Abstract
Liver X receptor (LXR)α and LXRβ play key roles in hepatic de novo lipogenesis through their regulation of lipogenic genes, including sterol regulatory element-binding protein (SREBP)-1c and carbohydrate responsive element-binding protein (ChREBP). LXRs activate lipogenic gene transcription in response to feeding, which is believed to be mediated by insulin. We have previously shown that LXRs are targets for glucose-hexosamine-derived O-linked β-N-acetylglucosamine (O-GlcNAc) modification enhancing their ability to regulate SREBP-1c promoter activity in vitro. To elucidate insulin-independent effects of feeding on LXR-mediated lipogenic gene expression in vivo, we subjected control and streptozotocin-treated LXRα/β+/+ and LXRα/β−/− mice to a fasting-refeeding regime. We show that under hyperglycemic and hypoinsulinemic conditions, LXRs maintain their ability to upregulate the expression of glycolytic and lipogenic enzymes, including glucokinase (GK), SREBP-1c, ChREBPα, and the newly identified shorter isoform ChREBPβ. Furthermore, glucose-dependent increases in LXR/retinoid X receptor-regulated luciferase activity driven by the ChREBPα promoter was mediated, at least in part, by O-GlcNAc transferase (OGT) signaling in Huh7 cells. Moreover, we show that LXR and OGT interact and colocalize in the nucleus and that loss of LXRs profoundly reduced nuclear O-GlcNAc signaling and ChREBPα promoter binding activity in vivo. In summary, our study provides evidence that LXRs act as nutrient and glucose metabolic sensors upstream of ChREBP by modulating GK expression, nuclear O-GlcNAc signaling, and ChREBP expression and activity.
Keywords: lipid metabolism, insulin, glucose, carbohydrate responsive element-binding protein α, carbohydrate responsive element-binding protein β, chromatin immunoprecipitation, O-linked β-N-acetylglucosamine, O-linked β-N-acetylglucosamine transferase
In mammals, excess dietary carbohydrates are converted into TGs through de novo lipogenesis (DNL) in liver and adipose tissue (1). Although insulin is a central regulator of hepatic DNL, glucose also plays a role in this process (2). The liver X receptors (LXRs) [LXRα (NR1H3) and LXRβ (NR1H2)] are oxysterol-activated nuclear receptors with important roles in cholesterol, glucose, and lipid metabolism (3, 4). LXRα is mainly expressed in liver, adipose, intestine, and macrophages, whereas LXRβ is ubiquitously expressed. Hepatic LXRs regulate the expression of sterol regulatory element-binding protein (SREBP)-1c (5) and carbohydrate responsive element-binding protein (ChREBP) (6), which, together with LXRs, are key regulators of many glycolytic and lipogenic genes, including glucokinase (GK) (7), acetyl-CoA carboxylase (ACC) (8), FAS (9), and stearoyl CoA desaturase (SCD)1 (10).
In addition to oxysterols and synthetic ligands, our laboratory and others have shown that insulin stimulates the lipogenic activity of LXRs in mouse liver (11, 12). The mechanisms involved are poorly understood, but may include posttranslational modifications [i.e., phosphorylation of LXR and/or synthesis of endogenous LXR ligand(s)]. Mitro et al. (13) reported that glucose bound to and activated LXR; however, these findings are controversial because the LXR ligand binding pocket accommodates small hydrophobic, rather than hydrophilic, compounds (14). We recently reported that LXRs were posttranslationally modified by O-linked β-N-acetylglucosamine (O-GlcNAc) in response to glucose, which in turn potentiated their transactivation of SREBP-1c (15). O-GlcNAc is a dynamically regulated posttranslational modification that is catalyzed by O-GlcNAc transferase (OGT). OGT uses UDP-N-acetylglucosamine (GlcNAc) as a substrate to add GlcNAc in O-glycosidic linkage to serine and threonine residues in target proteins, which are removed by O-GlcNAcase (OGA). The intracellular concentration of UDP-GlcNAc is controlled by the nutrient-responsive hexosamine biosynthetic pathway (HBP), a branch of glucose metabolism (16). Metabolic flux via the HBP is controlled by the rate-limiting enzyme glutamine:fructose-6-phosphate amidotransferase (GFAT). Concequently, UDP-GlcNAc pools are reduced when glucose uptake and glucose-6-phosphate (G-6-P) synthesis are limited (17). Because OGT activity is highly responsive to UDP-GlcNAc concentrations, OGT serves as a unique nutrient sensor (16). The activity of OGT and OGA are regulated by UDP-GlcNAc availability, protein-protein interactions, and posttranslational modifications, but the precise mechanisms involved are poorly understood (18, 19). Of note, the sensitivity of OGT to glucose increases with decreasing insulin signaling (20), whereas insulin mediates spatio-temporal activation of OGT, regulating substrate specificity of the enzyme at the plasma membrane (21).
Several studies have shown that ChREBP is activated by glucose through a variety of glucose metabolites generated downstream of G-6-P, including xylulose-5-phosphate, fructose-6-phosphate, and fructose-2,6-bisphosphate (22). The mechanisms through which these glucose metabolites modulate ChREBP activity are unclear, but are believed to involve allosteric regulation and dephosphorylation of the canonical isoform ChREBP at Ser196 and Thr666 leading to its nuclear translocation and regulation of target gene expression (23). Mutations of these residues, however, did not result in a constitutively active ChREBP (24), which suggests that ChREBPα is regulated by alternative means. In this respect, a recent study reported O-GlcNAcylation of ChREBP in response to hyperglycemia (25). Moreover, adenoviral overexpression of OGT in liver increased ChREBP O-GlcNAcylation, protein stability, and transactivation of liver pyruvate kinase (L-pk) and lipogenic genes (25). The lipogenic potential of LXRs was not investigated in that study.
Several reports show that LXR is essential for SREBP-1c activity in response to feeding, as SREBP-1c expression and hepatic DNL are drastically reduced in LXR-deficient mice (5, 26, 27). Although LXR clearly stimulates hepatic ChREBP expression after pharmacological activation (6), whether LXR regulates ChREBP under physiological conditions is less clear and may be dependent on insulin, dietary carbohydrate, and lipid content (26, 27). Notably, Herman et al. (28) recently identified a novel variant of ChREBP named ChREBPβ, which results from an alternative promoter located in exon1b of the ChREB gene. They further showed that glucose metabolism induced transcription of ChREBPα, promoting binding of ChREBPα to a carbohydrate response element (ChORE) identified in exon1b, inducing ChREBPβ transcription. Whether LXRs contribute to isoform-specific expression of ChREBP is currently not known.
To further explore the role of LXR and ChREBP in hepatic lipogenesis and to investigate insulin-independent effects of feeding on LXR activity, we subjected control and streptozotocin (STZ)-treated LXR+/+ and LXR−/− mice to a fasting-refeeding regime. Our results demonstrate that LXR regulates hepatic Gk expression and lipogenic genes also under hyperglycemic conditions independently of insulin. Furthermore, levels of O-GlcNAc-modified LXR were increased upon refeeding in both models, supporting the notion of LXR as a postprandial glucose-O-GlcNAc sensor. In support of our observations in vivo, LXR conferred high glucose responses on the ChREBPα promoter in Huh7 cells, an effect that was significantly inhibited by OGT depletion. We further show that nuclear, but not cytosolic, protein-O-GlcNAc levels are dramatically lower in LXR−/− compared with LXR+/+ mice. Accordingly, O-GlcNAcylation of ChREBP and the potential of ChREBPα to induce its target gene L-pk, was lower in refed LXR−/− compared with LXR+/+ mice. Combined, our work identifies LXR as an important integrator of hepatic glucose/O-GlcNAc metabolism and fatty acid synthesis upstream of ChREBP.
RESEARCH DESIGN AND METHODS
Materials
STZ (S0130), GW3965 (G6295), DMSO (D4540), formaldehyde (F1635), DMEM (D6046, D6546), fetal bovine serum (F7524), l-glutamine (G7513), 1% penicillin-streptomycin (P4458), and D-(+)-glucose solution (G8769) were purchased from Sigma-Aldrich. Glucose-free DMEM (11966-025), TRIzol® reagent (15596-018), UltraPure™ phenol:chloroform:isoamyl alcohol (15593-031), and isopropylthio-β-galactoside (IPTG) (15529-019) were from Invitrogen. Silencing RNA targeting OGT (ON-TARGET Plus SMART Pool, catalog number L-019111-00) and scrambled control siRNA (ON-TARGET Plus® Control Pool, catalog number D-001818-10-05) were from Thermo Scientific Dharmacon. Dual-Luciferase® reporter assay system (E194A) was from Promega. All other chemicals were of the highest quality available from commercial vendors.
Plasmids
LXRα/β cDNA in plasmids pcDNA3-FLAG-hLXRα and pcDNA3-FLAG-hLXRβ, previously described (15), were subcloned into pGEX-4T-1 or pEGFP vectors. The generation of pcDNA3-hRXRα has been described previously (29). pET-24b-His-ncOGT was a kind gift from Suzanne Walker (30). The pCAG-HA-OGT and the pCherry-OGT plasmids were kind gifts from Gerald Hart. The pGL3-basic and pGL3-basic-pChREBP (ChREBPα-prom) reporter plasmids were kindly provided by Karine Gauthier Vanacker (31). Sequences were verified by DNA sequencing.
Animals and treatment
Male LXRαβ-deficient (LXR−/−) mice and wild-type littermates (LXR+/+) were housed in a temperature-controlled (22°C) facility with a strict 12 h light/dark cycle. The mice had free access to food and water at all times. LXR−/− mice and corresponding controls had mixed genetic background based on 129/Sv and C57BL/6J strains, backcrossed in C57BL/6J for at least six generations. The generation of the LXR−/− mice has been described previously (32, 33). STZ was prepared in a sodium citrate buffer (50 mmol/l, pH 4.5) immediately before injections. STZ-treated mice were treated with two intraperitoneal injections of STZ (100 mg/kg) with a 1 day interval. Seven days after the first STZ injection, mice were included in a fasting-refeeding experiment with matched untreated control mice. Mice were fasted for 24 h or fasted for 24 h and refed for 12 h on a diet containing 64% carbohydrates, 31.5% protein, 4.5% fat, and no cholesterol (SDS RM no.1 maintenance, Special Diets Services, UK). The mice were euthanized by cervical dislocation at 8:00 AM and tissues were weighed and snap-frozen in liquid nitrogen and stored at −80°C until further analysis. All use of animals was approved and registered by the Norwegian Animal Research authority.
