sall T, Jakobsson M, Lind-Hallden C, Hallden C. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. Journal of Evolutionary Biology. 2003;16:1019–1029. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proceedings of the National Academy of Sciences of USA. 2014;111:585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics. 2005;169:1601–1615. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Rellan-Alvarez R, Fink-Straube C, Abadia J, Bauer P. Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell. 2012;24:2380–2400. doi: 10.1105/tpc.112.099077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annual Review of Plant Biology. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Molecular Ecology. 2000;9:2109–2118. doi: 10.1046/j.1365-294X.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. Arabidopsis, the Rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Sinclair SA, Sherson SM, Jarvis R, Camakaris J, Cobbett CS. The use of the zinc-fluorophore, Zinpyr-1, in the study of zinc homeostasis in Arabidopsis roots. The New Phytologist. 2007;174:39–45. doi: 10.1111/j.1469-8137.2007.02030.x. [DOI] [PubMed] [Google Scholar]
- Somerville C, Koornneef M. A fortunate choice: the history of Arabidopsis as a model plant. Nature Reviews Genetics. 2002;3:883–889. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates–gene discovery and beyond. Trends in Plant Science. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Streb S, Zeeman SC. The Arabidopsis Book. American Society of Plant Biologists; 2012. Starch metabolism in Arabidopsis; pp. 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Toomajian C, Sherman-Broyles S, Plagnol V, Guo YL, Hu TT, Clark RM, Nasrallah JB, Weigel D, Nordborg M. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- Tennstedt P, Peisker D, Bottcher C, Trampczynska A, Clemens S. Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiology. 2009;149:938–948. doi: 10.1104/pp.108.127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Trampczynska A, Bottcher C, Clemens S. The transition metal chelator nicotianamine is synthesized by filamentous fungi. FEBS Letters. 2006;580:3173–3178. doi: 10.1016/j.febslet.2006.04.073. [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Vorholt JA. Microbial life in the phyllosphere. Nature Reviews Microbiology. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- Wilczek AM, Cooper MD, Korves TM, Schmitt J. Lagging adaptation to warming climate in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of USA. 2014;111:7906–7913. doi: 10.1073/pnas.1406314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, Egan JF, Moyers BT, Petipas R, Giakountis A, Charbit E, Coupland G, Welch SM, Schmitt J. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufume S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, Parcy F, Jensen ST, Li H, Wagner D. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Developmental Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Wright SI, Lauga B, Charlesworth D. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Molecular Ecology. 2003;12:1247–1263. doi: 10.1046/j.1365-294X.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA. Natural enemies drive geographic variation in plant defenses. Science. 2012;338:116–119. doi: 10.1126/science.1226397. [DOI] [PubMed] [Google Scholar]