Abstract
The transcription factors hepatic nuclear factor (HNF)1α and HNF1β can bind to the HNF1 site on the proprotein convertase subtilisin/kexin type 9 (PCSK9) promoter to activate transcription in HepG2 cells. However, it is unknown whether one or both HNF1 factors are obligatory for transactivating hepatic PCSK9 gene expression in vivo. We developed shRNA adenoviral constructs (Ad-shHNF1α and Ad-shHNF1β) to examine the effects of knockdown of HNF1α or HNF1β on PCSK9 expression and its consequent impact on LDL receptor (LDLR) protein levels in cultured hepatic cells and liver tissue. We demonstrated that infection with Ad-shHNF1α, but not Ad-shHNF1β, markedly reduced PCSK9 mRNA expression in HepG2 cells with a concomitant increase in LDLR protein abundance. Injecting Ad-shHNF1α in mice fed a normal diet significantly (∼50%) reduced liver mRNA expression and serum concentration of PCSK9 with a concomitant increase (∼1.9-fold) in hepatic LDLR protein abundance. Furthermore, we observed a modest but significant reduction in circulating LDL cholesterol after knockdown of HNF1α in these normolipidemic mice. Consistent with the observation that knockdown of HNF1β did not affect PCSK9 mRNA or protein expression in cultured hepatic cells, Ad-shHNF1β infection in mice resulted in no change in the hepatic mRNA expression or serum content of PCSK9. Altogether, our study demonstrates that HNF1α, but not HNF1β, is the primary positive regulator of PCSK9 transcription in mouse liver.
Keywords: hepatic nuclear factor 1α, hepatic nuclear factor 1β, proprotein convertase subtilisin/kexin type 9, low density lipoprotein receptor, low density lipoprotein cholesterol
Elevated plasma cholesterol, specifically LDL cholesterol (LDL-C), is a significant risk factor for coronary heart disease and associated comorbidities (1). LDL-C is cleared from the circulation after binding to the extracellular domain of the transmembrane protein, LDL receptor (LDLR), causing its internalization in clathrin-coated vesicles (2, 3). Following processing in the endosome, where the lipoprotein particle dissociates in a pH-dependent fashion, LDLR is either recycled to the cell membrane or is targeted for lysosomal degradation (3). The endosomal sorting of LDLR is regulated by the protein, proprotein convertase subtilisin/kexin type 9 (PCSK9) (4, 5). PCSK9 is a liver-derived plasma protease which is initially synthesized as a ∼74 kDa precursor protein that undergoes autocatalytic cleavage of the prodomain to yield a ∼60 kDa mature form (6, 7). Circulating PCSK9 binds to the extracellular epidermal growth factor-like repeat A (EGF-A) domain of LDLR causing its internalization (8). The binding between PCSK9 and LDLR is enhanced at the acidic pH prevalent in the endosome-lysosomal compartment, which inhibits recycling of LDLR to the cell membrane and instead targets the PCSK9-LDLR complex for degradation in the lysosome (9). In human subjects, both gain- and loss-of-function mutations of PCSK9 have been described that result in chronically elevated (hypercholesterolemia) or suppressed (hypocholesterolemia) levels of circulating LDL-C, respectively (10, 11).
Although the liver is the primary source of secreted PCSK9 and is the most important organ for its function, PCSK9 is expressed in several tissues including intestine, kidney, and muscle (6). Analyses of the ∼650 bp proximal region of the PCSK9 promoter have thus far revealed three promoter elements that can modulate PCSK9 expression. Farthest upstream, a Sp1 site was implicated to play a modest role in PCSK9 transcription (12). A sterol-regulatory element (SRE), which forms the target site for sterol-regulatory element binding proteins (SREBPs), is present ∼330 bp upstream of the translation start site and functions as an important cis-element regulating PCSK9 transcription (13, 14). Cholesterol-lowering drugs, statins, inhibit hydroxymethylglutaryl-CoA, the rate-limiting enzyme in cholesterol biosynthesis and activate the SREBP pathway. Although this activation of SREBP results in induction of LDLR mRNA transcription, SREBP2 also coordinately induces PCSK9 via the SRE, thus diminishing the beneficial effects of statin treatment (15–17). Our laboratory has previously described a highly conserved hepatic nuclear factor (HNF)1 binding site present ∼380 bp upstream of the translation start site to be a critical regulator of PCSK9 expression. Additionally, in vitro knockdown of HNF1α represses PCSK9 protein levels in HepG2 cells (18).
HNF1α and HNF1β are homeobox proteins that belong to the HNF1 subfamily of transcription factors (19). HNF1α and HNF1β form homodimers that bind to GTTAATNATTAAC sites on the promoter of their target genes (20, 21). Although initially thought to be restricted to the liver, HNF1α and HNF1β have been reported to be expressed in a variety of other tissues, like pancreas, kidney, and intestine, where they play important roles in tissue development and differentiation (19, 22). While loss of HNF1β is developmentally lethal, germline deletion of HNF1α results in hepatic, pancreatic, and renal defects (22–25). A predominant phenotype of mono-allelic HNF1α and HNF1β mutations is the development of insulin-independent mature onset of diabetes (MODY type 3 and type 5, respectively) due to defective β-cell function (26, 27). Additionally, loss of HNF1α has also been associated with increased incidence of hepatocellular adenomas, as well as defective bile acid and cholesterol metabolism (28, 29).
HNF1α and HNF1β are paralogs with essentially identical in vitro binding properties (20). Although HNF1β is expressed at very low levels in adult hepatocytes, a recent report implicated that its expression is induced in HNF1α−/− livers (30). In vivo binding studies further indicated that loss of HNF1α triggers significantly higher HNF1β binding at several HNF1α target loci, including the PCSK9 promoter region (30). However, there is currently limited in vivo evidence linking the binding of these transcription factors on the PCSK9 promoter to the individual contributions of HNF1α and HNF1β in the regulation of PCSK9 expression in liver tissue, as well as on circulating cholesterol levels. In this current study, we thus investigated the effects that transient knockdown of HNF1α and HNF1β in mouse livers had on secreted PCSK9 and on hepatic LDLR expression under normal diet-fed conditions. Our results demonstrate for the first time that in vivo liver-specific knockdown of HNF1α, but not HNF1β, significantly decreases serum PCSK9 expression. This decrease in circulating PCSK9 is accompanied by increased expression of hepatic LDLR protein. Finally, we show that knockdown of HNF1α is associated with a modest reduction in circulating LDL-C in normolipidemic mice fed a normal chow diet.