Blood chemistries and liver lipid analyses
Blood glucose was measured at the time of euthanization with a handheld glucometer (Roche). Serum was separated from blood by centrifugation. Serum insulin was measured using the ultra-sensitive insulin kit from Mercodia (Mercodia AB, Uppsala, Sweden) according to the manufacturer’s instructions. Serum lipid quantification, using enzymatic in vitro tests, was done on a Roche Hitachi 917. TGs were determined with a TG GPO-PAP kit (11730711216; Roche). Liver samples were homogenized in cold sucrose buffer [0.25 M sucrose, 10 mM HEPES, 1 mM Na-EDTA (pH 7.4)] and centrifuged at 600 g for 10 min at 4°C and hepatic TGs were measured in the supernatant using the TG GPO-PAP kit according to the manufacturer’s protocol.
Cells cultures and transfections
Huh7 liver hepatoma cells were maintained in high glucose (25 mM) DMEM (D6546) or adapted to physiological glucose (5 mM) DMEM (D6046) supplemented with 10% fetal bovine serum, 4 mM l-glutamine, and 1% penicillin-streptomycin. Cells were transfected with Lipofectamine 2000 before luciferase reporter assay, immunofluorescence, and immunoprecipitation, as described in the sections below.
Luciferase reporter assay
Huh7 cells were transfected with 150 ng of pGL3-basic-pChREBP-Luc reporter plasmid (ChREBPα-prom), with 50 ng pcDNA3-FLAG-hLXRα, pcDNA3-FLAG-hLXRβ, pcDNA3-hRXRα, or empty pcDNA3 expression plasmid. pRL-CMV (Promega; 50 ng) was included as internal control to normalize for transfection efficiency. All transfections were performed using Lipofectamine 2000. Dual-Luciferase reporter assay was performed 24 h posttransfection as previously described (34). For siRNA experiments, Huh7 cells were transfected with 40 nM of siOGT or nontargeting siRNA. Posttransfection (24 or 48 h), the cells were further transfected with pChREBPα-prom and pRL-CMV with pcDNA3-FLAG-hLXRα, pcDNA3-hRXRα, and/or empty vector as described above in siRNA containing medium. Luciferase activity was measured after an additional 24 h. Transfections were verified by immunoblotting using 20 μl cell lysate and antibodies described below.
RNA extraction, cDNA synthesis, and quantitative RT-PCR
RNA was isolated with TRIZOL® reagent (Invitrogen) according to the manufacturer’s protocol, including high salt precipitation (0.8 M sodium acetate, 1.2 M NaCl) to avoid contaminating polysaccharides to coprecipitate with RNA. Isolated RNA was reverse transcribed into cDNA using a high-capacity cDNA Archive kit (Applied Biosystems). Analysis of mRNA expression was done by quantitative RT-PCR on a 7900HT instrument (Applied Biosystems). Gene expression was normalized against the expression of tata-binding protein (Tbp). Primers were as follows: TaqMan gene expression assays detecting mouse: Accα (Mm01304257_m1), Accβ (Mm01204678_m1), elongation of long-chain fatty acid family member 6 (Elovl6) (Mm00851223_s1), Fas (Mm00662319_m1), Gfat1 (Mm00600127_m1), Gfat2 (Mm00496565_m1), Gk (Mm00439129_m1), glycerol-3-phophate acyltransferase (Gpat) (Mm00833328_m1), L-pk (Mm00443090_m1), Ogt (Mm00507317_m1), Oga (Mm00452409_m1), Scd1 (Mm00772290_m1), tbp (Mm00446971_m1); or SYBR primers detecting mouse: Chrebpα (5′-CGACACTCACCCACCTCTTC, 5′-TTGTTCAGCCGGATCTTGTC) (28), Chrebpβ (5′-TCTGCAGATCGCGTGGAG, 5′-CTTGTCCCGGCATAGCAAC) (28), Lxrα (5′-GGAGTGTCGACTTCGCAAATG, 5′-CAGCACACACTCCTCCCTCA), Lxrβ (5′-GCTCTGCCTACATCGTGGTCA, 5′-TGCGCTCAGGCTCATCCT), Mlx (5′-GGAGCTCTCAGCTTGTGTCTTCA, 5′-CACCGATCACAATCTCTCGTAGAGT) (28), Srebp-1 (5′-GGAGCCATGGATTGCACATT, 5′-GCTTCCAGAGAGGAGGCCAG) (35).
Generation of LXR antibody
Rat LXRα and LXRβ proteins were purified as described previously (36). Rabbits were immunized at Agisera (Vännäs, Sweden) by a standard immunization program. Briefly, 0.25 mg of LXRα and LXRβ were injected four times and serum collected after 15 weeks. Purified LXRα and LXRβ protein (1.4 mg) in 0.2 M NaHCO3, 0.5 M NaCl (pH 8.3) buffer were coupled on a HiTrap NHS-activated matrix column (GE Healthcare). The immunized rabbit serum was added to the column and washed according to standard procedure. Elution of anti-LXRs was performed 10 times and eluates were pooled.
Liver extracts and immunoblot analysis
Nuclear and cytoplasmic proteins were prepared using the NE-PER extraction kit (Pierce Biotechnology) with the following inhibitors added to the buffers: 1 mM NaF, 1 mM Na3VO4, 1 mM β-glycerophosphate, 1 μM O-GlcNAcase inhibitor GlcNAc-thiazoline, and CompleteTM protease inhibitors (Roche Applied Science). Proteins were separated by SDS-PAGE (Bio-Rad) and blotted onto polyvinylidene difluoride membrane (Millipore). Primary antibodies used were rabbit anti-mouse LXR (1:500) (see generation of antibodies), retinoid X receptor (RXR)α (1:1,000) (sc-553; Santa Cruz Biotechnology), ChREBP (1:1,000) (NB400-135; Novus Biologicals), FAS (1:500) (sc-55580; Santa Cruz Biotechnology), SCD1 (1:2,000) (sc-14719; Santa Cruz Biotechnology), α-tubulin (1:20,000) (T5168; Sigma-Aldrich), lamin A (1:1,000) (L1293; Sigma-Aldrich), GFAT1/2 (1:3,000) (37), GFAT2 (1:1,000) (sc-134710; Santa Cruz Biotechnology), OGT (1:1,000) (AL28) (38), CTD110.6 (1:2,500) (MMS-248R; Covance), SREBP-1 (1:1,000) (39), L-PK (1:2,000) (MABS148; Merck Millipore), FLAG M2 (1:10,000) (F1804; Sigma-Aldrich), GK (1:2,000) (40), and glutathione-S-transferase (GST)-HRP (ab58626; Abcam). Secondary HRP-conjugated anti-mouse, 115-035-174; anti-rabbit, 211-032-171; anti-sheep, 713-035-003 (all from Jackson ImmunoResearch Laboratories); anti-goat, 605-4302 (Rockland) antibodies were used at 1:10,000 dilutions. Anti-mouse IgM (A8786; Sigma) was used at a 1:5,000 dilution. Clean-Blot IP detection reagent (HRP) (PIER21230, Pierce) was used at 1:1,000.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (41). Briefly, liver tissue was cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by 10 min incubation with 12.5 mM glycine. Samples were washed twice in cold PBS and harvested in lysis buffer [0.1% SDS, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, and 20 mM Tris (pH 8.0)]. Lysed tissue was sonicated using a Bioruptor (Diagenode) to an average size of 200–500 bp fragments. Chromatin was immunoprecipitated with 2 μg antibody against LXR, ChREBP (NB400-135; Novus Biologicals), or rabbit IgG (011-000-002; Jackson Laboratory). Sepharose beads (GE Healthcare) were washed three times in lysis buffer, incubated with BSA (1 μg/μl) for 3 h, added to the chromatin and left to rotate at 4°C overnight. Beads were washed twice with buffer 1 [0.1% SDS, 0.1% deoxycholic acid (DOC), 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 20 mM HEPES (pH 7.6)]; once in buffer 2 [0.1% SDS, 0.1% DOC, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, 20 mM HEPES (pH 7.6)], once in buffer 3 [0.25 M LiCl, 0.5% DOC, 0.5% Nonidet P-40, 1 mM EDTA, 20 mM HEPES (pH 7.6)], and twice in buffer 4 [1 mM EDTA and 20 mM HEPES (pH 7.6)]. All washing steps were done for 5 min at 4°C. DNA-protein complexes were eluted with 1% SDS and 0.1 M NaHCO3 and reverse cross-linked by adding 0.2 M NaCl and overnight incubation at 65°C. DNA was purified by phenol-chloroform extraction, precipitated in ethanol with sodium acetate, and dissolved in water. DNA enrichment was quantified by quantitative RT-PCR. ChIP PCR primers spanning mouse LXR response elements (LXREs) were designed to amplify known LXR-enriched regions (41) and were as follows: Srebp-1 LXRE (5′-CAGAACTTGCCTGGACCATT, 5′-AAATCTTGCTGCTGCCATTC), Chrebp LXRE (5′-CGTTCAATCTCTGTGGATCGT, 5′-AAGTGGCCTGATTCACCTCT), Gk LXRE (5′-CATTCTGGGCTCTTCTACGG, 5′-TGCAGAGTATGTTGGGGTCA), Fas LXRE (5′-GGGTTACTACCGGTCATCGT, 5′-CCGTCAGTGTTCCCTATCCT); Scd1 LXRE (5′-AAGTGCATTGCTGAGACTTCC, 5′-CCTCCACAGCCTCTTTGTCT). The binding of ChREBP to the ChORE in the promoter-proximal enhancer of L-pk has been described previously (25) and was as follows: L-pk ChORE (5′-GTCCCACACTTTGGAAGCAT, 5′-CCCAACACTGATTCTACCC).
Recombinant proteins and GST pulldown
pET-24b-His-ncOGT, pGEX-4T-1-hLXRα, and pGEX-4T-1-hLXRβ were expressed in Escherichia coli BL21 (DE3) Star (Invitrogen). Human His-ncOGT was grown and purified as previously described (30). GST fusion proteins were expressed as described (30). For GST pulldown assays, lysates were incubated with glutathione Sepharose 4B beads (GE Healthcare), washed six times [10 mM TrisHCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40], proteins were then eluted by boiling in 2× Laemmli buffer and separated by SDS-PAGE. Precipitated proteins were detected by immunoblotting.
