Abstract
The roundworm Caenorhabditis elegans has risen to the status of a top model organism for biological research in the last fifty years. Among laboratory animals, this tiny nematode is one of the simplest and easiest organisms to handle. And its life outside the laboratory is beginning to be unveiled. Like other model organisms, C. elegans has a boom-and-bust lifestyle. It feasts on ephemeral bacterial blooms in decomposing fruits and stems. After resource depletion, its young larvae enter a migratory diapause stage, called the dauer. Organisms known to be associated with C. elegans include migration vectors (such as snails, slugs and isopods) and pathogens (such as microsporidia, fungi, bacteria and viruses). By deepening our understanding of the natural history of C. elegans, we establish a broader context and improved tools for studying its biology.
DOI: http://dx.doi.org/10.7554/eLife.05849.001
Research organism: C. elegans
Introduction
The free-living nematode Caenorhabditis elegans is a major model species that is used in a range of biological research. After initial work by Emile Maupas (Maupas, 1900) and Victor Nigon (Nigon, 1949; Nigon et al., 1960) on its mode of reproduction, meiosis and development, experiments by Sydney Brenner and collaborators in the 1960s and 1970s raised C. elegans to the status of a premier model organism. Besides its genome, the first to be sequenced for a multicellular organism (The C. elegans Sequencing Consortium, 1998), an extensive body of knowledge is now available on the molecular, cellular, developmental and behavioral biology (The C. elegans Research Community) of this organism. A number of key discoveries have been made by studying C. elegans, including the molecular mechanisms of apoptosis (Conradt and Xue, 2005) and gene silencing by small RNAs (Grishok, 2013).
The C. elegans reference strain N2 is a laboratory animal. This strain, originally isolated in Bristol, England, was cultured in the laboratory for many years before it was first frozen. Its laboratory environment consisted of agar plates seeded with Escherichia coli as a food source. E. coli was used because, as another model organism, it was already available in many laboratories—not because it was originally associated with wild C. elegans. An independent subculture from the same wild Bristol isolate was maintained in liquid axenic culture, and by comparing both strains, we now know of several mutations that appeared and were fixed in the N2 lineage, some of which appear adaptive in the agar plate environment. These mutations pleiotropically affect many traits, such as behavior, reproduction, susceptibility to pathogens, body size and entry into the dauer stage (where C. elegans undergo developmental arrest at the third larval stage) (McGrath et al., 2009, 2011; Duveau and Félix, 2012; Andersen et al., 2014; Green et al., 2014). Like other model organisms, C. elegans N2 has thus been modified by domestication.
For a century, the only information available on the natural history of C. elegans was that it could be found in compost heaps and in rich humus (Hodgkin and Doniach, 1997). From 2000 onward, this began to change when researchers embarked on an extensive sampling of wild C. elegans populations. Their efforts were motivated by wanting to develop C. elegans into a model organism for evolutionary biology and ecology research, which could also leverage the tools and knowledge already acquired for this species. In turn, exploring the natural history of C. elegans provides a context for, and also informs, basic biological research conducted with this organism, such as studies of its genome, development, behavior and immune system. In this article, we summarize our current knowledge of the natural history of this C. elegans, show how the isolation of natural pathogens of C. elegans informed basic biological research, and discuss a number of open questions.
A rotting habitat and a boom-and-bust life cycle
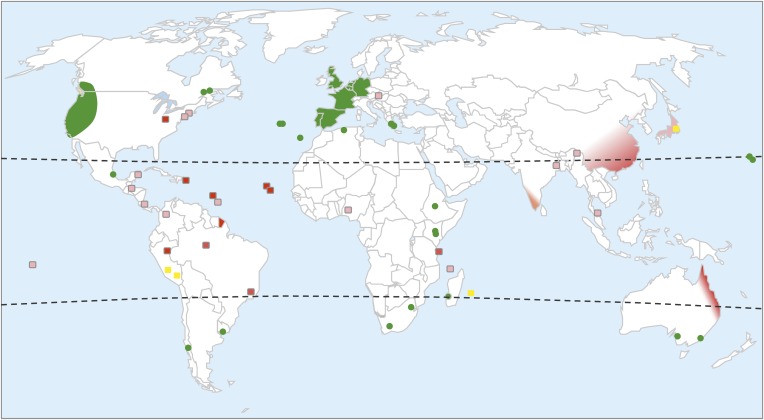
C. elegans is found worldwide, predominantly in humid temperate areas (Figures 1 and 2A-D) (Kiontke et al., 2011; Andersen et al., 2012). This species was originally isolated in rich soil or compost (Hodgkin and Doniach, 1997), where it is mostly found in a non-feeding stage called the dauer (Barrière and Félix, 2005a, 2007). More recently, feeding and reproducing stages of C. elegans have been found in decomposing plant material, such as fruits and thick herbaceous stems (Figure 2E–G) (Félix and Duveau, 2012). These rotting substrates in their late stages of decomposition provide abundant bacterial food for the nematode. Like other model organisms, C. elegans is thus partially associated with human activity (cultivated fruits and stems, compost), but the species is also commonly found on stems and fruits in wilder settings, such as woods (Félix and Duveau, 2012). New types of habitat and geographical locations may still be discovered.
Figure 1. Worldwide distribution of C. elegans.
Green shading highlights areas where C. elegans has been repeatedly collected. Green dots mark islands or locations where C. elegans has been collected at least once. Yellow squares represent areas where many Caenorhabditis species have been sampled and where C. elegans is present but rare (often found at altitude). Red shading highlights where C. elegans has never been collected despite the intensive sampling of many other Caenorhabditis species. Pink shading highlights where C. elegans has not been collected, despite the sampling of several other Caenorhabditis species. White represents areas that have never been sampled for C. elegans or very rarely. The distribution is inferred from published data (Abdul Kader and Côté, 1996; Barrière and Félix, 2005a, 2005b, 2007; Dolgin et al., 2008; Wang et al., 2010; Kiontke et al., 2011; Andersen et al., 2012; Félix and Duveau, 2012; Dey et al., 2013; Félix et al., 2013), WormBase, and our lab collection (http://www.justbio.com/worms/index.php).