MATERIALS AND METHODS
Animals
Experimental subjects were male C57BL/6J mice between 8 and 10 weeks of age (Jackson Laboratories). All mice were housed in the Veterans Affairs Palo Alto Health Care System Veterinary Medical Unit in a 12:12 light:dark cycle (lights on at 0600 hours) with a regular rodent chow diet and water provided ad libitum. All experimental analyses were conducted in accordance with animal use protocols approved by the Institutional Animal Care and Use Committee at the Veterans Affairs Palo Alto Health Care System. Starting 2 days prior to adenoviral injections, mice were housed three to four per cage and health parameters including body weight and food intake were monitored throughout the duration of the experiment. Food was removed from cages at 0800 hours and blood was collected from the retroorbital plexus, under anesthesia, at 1200 hours before injection (pre-treatment) and on days 4, 7, and 10 postinjection. On the day of the injection (day 0), all animals were retro-orbitally injected with 4–6 × 109 PFUs of virus particles. Animals were euthanized on day 10, and blood and liver were harvested for further analysis.
shRNA design and adenoviral vectors
HNF1α and HNF1β were transiently knocked down using shRNA-expressing plasmids and adenoviral vectors. A sequence spanning nucleotides 1004 to 1024 of the mouse HNF1α mRNA was selected as the target for RNAi. This sequence is identical among human, mouse, and rat species. Furthermore, it translates to amino acid positions 261 to 268, which form part of the homeodomain of HNF1α protein. For HNF1β, a sequence spanning nucleotides 1424 to 1444 of the mouse HNF1β mRNA transcript variant 1 was targeted for shRNA-mediated knockdown. This sequence is also identical among human, mouse, and rat species and forms part of the C-terminal domain, corresponding to amino acids 414 to 420 of the HNF1β protein sequence. To construct Ad-shHNF1α/Ad-shHNF1β adenoviral vectors, a U6 promoter-based shuttle vector (pSH-HNF1α or pSH-HNF1β) that expresses HNF1α or HNF1β shRNA was generated using Invitrogen’s BLOCK-iT™ U6 RNAi entry vector kit following the manufacturer’s instructions. The sequence identity and orientation of shRNA in the shuttle vector was confirmed by sequencing. The plasmid was then recombined with pAD/BLOCK-iT DEST vector to generate Ad-shHNF1α or Ad-shHNF1β viral vectors and transduced into HEK293A cells. The crude viral stocks were further amplified and purified according to the manufacturer’s guidelines. To account for effects of the RNAi expression vectors and adenoviral infections, Ad-shLacZ encoding a shRNA for the LacZ gene was used as control. Large scale virus preparation and purification for animal injections was performed at Vector Biolabs.
Cell culture and transfections
HEK293A cells were cotransfected with FLAG-tagged HNF1α or HNF1β and pENTRY plasmids for shLacZ, shHNF1α, or shHNF1β using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s guidelines. After 48 h posttransfection, cells were harvested and proteins were extracted in RIPA buffer [50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS (pH 7.4)] supplemented with 1 mM PMSF and protease inhibitor cocktail (Roche). The expression of HNF1α and HNF1β was estimated by immunoblotting with an anti-FLAG antibody (Sigma-Aldrich) as described later below. HepG2 cells were cultured in MEM supplemented with 10% FBS. The cells were seeded in 24- or 6-well plates at a density of 0.5 × 106 cells/ml and transduced with Ad-shLacZ, Ad-shHNF1α, or Ad-shHNF1β adenovirus particles at a multiplicity of infection (MOI) of 30 or 60. Following overnight incubation after adenoviral transduction, cells were washed and fresh growth medium was replaced. Cells were harvested in RIPA buffer or RNA lysis buffer 72 h later and stored at −80°C until further processing. Mouse primary hepatocytes were purchased from the University of California, San Francisco, Liver Center where the hepatocytes were isolated by perfusion from 6- to 8-week-old male C57BL/6J mice. The primary hepatocytes were initially cultured in plating medium (Williams E supplemented with hepatocyte plating supplement pack, Invitrogen) for 4 h, and later transferred to incubation medium (William E supplemented with hepatocyte maintenance supplement pack, Invitrogen) for the duration of the experiment.
Luciferase reporter assay
pLightSwitch_Promo vector containing the 1,192 bp promoter region of human LDLR was purchased from Switchgear Genomics. The 1,192 bp fragment was then sub-cloned into pGL3 basic promoter (Promega) and denoted pLDLR1192. The PCSK9-D4 promoter has been previously described (12, 18). HepG2 cells were cotransfected with pGL3-basic, pLDLR1192, or pPCSK9-D4 promoters and pENTR vectors for shLacZ, shHNF1α, or shHNF1β using FuGene 6 transfection reagent (Promega) according to manufacturer’s guidelines. To control for differences in transfection efficiency or efficacy across samples, pTK plasmid encoding Renilla luciferase gene was also transfected and was used to normalize firefly luciferase signal across all samples. Forty-eight hours after transfection, cells were harvested in 1× passive lysis buffer (Promega) and dual-luminescence signal was measured in all samples using the dual-luciferase reporter assay system (Promega).
Serum collection, PCSK9 ELISA, total cholesterol, and LDL-C estimation
Following collection using heparinized capillaries, blood samples (100 ul per animal) were allowed to coagulate at room temperature. The samples were centrifuged for 10 min at room temperature and separated serum was transferred to fresh tubes and frozen at −80°C. Levels of serum PCSK9 were measured using the Mouse Proprotein Convertase 9/PCSK9 Quantikine ELISA kit (R&D Systems) according to manufacturer’s guidelines. Briefly, serum samples were diluted 1:200 in Calibrator diluent and allowed to bind for 2 h onto microplate wells that were precoated with the capture antibody. Samples were then sequentially incubated with PCSK9 conjugate followed by the PCSK9 substrate solution with extensive intermittent washes between each step. The amount of PCSK9 in serum was estimated colorimetrically using a standard microplate reader (MDS Analytical Technologies). Total cholesterol and LDL-C were measured from serum samples using respective kits from StanBio Laboratories according to the manufacturer’s guidelines.
RNA isolation and real-time PCR
Total RNA was isolated from 20 mg of flash-frozen mouse liver tissue samples using the RNeasy PLUS mini kit (Qiagen). After RNA integrity was confirmed by gel electrophoresis, 1.5 μg of total RNA was reverse transcribed by random priming using the high-capacity cDNA reverse transcription kit (Life Technologies) according to the manufacturer’s guidelines. Real-time PCR was then performed in an ABI 7900HT sequence detection system using SyBr Green PCR Master Mix (Life Technologies) and PCR primers specific for each gene being amplified (Table 1). With duplicate measurements from each cDNA sample, the data were analyzed using the ΔΔCT method and relative expression of target mRNAs was normalized to that of GAPDH in each sample. For analyses from cultured HepG2 cells and mouse primary hepatocytes, the same procedure was followed with the exception that total RNA was extracted using Quick-RNA miniprep kit (Zymo Research) and each cDNA was measured in triplicate with species-specific primers.
TABLE 1.