Succinylated wheat germ agglutinin pulldown
Nuclear extracts (100 μg) were incubated with protein A/G-agarose beads (sc-2003; Santa Cruz Biotechnology) for 1 h at 4°C. Cleared extracts were transferred to new tubes and incubated with 30 μl of succinylated wheat germ agglutinin (sWGA)-agarose (Vector Lab) with or without 0.5 M GlcNAc overnight at 4°C. After four washes (PBS, 0.2% NP-40), proteins were eluted from the beads in 2× Laemmli buffer and separated by SDS-PAGE. The captured proteins were analyzed by immunoblotting.
Immunofluorescence
Huh7 cells were seeded on coverslips (2.5 × 105 cells/coverslip) and transfected with 200 ng pEGFP-LXRα or pEGFP-LXRβ and 800 ng pCherryOGT 24 h after seeding. Twenty-four hours after transfection, cells were treated with DMSO (0.1% final) or 1 μM GW3965 for 2 h by addition of ligand directly to culture media and cells were then fixed in 4% paraformaldehyde and permeabilized with 0.4% Triton X-100 for 20 min. Fixed cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector Laboratories). Images were acquired at 60× magnification using an Olympus FluoView 1000 confocal microscope.
Coimmunoprecipitation
Huh7 cells were transfected with 1 μg pCAG-HA-OGT and 0.5 μg pcDNA3-FLAG-LXRα or pcDNA3-FLAG-LXRβ. Cells were washed twice with cold PBS and harvested in lysis buffer [200 mM NaCl, 20 mM HEPES (pH 7.4), 1% NP-40] containing 1 mM NaF, 1 mM Na3VO4, 1 mM β-glycerophosphate, 1 μM O-GlcNAcase inhibitor GlcNAc-thiazoline, and CompleteTM protease inhibitors (Roche Applied Science). The lysate was cleared by centrifugation and the supernatants were immunoprecipitated with 2 μg FLAG M2, 2 μg OGT (AL28), and mouse IgG antibody (015-000-002; Jackson Laboratory) bound to protein G Dynabeads (Invitrogen) for 4 h at 4°C. Beads were washed three times in wash buffer [200 mM NaCl, 20 mM HEPES (pH 7.4), 0.1% NP-40] and proteins were eluted from the beads with 1× NuPAGE LDS sample buffer (Invitrogen) at 70°C for 10 min. Coimmunoprecipitated proteins were analyzed by immunoblotting with FLAG M2 or OGT antibodies. Liver was homogenized in lysis buffer with inhibitors and lysate was cleared by centrifugation for 10 min. Cleared lysates (500 μg) were immunoprecipitated with 3 μg antibody against LXR, OGT (AL28), or rabbit IgG (011-000-002; Jackson Laboratory) bound to protein G Dynabeads (Invitrogen). Coimmunoprecipitated proteins were analyzed by immunoblotting.
Statistical analysis
Statistical analyses were performed using SPSS 22 (IBM Corp.). All data are presented as means and SEM, and error bars for all results are derived from biological replicates rather than technical replicates. The homogeneity of variances between groups was evaluated using Levene’s test. If significant, a Welch test was applied to assess overall differences combined with a Dunnett’s T3 post hoc test to establish intergroup comparisons. If Levene’s test was not significant and homogeneity of variances could be assumed, statistical differences between groups were determined by one-way ANOVA followed by Tukey’s multiple comparison tests. P < 0.05 was considered statistically significant.
RESULTS
Feeding induces LXRα protein levels
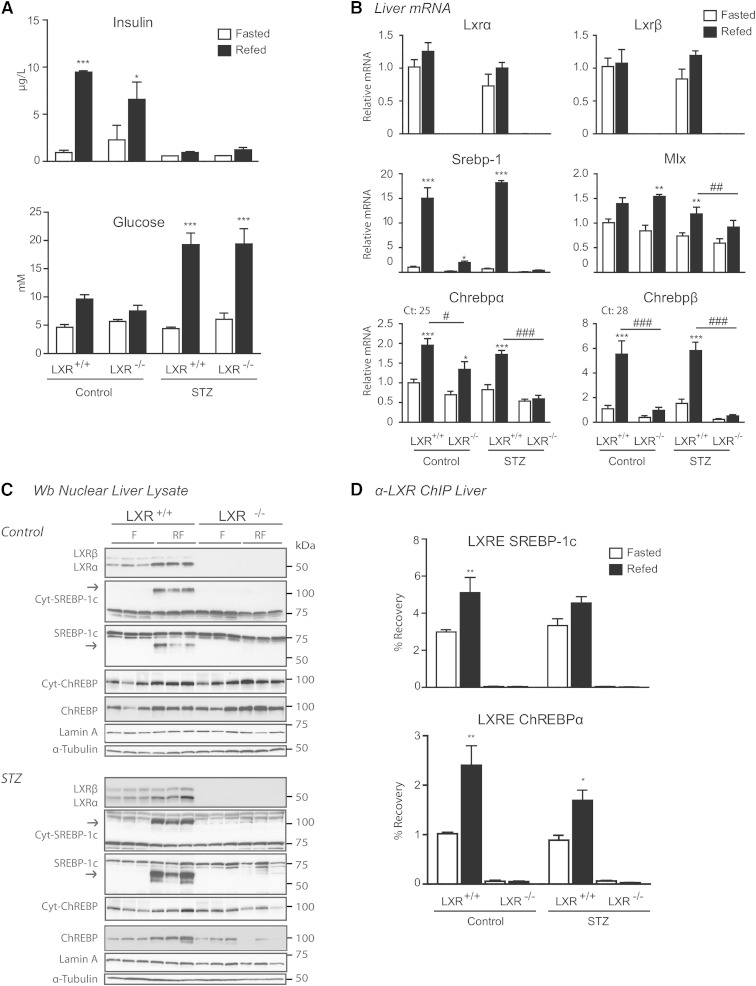
To examine the effects of insulin on LXR-regulated hepatic lipogenic target gene expression, LXR+/+ and LXR−/− control and STZ-treated mice were placed on a fasting-refeeding regime. STZ was used to destroy pancreatic β-cells and insulin production. Using this model, the effect of insulin could be distinguished from those of hyperglycemia in refed STZ-treated mice (low insulin, high glucose) compared with untreated control mice (high insulin, high glucose). In hepatocytes expressing the insulin-independent low-affinity glucose transporter, GLUT2, glycolysis is favored in the fed state when glucose is abundant (42, 43). LXR+/+ and LXR−/− refed control mice showed increased serum insulin and blood glucose levels, whereas refed STZ-treated mice were hyperglycemic with fasting insulin levels (Fig. 1A). Insulin levels were slightly, but not significantly, higher in fed LXR+/+ compared with LXR−/− control mice, supporting the known role of LXR in glucose homeostasis, insulin secretion, and pancreatic function (44, 45). No significant differences in body weight or liver weight were found between treatment-matched LXR+/+ and LXR−/− mice (supplementary Table 1). Lxrα and Lxrβ mRNA expression levels were unaffected in refed control and STZ-treated LXR+/+ mice when compared with treatment-matched fasted mice (Fig. 1B). Nuclear LXRα protein levels were 2.1- to 2.5-fold increased in refed control and STZ-treated mice (Fig. 1C; band intensity quantifications relative to loading control using Image J software), whereas LXRβ protein levels remained unchanged. These data suggest that feeding increases LXRα protein expression via posttranscriptional or posttranslational mechanisms in vivo.
Fig. 1.
LXRs confer differential regulation of hepatic ChREBP isoform expression. C57BL/6 control and STZ-treated male mice were fasted (white bars) or fasted-refed for 12 h (black bars). A: Insulin and glucose levels. B: Hepatic gene expression of Lxrα/β, Srebp-1, Mlx, and Chrebpα/β was analyzed by quantitative RT-PCR and normalized to Tbp. C: Nuclear lysates were subjected to SDS-PAGE and immunoblotted with antibodies against LXR, SREBP-1, and ChREBP. Cytosolic levels of SREBP-1 and ChREBP are also shown. α-Tubulin and lamin A were used as controls for cytosolic and nuclear fractions, respectively. Each lane represents independent mice from experimental groups. The samples for control and STZ-treated mice were loaded on separate gels and the films were developed with the same exposure time. One representative Western blot is shown (n = 2). D: LXR binding to LXRE-containing promoter regions of SREBP-1c and ChREBP were analyzed by ChIP with antibody recognizing LXRα/β. All values are mean ± SEM (n = 4–5). *P < 0.05, **P < 0.01, ***P < 0.001 compared with fasted within experimental groups. #P < 0.05, ##P < 0.01, ###P < 0.001 compared with refed within STZ treatment.
LXR confers differential regulation of hepatic ChREBP isoform expression
The expression and activity of SREBP-1c and ChREBP are regulated by insulin and glucose (43, 46). Because SREBP-1c and ChREBP can work in concert with LXRs to regulate hepatic lipogenic genes (5, 6, 47), we investigated the expression of SREBP-1 and ChREBP upon refeeding under normal and hyperglycemic insulin-independent conditions. In agreement with previous reports, SREBP-1c mRNA and protein levels were strongly induced in refed control and STZ-treated LXR+/+ mice (Fig. 1B, C) (15, 43) and almost completely abolished in LXR−/− mice (26, 47). Chrebpα mRNA expression was elevated (2-fold) by refeeding in LXR+/+ control and STZ-treated mice and this induction was significantly lower in LXR−/− mice. However, a 1.4-fold induction in Chrebpα mRNA levels was observed in control LXR−/− mice (no induction in STZ-treated LXR−/− mice) (Fig. 1B). Expression levels of the newly identified shorter isoform, Chrebpβ (28), were strongly induced by refeeding in both control and STZ-treated mice (6-fold), and almost abolished in LXR−/− mice. Interestingly, ChREBP cytosolic and nuclear protein levels, detected by immunoblotting with an antibody recognizing ChREBPα as well as ChREBPβ (our own observations in ChREBPβ overexpressing cells), were not statistically different between genotypes in refed control mice, but were almost abolished (70% lower in cytosolic, 90% lower in nuclear) in refed LXR−/− STZ-treated hyperglycemic mice compared with refed LXR+/+ mice treated with STZ (Fig. 1C; band intensity quantifications relative to loading control using Image J software). This suggests that transcriptional and posttranscriptional regulation of ChREBPα is dependent on LXR under hyperglycemic conditions lacking the postprandial insulin signal. ChREBPα is believed to be retained inactive in the cytoplasm during fasting (23). Our observed nuclear localization of ChREBP in the fasted state may reflect levels of the constitutively nuclear localized ChREBPβ (25). However, we were not able to distinguish between the two isoforms in Western blotting analysis (ChREBPα, 93 kDa; ChREBPβ, 90 kDa). Furthermore, we did not observe a significant fasting-refeeding shuttling of ChREBP between the cytosolic and nuclear compartments. Rather, cytosolic and nuclear ChREBP protein levels were comparable under each condition. ChIP assays revealed increased LXR recruitment to an LXRE in the ChREBPα promoter in refed control and STZ-treated mice. LXR recruitment to an LXRE in the SREBP-1 promoter was evident in refed control mice, but only a weak nonsignificant increase in LXR binding was observed in STZ-treated mice (Fig. 1D), which did not reflect the strong induction of Srebp-1c mRNA levels in these mice (Fig. 1B). This may be due to increased coactivator recruitment to this LXRE and/or increased SREBP-1c autoregulation under hyperglycemic conditions. Taken together, these data suggest that LXRs are essential regulators of SREBP-1 and ChREBP expression also in the absence of postprandial insulin.
