Abstract
Among all the metabolites present in the plasma, lipids, mainly triacylglycerol and diacylglycerol, show extensive circadian rhythms. These lipids are transported in the plasma as part of lipoproteins. Lipoproteins are synthesized primarily in the liver and intestine and their production exhibits circadian rhythmicity. Studies have shown that various proteins involved in lipid absorption and lipoprotein biosynthesis show circadian expression. Further, intestinal epithelial cells express circadian clock genes and these genes might control circadian expression of different proteins involved in intestinal lipid absorption. Intestinal circadian clock genes are synchronized by signals emanating from the suprachiasmatic nuclei that constitute a master clock and from signals coming from other environmental factors, such as food availability. Disruptions in central clock, as happens due to disruptions in the sleep/wake cycle, affect intestinal function. Similarly, irregularities in temporal food intake affect intestinal function. These changes predispose individuals to various metabolic disorders, such as metabolic syndrome, obesity, diabetes, and atherosclerosis. Here, we summarize how circadian rhythms regulate microsomal triglyceride transfer protein, apoAIV, and nocturnin to affect diurnal regulation of lipid absorption.
Keywords: lipoproteins, triglycerides, rhythms, clock genes, nocturnin, MTP, apoAIV
Several behavioral and physiologic activities show circadian rhythms that are attuned to changes in light within a 24 h day. Plasma triacylglycerols exhibit diurnal variations in humans and rodents (1–6). Due to their hydrophobic nature, these lipids are transported in the plasma as major core constituents of apoB-containing lipoproteins. These lipoproteins are assembled in the endoplasmic reticulum and Golgi of enterocytes and hepatocytes with the assistance of a dedicated chaperone, microsomal triglyceride transfer protein (MTP), to transport dietary and endogenous lipids, respectively (7–9). Therefore, it is possible that a major reason for the circadian regulation of plasma triglyceride depends on their assembly and secretion by the intestine and the liver.
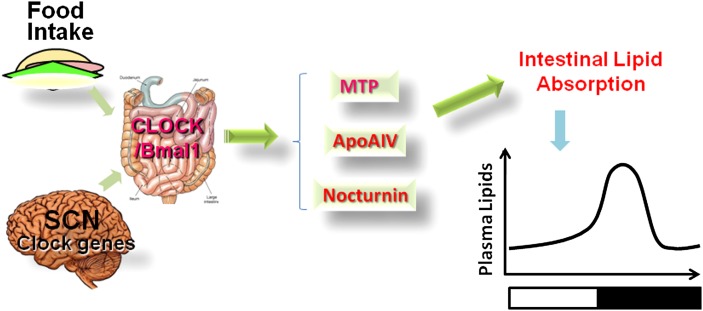
Circadian rhythms are centrally orchestrated by a set of clock genes expressed in the suprachiasmatic nuclei (SCN) of the brain. These genes are also expressed in other cells and their expression is entrained by various stimuli emanating from the SCN. Additionally, they are regulated by external stimuli such as food availability. Several intestinal functions, including lipid absorption, show diurnal variations. We will briefly introduce clock genes and their regulation and intestinal lipid absorption, and then discuss three proteins that have been shown to be involved in the diurnal regulation of lipid absorption (Fig. 1).
Fig. 1.
Diurnal regulators of intestinal lipid absorption. Both food- and light-entrained oscillators appear to affect the expression of clock genes in the intestine. Intestinal clock genes affect other genes to modulate intestinal lipid absorption. So far, at least three proteins, MTP, apoAIV, and nocturnin, that affect diurnal variations in intestinal lipid absorption have been identified. Changes in the diurnal expression of these genes appear to affect diurnal variations in plasma lipids.
LIGHT AND FOOD ENTRAINMENT OF CIRCADIAN RHYTHMS
Constant periodicity of sunrise and sunset affects every organism. Some wake up with the sunrise while others go to sleep. However, wakefulness is always associated with physical activity aimed at gathering food. In contrast, sleep is associated with low locomotor activity and fasting. The information about the sunlight is transmitted from the eye via retino-hypothalamic neurons to bilateral SCN in the brain (10–12). These photic signals are deciphered in the SCN resulting in the increased expression of a clock gene, brain and muscle arylhydrocarbon receptor nuclear translocator (ARNT)-like protein 1 (Bmal1). When the levels increase, Bmal1 forms heterodimers with another clock gene, circadian locomotor output cycles kaput (Clock). The Bmal1:Clock heterodimers interact with the E-boxes present in the promoter regions of period (Per) and cryptochrome (Cry) genes and increase their transcription (Fig. 2A). Per and Cry proteins also form heterodimers and act as repressors of the Bmal1:Clock heterodimers to reduce their own expression. This transcriptional auto-regulatory loop repeats with an approximate interval of 24 h and is further modulated by several posttranslational modifications, such as phosphorylation and acetylation (10–13). In addition to this major regulatory loop, expression of Bmal1 is regulated by other transcription factors that sense cellular energy levels and other environmental stimuli. These include retinoic acid receptor-related orphan receptor α (Rorα), PPARγ coactivator 1-α (PGC1α), and reverse erythroblastosis virus α (Rev-erbα). Rorα and PGC1α increase, while Rev-erbα suppresses, Bmal1 expression constituting a secondary regulatory loop (10–12). More molecular details about the regulation of these transcription factors and the regulation of circadian clock genes can be found in several excellent reviews (10–13). An important feature of these clock genes is that they need to be regularly entrained by light to maintain periodic rhythmicity. In the absence of regular exposure, the intensity of circadian response shortens with time and eventually disappears.
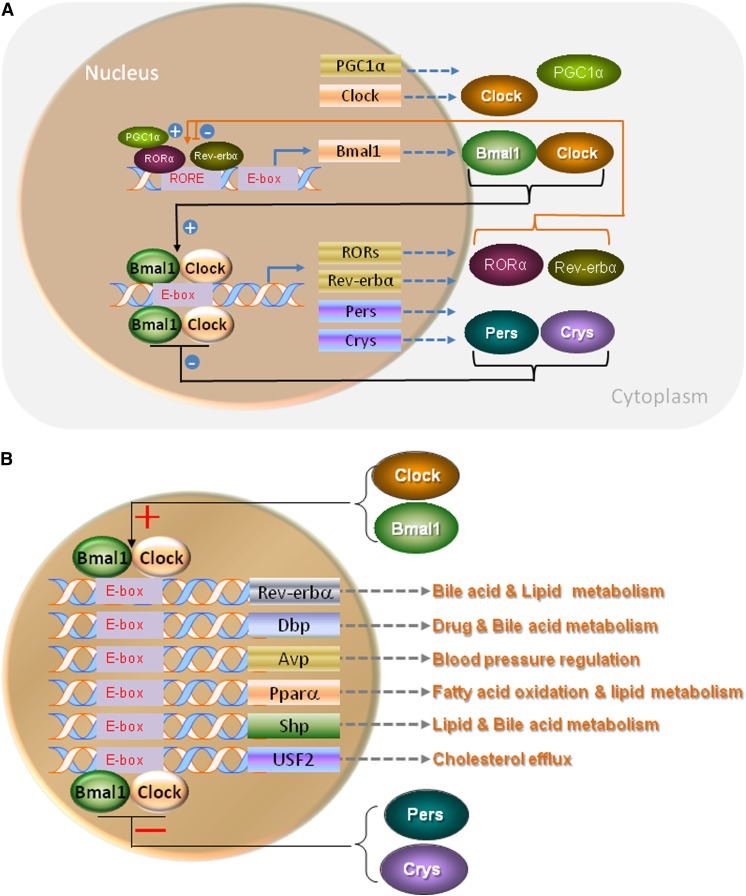
Fig. 2.