List of primers used for real-time PCR
| 5′–3′ | |
| Human | |
| HNF1α-forward | TGGCGCAGCAGTTCACCCAT |
| HNF1α-reverse | TGAAACGGTTCCTCCGCCCC |
| HNF1β-forward | GGCATTCCGGCAAAAGCTGGC |
| HNF1β-reverse | GGCTGTAGCGCACTCCTGACA |
| PCSK9-forward | AGGGGAGGACATCATTGGTG |
| PCSK9-reverse | CAGGTTGGGGGTCAGTACC |
| APOC3-forward | GAAGCACGCCACCAAGACCG |
| APOC3-reverse | AAGCCATCGGTCACCCAGCCC |
| PCK1-forward | GAGCTCAGAGGATGGGGAACC |
| PCK1-reverse | GGGAACACCTTCCGGAGACT |
| LIPC-forward | TGTGCAACTCTCTCGAAGCC |
| LIPC-reverse | ATCCAGCCCTGTGATTCTCC |
| NR1D1-forward | GGCGATCGCAACCTCTAGTT |
| NR1D1-reverse | ATGACGCCACCTGTGTTGTT |
| GAPDH-forward | ATGGGGAAGGTGAAGGTCG |
| GAPDH-reverse | GGGGTCATTGATGGCAACAATA |
| Mouse | |
| HNF1α-forward | CGCTCTGTACACCTGGTACG |
| HNF1α-reverse | CTTAGTTGGCAGCTCATCGC |
| HNF1β-forward | AGAGAGGGAGGCCTTAGTGG |
| HNF1β-reverse | CCTCCGTGACCAAGTTGGAG |
| PCSK9-forward | TTGCAGCAGCTGGGAACTT |
| PCSK9-reverse | CCGACTGTGATGACCTCTGGA |
| LDLR-forward | TCGTAGTGGACCCTGTGCAT |
| LDLR-reverse | GGAAAGATCTAGTGTGATGCCATT |
| APOC3-forward | TACAGGGCTACATGGAACAAGC |
| APOC3-reverse | CAGGGATCTGAAGTGATTGTCC |
| PCK1-forward | TCTTTGGTGGCCGTAGACCT |
| PCK1-reverse | GATGATCTTGCCCTTGTGTTCTG |
| HNF4α-forward | GCCTTCTGCGAACTCCTTCT |
| HNF4α-reverse | TCATTGCCTAGGAGCAGCAC |
| FABP1-forward | ACCATGAACTTCTCCGGCAA |
| FABP1-reverse | GACACCCCCTTGATGTCCTT |
| FABP5-forward | ACGGCTTTGAGGAGTACATGA |
| FABP5-reverse | GTTCATGACACACTCCACGATCA |
| GAPDH-forward | ATGGTGAAGGTCGGTGTGAA |
| GAPDH-reverse | ACTGGAACATGTAGACCATGTAGT |
Western blotting from mouse liver tissue
Total protein was extracted from flash-frozen mouse liver samples by homogenizing ∼50 mg of tissue in RIPA buffer supplemented with 1 mM PMSF and protease inhibitor cocktail (Roche). Protein content was quantified using BCA protein assay reagent (Pierce) and ∼75–100 μg of protein from individual samples was resolved by SDS-PAGE. Following transfer onto nitrocellulose membranes, LDLR, PCSK9, HNF1α, and β-actin proteins were detected by immunoblotting using rabbit anti-LDLR (Biovision), anti-PCSK9 (R&D Systems), goat anti-HNF1α (sc-6547, Santa Cruz Biotechnology), and monoclonal anti-β-actin (Clone AC-15, Sigma) antibodies. Immunoreactive bands of predicted molecular mass were visualized using SuperSignal West Femto chemiluminescent substrate (Pierce) and FluorChem E Western blot imaging system (Protein Simple). Background subtracted band densities were quantified using Alpha View SA imaging software (Protein Simple). Signal intensities for β-actin were used as normalization to control for protein loading differences between individual samples.
Statistical analysis
All values are expressed as mean ± SEM. For multiple group comparisons, one-way ANOVA with Student’s-Keuls pairwise post hoc test was performed using GraphPad Prism 5 software. For two group data analysis, unpaired two-tailed Student’s t-test was applied. A P value of <0.05 was considered statistically significant for all analyses.
RESULTS
HNF1α, but not HNF1β, regulates PCSK9 expression
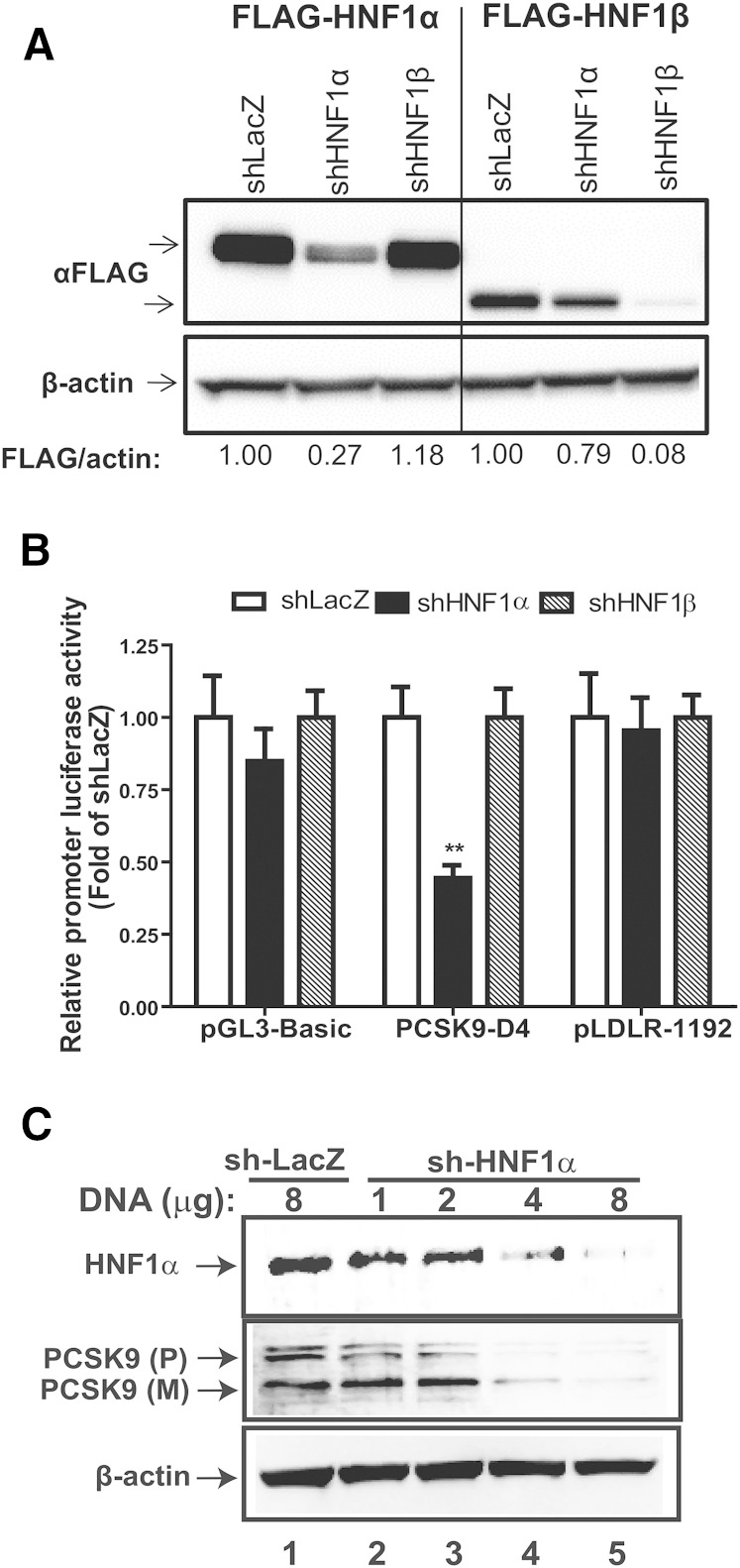
To better delineate the liver-specific roles of the HNF1 sub-family of transcription factors, we designed shRNAs against HNF1α and HNF1β and first tested them in cultured cells. For validation of knockdown and specificity, shRNA shuttle vectors were cotransfected into HEK293 cells with FLAG-tagged HNF1α or HNF1β expression plasmids. Western blot analysis with anti-FLAG antibody confirmed that, compared with cells cotransfected with a control (shLacZ) shuttle vector, both HNF1 shRNA constructs specifically reduced their target protein abundance by ∼70% and ∼90% for HNF1α and HNF1β, respectively (Fig. 1A).