Expression of hepatic de novo lipogenic genes and production of plasma TGs through LXR are not dependent on insulin signaling
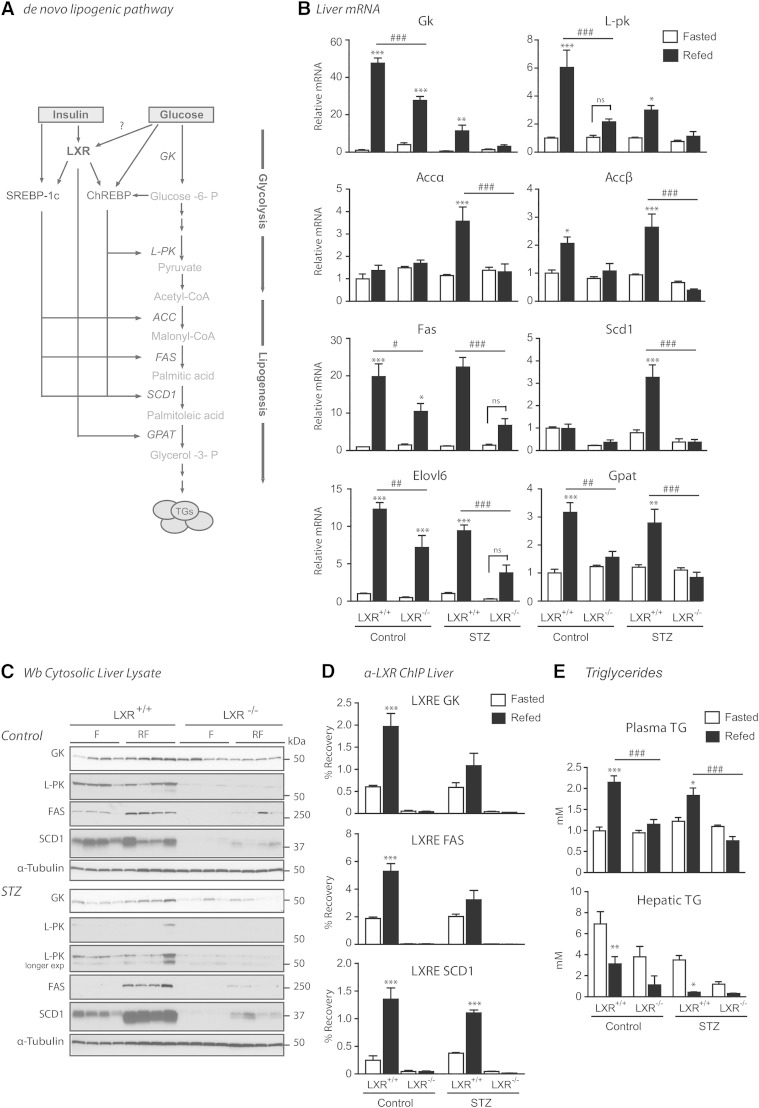
We next investigated the expression of genes downstream of the LXR lipogenic axis to further delineate the effects of hyperglycemic conditions on LXR activity. A schematic representation of DNL with the proteins involved is provided in Fig. 2A. Gk and L-pk mRNA levels were induced approximately 50-fold and 6-fold in refed control mice, respectively (Fig. 2B). These genes were also significantly upregulated in refed STZ-treated mice; however, the level of induction was less than that observed in control mice (10-fold for Gk, 3-fold for L-pk). Of note, expression of gluconeogenic genes, glucose-6-phosphatase and phosphoenolpyruvate carbokinase (PEPCK), and the gluconeogenic coactivator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC1α) were strongly downregulated in both control and STZ-treated mice (data not shown), supporting increased G-6-P synthesis above fasting levels in hyperglycemic STZ-treated mice. In support of our observations, others have shown downregulation of glucose-6-phosphatase and PEPCK in response to glucose and sucrose refeeding in control and STZ-treated mice (13, 43). Moreover, GLUT2 expression is increased in refed STZ-administered mice, ensuring increased hepatic glucose uptake in hyperglycemia (43). The mRNA levels of Fas, Elovl6, Accβ, and Gpat were elevated to a similar degree in both refed control and STZ-treated mice. Notably, the mRNA levels of Accα and Scd1 were highly induced in LXR+/+ STZ-treated mice, where SREBP-1c may be important (43). Interestingly, GK and L-PK protein levels did not mimic the induced mRNA expression levels in refed LXR+/+ mice, there were only small insignificant changes, which could be due to posttranslational regulation of the active proteins (Fig. 2C). GK protein levels were 1.4 times higher in refed control compared with fasted control LXR+/+ mice and 1.6 times higher in refed STZ compared with fasted STZ LXR+/+ mice. Fasting GK protein levels were 2.8 times higher in fasted control compared with fasted STZ mice and GK and L-PK levels were significantly lower in refed LXR−/− mice compared with LXR+/+ mice in both control (GK 50% lower, L-PK 92% lower) and STZ-treated mice (GK 65% lower, L-PK 77% lower) (Fig. 2C; band intensity quantifications relative to loading control using Image J software). LXR binding to the LXREs in the promoters of Gk, Fas, and Scd1 was significantly induced in refed control mice, but LXR binding was only significantly induced to the Scd1 LXRE in STZ-treated mice (Fig. 2D). Differences may be due to alternative LXREs in promoters or enhancers of these genes, and/or coregulator interactions. The induction of lipogenic genes was followed by increased plasma TGs in refed control and STZ-treated LXR+/+, but not LXR−/−, mice (Fig. 2E). Plasma nonesterified fatty acids and cholesterol were not statistically different between LXR+/+ and LXR−/− mice (data not shown). Similar to previous reports, LXR−/− mice accumulated fewer hepatic TGs compared with LXR+/+ littermates, and in STZ-treated compared with control mice (43, 48) (Fig. 2E). No differences in expression of hepatic fatty acid oxidation genes (Cpt1 and Acox1) or genes involved in endoplasmic reticulum stress (Hspa5, Hsp90b1, and Calr) were observed, suggesting that the lower hepatic TG content was independent of these pathways (data not shown). Taken together, our findings suggest that LXR regulates hepatic lipogenic genes and TG content under hyperglycemic conditions independently of insulin, suggestive of a role for glucose in regulating LXR activity as previously suggested (13, 15).
Fig. 2.
Expression of hepatic de novo lipogenic genes and production of plasma TGs through LXR are not dependent on insulin signaling. A: Simplified schematic overview of the genes involved in hepatic DNL. B: Hepatic gene expression was analyzed by quantitative RT-PCR and normalized to Tbp. C: Cytosolic lysates were immunoblotted with antibodies against GK, L-PK, FAS, and SCD1 with α-tubulin as loading control. Each lane represents independent mice from experimental groups. The samples for control and STZ-treated mice were loaded on separate gels and films were developed with the same exposure time, unless otherwise indicated. D: LXR binding at the LXRE-containing promoter regions of GK, FAS, and SCD1 were analyzed by ChIP experiments with antibody recognizing LXRα/β. E: Liver and plasma TGs. All values are given as mean ± SEM (n = 4–5). *P < 0.05, **P < 0.01, ***P < 0.001 compared with fasted within experimental group. #P < 0.05, ###P < 0.001 compared with refed within control or STZ treatment. ns, not significant.
