Abstract
Besides bulk amounts of SM, mammalian cells produce small quantities of the SM analog ceramide phosphoethanolamine (CPE). Little is known about the biological role of CPE or enzymes responsible for CPE production. Heterologous expression studies revealed that SM synthase (SMS)2 is a bifunctional enzyme producing both SM and CPE, whereas SMS-related protein (SMSr) serves as monofunctional CPE synthase. Acute disruption of SMSr catalytic activity in cultured cells causes a rise in endoplasmic reticulum (ER) ceramides, fragmentation of ER exit sites, and induction of mitochondrial apoptosis. To address the relevance of CPE biosynthesis in vivo, we analyzed the tissue-specific distribution of CPE in mice and generated mouse lines lacking SMSr and SMS2 catalytic activity. We found that CPE levels were >300-fold lower than SM in all tissues examined. Unexpectedly, combined inactivation of SMSr and SMS2 significantly reduced, but did not eliminate, tissue-specific CPE pools and had no obvious impact on mouse development or fertility. While SMSr is widely expressed and serves as the principal CPE synthase in the brain, blocking its catalytic activity did not affect ceramide levels or secretory pathway integrity in the brain or any other tissue. Our data provide a first inventory of CPE species and CPE-biosynthetic enzymes in mammals.
Keywords: brain lipids, enzyme inactivation, genetics, mass spectrometry, sterile α motif domain-containing protein 8, sphingomyelin synthase-related protein, sphingolipids, sphingomyelin synthase, transgenic mice
Sphingolipids are vital components of cellular membranes that contribute to mechanical stability, signaling, and molecular sorting (1–3). Ceramides are obligatory precursors for sphingolipid biosynthesis, but also potent mediators of various stress responses, cell cycle arrest, and apoptosis (2, 4, 5). Consequently, cells should monitor sphingolipid biosynthesis closely to avoid jeopardizing their growth and viability. How this works is largely unexplored. Ceramides are synthesized on the cytosolic surface of the endoplasmic reticulum (ER) and transported by ceramide transfer protein (CERT) to the Golgi for conversion to SM (6). SM production is catalyzed by a phosphatidylcholine (PC):ceramide choline phosphotransferase or SM synthase (SMS). Mammalian cells contain two SMS isoforms, namely, SMS1 responsible for bulk production of SM in the Golgi lumen and SMS2 likely serving a role in regenerating SM from ceramides liberated by SM phosphodiesterases on the exoplasmic surface of the plasma membrane (7, 8). Together with a closely related enzyme, SMS-related protein (SMSr), they form the SMS protein family (9).
Both SMS1 and SMS2 support cell growth, at least in certain types of cultured cancer cells (8, 10). Moreover, SMS1 depletion has been reported to enhance ceramide production and apoptosis after photodamage (11), while its contribution to the generation of plasma membrane-associated SM is critical for Fas-clustering and Fas-mediated apoptosis (12). SMS1 deficiency in mice causes moderate neonatal lethality, reduced body weight, loss of fat tissue mass, and white adipose tissue function associated with impaired insulin secretion (13, 14). It also decreases SM and elevates glycosphingolipid levels in plasma, liver, and macrophages, and attenuates macrophage activation by NFκB and MAPK in response to inflammatory stimuli (15). SMS2 deficiency, on the other hand, ameliorates high fat-induced obesity (16, 17), attenuates lipopolysaccharide-induced lung injury (18), and reduces atherosclerosis (19). A recent study investigating SMS2 deficiency in the brain reported decreased expression and altered function of drug transporters (20).