Fig. 1.
HNF1α, but not HNF1β, regulates PCSK9 expression. A: HEK293 cells were transfected with shLacZ, shHNF1α, or shHNF1β shuttle vector along with FLAG-tagged HNF1α or HNF1β expression vector and 48 h later extracted protein was probed with anti-FLAG and anti-β-actin antibodies. B: Relative luciferase activities from HepG2 cells cotransfected with shLacZ, shHNF1α, or shHNF1β shuttle vector along with either pGL3-basic, pPCSK9-D4, or pLDLR-1192 promoter luciferase plasmids. Data represent summarized results (mean ± SEM) of four to six replicates per treatment and are expressed as the ratio of firefly/Renilla activity from each sample where the relative luciferase activity for shLacZ transfected cells is set to 1. **P < 0.01 compared with shLacZ cotransfected samples. C: HepG2 cells were transfected with shLacZ or shHNF1α shuttle vectors at the indicated DNA amounts. Total protein from transfected cells was extracted in RIPA buffer supplemented with PMSF, protease inhibitor cocktail, and phosphatase inhibitor and was separated by SDS-PAGE. Western blotting was performed for detection of HNF1α, PCSK9 and β-actin.
The PCSK9 promoter contains a HNF1 binding site and is thus considered a target of HNF1α/1β regulated activation. To confirm whether knockdown of HNF1α or HNF1β affected PCSK9 promoter activity, we utilized the D4 promoter construct which contains the proximal PCSK9 promoter region (−440 to −94) including the Sp1, HNF1, and SRE sites (12, 18). We cotransfected HepG2 cells with the D4 plasmid along with shuttle vectors for shHNF1α or shHNF1β and monitored luciferase reported promoter activity. Compared with the control (shLacZ), knockdown of HNF1α significantly (P < 0.01) reduced PCSK9 promoter activity by ∼55% (Fig. 1B). However, knockdown of HNF1β did not produce any reduction in the PCSK9 D4 promoter activity (Fig. 1B). In addition to PCSK9, we also tested whether knockdown of HNF1α or HNF1β had any effects on LDLR transcription. We cotransfected HepG2 cells with either the control (shLacZ), shHNF1α, or shHNF1β shuttle vectors along with the LDLR1192 promoter plasmid containing the proximal −1192 bp region of the LDLR promoter. Contrary to the effects observed on PCSK9 D4 plasmid, knockdown of either HNF1α or HNF1β did not produce any significant change in LDLR1192 activity, confirming that LDLR transcription is not regulated by HNF1α/1β (Fig. 1B). Western blotting analysis of HNF1α and PCSK9 in HepG2 cells transfected with either the shLacZ or shHNF1α shuttle vectors was next used to further validate the luciferase reporter assay results and confirmed that knockdown of HNF1α resulted in dose-dependent decrease in PCSK9 protein levels (Fig. 1C).
Knockdown of HNF1α, but not HNF1β, results in elevated LDLR protein expression in cultured hepatic cells
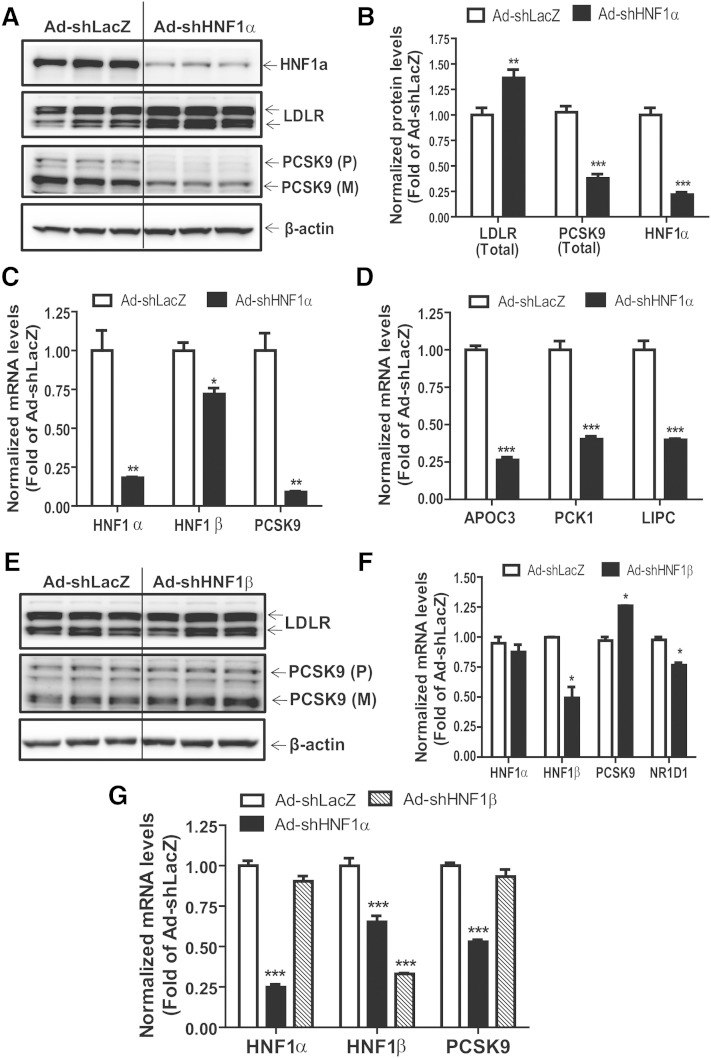
After subcloning into destination vectors and packaging in adenovirus particles, HepG2 cells were transduced with either control (Ad-shLacZ), shHNF1α (Ad-shHNF1α), or shHNF1β (Ad-shHNF1β) expressing virions. We first investigated the effects that HNF1α knockdown had on protein expression of candidate genes in HepG2 cells. Compared with Ad-shLacZ-transduced controls, we observed a robust knockdown of ∼80% (P < 0.001) in HNF1α protein expression from HepG2 cells transduced with the Ad-shHNF1α adenovirus (Fig. 2A, B). Western blot analysis confirmed that knockdown of HNF1α significantly reduced PCSK9 protein expression by ∼63% (P < 0.001) in the transduced cells (Fig. 2A, B). Because PCSK9 is responsible for LDLR protein degradation without affecting its steady state mRNA levels, we also assessed the effects of HNF1α knockdown on LDLR and observed a substantial ∼30% (P < 0.01) increase in LDLR protein expression in Ad-shHNF1α-transduced cells (Fig. 2A, B). To further confirm that the reduction in PCSK9 protein was a direct outcome of the shRNA, we profiled mRNA expression of HNF1α and corresponding target genes. Real-time PCR from cells collected 72 h posttransduction revealed significant (P < 0.01) knockdown of HNF1α mRNA by ∼80% in cells transduced with Ad-shHNF1α virus compared with those transduced with the control (Fig. 2C). Importantly, knockdown of HNF1α resulted in a significant (P < 0.01) ∼90% reduction in PCSK9 mRNA expression compared with that observed in the control-transduced HepG2 cells (Fig. 2C). Profiling of some canonical HNF1α target genes, APOC3, PCK1, and LIPC, further validated functional knockdown of HNF1α (Fig. 2D).