Regulation of clock and clock-controlled genes by Bmal1:Clock heterodimers. A: Regulation of clock genes: Bmal1:Clock heterodimers interact with E-box elements present in the promoters to increase the expression of different clock genes. When the levels of Per and Cry proteins increase, they form heterodimers and act as repressors reducing the expression of Bmal1 constituting the primary auto-regulatory transcriptional loop. Bmal1:Clock heterodimers also increase temporal expression of Rorα and Rev-erbα that act as activator and repressor, respectively, of Bmal1 expression by binding to a retinoic acid-related orphan receptor response element (RORE), and constitute a secondary regulatory loop that affects diurnal regulation. PGC1α can interact with Rorα to increase Bmal1 expression. These two complementary auto-regulatory loops are involved in the control of circadian rhythms. B: Regulation of clock-controlled genes: Bmal1:Clock heterodimers also interact with E-boxes present in the promoter regions of several other transcription factor genes to augment their expression. These transcription factors then regulate the expression of several genes involved different pathways to affect metabolism.
In addition to the clock genes described above, Bmal1:Clock heterodimers also interact with E-boxes present in the promoters of several “clock controlled genes” (Fig. 2B) and increase their expression. These transcription factors then modulate the expression of key proteins involved in different metabolic pathways. Thus, Bmal1:Clock heterodimers increase the expression of repressors to downregulate their own expression and also increase the expression of other transcription factors to modulate different metabolic pathways.
Besides SCN, clock genes are also expressed in almost all the cells. These cells are not synchronized by light; instead, they are synchronized by hormonal and neuronal signals emanating from the SCN. These peripheral clock genes are susceptible to changes in the environment, such as heat and food. Thus, while the regulation of clock genes in the SCN is mainly entrained by the light, the peripheral clock genes are regulated by various stimuli originating from the SCN, as well as other environmental (food, temperature) and physiologic (NAD+, hormones, nutrients, etc.) cues (14–20). Therefore, diurnal regulation of peripheral tissues, such as the intestine, is affected by several central, hormonal, and environmental stimuli.
Food is a potent synchronizer of peripheral clocks and entrains various behavioral and physiologic activities (14–16, 20–25). This can be easily demonstrated by providing food for a few days in the daytime to rodents that usually consume their meal in the nighttime with no disruptions in the light on/off schedule. Changes after food entrainment have characteristic features of circadian rhythms. These rhythms are sustained for some time after the food entrainment is discontinued. The identity of a center, if it exists, that controls and elucidates food-entrained oscillations is unknown. There is significant evidence to suggest that the food-entrained oscillator might be independent of the SCN. However, the clock genes involved in the light-induced oscillators may participate in the food-entrainment response. It is likely that the food-entrained oscillator is a network of several neural sites in the brain that cooperate to elicit a behavioral and physiologic response (20, 26).
EXPRESSION OF CLOCK GENES IN THE INTESTINE
Various functions of the intestine, such as motility, gastric emptying, DNA synthesis, epithelial cell renewal, food anticipatory activity, and nutrient absorption, exhibit circadian rhythms (14, 20, 27–29). It is well-known that the major complaints in shift workers and transcontinental travelers are related to gastrointestinal disturbances (30). Hence, it is possible that these activities are regulated by clock genes and disruptions in the diurnal expression of clock genes might be a cause for gastrointestinal discomforts. Indeed, the expression of various clock genes has been documented in various parts of the intestine. The colon exhibits the highest expression of clock genes (17, 31). We showed that the expression of these proteins increases from the duodenum to the colon (32). We also measured the expression of clock genes in intestinal mucosal and epithelial cells. Clock proteins were more abundant in the epithelial cells compared with the mucosal cells. Thus, expression of clock genes increases from duodenum to colon and from mucosal cells to epithelial cells.
Clock genes in the jejunum and colon show diurnal variations (17, 31, 32). The peaks and nadirs in the expression of these clock genes are in phase with their expression in the liver. However, the peak expressions of these genes are phase delayed compared with their temporal expression in the SCN (15). Thus, it is possible that a temporal delay in the rhythmicity of the expression of intestinal clock genes might be secondary to the neuronal and hormonal signals emanating from the SCN.
Circadian expression of clock genes in the intestine is altered with the feeding schedule (15, 32). After food entrainment, most of the clock genes are expressed at the time of food availability, instead of their normal peak expression at night. Thus, food availability can alter the expression pattern of clock genes in the intestine. However, the proper food-entrainment response is not seen in mice kept in constant dark or constant light. Thus, normal functioning of light-entrained response is necessary for the adaptation to food entrainment.
In short, intestinal cells express different clock genes and their function is likely modulated by these genes. Disruptions in circadian expression of clock genes in the intestine might contribute to gastrointestinal discomforts during trans-continental flights and while working at odd hours.
CIRCADIAN REGULATION OF INTESTINAL LIPID ABSORPTION
The major function of the small intestine is to digest and absorb food. Macronutrients, carbohydrates, lipids, and proteins are hydrolyzed in the lumen of the intestine and products are retrieved by enterocytes involving various transporters. Intestinal lipid absorption involves hydrolysis of dietary fat in the intestinal lumen, uptake of hydrolyzed products by the enterocytes, resynthesis of lipids, and assembly and secretion of chylomicrons. Several reviews have extensively discussed these steps in lipid absorption (7, 8, 14, 20, 33–42). Because lipids are water-insoluble, an important step in the digestion of these lipids is their emulsification with bile salts (39, 43). In this process, hydrophobic lipids are incorporated into bile salt micelles rendering them water-miscible. The digestion of triacylglycerols in the intestinal lumen produces unesterified fatty acids and monoacylglycerols (36). The digestion of phospholipids is carried out mainly by pancreatic phospholipases A2, yielding free fatty acids and lysophospholipids. Cholesterol esterase hydrolyzes cholesterol esters into free cholesterol and unesterified fatty acids. These hydrolyzed products are taken up by enterocytes (36, 38, 41, 44). Thus, the major steps involved in the transport of dietary lipids from the intestinal lumen to enterocytes are emulsification with bile, hydrolysis by esterases, and uptake by transporters.