High glucose regulates LXR transactivation of the ChREBPα promoter via O-GlcNAc
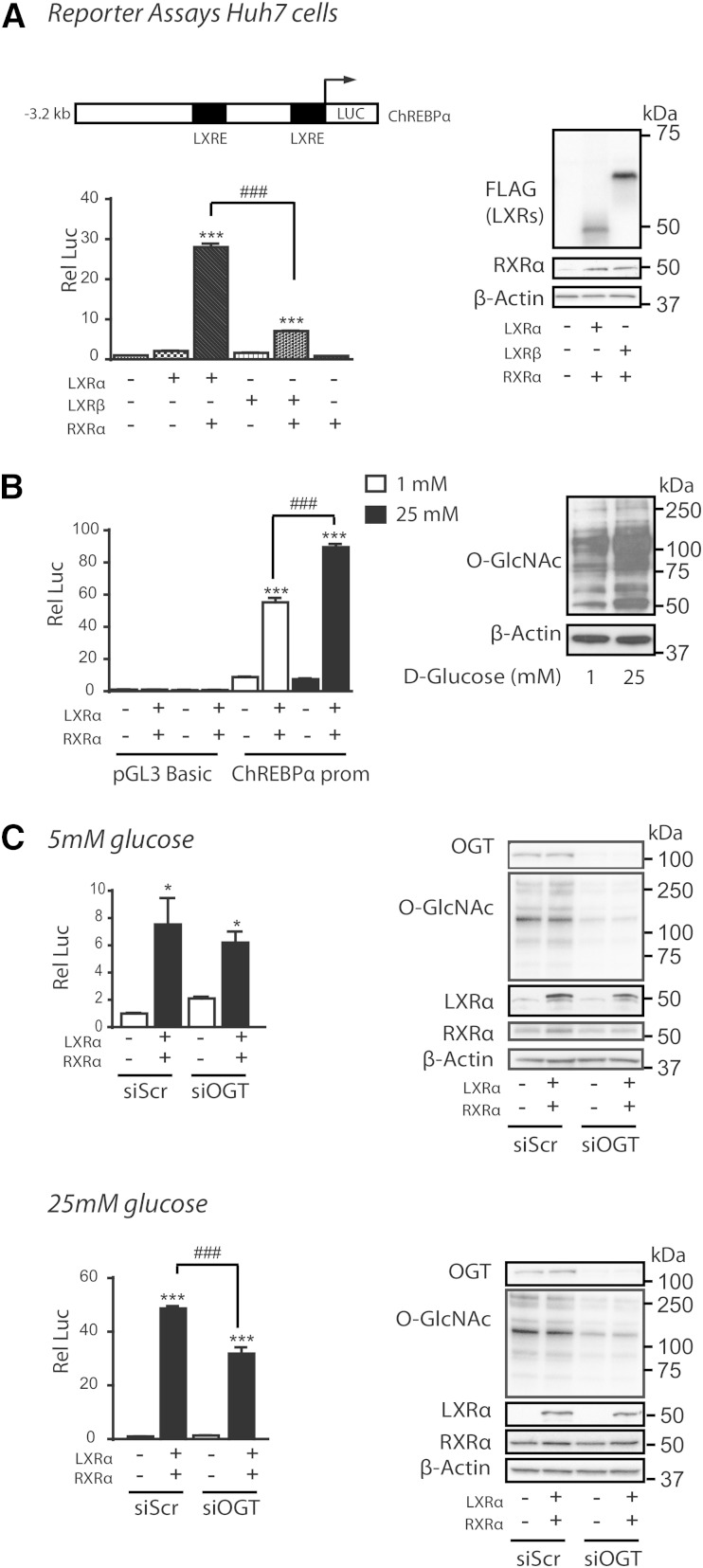
We have previously reported increased O-GlcNAcylation of LXR in vivo and in vitro in response to refeeding and high glucose treatment contributing to increased SREBP-1c promoter activation (15). To examine whether high glucose can regulate the ChREBPα promoter through LXRs, Huh7 cells were cotransfected with LXRα or LXRβ and/or RXRα expression vectors together with a luciferase reporter construct containing 3.2 kb of the 5′-flanking region of the mouse ChREBPα promoter (Fig. 3A, B). Cotransfected with RXRα, LXRα conferred an approximately 30-fold induction of ChREBPα promoter-luciferase activity as compared with 8-fold by LXRβ in Huh7 cells adapted to high glucose (25 mM) (Fig. 3A). Moreover, in cells adapted to physiological glucose levels (5 mM), we observed a 30% increase in ChREBPα promoter-luciferase activity in LXRα/RXRα transfected cells after high glucose (25 mM) compared with low glucose (1 mM) treatment for 24 h (Fig. 3B). To determine whether OGT-mediated O-GlcNAc signaling was conferring the glucose effect on ChREBPα expression through LXRα, Huh7 cells, cultured in physiological (5 mM) and high glucose, were transfected with scrambled siRNA or siRNA against OGT. One to two days after siRNA transfection, cells were transfected with LXRα/RXRα and ChREBPα promoter construct in siRNA-containing medium and cells were analyzed for luciferase activity the following day. The stimulating effect of high glucose through LXRα was approximately 30% reduced in siOGT-transfected cells, whereas no significant reduction was observed under normoglycemic (5 mM) conditions (Fig. 3C), arguing for a role for OGT signaling in high glucose/LXR-mediated transactivation of the ChREBPα promoter.
Fig. 3.
High glucose regulates LXR transactivation of the ChREBPα promoter via O-GlcNAc. A: Huh7 cells maintained in 25 mM glucose were transfected with ChREBPα-Firefly luciferase reporter (ChREBPα-prom), Renilla luciferase control plasmid (pRL-CMV) and RXRα expression vector and/or LXRα or LXRβ expression vectors or empty vector (pcDNA3.1) (n = 3). Dual luciferase assay was performed and lysates were immunoblotted with antibodies against FLAG, RXRα, and β-actin as loading control. A schematic figure of the ChREBPα reporter construct (ChREBPα-prom) including the two LXREs is shown. B: Huh7 cells maintained in 5 mM glucose were transfected with ChREBPα-prom or pGL3-Basic with empty expression vector or RXRα and LXRα expression vectors. All transfections contained pRL-CMV. After 6 h, the cells were treated with 1 mM or 25 mM glucose for 24 h (n = 3). Dual luciferase assay was performed and lysates were subjected to SDS-PAGE and immunoblotted with an antibody detecting O-GlcNAc epitopes on proteins (CTD 110.6) with β-actin as loading control. One representative Western blot is shown (n = 3). C: Huh7 cells maintained in 5 mM glucose (upper panel) and 25 mM glucose (lower panel) were transfected with 40 nM siRNA, nontargeting pool (siSrc), or human OGT (siOGT). After 24 h (25 mM glucose) and 48 h (5 mM glucose) the cells were further transfected with ChREBPα-prom, pRL-CMV with empty vector, or RXRα and LXRα expression vectors. Dual luciferase assay was performed after another 24 h. Lysates were subjected to SDS-PAGE and immunoblotted with antibodies against OGT, O-GlcNAc (CTD 110.6), LXRα, and RXRα and β-actin as loading control (n = 3). All values are given as mean ± SEM. Statistical differences are shown as *P < 0.05 or ***P < 0.001 between respective controls or as ###P < 0.001 between indicated groups.
LXR regulates nuclear O-GlcNAc signaling
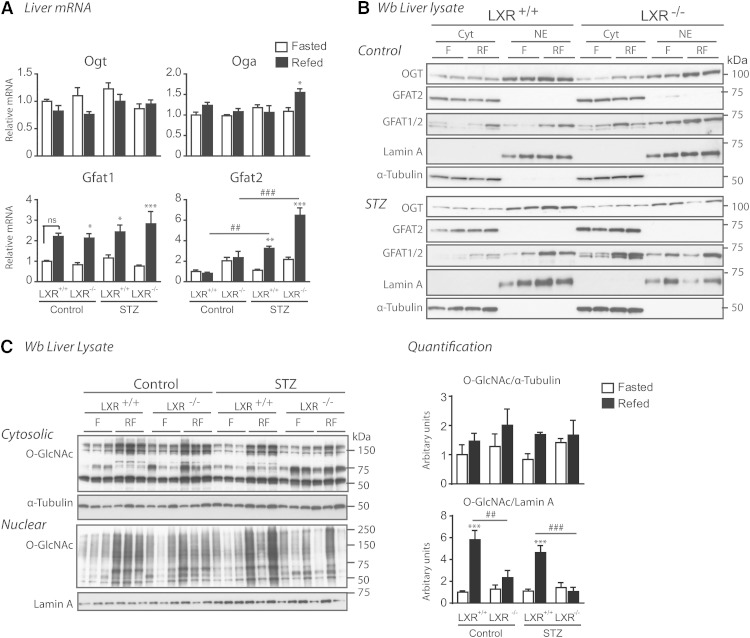
Having established a role for LXR in mediating glucose responses on the promoters of ChREBPα and SREBP-1c (15) via HBP/O-GlcNAc signaling, we next investigated the impact of feeding and LXR on expression of central genes in this pathway (Fig. 4A). Expression levels of GFAT1, the rate limiting enzyme of HBP, was upregulated by feeding in the absence and presence of insulin, whereas the less characterized isoform, GFAT2, was only upregulated in STZ-treated mice (3-fold) and more so in LXR−/− mice (6-fold), suggesting a possible negative feedback by LXRs on GFAT2 expression under hyperglycemic conditions. GFAT1, but not GFAT2, was localized in both cytoplasmic and nuclear fractions in LXR+/+ and LXR−/− mice, suggesting a possible nuclear production of glucosamine-6-phosphate, an intermediary product in the HBP generating UDP-GlcNAc, the substrate for OGT (Fig. 4B). Hepatic UDP-GlcNAc concentrations have been previously shown to be increased by approximately 60% in ad libitum-fed STZ-treated rats compared with controls (49). OGT and OGA expression were not regulated by feeding or LXR (Fig. 4A, B). Surprisingly, nucleocytoplasmic OGT protein expression was unaffected (Fig. 4B), but nuclear protein O-GlcNAcylation was significantly lower in refed LXR−/− mice under both control and hyperglycemic conditions (Fig. 4C), suggesting that LXR, by direct or indirect mechanisms, regulates nuclear OGT activity. In line with our observations in vivo, LXR potentiated nuclear, but not cytosolic, protein O-GlcNAcylation in LXR-tranfected Huh7 cells compared with empty vector controls (data not shown). Cytosolic protein O-GlcNAcylation was not affected by LXR deficiency as revealed by Western blotting analysis using a pan-O-GlcNAc-specific antibody (CTD 110.6), and total nuclear and cytosolic protein O-GlcNAcylation levels were similar in control and STZ-treated mice (Fig. 4C).
Fig. 4.
LXR regulates nuclear O-GlcNAc signaling. A: Hepatic gene expression of enzymes involved in HBP was analyzed in fasted (white bars) and refed (black bars) mice by quantitative RT-PCR and normalized to Tbp. B: Liver lysates from cytosolic (Cyt) and nuclear (NE) compartments were immunoblotted with antibodies against OGT, GFAT1/2, and GFAT2. α-Tubulin and lamin A were used as controls for Cyt and NE fractions, respectively. Each lane represents lysate from independent mice from each experimental group. A representative Western blot is shown (n = 2). C: Cytosolic and nuclear liver lysates were immunoblotted with anti-O-GlcNAc antibody (CTD110.6). α-Tubulin and lamin A were used as controls for Cyt and NE fractions, respectively. One representative Western blot is shown (n = 2). Quantification of total cytosolic and nuclear O-GlcNAc modified proteins was analyzed by Image J. All values are given as mean ± SEM (n = 4–5). *P < 0.05, **P < 0.01, ***P < 0.001 compared with fasted within experimental group. ##P < 0.01, ###P < 0.001 compared with refed between indicated groups. ns, not significant.