Fig. 2.
Knockdown of HNF1α results in elevated LDLR protein levels. HepG2 or mouse primary hepatocyte cells were seeded in 6-well or 24-well cell culture plates. After 24 h, cells were transduced with Ad-shLacZ (30 or 60 MOI), Ad-shHNF1α (30 MOI), or Ad-shHNF1β (60 MOI) adenoviruses. Fresh cell culture medium was replaced 6–8 h later and cells were incubated for a further 60–66 h. Total RNA and protein were collected from transduced cells for real-time PCR and Western blotting analyses. A, E: Western blotting with antibodies to LDLR, HNF1α, PCSK9, and β-actin was conducted by analyzing individual homogenates from Ad-shHNF1α- (A) or Ad-shHNF1β-transduced (E) HepG2 cells. B: Quantification of LDLR, PCSK9, and HNF1α from Ad-shLacZ- and Ad-shHNF1α-transduced HeG2 cells. C, D, F, G: Quantitative real-time PCR was conducted to determine the relative expression levels of mRNAs of interest from cultured HepG2 cells (C, D, F) or mouse primary hepatocytes (G). Data represent summarized result (mean ± SEM) of three independent replicates per treatment after normalization with GAPDH. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with Ad-shLacZ transduced control cells which was set at 1.
We also determined protein and mRNA expression of several genes from Ad-shHNF1β-transduced cells. While the Ad-shHNF1β adenovirus significantly knocked down HNF1β mRNA expression by ∼50% (P < 0.05), we did not observe any reduction in either PCSK9 mRNA or protein expression in these cells (Fig. 2E, F). We did observe a slight increase in steady-state PCSK9 mRNA expression in Ad-shHNF1β-transduced cells (Fig. 2F). Importantly, a canonical HNF1β target gene, NR1D1 did exhibit modest but significant reduction of ∼25% (P < 0.05) in Ad-shHNF1β-transduced cells compared with Ad-shLacZ-transduced controls (Fig. 2F). Additionally, LDLR protein expression in Ad-shHNF1β-transduced cells was comparable to the levels observed in Ad-shLacZ-transduced controls (Fig. 2E). These data confirmed that reducing HNF1β expression via shHNF1β transduction had no effect in PCSK9 transcription in HepG2 cells.
Although our shRNA sequences were designed to knockdown both human and mouse HNF1α and HNF1β, before proceeding with in vivo analyses, we validated their effects in the murine system in vitro by transducing mouse primary hepatocytes with Ad-shLacZ, Ad-shHNF1α, and Ad-shHNF1β adenoviruses. Whereas knockdown of HNF1β did not affect PCSK9 mRNA expression, real-time PCR analyses confirmed reduced HNF1α and a concurrent reduction in PCSK9 mRNA expression in cells transduced with the Ad-shHNF1α virus compared with controls (Fig. 2G), thus confirming that HNF1α and HNF1β shRNA vectors were able to target endogenous mouse HNF1α/β transcripts, respectively.
Decrease in circulating PCSK9 after liver-specific knockdown of HNF1α
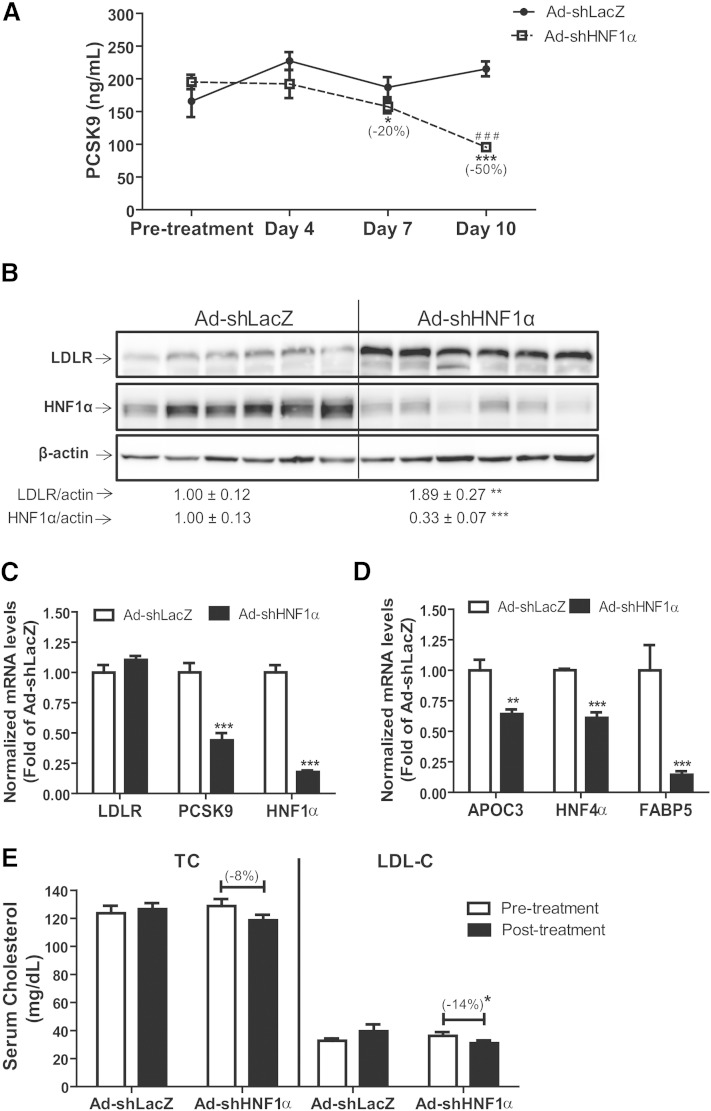
Expanding on our in vitro finding that knockdown of HNF1α resulted in decreased PCSK9 expression in cultured HepG2 cells and in mouse primary hepatocytes, in vivo functions of HNF1α were tested by injecting C57BL/6J mice with either Ad-shHNF1α or with Ad-shLacZ as control. Animals were fasted for 4 h and blood was drawn before injection, as well as on days 4, 7, and 10 after injection with the shRNA adenovirus. Using ELISA, we analyzed PCSK9 levels in serum collected at various time points from groups of mice. As shown in Fig. 3A, in Ad-shLacZ-injected mice, serum PCSK9 levels fluctuated during the 10 day course of study and a small but nonsignificant increase was observed at day 10 as compared with pretreatment values. In contrast, serum PCSK9 levels in Ad-shHNF1α-infected mice gradually decreased during the course of the entire study with significant reductions of ∼20 and ∼50% compared with pretreatment levels observed at days 7 (P < 0.05) and 10 (P < 0.001) postinjection, respectively.
Fig. 3.