Figure 2. The habitat of C. elegans at different scales.
(A–D) Landscapes that correspond to the macroscale C. elegans habitat; all are relatively humid areas where C. elegans has been found: (A) wet shrubland; (B) urban garden; (C) riverbank; and (D) fruit trees. (E–G) Bacteria-rich decomposing vegetal substrates, corresponding to the microscale C. elegans habitat: (E) Arum stem; (F) oranges and (G) plums. (H) Detail of a rotting apple at the stage where C. elegans proliferates. Springtails (white) and a mite are examples of animals that share the bacteria-rich habitat of C. elegans and that are potential carriers and/or predators (see also Table 1). (I) C. elegans nematodes on an E. coli lawn, just coming out of a rotten fruit. (J) Scanning electron micrograph of C. elegans infected with the fungus Drechmeria coniospora. Image credits: Marie-Anne Félix.
Two alternative life cycles have been described in the laboratory for C. elegans, depending on environmental conditions. If well fed, newly hatched individuals pass through four larval stages (L1, L2, L3, L4) and reach the adult stage after 3 days. Under stressful conditions (such as crowding, limited food supply, and heat stress), individuals can shift during the L1 stage to an alternative developmental route and enter a predauer stage (L2d), followed by the non-feeding diapause stage called dauer (an alternative L3 stage) (Figure 3). Dauer larvae are resistant to various stresses and can survive for several months without food. Upon their return to more favorable conditions, dauer larvae feed again and resume development.
Figure 3. A schematic of C. elegans lifecycle in the wild.
In bacteria-rich habitats (beige), the C. elegans life cycle begins with an embryonic (E) stage, followed by four larval (L1-L4) stages, and ends with an adult stage (Ad). Most animals are self-fertilizing hermaphrodites; males are rare and breeding with males therefore uncommon. Under suboptimal conditions (such as crowding and starvation), L1 larvae can enter a predauer stage (L2d) followed by the diapause stage (dauer). When better conditions arise, dauers develop into postdauer L3 larvae and re-enter the lifecycle at the L4 stage.
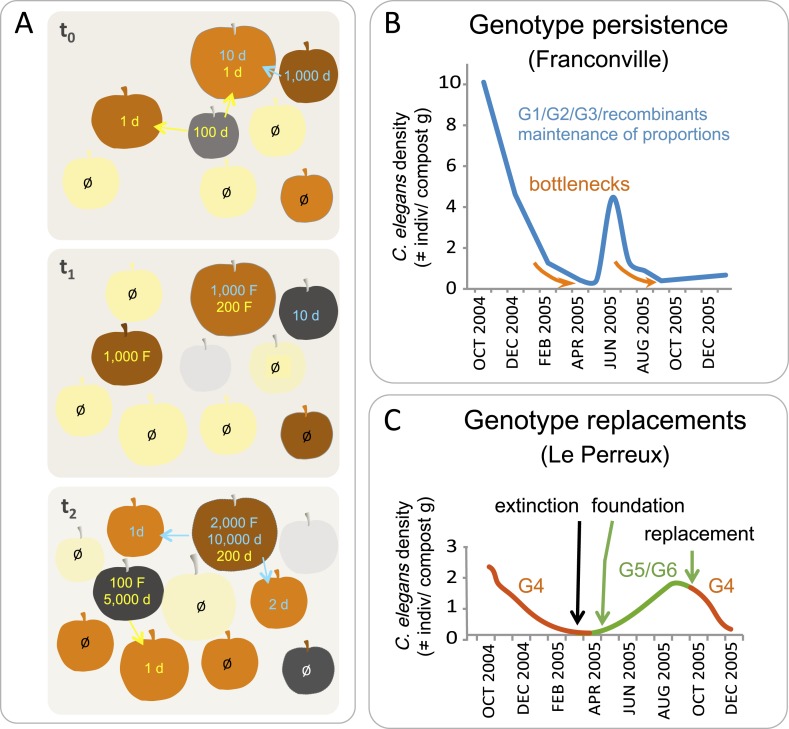
Population demographic surveys at the local scale in orchards and woods indicate that C. elegans has a boom-and-bust lifestyle (Félix and Duveau, 2012). C. elegans metapopulations evolve in a fluctuating environment where optimal habitats are randomly distributed in space and time (Figure 4A). A cycle of colonization of a food source likely begins when one to several dauer larvae discover a fruit or stem, exit the dauer stage and seed a growing population of up to 104 feeding nematodes at different life-cycle stages (Figure 3). Some moderate-sized populations found in rotting fruits and stems do not contain any dauer larvae, but larger ones always include only adults, L1, L2d and dauers (Félix and Duveau, 2012). As a food source runs low, dauers may leave it to explore the neighboring environment for new islands of resources. Most of them will fail.
Figure 4. C. elegans population dynamics in a natural habitat.
(A) A schematic of C. elegans population dynamics in an orchard. Population growth on a given apple has not been monitored to date and so is inferred here from data in (Félix and Duveau, 2012), based on many time points in an orchard and single time points on a given fruit. The fruits are shown at three time points (t) in identical positions on each panel, with t0 the first and t2 the last timepoint. Fruit colors indicate the degree of fruit decomposition, from early stages (yellow) to brown and dark grey (later stages), until disappearance (light grey). The number of feeding (F) and non-feeding dauer (d) individuals are indicated in color, with different colors representing different genotypes. Ø represents no colonization of a fruit by C. elegans. Arrows indicate dauer migration. How often different fruits or stems are colonized by several genotypes remains to be tested (Barrière and Félix, 2007; Andersen et al., 2012). (B–C) Actual population dynamics at the scale of a compost heap, from Barrière and Félix (2007). (B) In the first (Franconville) example, three main genotypes, G1, G2 and G3, persist in the heap at similar frequencies over the time period shown. In the second example (Le Perreux-sur-Marne), a single genotype, G4, was present, became extinct, then two new genotypes, G5 and G6, founded a new population. G4 reappeared in September, while G8 and G9 (not shown) disappeared. Genotypes were characterized using microsatellite markers.