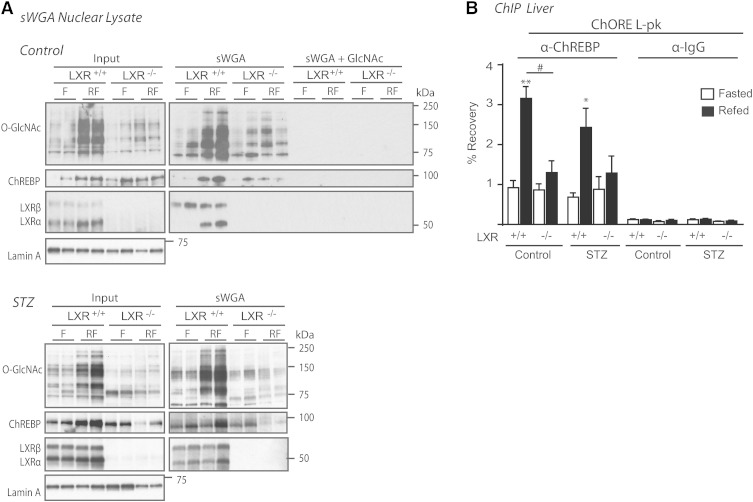
LXRs regulate O-GlcNAcylation of ChREBP and binding of ChREBPα to the L-pk promoter
Recently, O-GlcNAcylation of ChREBP was shown to potentiate its activity, stability, and binding to the promoter-proximal enhancer of L-pk (25). Based on that study and the reduced L-PK expression and protein O-GlcNAc levels observed in LXR−/− mice, we investigated whether LXR could regulate ChREBP activity through modulating O-GlcNAcylation of ChREBP. Using lectin (sWGA) pulldown experiments, we observed lower levels of O-GlcNAcylated proteins precipitated from nuclear lysates isolated from refed LXR−/− mice compared with LXR+/+ controls, which was in agreement with reduced total protein O-GlcNAc (Fig. 5A). Specifically, ChREBP O-GlcNAcylation was strongly reduced in refed LXR−/− mice compared with LXR+/+ mice as determined by 83 and 73% reduced recovery of ChREBP protein after sWGA pulldown in control and STZ-mice, respectively (Fig. 5A; band intensity quantification using Image J software). As expected and in line with our previous observations (15), LXRα was strongly recovered on sWGA beads in refed liver, whereas LXRβ was recovered under both fasted and refed conditions, suggestive of postprandial glucose-O-GlcNAc sensing via LXRα. SREBP-1 was not recovered on sWGA beads, suggesting that SREBP-1 is not a target for O-GlcNAc modification (data not shown). In ChIP assays, recruitment of ChREBP to its cognate DNA-binding site in the L-pk promoter was significantly higher in refed control compared with fasted mice (Fig. 5B), consistent with hepatic L-pk mRNA expression (Fig. 2B). Interestingly, ChREBP was not recruited to L-pk in control livers of LXR−/− mice despite high ChREBPα protein expression, suggesting that ChREBPβ is essential for ChREBP-dependent target gene activity in response to feeding in the presence of postprandial insulin signaling. Although ChREBP binding to the L-pk promoter was increased in the liver of refed STZ-treated mice (Fig. 5B), L-pk mRNA was only modestly increased (Fig. 2B), suggesting that the insulin signal is potentiating ChREBPα and/or ChREBPβ transactivation on the L-pk promoter.
Fig. 5.
LXR regulates O-GlcNAcylation of ChREBP and binding of ChREBPα to the L-PK promoter. A: Nuclear lysates were subjected to sWGA to precipitate GlcNAcylated proteins and immunoblotted with antibodies against O-GlcNAc (CTD 110.6), LXR, SREBP-1, and ChREBP (left panel). SREBP-1 was not recovered on sWGA beads (data not shown). Each lane represents independent mice from experimental groups. One representative Western blot is shown (n = 2). Specificity of the sWGA binding was confirmed by GlcNAc (0.5 M) competition (right, upper panel). sWGA from STZ-treated mice are shown in the lower panel. B: ChREBP binding to the ChORE containing region promoter of L-PK was detected by ChIP using antibodies against ChREBP or IgG as a control. Values are shown as mean ± SEM (n = 4–5). *P < 0.05, **P < 0.01 compared with fasted within experimental groups. #P < 0.05 compared with refed within control or STZ treatment.
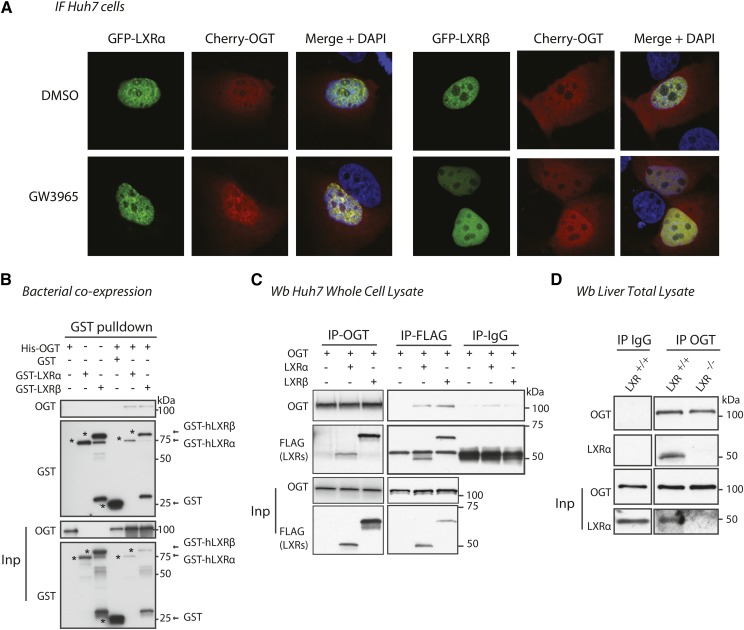
LXR interacts and colocalizes with OGT in vitro and in vivo
Prompted by our observations that LXRs are targets for O-GlcNAc modification (15) and appear to potentiate nuclear O-GlcNAc signaling without affecting expression of GFAT1/2, OGT, or OGA, we performed immunofluorescence and coimmunoprecipitation experiments to determine a possible colocalization and/or interaction of LXRs and OGT. Ectopically expressed cherry-OGT and EGFP-LXRα or EGFP-LXRβ colocalized in the nucleus of Huh7 cells treated with DMSO or GW3965, a synthetic LXR agonist (Fig. 6A). However, colocalization of LXRs and OGT was independent of ligand treatment (GW3965) or glucose concentration in the culture medium (5 mM vs. 25 mM) (data not shown). By GST pulldown experiments, we show that OGT and LXRs also interacted in a bacterial GST-LXRα/β/His-OGT coexpressed system, suggesting a direct interaction between OGT and LXRs (Fig. 6B). Finally, using a reciprocal coimmunoprecipitation strategy, we observed coimmunoprecipitation of OGT and LXRs in Huh7 cells cotransfected with HA-OGT and FLAG-LXRα or FLAG-LXRβ (Fig. 6C) and in the liver of fed wild-type mice (Fig. 6D). The interaction between OGT and LXRs in Huh7 cells was not affected by GW3965 (data not shown), suggesting that LXR interacts with OGT, indirectly or directly, via its N-terminal domain.
Fig. 6.
LXR and OGT colocalize and interact. A: Colocalization of overexpressed Cherry-OGT and GFP-LXRα or GFP-LXRβ in Huh7 cells under high glucose conditions (25 mM). Twenty-four hours after transfection, cells were treated for 2 h with DMSO or 1 μM of GW3965. B: GST pulldown of bacterial lysates coexpressing His-OGT and GST, GST-LXRα or GST-LXRβ. Precipitated proteins were subjected to SDS-PAGE and immunoblotted with antibodies against GST or OGT (AL28). C: Huh7 cells were cotransfected with HA-OGT and FLAG-LXRα or FLAG-LXRβ plasmids and incubated for 24 h under high glucose conditions. Immunoprecipitations (IP) of OGT and FLAG-LXRs were blotted with anti-FLAG and anti-OGT antibodies. Mouse IgG IP was performed as control. Input lysates (Inp, 5%) and immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-FLAG and anti-OGT antibodies. D: Liver lysates pooled from three fed wild-type (WT) and LXR−/− male mice were immunoprecipitated with anti-OGT or rabbit IgG antibody. Immunoprecipitated proteins were subject to SDS-PAGE and blotted with anti-OGT and anti-LXR antibodies.
DISCUSSION
LXRs regulate genes involved in hepatic fatty acid synthesis in response to synthetic and endogenous ligands and insulin (50). In addition to being a substrate for lipogenesis, glucose has emerged as a lipogenic signal (2) that activates LXR (13, 15), ChREBP (22), and SREBP-1c (43). In contrast to SREBP-1c, ChREBP expression and activity is seemingly unaffected by LXR deficiency in refed mice when both postprandial insulin and glucose signals are present (26). Pharmacological activation of LXR induces hepatic ChREBP expression (6, 41); however, whether LXR regulates ChREBP and de novo lipogenic gene expression in vivo under physiological conditions has not been firmly established (26, 27).
To discriminate between insulin and dietary activation of LXR in vivo, we subjected LXR+/+ and LXR−/− control and STZ-treated mice to a fasting-refeeding regime. The mice were fed a diet containing no cholesterol, omitting effects of dietary oxysterols on LXR activity. We show that LXRs regulate essential genes in the glycolytic and fatty acid synthesis pathway; Gk, L-pk Srebp-1, Chrebpα/β, Fas, Accα, Accβ, Scd1, and Gpat in response to feeding also in the absence of insulin. The refeeding-induction of these genes was blunted in LXR−/− control and hyperglycemic mice and correlated with lower protein levels of GK, L-PK, FAS, and SCD1 and serum TGs in LXR−/− mice, supporting the notion that LXRs are important glucose sensors of hepatic lipogenesis (13, 15), as glycolysis is dominant in the fed state when glucose is abundant (42). Recent ChIP-Sequencing analyses of LXR and SREBP-1 binding profiles in mouse liver and ChREBP binding in human HepG2 cells have revealed that their chromatin binding occurs at many genes involved in lipid metabolism (39, 41, 51). Moreover, mice deficient in LXR (5, 52), ChREBP (53), or SREBP-1c (54) all exhibit reduced expression of lipogenic enzymes, indicating that they each have prominent roles in the lipogenic pathway. The expression levels of lipogenic genes observed in LXR−/− mice are not expected to be mediated by SREBP-1, because its expression is dependent on LXR (5, 47). Furthermore, we performed a liftover of rat ChREBP ChIP-Sequencing profiles to mouse DNA and designed ChIP primers corresponding to ChREBP binding peaks in rat Fas, Scd1, and Acc promoters, but we were unable to detect ChREBP recruitment to the corresponding genomic regions in mouse liver extracts from control or STZ-treated wild-type or LXR−/− mice (Mandrup et al., unpublished observations). Future studies designed to delineate the relative role of LXR, SREBP-1c, and ChREBP in response to different diets and glucose levels will be important to further address their cooperative or independent regulation of lipogenic genes.
In the present study, we show that Chrebpα expression was blunted in STZ-treated LXR−/− mice, which is consistent with Chrebpα being an LXR target gene in liver (6, 13, 27). However, ChREBPα mRNA and protein levels were minimally affected by LXR deficiency in refed control mice, suggesting that ChREBPα expression becomes highly dependent on LXR in the absence of insulin. In line with these observations, we show that high glucose induced ChREBPα promoter activity through LXR in vitro, an effect that was inhibited by OGT depletion. Guinez et al. (25), reported increased ChREBP protein expression, but no increase in total Chrebp mRNA expression in liver and primary hepatocytes isolated from mice with adenoviral overexpression of OGT, arguing against a role for LXR in mediating a transcriptional glucose-response via O-GlcNAc on ChREBPα. However, these experiments were performed in the presence of insulin, and relative expression of ChREBPα and ChREBPβ isoforms were not investigated.