Decrease in circulating PCSK9 and increased hepatic LDLR after liver-specific knockdown of HNF1α. Wild-type 8–10-week-old male C57BL/6J mice (N = 6 animals per treatment) were retro-orbitally injected with 6 × 109 pfu/mouse of Ad-shLacZ or Ad-shHNF1α adenovirus particles. Four hour-fasted serum samples were collected prior to injection and on days 4, 7, and 10 after infection. A: Serum PCSK9 was measured using ELISA from all serum samples. Data represent (mean ± SEM) PCSK9 concentrations at each time point. *P < 0.05, ***P < 0.001 compared with pretreatment expression levels, and ###P < 0.001 compared with PCSK9 expression in Ad-shLacZ-infected animals at the same time point. B: Western blotting with antibodies to LDLR, HNF1α, and β-actin was conducted. Values are the mean ± SEM of six samples per group. **P < 0.01, ***P < 0.001 compared with the Ad-shLacZ group. C, D: Quantitative real-time PCR was used to determine the relative expression levels (mean ± SEM) of mRNAs of interest. Data represent hepatic mRNA levels of indicated genes after normalization with GAPDH mRNA levels. **P < 0.01, ***P < 0.001 compared with the Ad-shLacZ group which was set at 1. E: Total cholesterol and LDL-C estimations (mean ± SEM) from serum samples. * P < 0.05 compared with the pretreatment levels or Ad-shLacZ group at same time points.
We next quantified the hepatic expression of LDLR and HNF1α protein in control and Ad-shHNF1α-injected mice. Western blot analysis indicated that HNF1α protein levels were reduced by ∼67% (P < 0.001), while LDLR protein levels were significantly (P < 0.01) increased by ∼1.9-fold in Ad-shHNF1α-infected mouse liver tissues compared with control animals (Fig. 3B). Levels of PCSK9 protein in liver homogenates were not different (supplementary Fig. 1).
To confirm that the observed reduction in serum PCSK9 and corresponding increase in LDLR protein was an outcome of diminished hepatic HNF1α-mediated PCSK9 gene transcription, we quantified mRNA expression of PCSK9, HNF1α, and LDLR mRNA from mouse liver tissues. Real-time PCR analysis revealed that compared with Ad-shLacZ-infected control animals, HNF1α mRNA expression was reduced by ∼80% (P < 0.001) in mice infected with the Ad-shHNF1α adenovirus (Fig. 3C). Similar to results observed in cultured cells, this reduction in HNF1α was accompanied by a ∼50% reduction in PCSK9 mRNA expression (P < 0.001) in Ad-HNF1α-infected mice (Fig. 3C). While LDLR mRNA expression remained unchanged, the mRNA level of HNF1α target genes APOC3, HNF4α, and FABP5 was significantly (P < 0.05) reduced in Ad-shHNF1α-infected animals compared with controls (Fig. 3D).
We analyzed total cholesterol and LDL-C in pretreatment and day 10 posttreatment samples (Fig. 3E). We observed a slight, albeit nonsignificant, reduction of ∼8% in serum total cholesterol levels of Ad-shHNF1α-infected samples at 10 days posttreatment. Importantly, serum LDL-C levels were significantly (∼14%, P < 0.05) lower in the Ad-shHNF1α-infected animals at the time of termination of the experiment (day 10) (Fig. 3E).
Hepatic HNF1β knockdown increased serum PCSK9 levels in mice
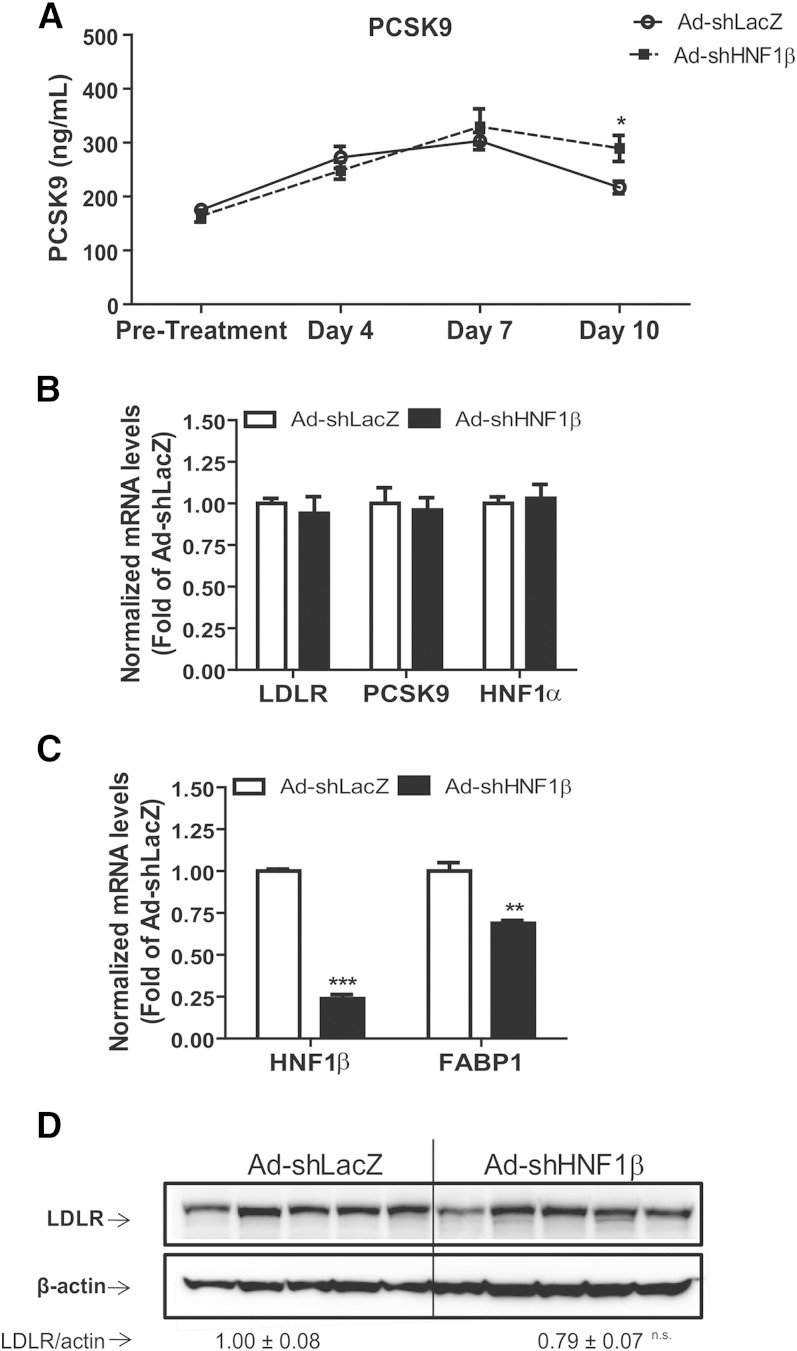
Our in vitro results demonstrated that HNF1α was the primary regulator of PCSK9 and HNF1β knockdown had no effect on PCSK9 expression. To verify whether the in vivo effects of HNF1β were similar to those observed in cultured cells, normal diet-fed mice were injected either with Ad-shLacZ or Ad-shHNF1β adenovirus. ELISA of serum samples collected at various time points revealed that circulating PCSK9 expression in Ad-shHNF1β-infected animals was comparable to that observed in Ad-shLacZ controls (Fig. 4A). There was no statistical difference in PCSK9 serum concentrations between the Ad-shHNF1β-treated and control animals in the pretreatment, day 4, or day 7 samples, while a slight increase in the serum PCSK9 level of the Ad-shHNF1β group as compared with the control group was observed at day 10 of postinfection (Fig. 4A). Gene expression analysis of liver tissue by real-time PCR showed that LDLR, PCSK9, and HNF1α mRNA expression remained unchanged between the treatment groups, whereas mRNA levels of HNF1β and its target gene FABP1 were markedly reduced in the Ad-sHNF1β-infected animals compared with Ad-shLacZ controls (Fig. 4B, C). Western blot analysis revealed that knockdown of HNF1β did not result in a significant reduction in hepatic LDLR protein (Fig. 4D). Furthermore, measurements of total cholesterol and LDL-C levels also indicated that there was no difference in serum cholesterol levels in Ad-shHNF1β mice compared with Ad-shLacZ-infected controls at the time of termination of the experiment (day 10, data not shown). Altogether, these data suggest that HNF1β knockdown did not affect PCSK9 gene expression in murine liver tissue.