Developmental regulation and the behavior of dauer larvae are central to the C. elegans lifestyle. Dauer larvae display active locomotion and a specific behavior called nictation, where they stand on their tail and wave their body in the air. Remarkably, dauers may also congregate to form a column and nictate as a group (Félix and Duveau, 2012) (see the video; http://www.wormatlas.org/dauer/behavior/Images/DBehaviorVID4.mov). These behaviors are thought to help dauers to find passing invertebrate hosts that they can use for their dispersal, such as isopods, snails and slugs. Together, dauer physiology and behavior suggest that this developmental stage plays a key role in C. elegans' stress resistance, long-distance dispersal, and possibly its overwintering capacity.
Over the year, in surveys performed in France and Germany, C. elegans populations in rotting fruits typically peak in the fall, with proliferation possible in spring through to early winter (Félix and Duveau, 2012; Petersen et al., 2014). This seasonal dynamic is consistent with that observed in a semi-natural habitat, such as a compost heap, where C. elegans population size varies over the year, from a dense population in autumn to one that declines and undergoes bottlenecks in winter (Figure 4B) (Barrière and Félix, 2007; Petersen et al., 2014). These bottlenecks can lead to local extinctions. Where this happens, a new genotype may colonize the compost heap when favorable conditions return (Figure 4C).
The spatio-temporal distribution of C. elegans genetic diversity reflects this boom-and-bust lifecycle coupled to active migration. At the global scale, C. elegans genetic diversity is low and displays little geographical structure, which must be due to long-distance migration, perhaps aided by larger hosts, such as birds, rodents or humans (see Box 1). In contrast, at a local scale, founder effects have a huge impact, with low diversity in a given sample contrasting with high molecular diversity but weak haplotype diversity at the scale of a square kilometer (Barrière and Félix, 2005a; Haber et al., 2005; Sivasundar and Hey, 2005; Andersen et al., 2012).
Box 1. Outstanding questions about the natural history of C. elegans.
- What is the pattern of C. elegans migration at different geographical distances? What are the relevant vectors? Are humans now major vectors over large scales?
Over a small scale, ongoing local population surveys will provide insights into migration patterns. Over large scales, selective sweeps estimated to date from 100–200 years ago have spread to different continents: their migration and selection might thus relate to human activity (e.g., via human travelling and the trafficking of goods, and via selection, perhaps by chemical compounds) (Andersen et al., 2012).
- What is C. elegans geographical center of origin? Are there species that are very closely related to C. elegans (see phylogeny in Box 2)?
Increased sampling in poorly sampled and isolated geographical regions might provide access to divergent populations that did not undergo the recent selective sweeps and so inform us of C. elegans' older history and habitats.
- What does C. elegans eat?
C. elegans likely feeds mostly on bacteria, but we do not know which bacteria. The sterols it requires might come from yeast or from the rotting plant substrate itself.
- What is the generation time and the number of generations per year in the wild?
The latter is surely far less than in the laboratory (ca. 150), but is it by one or two orders of magnitude?
- What are the relevant dauer entry and exit cues?
Whether associated organisms have an influence on dauer formation has not been investigated.
- Where does C. elegans spend its winter? What are the source populations?
In temperate regions, C. elegans is most often sampled in autumn, when it is found at high density. We ignore how and where this organism survives winter, for example, whether it becomes associated with a carrier organism.
C. elegans sex life
C. elegans has a very peculiar mode of reproduction called androdioecy. It can reproduce either by self-fertilizing (selfing) hermaphrodites or by hermaphrodites (XX) breeding with males (X0). Hermaphrodites develop a female soma, and early in adulthood produce sperm in limited quantities (ca. 200–300 sperm cells). Males occur by non-disjunction of the X chromosomes at meiosis or in the progeny of male-hermaphrodite crosses. The non-disjunction of X chromosomes occurs overall at a low frequency (e.g., 0.1%) in the laboratory, and this frequency varies with genotype and the environment (Hodgkin and Doniach, 1997; Teotonio et al., 2006). In natural populations, outcrossing does occur but the overall proportion of males is low, generally similar to the estimated rate of chromosomal non-disjunction measured in the lab (Barrière and Félix, 2007; Cutter et al., 2009; Rockman and Kruglyak, 2009; Andersen et al., 2012; Félix and Duveau, 2012). In rare instances, outcrossing is evident from a high proportion of males or of heterozygotes in a given sample (Sivasundar and Hey, 2005; Barrière and Félix, 2007; Félix and Duveau, 2012).
This combination of metapopulation dynamics, a very low outcrossing rate and selection has a major impact on the C. elegans genome (Barrière and Félix, 2005b, 2007; Cutter, 2006; Cutter et al., 2009; Rockman and Kruglyak, 2009; Andersen et al., 2012). These three factors reduce the effective population size, and thus genetic diversity, and can wipe out diversity in a large genomic region. Indeed, positively selected alleles carry with them a large part of the genome by linkage, at the scale of megabases, particularly in the center of chromosomes where recombination is low. For example, most C. elegans isolates carry a recently swept genotype spanning a large part of chromosome V; several other chromosomes have also experienced detectable sweeps (Andersen et al., 2012). Because of high linkage disequilibrium due to selfing, selective sweeps may cover large fractions of chromosomes and have erased much genetic diversity. Background selection (the loss of non-deleterious variants due to selection against linked deleterious alleles) may further reduce linked genetic diversity in this selfing species (Rockman et al., 2010). As a consequence, genome-wide pairwise genetic diversity is low (ca. 10−3/bp), especially compared to outcrossing relatives (Jovelin et al., 2003; Cutter et al., 2009; Wang et al., 2010; Dey et al., 2013; Gimond et al., 2013). Overall, this means that much of the C. elegans genome has been shaped by selection at linked sites, not directly by selection at the locus of interest. These extreme sweeps also mean that recent events in historical times, perhaps mediated by humans, may have had a significant impact on C. elegans.