Besides bulk amounts of SM, mammalian cells also produce small quantities of the SM analog, ceramide phosphoethanolamine (CPE) (21, 22), a widespread but largely unexplored sphingolipid. We recently demonstrated that SMS2 is, in fact, a bifunctional enzyme that produces both SM and CPE at the plasma membrane (23). The biological relevance of SMS2-catalyzed CPE production remains to be established. In addition, we found that SMSr functions as monofunctional CPE synthase in the ER (24). SMSr is the best-conserved member of the SMS protein family, with homologs in organisms such as Drosophila, which lacks SM and synthesizes CPE as the principal sphingolipid. While Drosophila SMSr exhibits CPE synthase activity (24), bulk production of CPE in this organism is mediated by an insect-specific enzyme that belongs to the CDP-alcohol phosphotransferase superfamily (25). Remarkably, acute disruption of SMSr catalytic activity in cultured mammalian and insect cells causes a substantial rise in ER ceramides, leading to a structural collapse of ER exit sites and fragmentation of the Golgi complex (24). Subsequent work revealed that SMSr acts as a suppressor of ceramide-induced mitochondrial apoptosis in various human carcinoma cell lines, including HeLa (26). We also found that SMSr-catalyzed CPE production, although required, is not sufficient to suppress ceramide-induced cell death and that SMSr-mediated control over ER ceramides is critically dependent on the enzyme’s N-terminal sterile α motif (SAM) domain (26). Based on these results, we postulated a primary role of SMSr in monitoring ER ceramide levels to prevent inappropriate cell death during sphingolipid biosynthesis.
In the current study, we generated mouse lines lacking SMSr catalytic activity and crossbred these with SMS2-null mice (27) in order to investigate the impact of disrupting SMS-mediated CPE production in vivo. In one mouse line, we introduced a point mutation in the active site of SMSr, creating a catalytically inactive enzyme (SMSrD348E). To circumvent potential embryonic lethality, we generated a mouse line for conditional depletion of smsr exon 6, encoding two catalytically essential residues. As SMSrD348E mice proved viable, we ubiquitously deleted exon 6 in these mice (SMSrdelEx6). Next, we analyzed the tissue distribution and abundance of CPE in wild-type, SMSrD348E, SMS2gt, and SMSrD348E×SMS2gt double mutant mice. We report on the consequences of disrupting SMSr- and SMS2-catalyzed CPE production on the lipid composition, cell survival, and integrity in a variety of mouse tissues.
MATERIAL AND METHODS
Generation of the SMSrD348E vector
In the first step, flanking regions of the 5′ and 3′ homologous regions (HRs) (5′HR and 3′HR) for homologous recombination in embryonic stem (ES) cells were amplified via PCR and cloned into the pDTA-vector, which contained a MC1-diphtheria toxin A (DTA)-negative selection cassette (28). This vector then served as a retrieval vector in a gap repair cloning step on a bacterial artificial chromosome (#bMQ 369P09; Sanger Institute, UK) to insert the genomic sequence of exon 4 to exon 6 (29). The point mutation D348E and insertion of the MfeI restriction site in exon 5 of the smsr gene and a part of the 3′ region of smsr were generated by PCR mutagenesis, and the resulting fragment was cloned into the pBluescript vector. Between exon 6 and the untranslated region (UTR) of smsr, a β-galactosidase gene (lacZ) reporter gene with an internal ribosomal entry site was inserted into the vector mentioned above. In the next step we inserted the neomycin resistance gene, driven by the Simian virus 40 promoter and flanked by Flp recognition target (frt) sites into the vector described above between exon 5 and 6 of the smsr gene. To obtain the final SMSrD348E vector, we combined both vectors via a gap repair cloning step, which resulted in a vector containing the DTA selection cassette followed by the genomic sequences of exon 4 to exon 5 and the neomycin resistance gene downstream of exon 5 followed by the D348E mutation in exon 6 and the nuclear localization signal-lacz reporter gene with internal ribosomal entry site downstream of exon 6 and part of the 3′UTR. The final nonconditional SMSrD348E vector was analyzed by restriction mapping and partial sequencing (GATC Biotech, Germany). The functionality of the frt sites was tested by transformation of the targeting vector into Flp recombinase expressing Escherichia coli bacteria (30).