Fig. 4.
Knockdown of hepatic HNF1β did not reduce circulating PCSK9. Wild-type 8–10-week-old male C57BL/6J mice (N = 5 animals per treatment) were retro-orbitally injected with 4 × 109 pfu/mouse of Ad-shLacZ or Ad-shHNF1β adenovirus particles. Four hour-fasted serum samples were collected prior to injection and on days 4, 7, and 10 after infection. A: Serum PCSK9 was measured using ELISA from all serum samples. Data represent (mean ± SEM) PCSK9 concentrations at each time point. *P < 0.05 compared with PCSK9 expression in Ad-shLacZ infected animals at the same time point. B, C: Quantitative real-time PCR was used to determine the relative expression levels (mean ± SEM) of mRNAs of interest. Data represent hepatic mRNA levels of indicated genes after normalization with GAPDH mRNA levels. **P < 0.01, ***P < 0.001 compared with the Ad-shLacZ group which was set at 1. D: Western blotting with antibodies to LDLR and β-actin was conducted. Values are the mean ± SEM of six samples per group.
DISCUSSION
In the present study, we analyzed the effects of liver-specific knockdown of HNF1α and its paralog HNF1β in adult wild-type mice. We show that HNF1α, but not HNF1β, is the primary positive regulator of PCSK9 transcription in liver tissue in vivo. Our in vitro studies first confirmed that loss of HNF1α reduced the promoter activity and mRNA expression of PCSK9, as well as resulted in increased expression of LDLR protein. Using adenoviral delivery of shRNA targeted against HNF1α or HNF1β, we show that knockdown of HNF1α caused significant reduction in hepatic mRNA expression and serum concentrations of PCSK9. While the expression of LDLR mRNA remains unchanged in the liver, LDLR protein levels were significantly increased after knockdown of HNF1α. In contrast to HNF1α, HNF1β knockdown did not affect PCSK9 expression either in HepG2 cells, mouse primary hepatocytes, or mouse liver tissue. To our knowledge, this is the first study addressing the in vivo liver-specific function of HNF1α and its paralog HNF1β in the regulation of PCSK9, LDLR, and serum cholesterol in adult animals.
Although a recent report indicated that HNF1α acts downstream of the insulin receptor (InsR) such that shRNA knockdown of InsR leads to increased expression of HNF1α, increased HNF1α binding on PCSK9 promoter, and increased PCSK9 expression (31), current in vivo understanding of the role of HNF1α in regulating plasma lipid metabolism is based primarily on studies involving HNF1α knockout transgenic animals. Because HNF1α is an important transcription factor controlling gene expression programs in multiple tissues, developmental loss of HNF1α is associated with various pleiotropic effects and abnormalities associated with lipid metabolism that may be potentially independent of its role in regulation of PCSK9 (29). For example, HNF1α plays a crucial role in regulation of β-cell transcriptional networks involved in glucose homeostasis and is responsible for a sub-type of maturity-onset diabetes (MODY3), as well as other deficiencies in β-cell proliferation and function (26, 32). Thus, in order to address the specific function of HNF1 factors in regulating plasma cholesterol metabolism through the hepatic PCSK9-LDLR axis, we employed a shRNA-based transient knockdown model and adenovirus delivery system for studying the in vivo effects of HNF1α/HNF1β knockdown on PCSK9 expression. Previous in vitro data from our laboratory indicate that both HNF1α and HNF1β bind to the PCSK9 promoter in HepG2 cells that overexpressed HNF1α or HNF1β (18). A recent study investigating in vivo promoter occupancy supported this observation and further indicated that in liver and islet tissues from HNF1α−/− transgenic animals the occupancy of PCSK9 promoter by HNF1β is dramatically increased by up to 20-fold (30). These studies pointed to the possibility that, like some other HNF1α target genes, the in vivo regulation of PCSK9 may exhibit redundancy in the ability of HNF1β to complement absence of HNF1α. We thus tested the effects of knockdown of not only HNF1α but also of HNF1β in mouse liver tissues. We observed >80% reduction in mRNA levels of hepatic HNF1α and HNF1β after 10 days postinfection. However, significant reductions in hepatic PCSK9 mRNA expression and serum PCSK9 concentrations were only observed in the Ad-shHNF1α virus-infected animals. Of note is that the infection dose of 4–6 × 109 PFU/animal and timeline (7–10 days postinfection) of analysis is consistent with other reports using adenoviral delivery vectors to knockdown gene expression in murine livers (33–35). Consistent with in vitro results, in vivo knockdown of HNF1β did not result in repression of hepatic PCSK9 mRNA levels. Thus, although both HNF1 transcription factors are capable of binding to the PCSK9 promoter with similar affinities, our results suggest that HNF1α and HNF1β play distinct roles in vivo. Whereas HNF1β is ineffective in modulating PCSK9 expression, loss of HNF1α results in a marked inhibition of PCSK9 mRNA transcription.
The experimental paradigm in our study used normal diet-fed adult wild-type mice. These mice are normolipidemic and yet, knockdown of HNF1α resulted in a significant (∼14%) reduction in circulating LDL-C levels at day 10 postinfection with the Ad-shHNF1α adenovirus. It is known that HNF1α is a critical transcription factor regulating various gene networks, including those involved in glucose and lipid metabolism (30, 36). The current experiments build upon recent studies elucidating the central role of HNF1α in regulation of PCSK9. Studies from our laboratory have indicated that the lipid lowering drug, berberine, partially exerts its effects in repressing PCSK9 transcription via reduction in HNF1α (18). Conversely we have also shown that in hamsters treated with rosuvastatin, in addition to the well-established pathway involving induction of PCSK9 by SREBP2, rosuvastatin also caused induction of HNF1α, which partially contributed to the increase in PCSK9 after treatment with the drug (37). Although it is yet unclear whether the statin-mediated induction of HNF1α occurs in vivo in humans, our studies suggest that liver-specific knockdown of HNF1α or utilizing agents that downregulate hepatic HNF1α expression may be an ancillary consideration when validating or exploring therapeutic regimens targeted toward lowering of LDL-C.