The selfing lifestyle of C. elegans also has an impact on phenomena such as inbreeding and outbreeding depression. Most animal species that reproduce by outcrossing suffer from inbreeding depression through the accumulation of deleterious recessive alleles. In a predominantly self-fertilizing organism, such as C. elegans, alleles are most often in a homozygous state due to repeated selfing, which allows deleterious recessive alleles to be purged by natural selection. Instead of inbreeding depression, wild C. elegans suffer from outbreeding depression (Dolgin et al., 2007). This phenomenon is best explained by the fact that selfing creates and maintains beneficial gene combinations, while outcrossing favors their disruption. Thus, the progeny of outcrossing are less fit than are the progeny of selfing. However, at least under certain circumstances of experimental evolution, outbreeding may contribute positively to C. elegans adaptation and inbreeding depression may reappear (Morran et al., 2009; Teotonio et al., 2012; Carvalho et al., 2014; Chelo et al., 2014).
Contributing also to a reduction in effective outcrossing, C. elegans possesses a widespread genetic incompatibility system, whereby a sperm poison (PEEL-1) arrests the development of embryos that do not synthesize the antidote (ZEEL-1) (Seidel et al., 2008, 2011). The zeel-1 and peel-1 genes are closely linked on chromosome I. The two allelic combinations, with both genes or without both genes, co-exist in the same geographical location, and the rest of the genome shows evidence of mixing. Yet this system, and other incompatibilities, may further contribute to lowering the effective outcrossing rate.
C. elegans has evolved selfing from an ancestor that reproduced through females and males (see Box 2). Male-specific genes are overall conserved yet appear to be subject to relaxed selection (Cutter, 2008; Thomas et al., 2012). Although C. elegans males still sense female pheromones from closely related species, hermaphrodites no longer produce pheromones to attract males (Chasnov and Chow, 2002; Chasnov et al., 2007), suggesting a partial degeneration of hermaphrodite outcrossing. On the male side, some aspects of mating behavior also appear to have degenerated, such as the ability to deposit a copulatory plug (Palopoli et al., 2008). The effect, and the future, of males in C. elegans natural populations remains an intriguing question.
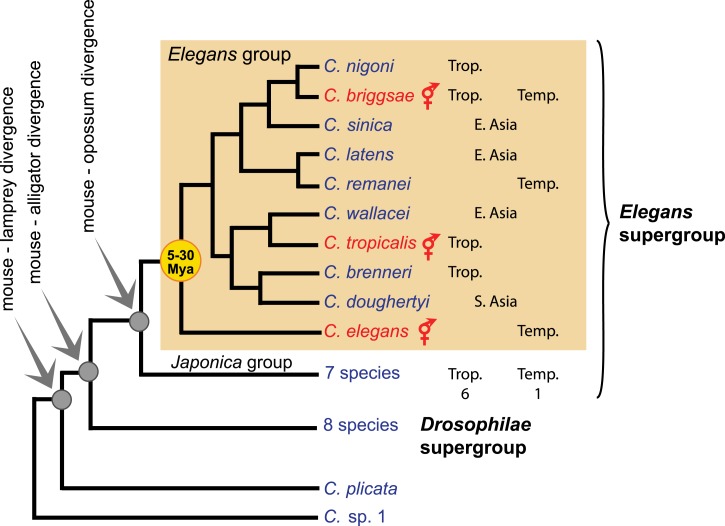
Box 2. The discovery and diversity of new Caenorhabditis species.
Within the last 5 years, many Caenorhabditis species have been newly discovered, due to the collaborative, worldwide sampling of rotting fruits, flowers and stems for new C. elegans isolates and Caenorhabditis species (Kiontke et al., 2011). Today, 27 species are available in culture and many more are being isolated.
The genus is presently composed of two basal species, C. plicata and C. sp. 1, and two species supergroups, named Elegans and Drosophilae, which are further divided into species groups (Kiontke et al., 2011; Sudhaus, 2011; Félix et al., 2014; Huang et al., 2014) (see accompanying Box figure). The genus is globally distributed, with a higher species diversity in the tropics. C. briggsae is the only species that is abundant in both tropical and temperate climates. The three selfing species (shown in red on the accompanying figure) are spread over several continents, while some male-female species have regional distributions.
Most species were collected in microhabitats similar to those described for C. elegans. However, given the unavoidable bias in sampling, some species may have unexplored preferred microhabitats and animal vectors (Kiontke and Sudhaus, 2006). For example, Caenorhabditis japonica appears to be specifically associated with the hemipteran bug Parastrachia japonensis, which feeds in summer on fruits of Schoepfia jasminodora (Kiontke et al., 2002; Okumura et al., 2013a, 2013b).
Box 2—Figure 1.
Position in the food chain
C. elegans shares its microhabitat with arthropods and with other microorganisms (bacteria and fungi) and invertebrates (Table 1), including other nematodes such as Oscheius, Pristionchus, Panagrellus and other Caenorhabditis species, such as Caenorhabditis briggsae (Kiontke et al., 2011; Félix and Duveau, 2012). If not with E. coli, it is noteworthy that C. elegans shares its rotting fruit habitat with two other top model organisms, Drosophila melanogaster and Saccharomyces cerevisiae (Fay and Benavides, 2005; Diezmann and Dietrich, 2009; Schacherer et al., 2009; Charron et al., 2014). A specific association is actually found between another Caenorhabditis species and another Drosophila species: this nematode species, C. drosophilae, feeds on rotting cactus in desert areas and its dauer juveniles use a local Drosophila species as a vector to move between cacti (Kiontke, 1997).