In agreement with earlier observations (28, 55), feeding in both mouse models strongly induced mRNA expression of Chrebpβ, which is believed to have higher activity than ChREBPα due to a lack of the N-terminal low glucose inhibitory domain and constitutive nuclear localization (28). Feeding-regulation of Chrebpβ mRNA expression was dependent on LXRs in both control and STZ-mice, which suggests a role for LXRs in regulating ChREBPα DNA binding and/or its transactivating activity on the ChREBPβ promoter under both physiological and hyperglycemic conditions because ChREBPα has been shown to regulate the ChREBPβ promoter in response to postprandial glucose levels (28).
ChREBP activity has been reported to be dependent on glucose metabolites downstream of GK (22). In our study, feeding induced ChREBPα activity (increased ChREBPα-specific target gene expression of Chrebpβ and L-pk and increased ChREBP promoter binding to the L-Pk promoter) in both control and STZ-treated mice, supporting increased GK-mediated G-6-P synthesis above fasting levels under hyperglycemic and hypoinsulinemic conditions. Furthermore, feeding-induced ChREBP activity appeared to be dependent on LXRs, as reflected by decreased mRNA expression of L-pk and Chrebpβ and no induction of ChREBP L-pk promoter binding in refed LXR−/− control mice despite high ChREBPα protein expression (low Chrebpβ expression). These findings are supported by reduced RNA polymerase II recruitment to L-pk in LXR−/− compared with LXR+/+ mice treated with pharmacological LXR agonists (41). In contrast to our results, one study reported that LXR is dispensable for ChREBP-activated DNL gene expression in high carbohydrate refed mice (26). However, in that study, the transcriptional level of Gk was unaffected by the loss of LXR, suggesting different responses due to different diets and/or LXR mouse models. We observed lower GK mRNA and protein levels in LXR−/− mice. The role of LXR in regulating GK is supported by reduced GK expression in LXR−/− mice (27, 45), reduced glucose phosphorylation to G-6-P by GK in LXRα−/− mice (49), elevated GK transcription in mice treated with pharmacological LXR agonists (56), and LXR binding to an LXRE in Gk promoter in rat liver extracts (7). Moreover, we report induced binding of LXR to a mouse LXRE in the GK promoter upon refeeding, with a similar trend in STZ-treated mice. Elevated Gk transcription in STZ-treated mice may be mediated by SREBP-1c (43), and possibly other transcription factors or coactivator complexes recruited to LXR at the GK promoter. We have previously shown that binding of LXR to the LXRE in the SREBP-1c promoter was unaffected by LXR agonist treatment, whereas RNA polymerase II was recruited to the Srebp-1c gene upon LXR activation, supporting that LXR activation of target genes may be mediated by coregulator recruitment to some genes (41).
Surprisingly, we observed a strong reduction in nuclear O-GlcNAc signaling in the liver of control and STZ-treated refed LXR−/− mice. Despite lower GK expression in LXR−/− mice, cytosolic protein O-GlcNAc levels were not reduced, suggesting that the effect of LXR on nuclear O-GlcNAc levels is not due to decreased flux through the HBP. Instead, we believe this is due to LXR-dependent expression of a protein(s) modulating HBP flux or OGT activity and/or that LXR may influence nuclear substrate specificity of OGT, which has been suggested for other OGT-interacting proteins (57). In support of the latter hypothesis, we observed that OGT and LXR interact and colocalize in the nucleus. Because OGT has been shown to be an important coactivator of nutrient-responsive gene transcription (59–61), we hypothesize a role for LXR in regulating substrate specificity of OGT on specific target gene promoters. OGT has previously been shown to target and activate ChREBPα, leading to increased ChREBP protein stabilization and binding to the L-pk promoter concomitant with increased DNL gene expression (25). This model is in agreement with our results showing reduced nuclear O-GlcNAc modified ChREBP, L-PK expression, and binding of ChREBP to the L-pk promoter in LXR−/− mice, suggesting that LXRs potentiate ChREBP activity via regulation of Gk expression and nuclear O-GlcNAc signaling.
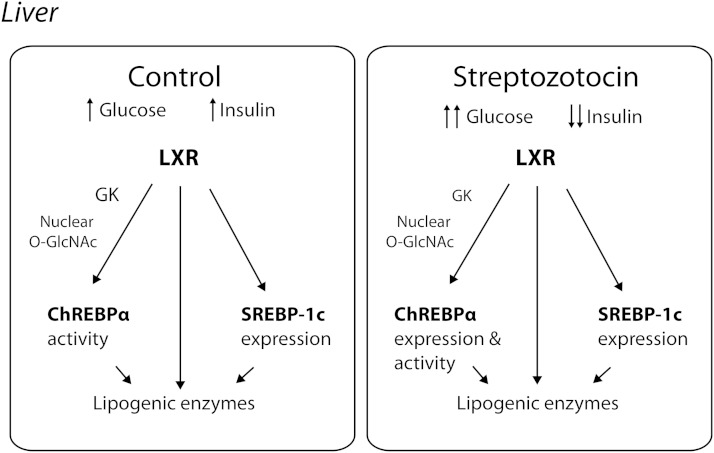
In summary, we have found that LXRs act as postprandial nutrient and glucose metabolic sensors upstream of ChREBP in vivo based on the following observations: 1) LXRs do not require insulin for activation of Gk and lipogenic gene expression in response to refeeding, specifically, both SREBP-1c and ChREBPβ expression is completely dependent on LXRs under hyperglycemic insulin-independent conditions. 2) LXRs interact with and modify the activity of nuclear OGT, a sensor of glucose metabolic flux via the HBP. 3) Transactivation of the L-PK promoter by glucose sensing ChREBPα is dependent on LXR. 4) In the absence of insulin, feeding-induced ChREBPα expression is dependent on LXR (in vivo), in part via glucose-O-GlcNAc-dependent signaling (in vitro) (summarized in Fig. 7). Further characterization of the LXR-OGT interaction and how this is related to LXR, OGT, and ChREBPα/β activity, as well as mass-spectrometry studies to identify O-GlcNAc site(s) on LXRs are currently ongoing in our laboratory. These studies will be important in determining the potential cross-talk among OGT, LXR, and ChREBPα/β and their relative activity in response to glycemia, insulin, and oxysterols.
Fig. 7.
LXR regulates ChREBPα activity in response to feeding. LXR regulates the hepatic expression of lipogenic enzymes directly and through SREBP-1c expression and ChREBPα activity in response to elevated postprandial glucose and insulin levels (Control). LXR maintains its ability to induce the expression of lipogenic target genes under hyperglycemic and hypoinsulinemic conditions (STZ), by regulating the expression of SREBP-1c and expression and activity of ChREBPα.
Supplementary Material
Acknowledgments
The authors thank Prof. Jan-Åke Gustafsson (Karolinska Insitute) for the original LXR-null mice. The authors acknowledge Christin Zwafink, Borghild Arntsen, and Sverre Holm (University of Oslo) for excellent technical assistance. They thank Knut Thomas Dalen and Elin Holter Anthonisen (University of Oslo) for helpful discussions. The authors are grateful to members of the Mandrup (University of Southern Denmark), Nebb (University of Oslo), and the Matthews (University of Toronto) laboratories for experimental input and discussions. The authors are grateful to Prof. Gerald Hart (Johns Hopkins University) for insightful discussions, providing the OGT antibody (AL28), and the pCherry-OGT and pCAG-OGT plasmids. They also thank Dr. Timothy F. Osborne (University of California) for providing the SREBP-1 antibody, Prof. Catherine Postic (Université Paris Descartes) for providing the GK antibody, Prof. Cora Weigert (University of Tübingen) for the GFAT1/2, Prof. Suzanne Walker (Harvard Medical Scool) for the His-ncOGT plasmid, and Karine Gauthier Vanacker (Institut de Génomique Fonctionnelle de Lyon) for the ChREBPα-prom plasmid.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- ChIP
- chromatin immunoprecipitation
- ChORE
- carbohydrate response element
- ChREBP
- carbohydrate responsive element-binding protein
- DNL
- de novo lipogenesis
- DOC
- deoxycholic acid
- Elovl6
- elongation of long-chain fatty acid family member 6
- G-6-P
- glucose-6-phosphate
- GFAT
- glutamine:fructose-6-phosphate amidotransferase
- GK
- glucokinase
- GlcNAc
- N-acetylglucosamine
- GPAT
- glycerol-3-phophate acyltransferase
- GST
- glutathione-S-transferase
- HBP
- hexosamine biosynthetic pathway
- L-PK
- liver pyruvate kinase
- LXR
- liver X receptor
- LXRE
- liver X receptor response element
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- OGA
- O-linked β-N-acetylglucosaminase
- OGT
- O-linked β-N-acetylglucosamine transferase
- RXR
- retinoid X receptor
- SCD
- stearoyl CoA desaturase
- SREBP
- sterol regulatory element-binding protein
- STZ
- streptozotocin
- sWGA
- succinylated wheat germ agglutinin
- Tbp
- tata-binding protein
- UDP-GlcNAc
- UDP-N-acetylglucosamine
This work was funded by grants to H.I.N. and L.M.G-W. from the University of Oslo, the Johan Throne Holst Foundation, the Novo Nordisk Foundation, Anders Jahre Foundation, the Research Council of Norway, Nordic Academy for Advanced Study (NorFA), KaroBio AB, and the Swedish Science Council; grants to S.M. from the Danish Independent Research Council, Natural Science, and the Danish Independent Research Council, and Health Science; and a grant from the Canadian Institutes of Health Research (MOP-125919) to J.M. No potential conflicts of interest relevant to this article were reported.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
- 1.Strable M. S., Ntambi J. M. 2010. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45: 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oosterveer M. H., Schoonjans K. 2014. Hepatic glucose sensing and integrative pathways in the liver. Cell. Mol. Life Sci. 71: 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. 1999. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA. 96: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkin A. C., Tontonoz P. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha J. Y., Repa J. J. 2007. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 282: 743–751. [DOI] [PubMed] [Google Scholar]
- 7.Kim T. H., Kim H., Park J. M., Im S. S., Bae J. S., Kim M. Y., Yoon H. G., Cha J. Y., Kim K. S., Ahn Y. H. 2009. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J. Biol. Chem. 284: 15071–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talukdar S., Hillgartner F. B. 2006. The mechanism mediating the activation of acetyl-coenzyme A carboxylase-alpha gene transcription by the liver X receptor agonist T0-901317. J. Lipid Res. 47: 2451–2461. [DOI] [PubMed] [Google Scholar]
- 9.Joseph S. B., Laffitte B. A., Patel P. H., Watson M. A., Matsukuma K. E., Walczak R., Collins J. L., Osborne T. F., Tontonoz P. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277: 11019–11025. [DOI] [PubMed] [Google Scholar]
- 10.Chu K., Miyazaki M., Man W. C., Ntambi J. M. 2006. Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol. Cell. Biol. 26: 6786–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobin K. A., Ulven S. M., Schuster G. U., Steineger H. H., Andresen S. M., Gustafsson J. A., Nebb H. I. 2002. Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J. Biol. Chem. 277: 10691–10697. [DOI] [PubMed] [Google Scholar]