After uptake, fatty acids are transported in the cells by fatty acid binding proteins. These proteins deliver fatty acids to various organelles. In the endoplasmic reticulum (ER), fatty acids are used for the synthesis of triacylglycerols, phospholipids, and cholesterol esters. These lipids are then packaged into lipoproteins called chylomicrons (8, 45). Chylomicrons are very large spherical triacylglycerol-rich particles that also contain phospholipids and cholesterol. The surface of these particles is covered with a phospholipid monolayer and the core is enriched in triacylglycerols and cholesteryl esters. The surface of these particles also contains apoB48 that acts as a scaffolding protein. Triacylglycerol absorption is dependent on the assembly and secretion of these particles. In contrast, phospholipids and cholesterol are absorbed via chylomicrons and the HDL pathway (8, 46). In the HDL pathway, ABCA1 and apoAI play a role (46–48). ABCA1 and apoAI deficiencies reduce cholesterol transport via the HDL pathway with no effect on cholesterol secretion via the chylomicron pathway. In short, fatty acids are used for the synthesis of lipids for packaging with lipoproteins and secretion. All triglycerides are transported via chylomicrons, while cholesterol and phospholipids are transported with both chylomicrons and HDLs.
Lipid absorption studies have been performed in animals, jejunal loops, and isolated primary enterocytes. These studies have revealed that lipid absorption is maximal at night and lowest in the day (5). Because lipid absorption involves uptake of hydrolyzed lipid products followed by their secretion with lipoproteins (7, 14, 33–42), we studied both the uptake and secretion of fatty acids and cholesterol by primary enterocytes (5, 32, 49). These studies showed that enterocytes exhibit diurnal variations with respect to their capacity to take up and secrete fat. Thus, various steps in the uptake, packaging, and secretion are probably controlled by clock genes.
To understand the molecular basis for diurnal lipid absorption, we looked for changes in different proteins involved in the uptake and packaging of fatty acids and cholesterol. Most of the genes examined showed diurnal changes in their expression (32). These include apoB, MTP, apoAIV, diacylglycerol O-acyltransferase 2 (DGAT2), fatty acid synthase, and stearoyl-CoA desaturase-1 (SCD-1). Thus, at least some of the genes involved in the synthesis and secretion of triglycerides are regulated by circadian rhythms. More studies are needed to establish that the individual steps and proteins involved in lipid uptake and secretion are indeed regulated by circadian clock genes. This can be achieved by measuring temporal changes in different processes and proteins involved in lipid absorption in mice deficient in specific clock genes.
Clock
The role of Clock in circadian control of lipid absorption has largely been derived from studies in mice that express a dominant negative Clock protein (ClockΔ19) in C57Bl/6J background. These mice are arrhythmic and show longer periodicity (26–29 h instead of 23–24 h) in their locomotor activity (50). The Clock mutant allele encodes a protein with 51 amino acid deletion in its putative transcriptional regulatory domain. It interacts with Bmal1, binds to E-box enhancer sequences, acts in a dominant negative fashion (51), decreases the transcription of Per and other circadian clock genes, and disables the negative feedback loop of circadian rhythm. These Clock mutant (ClkΔ19/Δ19) mice are entrainable during a normal light/dark cycle, but lose this ability when placed in the dark (50). In addition, they show physiologic abnormalities such as reduced fertility, obesity, hyperleptinemia, hyperlipidemia, hepatic steatosis, hyperglycemia, and metabolic syndrome (52, 53). They have been extensively used to study the role of circadian rhythms in metabolism.
We studied the expression of clock genes and different nutrient transporters in mice expressing normal and dominant negative Clock protein (32, 49). Our data show that normal intestinal cells express canonical clock genes in a circadian manner and are susceptible to attunement by food. Normal Clock expression is important for the circadian and food-entrained expression of nutrient transport proteins as well as in the absorption of macronutrients, as Clock mutant mice do not show circadian expression of genes involved in lipid absorption and do not respond to food entrainment (32). Thus, circadian rhythms and Clock protein are important in macronutrient absorption by the intestine.
Lipid absorption studies in Clock mutant mice involving in situ intestinal loops and isolated enterocytes showed that uptake of fatty acid and triglyceride secretion are significantly altered in these mice. As opposed to wild-type mice, ClkΔ19/Δ19 mice did not show significant differences in the uptake of fatty acids or triglyceride secretion by intestinal loops or enterocytes at midnight or midday, suggesting that diurnal variations in lipid uptake were lost in the mutant mice. Thus, these mutant mice were absorbing lipids throughout the day, and this sustained high lipid absorption might be a reason for the hypertriglyceridemia observed in these mice.
To understand the molecular basis for defects in circadian regulation of lipid absorption in clock mutant mice, we measured expression levels of different genes involved in lipid absorption in wild-type and ClkΔ19/Δ19 mice (49). The expression of several of the studied genes in the ClkΔ19/Δ19 mice was altered compared with their wild-type siblings. In wild-type mice, most of the genes exhibited diurnal expression; however, they did not show diurnal variations in ClkΔ19/Δ19 mice, suggesting that the absence of circadian variations in lipid absorption might be due to alterations in molecular events that control the expression of genes involved in lipid absorption.
Because Clock mutant mice develop hyperlipidemia, we reasoned that they may be more susceptible to atherosclerosis. To test this hypothesis, Clock mutant mice were crossed to Apoe−/− and Ldlr−/− mice. ClkΔ19/Δ19Apoe−/− and ClkΔ19/Δ19Ldlr−/− mice showed higher atherosclerosis (54). Physiologic studies indicated that these mice were assembling and secreting more chylomicrons and had higher lipid levels. In addition, macrophages from ClkΔ19/Δ19Apoe−/− mice were defective in cholesterol efflux. Therefore, these studies suggested that Clock reduces atherosclerosis by reducing intestinal lipid absorption and enhancing cholesterol efflux from macrophages.
MTP
MTP is a chaperone that is crucial for the biosynthesis of apoB-containing lipoproteins (9, 55–57). It transfers lipids, mainly neutral lipids, in vitro between membrane vesicles, as well as physically interacts with apoB. Thus, by interacting with apoB and lipidating this peptide during translation, MTP assists in the formation of primordial apoB-containing lipoproteins. Formation of these particles prevents proteasomal degradation of apoB that occurs in the absence of lipid supply or MTP deficiency.
The major organs expressing MTP are the intestine and the liver (9, 55–58). Mechanisms controlling different levels of MTP in tissues have not been explained. MTP expression is modulated by macronutrients (59). Recently, microRNA-30c has been shown to regulates its expression and reduce hyperlipidemia and atherosclerosis (60). In general, there is a good agreement between cellular levels of MTP mRNA, protein, and activity suggesting that MTP is mainly regulated at the transcriptional level. Various transcription factors and cis-elements involved in the regulation of MTP have been reviewed (59). A short promoter with few cis-elements appears sufficient for its expression in cells. The promoter contains positive regulatory hepatic nuclear factor (HNF)-1, HNF-4, Fox, and negative sterol/insulin regulatory elements that bind to HNF-1α, HNF-4α, FoxO1/A2, and SREBP transcription factors, respectively. Thus, MTP expression is controlled by various transcription factors that interact with specific elements present in the promoter.
Intestinal and hepatic MTP expression shows modest diurnal variations with highest levels found at midnight (5, 32, 49). These changes are not seen when mice are placed in constant dark for 5 days, indicating that light entrainment might be needed for proper diurnal expression of the MTP gene. Further, expression of intestinal and hepatic MTP was altered when mice were subjected to food entrainment, indicating that MTP responds to food-entrained oscillator. Thus, diurnal variations in the expression of MTP gene are under the control of both light- and food-entrained oscillators. The regulation of MTP by light-entrained oscillators was further supported by the observations that diurnal variations in MTP expression were absent in ClkΔ19/Δ19 mice (5). Normal Clock expression was also needed for food-entrained changes in MTP expression as ClkΔ19/Δ19 mice were unable to increase MTP expression at the time of food availability after food entrainment compared with wild-type mice. Thus, Clock appears to be necessary for both light- and food-entrained oscillatory changes in MTP expression.