Table 1.
The biotic environment of C. elegans
Some organisms may act as both food and pathogen, or as both vector and predator. Other nematodes may be competitors for food but also predators. The predatory relationships are inferred from the co-occurrence of the two species in the wild, from laboratory predation assays, and, in the case of mites and fungi, by observation on the laboratory isolation plate; specific studies would be needed to observe them directly in a natural setting.
and our unpublished observations.
The C. elegans diet mainly consists of bacteria and small eukaryotes (Félix and Braendle, 2010; Félix and Duveau, 2012), but has not been further characterized. The trophic interaction between the nematode and its prey is not univocal: as C. elegans adults grow old, they can serve as food to the same microorganisms.
Predators of C. elegans are also little studied. From co-occurrence in the wild and from laboratory experiments, possible natural predators include small arthropods, such as mites or springtails (Figure 2H), other nematodes, such as Pristionchus spp., and trapping nematophagous fungi (Lee and Widden, 1996; Kiontke and Sudhaus, 2006; Bento et al., 2010; Maguire et al., 2011; Félix and Duveau, 2012) (Table 1).
C. elegans also constantly interacts with a variety of obligate and non-obligate parasites, such as fungi, microsporidia, bacteria and viruses (Troemel et al., 2008; Félix et al., 2011; Félix and Duveau, 2012; Hodgkin et al., 2013) (Table 1). These parasites infect their host via the two most exposed parts of the nematode, the cuticle and the intestine. Some non-invasive bacteria form a biofilm along the nematode's cuticle or directly stick to it (Hodgkin et al., 2013). Other bacteria proliferate in the nematode gut, which may induce constipation and likely impairs nutrient uptake (Félix and Duveau, 2012). The most intrusive parasites enter and proliferate inside the nematode body. Some pierce the cuticle (e.g., Drechmeria coniospora [Couillault et al., 2004], Figure 2J), while others enter intestinal cells via the apical membrane (e.g., microsporidia and Orsay virus [Troemel et al., 2008; Félix et al., 2011]).
A model organism strengthened by its natural history
Numerous characteristics of C. elegans biology arise from its natural history and explain why this animal has become such an emblematic species for scientists. While some of these characteristics were clear for the early pioneers of C. elegans research, others were only discovered later.
First, like other model organisms, C. elegans rapidly proliferates in the wild upon encountering a food source, and can remain in a resting stage for long periods of time. In the lab, these features translate into organisms with short generation times (for their group) and good storage ability. Indeed, C. elegans displays a short generation time compared with many other nematodes and can simply be maintained on cholesterol-supplemented agar plates and fed E. coli. The natural ability of dauer larvae to live for several months offers a convenient option for short-term storage. C. elegans survives freezing, which enables their long-term storage at −80°C or in liquid nitrogen—an extremely useful feature for genotype storage and experimental design.
Second, the natural mode of reproduction of C. elegans makes this nematode a dream come true for geneticists. Selfing permits the generation of homozygous stocks, without inbreeding depression. The ability to induce males makes genetic crosses easy.
Third, C. elegans egg shells are naturally resistant to many environmental stresses, allowing the use of bleach. Bleaching is widely employed in the laboratory: in a population, only embryos will survive bleaching, thus allowing synchronization and removal of microbial contaminants.
Finally, a convenient feature of the reference C. elegans strain, N2, that is found in some but not all C. elegans wild isolates or Caenorhabditis species is an active RNA interference pathway. This pathway was perhaps selected in response to encounters with viruses and transposons (Tijsterman et al., 2002; Félix, 2008; Félix et al., 2011).
Recent findings informed by natural history
The exploration of the natural habitat of C. elegans opens new doors to study its biology, for example, its development, physiology and behavior. Strikingly, for a top model organism such as C. elegans, a large proportion of its genes are, as yet, not associated with any mutant phenotype. Our exploration of its natural history advances biological research in two key respects. First, the quest for wild isolates of C. elegans has yielded deletion mutants for many genes (Maydan et al., 2007; Thompson et al., 2013). Second, our improved knowledge of its wild habitat gives us new environmental parameters to test for mutant phenotypes.
As an example, an obvious ecological parameter is the interaction between C. elegans and its natural parasites, which have likely had a considerable impact on its recent evolution. From genomic analyses, expanded gene families, such as ubiquitin-adaptor genes, were hypothesized to have evolved under diversifying selection provided by pathogens or toxins (Thomas, 2006). Recently, a ubiquitin-mediated response was indeed shown to take part in the C. elegans defense against two wild parasites, Nematocida parisii (microsporidia) and the Orsay virus (Bakowski et al., 2014), as well as in behavioral avoidance of a bacterial pathogen (Chang et al., 2011).
The isolation of the Orsay virus, an RNA virus, was also instrumental in demonstrating in C. elegans the antiviral role of genes involved in RNA interference (so far studied in response to artificially administered double-stranded RNAs) (Félix et al., 2011). Moreover, an exploration of natural variation in C. elegans identified, by a genome-wide association study, a locus called drh-1. This locus was shown to carry in many C. elegans isolates a deletion that renders them permissive for Orsay virus replication. The drh-1 gene is homologous to the RIG-I family of genes that encode mammalian viral sensors. In C. elegans, this likely upstream sensor of viral RNAs was shown to activate an antiviral small RNA pathway (Ashe et al., 2013). Further molecular immunity pathways will likely be unraveled by studying the interaction of C. elegans with its natural associates (Liu et al., 2014) and its corresponding natural phenotypic variation.