- 12.Chen G., Liang G., Ou J., Goldstein J. L., Brown M. S. 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. USA. 101: 11245–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitro N., Mak P. A., Vargas L., Godio C., Hampton E., Molteni V., Kreusch A., Saez E. 2007. The nuclear receptor LXR is a glucose sensor. Nature. 445: 219–223. [DOI] [PubMed] [Google Scholar]
- 14.Lazar M. A., Willson T. M. 2007. Sweet dreams for LXR. Cell Metab. 5: 159–161. [DOI] [PubMed] [Google Scholar]
- 15.Anthonisen E. H., Berven L., Holm S., Nygard M., Nebb H. I., Gronning-Wang L. M. 2010. Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J. Biol. Chem. 285: 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J., Hart G. W. 2013. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics. 10: 365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellen K. E., Lu C., Mancuso A., Lemons J. M., Ryczko M., Dennis J. W., Rabinowitz J. D., Coller H. A., Thompson C. B. 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 24: 2784–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vocadlo D. J. 2012. O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol. 16: 488–497. [DOI] [PubMed] [Google Scholar]
- 19.Ruan H. B., Singh J. P., Li M. D., Wu J., Yang X. 2013. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 24: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondoux M. A., Love D. C., Ghosh S. K., Fukushige T., Bond M., Weerasinghe G. R., Hanover J. A., Krause M. W. 2011. O-linked-N-acetylglucosamine cycling and insulin signaling are required for the glucose stress response in Caenorhabditis elegans. Genetics. 188: 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., et al. 2008. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 451: 964–969. [DOI] [PubMed] [Google Scholar]
- 22.Poupeau A., Postic C. 2011. Cross-regulation of hepatic glucose metabolism via ChREBP and nuclear receptors. Biochim. Biophys. Acta. 1812: 995–1006. [DOI] [PubMed] [Google Scholar]
- 23.Kabashima T., Kawaguchi T., Wadzinski B. E., Uyeda K. 2003. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. USA. 100: 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsatsos N. G., Towle H. C. 2006. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem. Biophys. Res. Commun. 340: 449–456. [DOI] [PubMed] [Google Scholar]
- 25.Guinez C., Filhoulaud G., Rayah-Benhamed F., Marmier S., Dubuquoy C., Dentin R., Moldes M., Burnol A. F., Yang X., Lefebvre T., et al. 2011. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes. 60: 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denechaud P. D., Bossard P., Lobaccaro J. M., Millatt L., Staels B., Girard J., Postic C. 2008. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J. Clin. Invest. 118: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalaany N. Y., Gauthier K. C., Zavacki A. M., Mammen P. P. A., Kitazume T., Peterson J. A., Horton J. D., Garry D. J., Bianco A. C., Mangelsdorf D. J. 2005. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1: 231–244. [DOI] [PubMed] [Google Scholar]
- 28.Herman M. A., Peroni O. D., Villoria J., Schon M. R., Abumrad N. A., Bluher M., Klein S., Kahn B. B. 2012. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 484: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weedon-Fekjaer M. S., Dalen K. T., Solaas K., Staff A. C., Duttaroy A. K., Nebb H. I. 2010. Activation of LXR increases acyl-CoA synthetase activity through direct regulation of ACSL3 in human placental trophoblast cells. J. Lipid Res. 51: 1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross B. J., Kraybill B. C., Walker S. 2005. Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 127: 14588–14589. [DOI] [PubMed] [Google Scholar]
- 31.Gauthier K., Billon C., Bissler M., Beylot M., Lobaccaro J. M., Vanacker J. M., Samarut J. 2010. Thyroid hormone receptor beta (TRbeta) and liver X receptor (LXR) regulate carbohydrate-response element-binding protein (ChREBP) expression in a tissue-selective manner. J. Biol. Chem. 285: 28156–28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti S., Schuster G., Parini P., Feltkamp D., Diczfalusy U., Rudling M., Angelin B., Bjorkhem I., Pettersson S., Gustafsson J. A. 2001. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRbeta-deficient mice. J. Clin. Invest. 107: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster G. U., Parini P., Wang L., Alberti S., Steffensen K. R., Hansson G. K., Angelin B., Gustafsson J. A. 2002. Accumulation of foam cells in liver X receptor-deficient mice. Circulation. 106: 1147–1153. [DOI] [PubMed] [Google Scholar]
- 34.Bindesbøll C., Berg O., Arntsen B., Nebb H. I., Dalen K. T. 2013. Fatty acids regulate perilipin5 in muscle by activating PPARdelta. J. Lipid Res. 54: 1949–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsson T., Venteclef N., Toresson G., Damdimopoulos A. E., Ehrlund A., Lou X., Sanyal S., Steffensen K. R., Gustafsson J. A., Treuter E. 2009. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol. Cell. 34: 510–518. [DOI] [PubMed] [Google Scholar]
- 36.Toresson G., Schuster G. U., Steffensen K. R., Bengtsson M., Ljunggren J., Dahlman-Wright K., Gustafsson J. A. 2004. Purification of functional full-length liver X receptor beta produced in Escherichia coli. Protein Expr. Purif. 35: 190–198. [DOI] [PubMed] [Google Scholar]
- 37.Nerlich A. G., Sauer U., Kolm-Litty V., Wagner E., Koch M., Schleicher E. D. 1998. Expression of glutamine:fructose-6-phosphate amidotransferase in human tissues: evidence for high variability and distinct regulation in diabetes. Diabetes. 47: 170–178. [DOI] [PubMed] [Google Scholar]
- 38.Iyer S. P., Akimoto Y., Hart G. W. 2003. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J. Biol. Chem. 278: 5399–5409. [DOI] [PubMed] [Google Scholar]
- 39.Seo Y. K., Chong H. K., Infante A. M., Im S. S., Xie X., Osborne T. F. 2009. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc. Natl. Acad. Sci. USA. 106: 13765–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dentin R., Benhamed F., Pegorier J. P., Foufelle F., Viollet B., Vaulont S., Girard J., Postic C. 2005. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Invest. 115: 2843–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boergesen M., Pedersen T. A., Gross B., van Heeringen S. J., Hagenbeek D., Bindesboll C., Caron S., Lalloyer F., Steffensen K. R., Nebb H. I., et al. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32: 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rui L. 2014. Energy metabolism in the liver. Compr. Physiol. 4: 177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuzaka T., Shimano H., Yahagi N., Amemiya-Kudo M., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Tomita S., Sekiya M., et al. 2004. Insulin-independent induction of sterol regulatory element-binding protein-1c expression in the livers of streptozotocin-treated mice. Diabetes. 53: 560–569. [DOI] [PubMed] [Google Scholar]
- 44.Efanov A. M., Sewing S., Bokvist K., Gromada J. 2004. Liver X receptor activation stimulates insulin secretion via modulation of glucose and lipid metabolism in pancreatic beta-cells. Diabetes. 53(Suppl 3): S75–S78. [DOI] [PubMed] [Google Scholar]
- 45.Beaven S. W., Matveyenko A., Wroblewski K., Chao L., Wilpitz D., Hsu T. W., Lentz J., Drew B., Hevener A. L., Tontonoz P. 2013. Reciprocal regulation of hepatic and adipose lipogenesis by liver X receptors in obesity and insulin resistance. Cell Metab. 18: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA. 98: 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oosterveer M. H., van Dijk T. H., Grefhorst A., Bloks V. W., Havinga R., Kuipers F., Reijngoud D. J. 2008. Lxralpha deficiency hampers the hepatic adaptive response to fasting in mice. J. Biol. Chem. 283: 25437–25445. [DOI] [PubMed] [Google Scholar]
- 49.Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., Hart G. W. 2008. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283: 16283–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grønning-Wang L. M., Bindesbøll C., Nebb H. I. 2013. The role of liver X receptor in hepatic de novo lipogenesis and cross-talk with insulin and glucose signaling. In InTech. R. V. Baez, editor. Accessed January 23, 2013, at http://www.intechopen.com/books/lipid-metabolism/the-role-of-liver-x-receptor-in-hepatic-de-novo-lipogenesis-and-cross-talk-with-insulin-and-glucose-. [Google Scholar]
- 51.Jeong Y. S., Kim D., Lee Y. S., Kim H. J., Han J. Y., Im S. S., Chong H. K., Kwon J. K., Cho Y. H., Kim W. K., et al. 2011. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE. 6: e22544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steffensen K. R., Gustafsson J. A. 2004. Putative metabolic effects of the liver X receptor (LXR). Diabetes. 53(Suppl 1): S36–S42. [DOI] [PubMed] [Google Scholar]
- 53.Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA. 101: 7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. 2002. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 277: 9520–9528. [DOI] [PubMed] [Google Scholar]
- 55.Eissing L., Scherer T., Todter K., Knippschild U., Greve J. W., Buurman W. A., Pinnschmidt H. O., Rensen S. S., Wolf A. M., Bartelt A., et al. 2013. De novo lipogenesis in human fat and liver is linked to ChREBP-beta and metabolic health. Nat. Commun. 4: 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laffitte B. A., Chao L. C., Li J., Walczak R., Hummasti S., Joseph S. B., Castrillo A., Wilpitz D. C., Mangelsdorf D. J., Collins J. L., et al. 2003. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA. 100: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. 2008. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283: 33935–33941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bond M. R., Hanover J. A. 2013. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33: 205–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis B. A., Hanover J. A. 2014. O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289: 34440–34448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harwood K. R., Hanover J. A. 2014. Nutrient-driven O-GlcNAc cycling - think globally but act locally. J. Cell Sci. 127: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.