Evidence that Clock regulates MTP comes from studies in ClkΔ19/Δ19 mice (49). MTP expression did not show diurnal variations in the intestine and livers of ClkΔ19/Δ19 mice. The role of Clock in MTP regulation was further demonstrated by reducing the expression using siRNA for Clock. Knockdown of Clock increased MTP expression, indicating a reciprocal relationship. We observed that MTP lacks an E-box in its promoter that is recognized by Bmal1:Clock heterodimers. Hence, we reasoned that increases in activators or reductions in repressors might explain increases in MTP expression in ClkΔ19/Δ19 mice. siRNA-mediated knockdown of Clock in cells reduced or had no effect on activators, however, one of the repressors, small heterodimer partner (Shp), was reduced (52). Further, knockdown of Shp increased MTP expression. These studies suggested that Clock might regulate MTP expression by modulating Shp expression.
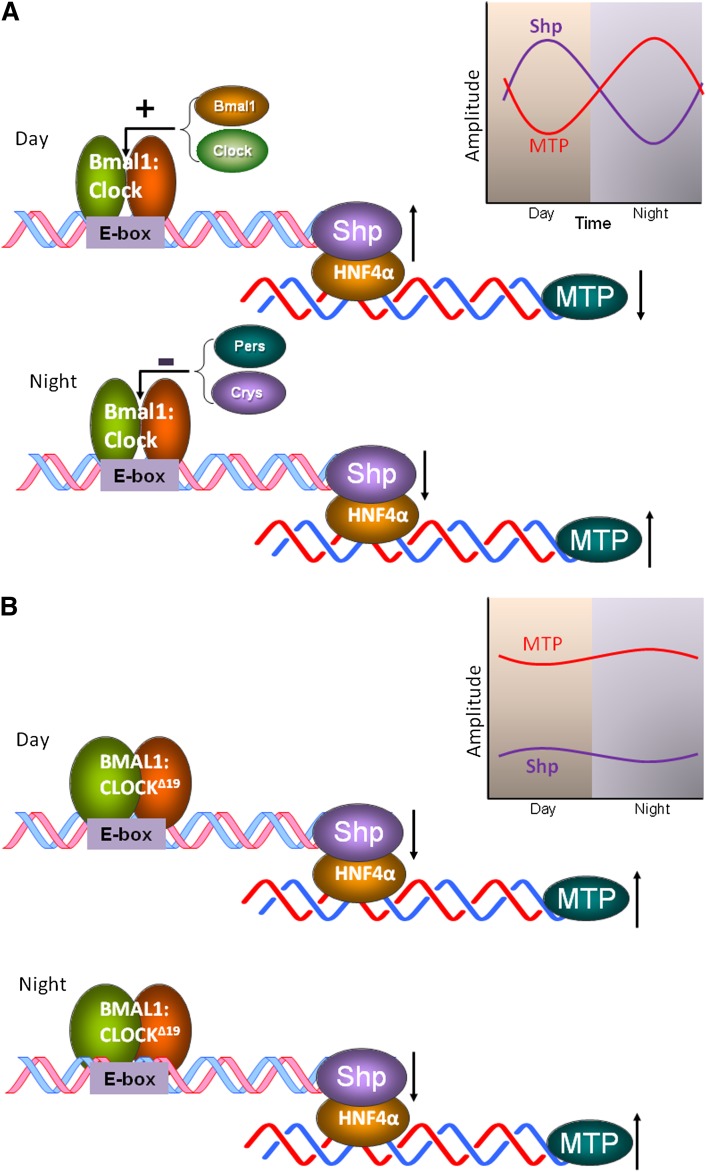
The Shp gene contains an E-box. In wild-type mice, Bmal1:Clock heterodimers interact with the E-box enhancer elements in the promoter of the Shp gene increasing its expression at dawn (Fig. 3A) (61). When levels increase, Shp interacts with several transcription factors that activate MTP gene expression (49). These include HNF-4α, HNF-1α, and liver receptor homolog-1 (LRH-1). By binding to these transcription factors, Shp represses the expression of MTP. The maximum association of Shp with MTP promoter occurs in the midday and is correlated with low levels of MTP in the daytime. Shp levels reduce at the end of the day and this derepresses MTP expression in the early hours of the night to maximize lipid absorption and transport. This is a simplistic picture about the regulation of MTP by Clock. Other clock genes might also be involved in the regulation of MTP. Hence, more studies are needed to understand how other clock genes regulate MTP to modulate intestinal lipid absorption and plasma lipid levels.
Fig. 3.
Regulation of MTP by Clock involving Shp. A: In wild-type mice, Bmal1:Clock heterodimers interact with the E-box of Shp in the day to increase expression. When Shp levels increase, they interact with different activators that are bound to the MTP promoter, such as HNF-4α, to reduce expression. At night, when concentrations of Per and Cry proteins increase, the activity of Baml1:Clock is repressed resulting in reduced expression of Shp and increased expression of MTP. B: In ClockΔ19/Δ19 mice, binding of Bmal1:ClockΔ19 heterodimers to the E-box reduces expression of Shp. Further, this binding is perhaps not affected by Per:Cry heterodimers at night. Therefore, expression of Shp is low at all times and does not show diurnal variations. Reduced levels and loss of circadian expression of Shp might contribute to high MTP expression throughout the day.
In ClkΔ19/Δ19 mice (Fig. 3B), Bmal1:ClockΔ19 heterodimer probably interacts with the E-box resulting in reduced Shp expression and increased MTP expression. Because the activity of the Bmal1:ClockΔ19 heterodimer does not show diurnal variations, the levels of Shp do not change throughout the day in ClkΔ19/Δ19 mice. Therefore, MTP levels are perhaps high at all times, contributing to sustained hypertriglyceridemia.
apoAIV
apoAIV is a 46 kDa protein found associated with chylomicrons and HDLs, as well as in lipid-free form in the plasma (62, 63). In humans, it is mainly synthesized by the intestine; but rodent livers also synthesize apoAIV (64). apoAIV is incorporated on the surface of chylomicrons in the early stages of their biogenesis in the ER (65) and is secreted from the basolateral side of enterocytes. There is considerable evidence that dietary fat increases apoAIV expression; fat ingestion and intestinal perfusion of lipids increase synthesis and secretion of apoAIV in rodents (66–68). Lu et al. (69) have demonstrated that a high-fat diet induces apoAIV expression by 7-fold in newborn swine jejunum. Similarly, a 3-fold increase in apoAIV mRNA levels was observed in Caco-2 cells supplemented apically with lipid micelles (70). Fat feeding increases transcription of apoAIV. Mechanistic studies indicate that fat feeding increases the binding of HNF-4α to the apoAIV promoter (70, 71). Thus, fat feeding may induce apoAIV synthesis by increasing gene transcription.