How C. elegans development and behavior are modulated by the environment will also be greatly enriched by a better knowledge of its natural history. A key component of its population cycles that remains to be explored is the regulation of dauer entry and exit by relevant environmental factors, besides crowding and high temperature. Behaviorally, the dauer larva has been little studied; for example, we still know little about its olfactory responses. Recent work has demonstrated the importance of a specific set of neurons (called IL2 cells) and of cholinergic transmission in controlling dauer nictation (Lee et al., 2011). When present at high density, the dauers can nictate in large groups (Félix and Duveau, 2012). The mechanistic basis and environmental regulation of this host-seeking behavior are still poorly understood.
Another striking resource provided by the exploration of C. elegans natural history is the phenotypic diversity at the intra- and inter-specific level (see Box 2). As an example, the mode of reproduction has been modified multiple times in the Caenorhabditis genus. While most species reproduce through males and females, C. elegans, C. briggsae and Caenorhabditis tropicalis are exceptions by having evolved spermatogenesis in a female soma. Phylogenetic studies and molecular dissection of germ line sex determination argue for a convergent evolution of androdioecy, three times in the Caenorhabditis genus (Guo et al., 2009; Kiontke et al., 2011; Thomas et al., 2012). Another opening provided by the huge rise in Caenorhabditis species and strain collections is the ability to study reproductive isolation and speciation mechanisms in this group (Woodruff et al., 2010; Kiontke et al., 2011; Dey et al., 2012).
Conclusion
Despite intense laboratory work on C. elegans, many important features of this species in the wild remain mysterious (Box 1). We still know very little concerning this nematode's feeding, dauer entry and exit cues, migration, overwintering, and we have only just started to glimpse organisms associated with it, be they food or pathogens. A greater understanding of its natural history could greatly inform our studies of its physiology, behavior and immunity. Comparative studies between Caenorhabditis species also promise to provide a better understanding of the role and the evolutionary history of specific gene families and other orphan genes.
In addition, improving our understanding of C. elegans' natural history will help establish it as a model species for evolutionary and ecological studies. The diversity of modes of reproduction in the Caenorhabditis genus makes them attractive models for studies of the causes and consequences of such variation. Its boom-and-bust lifestyle is favorable for studies of C. elegans metapopulation dynamics in different environments.
To conclude, knowing more about C. elegans' life outside of the Petri dish is opening new avenues to laboratory research. As for other model organisms, it also helps us to realize how idiosyncratic some features of the C. elegans N2 reference strain are—only a snapshot in laboratory evolution.
Acknowledgements
We thank our colleagues from the Caenorhabditis evolution and ecology community for fruitful discussions and sharing. We are funded by a grant from the Agence Nationale pour la Recherche 11 BSV3 01301 and a Coup d'Elan from the Bettencourt-Schueller Foundation.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Competing interests:
The authors declare that no competing interests exist.
Author contributions
LF, Drafting or revising the article.
M-AF, Drafting or revising the article.
Funding Information
This paper was supported by the following grants:
Agence Nationale de la Recherche (ANR) 11 BSV3 01301 to Lise Frézal, Marie-Anne Félix.
Fondation Bettencourt Schueller Coup d'Elan 2012 to Lise Frézal, Marie-Anne Félix.
References
- Abdul Kader N, Côté MG. Isolement, identification et caractérisation de souches québécoises du nématode Caenorhabditis elegans. Fundamental and Applied Nematology. 1996;19:381–389. [Google Scholar]
- Andersen EC, Bloom JS, Gerke JP, Kruglyak L. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLOS Genetics. 2014;10:e1004156. doi: 10.1371/journal.pgen.1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nature Genetics. 2012;45:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Bélicard T, Le Pen J, Sarkies P, Frézal L, Lehrbach NJ, Félix MA, Miska EA. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife. 2013;2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLOS Pathogens. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi C, Cho S, Ellis RE. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science. 2009;326:1002–1005. doi: 10.1126/science.1176013. [DOI] [PubMed] [Google Scholar]
- Barrière A, Félix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Current Biology. 2005a;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Barrière A, Félix MA. Natural variation and population genetics of C. elegans. In: The C. elegans Research Community, editor. Wormbook. 2005b. http://www.wormbook.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Félix MA. Temporal dynamics and linkage disequilibrium in natural C. elegans populations. Genetics. 2007;176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Félix MA. Isolation of C. elegans and related nematodes. In: The C. elegans Research Community, editor. Wormbook. 2014. http://www.wormbook.org/ [DOI] [PubMed] [Google Scholar]
- Barrière A, Ruvinsky I. Pervasive divergence of transcriptional gene regulation in Caenorhabditis nematodes. PLOS Genetics. 2014;10:e1004435. doi: 10.1371/journal.pgen.1004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G, Ogawa A, Sommer RJ. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature. 2010;466:494–497. doi: 10.1038/nature09164. [DOI] [PubMed] [Google Scholar]
- Carvalho S, Phillips PC, Teotónio H. Hermaphrodite life history and the maintenance of partial selfing in experimental populations of Caenorhabditis elegans. BMC Evolutionary Biology. 2014;14:117. doi: 10.1186/1471-2148-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell-Chen EP, Chen J, Lewis EE, Douhan GW, Nadler SA, Carey JR. Revising the standard wisdom of C. elegans natural history: ecology of longevity. Science of Aging Knowledge Environment. 2005;40:pe30. doi: 10.1126/sageke.2005.40.pe30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Paek J, Kim DH. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480:525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Leducq JB, Bertin C, Dubé AK, Landry CR. Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Saccharomyces paradoxus in North America. FEMS Yeast Research. 2014;14:281–288. doi: 10.1111/1567-1364.12100. [DOI] [PubMed] [Google Scholar]