In summary, utilizing adenovirus-mediated transient expression of shRNAs that target HNF1α and HNF1β, we demonstrate for the first time that downregulation of HNF1α in liver tissue led to significant reduction of serum PCSK9 and LDL-C levels through upregulation of liver LDLR protein. While HNF1β is a coregulator of HNF1α in transactivating several genes containing HNF1 binding sites in their promoters, it is expendable for PCSK9 transcription in liver tissue. This work underscores the importance of HNF1α in the control of hepatic PCSK9 expression and as a potential therapeutic target for lowering plasma cholesterol.
Supplementary Material
Footnotes
Abbreviations:
- HNF
- hepatic nuclear factor
- LDL-C
- LDL-cholesterol
- LDLR
- LDL receptor
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- SRE
- sterol-regulatory element
- SREBP
- SRE binding protein
This work was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service) and by grant (1R01AT006336-01A1) from the National Center of Complementary and Alternative Medicine. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Grundy S. M., Cleeman J. I., Merz C. N., Brewer H. B., Jr, Clark L. T., Hunninghake D. B., Pasternak R. C., Smith S. C., Jr, Stone N. J. 2004. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. J. Am. Coll. Cardiol. 44: 720–732. [DOI] [PubMed] [Google Scholar]
- 2.Spady D. K. 1992. Hepatic clearance of plasma low density lipoproteins. Semin. Liver Dis. 12: 373–385. [DOI] [PubMed] [Google Scholar]
- 3.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell K. N., Breslow J. L. 2004. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA. 101: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian Y-W., Schmidt R. J., Zhang Y., Chu S., Lin A., Wang H., Wang X., Beyer T. P., Bensch W. R., Li W., et al. 2007. Secreted PCSK9 downregulates low density lipoprotein receptor through receptor-mediated endocytosis. J. Lipid Res. 48: 1488–1498. [DOI] [PubMed] [Google Scholar]
- 6.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Bélanger J., Stifani S., Basak A., Prat A., Chrétien M. 2003. The secretory proprotein convertase neutal apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279: 48865–48875. [DOI] [PubMed] [Google Scholar]
- 8.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 9.Fisher T. S., Lo S. P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., et al. 2007. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 282: 20502–20512. [DOI] [PubMed] [Google Scholar]
- 10.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 11.Benjannet S., Hamelin J., Chretien M., Seidah N. G. 2012. Loss- and gain-of-function PCSK9 variants: cleavage specificity, dominant negative effects, and low density lipoprotein receptor (LDLR) degradation. J. Biol. Chem. 287: 33745–33755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong H. J., Lee H-S., Kim K-S., Kim Y-K., Yoon D., Park S. W. 2008. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 49: 399–409. [DOI] [PubMed] [Google Scholar]
- 13.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell K.N., Soccio R.E., Duncan E.M., Sehayek E., Breslow J.L. 2003. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 44: 2109–2119. [DOI] [PubMed] [Google Scholar]
- 15.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., Prat A. 2004. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 24: 1454–1459. [DOI] [PubMed] [Google Scholar]
- 16.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y-A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 102: 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attie A. D., Seidah N. G. 2005. Dual regulation of the LDL receptor-some clarity and new questions. Cell Metab. 1: 290–292. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Dong B., Park S. W., Lee H-S., Chen W., Liu J. 2009. Hepatocyte nuclear factor 1alpha plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J. Biol. Chem. 284: 28885–28895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cereghini S. 1996. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 10: 267–282. [PubMed] [Google Scholar]
- 20.Mendel D. B., Hansen L. P., Graves M. K., Conley P. B., Crabtree G. R. 1991. HNF-1α and HNF-1 β (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev. 5: 1042–1056. [DOI] [PubMed] [Google Scholar]
- 21.Mendel D. B., Crabtree G. R. 1991. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J. Biol. Chem. 266: 677–680. [PubMed] [Google Scholar]
- 22.Pontoglio M., Barra J., Hadchouel M., Doyen A., Kress C., Bach J. P., Babinet C., Yaniv M. 1996. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 84: 575–585. [DOI] [PubMed] [Google Scholar]
- 23.Pontoglio M., Sreenan S., Roe M., Pugh W., Ostrega D., Doyen A., Pick A. J., Baldwin A., Velho G., Froguel P., et al. 1998. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J. Clin. Invest. 101: 2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffinier C., Thepot D., Babinet C., Yaniv M., Barra J. 1999. Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development. 126: 4785–4794. [DOI] [PubMed] [Google Scholar]
- 25.Barbacci E., Reber M., Ott M. O., Breillat C., Huetz F., Cereghini S. 1999. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 126: 4795–4805. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell S. M., Frayling T. M. 2002. The role of transcription factors in maturity-onset diabetes of the young. Mol. Genet. Metab. 77: 35–43. [DOI] [PubMed] [Google Scholar]
- 27.Ryffel G. U. 2001. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 27: 11–29. [DOI] [PubMed] [Google Scholar]
- 28.Pelletier L., Rebouissou S., Paris A., Rathahao-Paris E., Perdu E., Bioulac-Sage P., Imbeaud S., Zucman-Rossi J. 2010. Loss of hepatocyte nuclear factor 1alpha function in human hepatocellular adenomas leads to aberrant activation of signaling pathways involved in tumorigenesis. Hepatology. 51: 557–566. [DOI] [PubMed] [Google Scholar]
- 29.Shih D. Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B. L., Suchy F. J., Shefer S., Bollileni J. S., Gonzalez F. J., Breslow J. L., et al. 2001. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27: 375–382. [DOI] [PubMed] [Google Scholar]
- 30.Servitja J-M., Pignatelli M., Maestro M. A., Cardalda C., Boj S. F., Lozano J., Blanco E., Lafuente A., McCarthy M. I., Sumoy L., et al. 2009. HNF1α (MODY3) controls tissue-specific transcription programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol. Cell. Biol. 29: 2945–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ai D., Chen C., Han S., Ganda A., Murphy A. J., Haeusler R., Thorp E., Accili D., Horton J. D., Tall A. R. 2012. Regulation of hepatic LDL receptors by mTORC1 and PCSK9 in mice. J. Clin. Invest. 122: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih D. Q., Screenan S., Munoz K. N., Philipson L., Pontoglio M., Yaniv M., Polonsky K. S., Stoffel M. 2001. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 50: 2472–2480. [DOI] [PubMed] [Google Scholar]
- 33.Ragozin S., Niemeier A., Laatsch A., Loeffler B., Merkel M., Beisiegel U., Heeren J. 2005. Knockdown of hepatic ABCA1 by RNA interference decreases plasma HDL cholesterol levels and influences postprandial lipemia in mice. Arterioscler. Thromb. Vasc. Biol. 25: 1433–1438. [DOI] [PubMed] [Google Scholar]
- 34.Jin W., Wang X., Millar J. S., Quertermous T., Rothblat G. H., Glick J. M., Rader D. J. 2007. Hepatic proprotein convertases modulate HDL metabolism. Cell Metab. 6: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A. B., Kan C. F., Shende V., Dong B., Liu J. 2014. A novel posttranscriptional mechanism for dietary cholesterol-mediated suppression of liver LDL receptor expression. J. Lipid Res. 55: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., et al. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science. 303: 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong B., Wu M., Li H., Kraemer F. B., Adeli K., Seidah N. G., Park S. W., Liu J. 2010. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 51: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.