Recently it has been shown that cAMP responsive element-binding protein, hepatocyte specific (CREBH) regulates apoAIV expression by binding to two CREBH elements present in the apoAIV promoter (72). CREBH is an ER membrane anchored bZIP transcription factor that is mainly expressed in the liver and intestine (72, 73). CREBH deficiency reduces apoAIV expression in the liver and intestine. apoAIV expression is increased after the overexpression of CREBH in the liver and in cultured cells. High-fat diet induces steatosis in the liver leading to increased expression of CREBH and apoAIV (72, 73). Fasting is known to induce CREBH and apoAIV expression in the liver. However, it is not clear whether prolonged fasting increases CREBH and apoAIV in the intestine. It remains to be determined whether CREBH is involved in diurnal regulation of apoAIV expression.
Transgenic overexpression of human apoAIV in mice increases serum VLDL cholesterol and triacylglycerols in the fed state (68). An approximately 50-fold enhanced expression of apoAIV in a newborn swine enterocyte cell line, IPEC-1, increased secretion of nascent triacylglycerols and phospholipids with chylomicrons by 2- to 3-fold (69, 74). VerHague et al. (75) have shown that apoAIV enhances VLDL triglyceride production by enhancing core expansion, not particle number, in steatotic livers. Molecular mechanisms involved in this process are not fully understood, but Weinberg et al. (76) have suggested that apoAIV may help in the expansion of nascent lipoproteins into larger lipoproteins by maintaining interfacial tension and elasticity of the larger particles. Besides these biophysical mechanisms, Yao et al. (77) showed that overexpression of apoAIV increases MTP mRNA protein and activity involving pretranslational mechanisms. Pan et al. (78) showed that this increase in MTP might involve transcription factors FoxO1 and FoxA2. In contrast to these studies, other studies suggest no increase in MTP after apoAIV overexpression (75, 79). Thus, how apoAIV assists in fat absorption at the molecular and biochemical levels needs further exploration.
Apart from these changes at the intestinal level, apoAIV might regulate lipid absorption involving central neuronal controls. Liu et al. (80) showed that apoAIV is expressed in the hypothalamus, and the hypothalamic apoAIV gene expression is reduced by food deprivation and restored by lipid refeeding. Blocking the action of endogenous apoAIV with its antibody increases meal size, implying that endogenous apoAIV exerts an inhibitory tone on feeding (81). Therefore, it is possible that hypothalamic apoAIV might control lipid absorption. In fact, it has been suggested that apoAIV affects food intake and acts as a satiety factor in rats (82). However, genetic ablation and overexpression studies in mice showed no influence on dietary lipid absorption and feeding behavior (68, 83). However, Weinstock et al. (83) did observe that apoAIV knockout male mice took up more food after a long fast and suggested that under certain conditions apoAIV might serve as a satiety signal.
Another feature of apoAIV that is pertinent to lipid absorption is its circadian expression (13, 38, 39). Serum apoAIV levels exhibit circadian rhythms. Further, intestinal apoAIV protein and mRNA levels are higher in the dark. We have shown that apoAIV mRNA levels show circadian expression in ad libitum-fed mice subjected to a 12 h light/dark cycle. Further, its expression is induced by food entrainment (13). Inductions in mRNA after food entrainment are suppressed when animals are placed in continuous dark or light. Diurnal changes and response to food entrainment were not seen in ClkΔ19/Δ19 mice. Therefore, it was concluded that apoAIV belongs to a family of genes that is regulated by both light and food (13). These changes in apoAIV expression are similar to those observed for MTP and plasma lipid levels (12, 14). Hence, we hypothesized that apoAIV might play a role in the diurnal regulation of lipid absorption and plasma lipid levels. To test this hypothesis, we studied diurnal changes in plasma lipids, lipid absorption, and intestinal MTP in apoAIV knockout mice and compared them with wild-type controls. Increases in intestinal MTP, intestinal triacylglycerol absorption, and in plasma triacylglycerols were reduced at midnight in apoAIV knockout mice compared with wild-type controls, indicating that diurnal increases in apoAIV might optimize lipid absorption in the postprandial state (78). Mechanistic studies showed that apoAIV might contribute to optimum lipid absorption at mealtime by enhancing the expression of FoxO1 and FoxA2 transcription factors, as well as MTP. Peaks in the expression of FoxO1 and FoxA2 occurred before the maximum expression of MTP in mice fed during the day. Further, these transcription factors bound to the MTP promoter and enhanced its expression. Therefore, temporal increases in apoAIV expression at night or at mealtime might increase the expression of FoxO1 and FoxA2, enhance the binding of these transcription factors to the Mttp promoter, and elevate MTP expression. Higher amounts of MTP in congruence with augmentations in apoAIV after a fatty meal might optimize packaging and secretion of lipids with chylomicrons by enterocytes, contributing to increases in plasma triacylglycerol.
Nocturnin
Nocturnin was identified as a RNA transcript that shows significant circadian expression in the early part of the night in the retina of Xenopus (84, 85). Nocturnin is an exoRNase that deadenylates mRNA with specificity toward poly(A) nucleotides. Deadenylation of mRNA results in rapid degradation and inhibition of their translation. Inhibition of translation probably occurs because proteins that interact with poly(A) tails are also known to interact with mRNA caps to form translationally optimum loops. Truncations of poly(A) tails might disrupt the binding of proteins and formation of translationally active loops. Both of these mechanisms reduce the amounts of protein translated from deadenylated mRNAs. Kojima, Sher-Chen, and Green (86) sequenced hepatic poly(A) mRNAs with different poly(A) tail lengths and showed that ∼2.5% of the total mRNA contained different lengths of poly(A) tails and their levels exhibit circadian variations. More importantly, rhythmicity in poly(A) tail length was correlated with rhythmic expression of proteins. Mechanisms for the origin of rhythmicity in the tail length of mRNA are not known, but it is possible that enzymes involved in the processing of poly(A) tails might play a role in imparting circadian regulation. Thus, nocturnin could potentially regulate protein levels involving posttranscriptional mRNA degradation or by inhibiting mRNA translation. Due to its circadian expression, nocturnin could alter mRNA stability and protein translation in a temporal fashion and cause specific time-dependent changes in protein levels. Specific transcripts that are temporally regulated by nocturnin have not been identified.
In mice, nocturnin is expressed in a wide variety of tissues. It is expressed throughout the small intestine with highest levels in the proximal portion (87). Jejunal nocturnin mRNA levels exhibit circadian changes with peak levels present at dark onset. Expression of nocturnin is increased within 2 h of an olive oil gavage, suggesting that it is induced after fat feeding. Thus, nocturnin is likely regulated by both light- and food-entrained oscillators.