- Chasnov JR, Chow KL. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics. 2002;160:983–994. doi: 10.1093/genetics/160.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov JR, So WK, Chan CM, Chow KL. The species, sex and stage specificity of a Caenorhabditis sex pheromone. Proceedings of the National Academy of Sciences of USA. 2007;104:6730–6735. doi: 10.1073/pnas.0608050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelo IM, Carvalho S, Roque M, Proulx SR, Teotonio H. The genetic basis and experimental evolution of inbreeding depression in Caenorhabditis elegans. Heredity. 2014;112:248–254. doi: 10.1038/hdy.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Xue D. In: Wormbook. The C. elegans Research Community, editor. 2005. http://www.wormbook.org/ [DOI] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nature Immunology. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila, inferred from direct estimates of the neutral mutation rate. Molecular Biology and Evolution. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Dey A, Murray RL. Evolution of the Caenorhabditis elegans genome. Molecular Biology and Evolution. 2009;26:1199–1234. doi: 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- Dey A, Jeon Y, Wang GX, Cutter AD. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics. 2012;191:1257–1269. doi: 10.1534/genetics.112.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chan CK, Thomas CG, Cutter AD. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proceedings of the National Academy of Sciences of the USA. 2013;110:11056–11060. doi: 10.1073/pnas.1303057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezmann S, Dietrich FS. Saccharomyces cerevisiae: population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLOS ONE. 2009;4:e5317. doi: 10.1371/journal.pone.0005317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin ES, Charlesworth B, Baird SE, Cutter AD. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution. 2007;61:1339–1352. doi: 10.1111/j.1558-5646.2007.00118.x. [DOI] [PubMed] [Google Scholar]
- Dolgin ES, Félix M-A, Cutter AD. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 2008;100:304–315. doi: 10.1038/sj.hdy.6801079. [DOI] [PubMed] [Google Scholar]
- Duveau F, Félix MA. Role of pleiotropy in the evolution of a cryptic developmental variation in C. elegans. PLOS Biology. 2012;10:e1001230. doi: 10.1371/journal.pbio.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLOS Genetics. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Current Biology. 2007;17:103–114. doi: 10.1016/j.cub.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Félix MA. RNA interference in nematodes and the chance that favored Sydney Brenner. Journal of Biology. 2008;7:34. doi: 10.1186/jbiol97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLOS Biology. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Braendle C. The natural history of Caenorhabditis elegans. Current Biology. 2010;20:R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- Félix MA, Braendle C, Cutter AD. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLOS ONE. 2014;9:e94723. doi: 10.1371/journal.pone.0094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biology. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Jovelin R, Ferrari C, Han S, Cho YR, Andersen EC, Cutter AD, Braendle C. Species richness, distribution and genetic diversity of Caenorhabditis nematodes in a remote tropical rainforest. BMC Evolutionary Biology. 2013;13:10. doi: 10.1186/1471-2148-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimond C, Jovelin R, Han S, Ferrari C, Cutter AD, Braendle C. Outbreeding depression with low genetic variation in selfing Caenorhabditis nematodes. Evolution. 2013;67:3087–3101. doi: 10.1111/evo.12203. [DOI] [PubMed] [Google Scholar]
- Green JW, Stastna JJ, Orbidans HE, Harvey SC. Highly polygenic variation in environmental perception determines dauer larvae formation in growing populations of Caenorhabditis elegans. PLOS ONE. 2014;9:e112830. doi: 10.1371/journal.pone.0112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A. Biology and mechanisms of short RNAs in Caenorhabditis elegans. Advances in Genetics. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- Guo Y, Lang S, Ellis RE. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Current Biology. 2009;19:1853–1860. doi: 10.1016/j.cub.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Haber M, Schüngel M, Putz A, Müller S, Hasert B, Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Molecular Biology and Evolution. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Félix MA, Clark LC, Stroud D, Gravato-Nobre MJ. Two Leucobacter strains exert complementary virulence on Caenorhabditis including death by worm-star formation. Current Biology. 2013;23:2157–2161. doi: 10.1016/j.cub.2013.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RE, Ren X, Qiu Y, Zhao Z. Description of Caenorhabditis sinica sp. n. (Nematoda: Rhabditidae), a nematode species used in comparative biology for C. elegans. PLOS ONE. 2014;9:e110957. doi: 10.1371/journal.pone.0110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson HB, Jeyaprakash A, Zuckerman BM. Differential adhesion and infection of nematodes by the endoparasitic fungus Meria coniospora (Deuteromycetes) Applied and Environmental Microbiology. 1985;49:552–555. doi: 10.1128/aem.49.3.552-555.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Ajie BC, Phillips PC. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Molecular Ecology. 2003;12:1325–1337. doi: 10.1046/j.1365-294X.2003.01805.x. [DOI] [PubMed] [Google Scholar]
- Kiontke K. Description of Rhabditis (Caenorhabditis) drosophilae n. sp. and R. (C.) sonorae n. sp. (Nematoda: Rhabditida) from saguaro cactus rot in Arizona. Fundamental and Applied Nematology. 1997;20:305–315. [Google Scholar]
- Kiontke K, Félix MA, Ailion M, Rockman MV, Braendle C, Pénigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evolutionary Biology. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Hironaka M, Sudhaus W. Description of Caenorhabditis japonica n. sp. (Nematoda: Rhabditida) associated with the burrower bug Parastrachia japonensis (Heteroptera: Cynidae) in Japan. Nematology. 2002;4:933–941. doi: 10.1163/156854102321122557. [DOI] [Google Scholar]
- Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. In: The C. elegans Research Community, editor. Wormbook. 2006. http://www.wormbook.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Q, Widden P. Folsomia candida, a ‘fungivorous’ collembolan, feeds preferentially on nematodes rather than soil fungi. Soil Biology & Biochemistry. 1996;28:689–690. doi: 10.1016/0038-0717(95)00158-1. [DOI] [Google Scholar]
- Lee H, Choi MK, Lee D, Kim HS, Hwang H, Kim H, Park S, Paik YK, Lee J. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nature Neuroscience. 2011;15:107–112. doi: 10.1038/nn.2975. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SM, Clark CM, Nunnari J, Pirri JK, Alkema MJ. The C. elegans touch response facilitates escape from predacious fungi. Current Biology. 2011;21:1326–1330. doi: 10.1016/j.cub.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupas E. Modes et formes de reproduction des nématodes. Archives de Zoologie Expérimentale et Générale. 1900;8:463–624. [Google Scholar]
- Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Research. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigon V. Modalités de la reproduction et déterminisme du sexe chez quelques nématodes libres. Annales Des Sciences Naturelles, Zoologie. 1949;11:1–132. [Google Scholar]
- Nigon V, Guerrier P, Monin H. L'architecture polaire de l'œuf et les mouvements des constituants cellulaires au cours des premières étapes du développement chez quelques Nématodes. Bulletin Biologique De La France Et De La Belgique. 1960;93:131–202. [Google Scholar]
- Okumura E, Ishikawa Y, Tanaka R, Yoshiga T. Propagation of Caenorhabditis japonica in the nest of its carrier insect, Parastrachia japonensis. Zoological Science. 2013a;30:174–177. doi: 10.2108/zsj.30.174. [DOI] [PubMed] [Google Scholar]
- Okumura E, Tanaka R, Yoshiga T. Species-specific recognition of the carrier insect by dauer larvae of the nematode Caenorhabditis japonica. Journal of Experimental Biology. 2013b;216:568–572. doi: 10.1242/jeb.073593. [DOI] [PubMed] [Google Scholar]
- Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénigault JB, Félix MA. Evolution of a system sensitive to stochastic noise: P3.p cell fate in Caenorhabditis. Developmental Biology. 2011;357:419–427. doi: 10.1016/j.ydbio.2011.05.675. [DOI] [PubMed] [Google Scholar]
- Petersen C, Dirksen P, Prahl S, Strathmann EA, Schulenburg H. The prevalence of Caenorhabditis elegans across 1.5 years in selected North German locations: the importance of substrate type, abiotic parameters, and Caenorhabditis competitors. BMC Ecology. 2014;14:4. doi: 10.1186/1472-6785-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proceedings of the National Academy of Sciences of USA. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riche S, Zouak M, Argoul F, Arneodo A, Pecreaux J, Delattre M. Evolutionary comparisons reveal a positional switch for spindle pole oscillations in Caenorhabditis embryos. Journal of Cell Biology. 2013;201:653–662. doi: 10.1083/jcb.201210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLOS Genetics. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLOS Biology. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A, Hey J. Sampling from natural populations with RNAI reveals high outcrossing and population structure in Caenorhabditis elegans. Current Biology. 2005;15:1598–1602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Sudhaus W. Phylogenetic systematisation and catalogue of paraphyletic ‘Rhabditidae’ (Secernentea, Nematoda) Journal of Nematode Morphology and Systematics. 2011;14:113–178. [Google Scholar]
- Sudhaus W, Kiontke K. Phylogeny of Rhabditis subgenus Caenorhabditis (Rhabditidae, Nematoda) Journal of Zoological Systematics and Evolutionary Research. 1996;34:217–233. doi: 10.1111/j.1439-0469.1996.tb00827.x. [DOI] [Google Scholar]
- Sudhaus W, Kühne R. Nematodes associated with Psychodidae: description of Rhabditis berolina sp. n. and redescription of R. dubia Bovien, 1937 (Nematoda: Rhabditidae), with biological and ecological notes, and a phylogenetic discussion. Nematologica. 1989;35:305–320. doi: 10.1163/002825989X00412. [DOI] [Google Scholar]
- Teotonio H, Carvalho S, Manoel D, Roque M, Chelo IM. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLOS ONE. 2012;7:e35811. doi: 10.1371/journal.pone.0035811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotonio H, Manoel D, Phillips PC. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution. 2006;60:1300–1305. doi: 10.1111/j.0014-3820.2006.tb01207.x. [DOI] [PubMed] [Google Scholar]
- The C. elegans research community . Wormbook: the online review of C. elegans biology. 2005/2014. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Thomas CG, Li R, Smith HE, Woodruff GC, Oliver B, Haag ES. Simplification and desexualization of gene expression in self-fertile nematodes. Current Biology. 2012;22:2167–2172. doi: 10.1016/j.cub.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Woodruff GC, Haag ES. Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends in Genetics. 2012;28:213–220. doi: 10.1016/j.tig.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Research. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, Kieffer A, Lau J, Lee N, Miller A, Raymant G, Shen B, Shendure J, Taylor J, Turner EH, Hillier LW, Moerman DG, Waterston RH. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Research. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterl RH. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Genome Research. 2002;12:1535–1540. doi: 10.1016/S0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Félix M-A, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode C. elegans. PLOS Biology. 2008;6:e309. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Ramani AK, McKay SJ, Fraser AG. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLOS Genetics. 2014;10:e1004077. doi: 10.1371/journal.pgen.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GX, Ren S, Ren Y, Ai H, Cutter AD. Extremely high molecular diversity within the East Asian nematode Caenorhabditis sp. 5. Molecular Ecology. 2010;19:5022–5029. doi: 10.1111/j.1365-294X.2010.04862.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Chamberlin H. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-28 ovo gene. Genes & Development. 2002;16:2345–2348. doi: 10.1101/gad.996302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff GC, Eke O, Baird SE, Félix M-A, Haag ES. Fertile interspecies hybrids in Caenorhabditis nematodes and their implications for the evolution of hermaphroditism. Genetics. 2010;186:997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]