Ablation of nocturnin in mice is not correlated with any obvious developmental, reproductive, and circadian abnormalities. However, these mice remain lean on a high-fat diet and do not develop hepatosteatosis (88). Nocturnin-deficient mice absorb fewer triacylglycerols after an oral fat gavage (87). Further, they absorb lesser amounts of radiolabeled triacylglycerols and cholesterol and retain more radiolabeled lipids in the intestinal segments (87). In these mice, more lipid droplets are present in the perinuclear region of the enterocyte nuclei toward the apical side. Experiments in isolated enterocytes corroborated observations made in nocturnin-deficient mice. Nocturnin-deficient enterocytes retain more and secrete fewer radiolabeled lipids (87). Analysis of secreted lipoproteins revealed that triacylglycerol secretion with chylomicrons was significantly reduced. Secretion of cholesterol was less with both chylomicrons and HDLs. Reductions in the secretion of lipids with chylomicrons were not due to reduced expression of MTP. In contrast, MTP levels were increased in the intestine, but not in the liver, of nocturnin-deficient mice (87). Thus, nocturnin-deficient mice fed a high-fat diet do not gain weight like the wild-type controls, most likely secondary to reduced fat absorption that is independent of changes in MTP activity. The importance of reduced fat absorption in nocturnin-deficient mice is supported further by the observations that feeding high carbohydrate and low fat diets has no discriminatory effect on weight gain in these mice. Thus, it is likely that the lean phenotype of nocturnin-deficient mice fed a high-fat diet might be secondary to lower fat absorption and inhibition of nocturnin might be a way to reduce hyperlipidemia and hepatosteatosis.
FUTURE DIRECTIONS AND PERSPECTIVES
Most of the studies described above about the circadian regulation of intestinal lipid absorption have been performed using Clock mutant mice. The expression of dominant negative mutant protein may or may not reflect the actual mode of action of Clock itself. Therefore, similar studies are needed in Clock−/− mice. Further, the role of Clock can be supplemented with studies in Bmal1−/− mice. Studies in Clock−/− and Bmal1−/− mice can provide substantial supportive evidence for the regulation of intestinal lipid absorption by the positive regulators of circadian rhythms. Complementary studies in mice deficient in circadian repressors, Pers and Crys, are also needed to identify whether circadian activators and repressors have similar or opposite effects on intestinal lipid absorption.
Some progress has been made to understand the circadian regulation of proteins involved in lipid absorption by the Clock protein. However, there is a need for a comprehensive understanding of the different steps involved in lipid absorption and their regulation by various clock genes. In this regard, studies in mice deficient in different clock genes might provide invaluable information.
Nocturnin is not directly involved in lipid absorption but appears to modulate lipid absorption by temporally regulating unknown intestinal proteins involved in lipid absorption and transport. Identification of these proteins may provide valuable clues to the mechanisms that contribute to circadian regulation of intestinal lipid absorption during high fat consumption. Furthermore, these studies could identify new targets that might be amenable to control lipid absorption.
In summary, intestinal lipid absorption is a diurnally regulated process. The mechanisms involved in the diurnal regulation of different genes involved in intestinal lipid absorption have not been elucidated. Such knowledge might be useful in minimizing gastrointestinal adverse events associated with delivery of some drugs, and in identifying new targets and/or modalities to avoid intestinal discomforts associated with abnormal working schedules.
Footnotes
Abbreviations:
- ARNT
- arylhydrocarbon receptor nuclear translocator
- Bmal1
- brain and muscle ARNT-like protein 1
- Clock
- circadian locomotor output cycles kaput
- CREBH
- cAMP responsive element-binding protein H
- Cry
- cryptochrome
- ER
- endoplasmic reticulum
- HNF
- hepatic nuclear factor
- MTP
- microsomal triglyceride transfer protein
- Per
- period
- PGC1α
- PPARγ coactivator 1-α
- Rev-erbα
- reverse erythroblastosis virus α
- Rorα
- retinoic acid receptor-related orphan receptor α
- SCN
- suprachiasmatic nuclei
- Shp
- small heterodimer partner
This work was supported in part by National Institutes of Health Grant DK-81879 and VA Merit Award BX001728.
REFERENCES
- 1.Schlierf G., Dorow E. 1973. Diurnal patterns of triglycerides, free fatty acids, blood sugar, and insulin during carbohydrate-induction in man and their modification by nocturnal suppression of lipolysis. J. Clin. Invest. 52: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maillot F., Garrigue M. A., Pinault M., Objois M., Theret V., Lamisse F., Hoinard C., Antoine J. M., Lairon D., Couet C. 2005. Changes in plasma triacylglycerol concentrations after sequential lunch and dinner in healthy subjects. Diabetes Metab. 31: 69–77. [DOI] [PubMed] [Google Scholar]
- 3.Mondola P., Gambardella P., Santangelo F., Santillo M., Greco A. M. 1995. Circadian rhythms of lipid and apolipoprotein pattern in adult fasted rats. Physiol. Behav. 58: 175–180. [DOI] [PubMed] [Google Scholar]
- 4.Bruckdorfer K. R., Kang S. S., Khan I. H., Bourne A. R., Yudkin J. 1974. Diurnal changes in the concentrations of plasma lipids, sugars, insulin and corticosterone in rats fed diets containing various carbohydrates. Horm. Metab. Res. 6: 99–106. [DOI] [PubMed] [Google Scholar]
- 5.Pan X., Hussain M. M. 2007. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J. Biol. Chem. 282: 24707–24719. [DOI] [PubMed] [Google Scholar]
- 6.Chua E. C., Shui G., Lee I. T., Lau P., Tan L. C., Yeo S. C., Lam B. D., Bulchand S., Summers S. A., Puvanendran K., et al. 2013. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA. 110: 14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain M. M., Fatma S., Pan X., Iqbal J. 2005. Intestinal lipoprotein assembly. Curr. Opin. Lipidol. 16: 281–285. [DOI] [PubMed] [Google Scholar]
- 8.Hussain M. M. 2014. Intestinal lipid absorption and lipoprotein formation. Curr. Opin. Lipidol. 25: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain M. M., Rava P., Walsh M., Rana M., Iqbal J. 2012. Multiple functions of microsomal triglyceride transfer protein. Nutr. Metab. (Lond). 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green C. B., Takahashi J. S., Bass J. 2008. The meter of metabolism. Cell. 134: 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levi F., Schibler U. 2007. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 47: 593–628. [DOI] [PubMed] [Google Scholar]
- 12.Partch C. L., Green C. B., Takahashi J. S. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bass J. 2012. Circadian topology of metabolism. Nature. 491: 348–356. [DOI] [PubMed] [Google Scholar]
- 14.Hussain M. M., Pan X. 2009. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol. Metab. 20: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., Menaker M. 2001. Entrainment of the circadian clock in the liver by feeding. Science. 291: 490–493. [DOI] [PubMed] [Google Scholar]
- 16.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14: 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoogerwerf W. A., Hellmich H. L., Cornelissen G., Halberg F., Shahinian V. B., Bostwick J., Savidge T. C., Cassone V. M. 2007. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 133: 1250–1260. [DOI] [PubMed] [Google Scholar]
- 18.Morf J., Rey G., Schneider K., Stratmann M., Fujita J., Naef F., Schibler U. 2012. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 338: 379–383. [DOI] [PubMed] [Google Scholar]
- 19.Peek C. B., Affinati A. H., Ramsey K. M., Kuo H. Y., Yu W., Sena L. A., Ilkayeva O., Marcheva B., Kobayashi Y., Omura C., et al. 2013. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 342: 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain M. M. 2014. Regulation of intestinal lipid absorption by clock genes. Annu. Rev. Nutr. 34: 357–375. [DOI] [PubMed] [Google Scholar]
- 21.Hara R., Wan K., Wakamatsu H., Aida R., Moriya T., Akiyama M., Shibata S. 2001. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 6: 269–278. [DOI] [PubMed] [Google Scholar]
- 22.Stephan F. K. 2002. The “other” circadian system: food as a Zeitgeber. J. Biol. Rhythms. 17: 284–292. [DOI] [PubMed] [Google Scholar]
- 23.Mendoza J. 2007. Circadian clocks: setting time by food. J. Neuroendocrinol. 19: 127–137. [DOI] [PubMed] [Google Scholar]
- 24.Feillet C. A., Albrecht U., Challet E. 2006. “Feeding time” for the brain: a matter of clocks. J. Physiol. Paris. 100: 252–260. [DOI] [PubMed] [Google Scholar]
- 25.Patton D. F., Mistlberger R. E. 2013. Circadian adaptations to meal timing: neuroendocrine mechanisms. Front. Neurosci. 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechtold D. A., Loudon A. S. 2013. Hypothalamic clocks and rhythms in feeding behaviour. Trends Neurosci. 36: 74–82. [DOI] [PubMed] [Google Scholar]
- 27.Hoogerwerf W. A. 2006. Biologic clocks and the gut. Curr. Gastroenterol. Rep. 8: 353–359. [DOI] [PubMed] [Google Scholar]
- 28.Soták M., Polidarová L., Musilková J., Hock M., Sumová A., Pácha J. 2011. Circadian regulation of electrolyte absorption in the rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 301: G1066–G1074. [DOI] [PubMed] [Google Scholar]
- 29.Scheving L. A. 2000. Biological clocks and the digestive system. Gastroenterology. 119: 536–549. [DOI] [PubMed] [Google Scholar]
- 30.Vener K. J., Szabo S., Moore J. G. 1989. The effect of shift work on gastrointestinal (GI) function: a review. Chronobiologia. 16: 421–439. [PubMed] [Google Scholar]
- 31.Sládek M., Rybová M., Jindráková Z., Zemanová Z., Polidarová L., Mrnka L., O’Neill J., Pácha J., Sumová A. 2007. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 133: 1240–1249. [DOI] [PubMed] [Google Scholar]
- 32.Pan X., Hussain M. M. 2009. Clock is important for food and circadian regulation of macronutrient absorption in mice. J. Lipid Res. 50: 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain M. M., Kancha R. K., Zhou Z., Luchoomun J., Zu H., Bakillah A. 1996. Chylomicron assembly and catabolism: role of apolipoproteins and receptors. Biochim. Biophys. Acta. 1300: 151–170. [DOI] [PubMed] [Google Scholar]
- 34.Hussain M. M. 2000. A proposed model for the assembly of chylomicrons. Atherosclerosis. 148: 1–15. [DOI] [PubMed] [Google Scholar]
- 35.Hussain M. M., Kedees M. H., Singh K., Athar H., Jamali N. Z. 2001. Signposts in the assembly of chylomicrons. Front. Biosci. 6: D320–D331. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal J., Hussain M. M. 2009. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 296: E1183–E1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain M. M., Leung T. M., Zhou L., Abu-Merhi S. 2013. Regulating intestinal function to reduce atherogenic lipoproteins. Clin. Lipidol. 8: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X., Hussain M. M. 2012. Gut triglyceride production. Biochim. Biophys. Acta. 1821: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan C. T., Tso P. 2001. Intestinal lipid absorption and transport. Front. Biosci. 6: D299–D319. [DOI] [PubMed] [Google Scholar]
- 40.Tso P. 1985. Gastrointestinal digestion and absorption of lipid. Adv. Lipid Res. 21: 143–186. [DOI] [PubMed] [Google Scholar]
- 41.Abumrad N. A., Davidson N. O. 2012. Role of the gut in lipid homeostasis. Physiol. Rev. 92: 1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansbach C. M., Gorelick F. 2007. Development and physiological regulation of intestinal lipid absorption. II. Dietary lipid absorption, complex lipid synthesis, and the intracellular packaging and secretion of chylomicrons. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G645–G650. [DOI] [PubMed] [Google Scholar]
- 43.Mu H., Hoy C. E. 2004. The digestion of dietary triacylglycerols. Prog. Lipid Res. 43: 105–133. [DOI] [PubMed] [Google Scholar]
- 44.Abumrad N., Harmon C., Ibrahimi A. 1998. Membrane transport of long-chain fatty acids: evidence for a facilitated process. J. Lipid Res. 39: 2309–2318. [PubMed] [Google Scholar]
- 45.Mansbach C. M., Siddiqi S. A. 2010. The biogenesis of chylomicrons. Annu. Rev. Physiol. 72: 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iqbal J., Anwar K., Hussain M. M. 2003. Multiple, independently regulated pathways of cholesterol transport across the intestinal epithelial cells. J. Biol. Chem. 278: 31610–31620. [DOI] [PubMed] [Google Scholar]
- 47.Brunham L. R., Kruit J. K., Iqbal J., Fievet C., Timmins J. A., Pape T. D., Coburn B. A., Bissada N., Staels B., Groen A. K., et al. 2006. Intestinal ABCA1 directly contributes to HDL biogenesis in vivo. J. Clin. Invest. 116: 1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal J., Hussain M. M. 2005. Evidence for multiple complementary pathways for efficient cholesterol absorption in mice. J. Lipid Res. 46: 1491–1501. [DOI] [PubMed] [Google Scholar]
- 49.Pan X., Zhang Y., Wang L., Hussain M. M. 2010. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 12: 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., Takahashi J. S. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 264: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darlington T. K., Wager-Smith K., Ceriani M. F., Staknis D., Gekakis N., Steeves T. D., Weitz C. J., Takahashi J. S., Kay S. A. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 280: 1599–1603. [DOI] [PubMed] [Google Scholar]
- 52.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 308: 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 466: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan X., Jiang X. C., Hussain M. M. 2013. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation. 128: 1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain M. M., Rava P., Pan X., Dai K., Dougan S. K., Iqbal J., Lazare F., Khatun I. 2008. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr. Opin. Lipidol. 19: 277–284. [DOI] [PubMed] [Google Scholar]
- 56.Berriot-Varoqueaux N., Aggerbeck L. P., Samson-Bouma M., Wetterau J. R. 2000. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu. Rev. Nutr. 20: 663–697. [DOI] [PubMed] [Google Scholar]
- 57.Hussain M. M., Iqbal J., Anwar K., Rava P., Dai K. 2003. Microsomal triglyceride transfer protein: a multifunctional protein. Front. Biosci. 8: s500–s506. [DOI] [PubMed] [Google Scholar]
- 58.Hussain M. M., Shi J., Dreizen P. 2003. Microsomal triglyceride transfer protein and its role in apolipoprotein B-lipoprotein assembly. J. Lipid Res. 44: 22–32. [DOI] [PubMed] [Google Scholar]
- 59.Hussain M. M., Nijstad N., Franceschini L. 2011. Regulation of microsomal triglyceride transfer protein. Clin. Lipidol. 6: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., Hussain M. M. 2013. MicroRNA-30c reduces hyperlipidemia and atherosclerosis by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 19: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oiwa A., Kakizawa T., Miyamoto T., Yamashita K., Jiang W., Takeda T., Suzuki S., Hashizume K. 2007. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem. Biophys. Res. Commun. 353: 895–901. [DOI] [PubMed] [Google Scholar]
- 62.Bisgaier C. L., Sachdev O. P., Megna L., Glickman R. M. 1985. Distribution of apolipoprotein A-IV in human plasma. J. Lipid Res. 26: 11–25. [PubMed] [Google Scholar]
- 63.Green P. H., Glickman R. M., Saudek C. D., Blum C. B., Tall A. R. 1979. Human intestinal lipoproteins. Studies in chyluric subjects. J. Clin. Invest. 64: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elshourbagy N. A., Boguski M. S., Liao W. S., Jefferson L. S., Gordon J. I., Taylor J. M. 1985. Expression of rat apolipoprotein A-IV and A-I genes: mRNA induction during development and in response to glucocorticoids and insulin. Proc. Natl. Acad. Sci. USA. 82: 8242–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar N. S., Mansbach C. M., II 1999. Prechylomicron transport vesicle: isolation and partial characterization. Am. J. Physiol. 276: G378–G386. [DOI] [PubMed] [Google Scholar]
- 66.Apfelbaum T. F., Davidson N. O., Glickman R. M. 1987. Apolipoprotein A-IV synthesis in rat intestine: regulation by dietary triglyceride. Am. J. Physiol. 252: G662–G666. [DOI] [PubMed] [Google Scholar]
- 67.Weinberg R. B., Dantzker C., Patton C. S. 1990. Sensitivity of serum apolipoprotein A-IV levels to changes in dietary fat content. Gastroenterology. 98: 17–24. [DOI] [PubMed] [Google Scholar]
- 68.Aalto-Setälä K., Bisgaier C. L., Ho A., Kieft K. A., Traber M. G., Kayden H. J., Ramakrishnan R., Walsh A., Essenburg A. D., Breslow J. L. 1994. Intestinal expression of human apolipoprotein A-IV in transgenic mice fails to influence dietary lipid absorption or feeding behavior. J. Clin. Invest. 93: 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu S., Yao Y., Meng S., Cheng X., Black D. D. 2002. Overexpression of apolipoprotein A-IV enhances lipid transport in newborn swine intestinal epithelial cells. J. Biol. Chem. 277: 31929–31937. [DOI] [PubMed] [Google Scholar]
- 70.Carrière V., Vidal R., Lazou C., Lacasa M., Delers F., Ribeiro A., Rousset M., Chambaz J., Lacorte J. M. 2005. HNF-4-dependent induction of apolipoprotein A-IV gene transcription by an apical supply of lipid micelles in intestinal cells. J. Biol. Chem. 280: 5406–5413. [DOI] [PubMed] [Google Scholar]
- 71.Leng S., Lu S., Yao Y., Kan Z., Morris G. S., Stair B. R., Cherny M. A., Black D. D. 2007. Hepatocyte nuclear factor-4 mediates apolipoprotein A-IV transcriptional regulation by fatty acid in newborn swine enterocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G475–G483. [DOI] [PubMed] [Google Scholar]
- 72.Xu X., Park J. G., So J. S., Hur K. Y., Lee A. H. 2014. Transcriptional regulation of apolipoprotein A-IV by the transcription factor CREBH. J. Lipid Res. 55: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J. H., Giannikopoulos P., Duncan S. A., Wang J., Johansen C. T., Brown J. D., Plutzky J., Hegele R. A., Glimcher L. H., Lee A. H. 2011. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med. 17: 812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu S., Yao Y., Cheng X., Mitchell S., Leng S., Meng S., Gallagher J. W., Shelness G. S., Morris G. S., Mahan J., et al. 2006. Overexpression of apolipoprotein A-IV enhances lipid secretion in IPEC-1 cells by increasing chylomicron size. J. Biol. Chem. 281: 3473–3483. [DOI] [PubMed] [Google Scholar]
- 75.VerHague M. A., Cheng D., Weinberg R. B., Shelness G. S. 2013. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arterioscler. Thromb. Vasc. Biol. 33: 2501–2508. [DOI] [PubMed] [Google Scholar]
- 76.Weinberg R. B., Cook V. R., DeLozier J. A., Shelness G. S. 2000. Dynamic interfacial properties of human apolipoproteins A-IV and B-17 at the air/water and oil/water interface. J. Lipid Res. 41: 1419–1427. [PubMed] [Google Scholar]
- 77.Yao Y., Lu S., Huang Y., Beeman-Black C. C., Lu R., Pan X., Hussain M. M., Black D. D. 2011. Regulation of microsomal triglyceride transfer protein by apolipoprotein A-IV in newborn swine intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 300: G357–G363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan X., Munshi M. K., Iqbal J., Queiroz J., Sirwi A. A., Shah S., Younus A., Hussain M. M. 2013. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J. Biol. Chem. 288: 20464–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinberg R. B., Gallagher J. W., Fabritius M. A., Shelness G. S. 2012. ApoA-IV modulates the secretory trafficking of apoB and the size of triglyceride-rich lipoproteins. J. Lipid Res. 53: 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu M., Shen L., Liu Y., Tajima D., Sakai R., Woods S. C., Tso P. 2004. Diurnal rhythm of apolipoprotein A-IV in rat hypothalamus and its relation to food intake and corticosterone. Endocrinology. 145: 3232–3238. [DOI] [PubMed] [Google Scholar]
- 81.Fujimoto K., Cardelli J. A., Tso P. 1992. Increased apolipoprotein A-IV in rat mesenteric lymph after lipid meal acts as a physiological signal for satiation. Am. J. Physiol. 262: G1002–G1006. [DOI] [PubMed] [Google Scholar]
- 82.Qin X., Tso P. 2005. The role of apolipoprotein AIV on the control of food intake. Curr. Drug Targets. 6: 145–151. [DOI] [PubMed] [Google Scholar]
- 83.Weinstock P. H., Bisgaier C. L., Hayek T., Aalto-Setala K., Sehayek E., Wu L., Sheiffele P., Merkel M., Essenburg A. D., Breslow J. L. 1997. Decreased HDL cholesterol levels but normal lipid absorption, growth, and feeding behavior in apolipoprotein A-IV knockout mice. J. Lipid Res. 38: 1782–1794. [PubMed] [Google Scholar]
- 84.Douris N., Green C. B. 2008. NOC out the fat: a short review of the circadian deadenylase Nocturnin. Ann. Med. 40: 622–626. [DOI] [PubMed] [Google Scholar]
- 85.Stubblefield J. J., Terrien J., Green C. B. 2012. Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol. Metab. 23: 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kojima S., Sher-Chen E. L., Green C. B. 2012. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26: 2724–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Douris N., Kojima S., Pan X., Lerch-Gaggl A. F., Duong S. Q., Hussain M. M., Green C. B. 2011. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 21: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Green C. B., Douris N., Kojima S., Strayer C. A., Fogerty J., Lourim D., Keller S. R., Besharse J. C. 2007. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. USA. 104: 9888–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]