Generation of the conditional SMSrdelEx6 vector
Using mutagenesis PCR with the previously mentioned bacterial artificial chromosome (#bMQ 369P09; Sanger Institute) flanking regions of the 5′HR were generated and cloned into the vector ploxP-frt-dualneo-frt-loxP. The vector provides loxP sequences for conditional deletion of exon 6 surrounding a Simian virus 40 promoter-driven neomycin resistance gene flanked by frt sites. Insertion took place upstream of the 5′loxP site and the 5′HR was extended to its full size (a 4 kb part of intron 4) by gap repair cloning in the last step. In the next step, flanking regions of an internal HR downstream of the 5′loxP site (5′ flanking region, part of exon 5; 3′ flanking region, part of 3′UTR) were produced by mutagenesis PCR. The region ranging from exon 5 to the 3′UTR was completed in the second to last step by gap repair cloning. The 3′HR representing the 1.8 kb 5′ region of the 3′UTR was generated by PCR and cloned into the vector pMJ_SMSrbisEx5-eGFP (containing the SMSr cDNA sequence up to exon 5 followed by an in-frame egfp sequence) downstream of the egfp sequence. From this vector, the resulting sequence consisting of the 3′ region of intron 4, exon 5, egfp, and the 3′HR was first cloned into the pDTA vector harboring the MC1-DTA-negative selection cassette (28). Then the sequence, together with the downstream DTA-cassette, was inserted downstream of the 3′ loxP site into the initially produced vector containing the 5′HR. The targeting vector was finished by two independent gap repair cloning steps as mentioned above. The SMSrD348E targeting vector identity was verified by restriction mapping and partial sequencing. Functionality of the loxP or frt sites was proved by transformation of E. coli bacteria expressing the Cre or Flp recombinase, respectively (30). The conditional SMSrdelEx6 vector allows the cell type- or time-specific expression of the generated mutation. In contrast, the mutation in SMSrD348E mice is already present in the zygote and hence in all descendent cells.
Screening of ES cell clones
For transfection of HM1 ES cells (31) via electroporation (0.8 kV, 3 μF; Gene Pulser, Bio-Rad), 300 μg of DNA of the corresponding vector were linearized by NotI digestion. Selection of transfected ES cells was carried out with 350 μg/μl G418-neomycin (Invitrogen, Germany). Resulting ES cell clones were tested by Southern blot analyses for recombination at the 3′HP and 5′HR (external probes), as well as for single integration of the vector construct (internal probe; data not shown). The transfection of cells with the SMSrD348E vector yielded 22% stably transfected cell clones that carried the vector integrated by homologous recombination. Transfection with the SMSrdelEx6 vector yielded 21.5% stably transfected cells.
Generation of SMSrD348E and SMSrdelEx6 mice
Homologously recombined ES cells were injected into C57BL/6 mouse blastocysts (32). Blastocysts were used to generate germ-line transmission chimeras. These chimeras were mated with C57BL/6 mice and the homologous recombination was verified by PCR from tail tip DNA of the agouti-colored offspring. For the SMSrD348E mice, we used an exon-specific sense primer (SMSr_1, 5′-GAA ACT ACG ACG GGC ATT-3′) together with an intron-specific antisense primer (SMSr_3b, 5′-TTC AGC TCT GTC TCA TGT GGC-3′) and a neomycin cassette-specific antisense primer (SMSr_2b, 5′-CCT TCC GTG TTT CAG TTA GCC-3′).
For the SMSrdelEx6 mice, we used an intron-specific sense primer (SMSr_loxP_for 5′-AGC TCT GGT AAT TCT CTG GGC-3′) combined with an eGFP-specific antisense primer (SMSr_eGFP_rev, 5′-AAG TCG TGC TGC TTC ATG TGG-3′) and an intron-specific antisense primer (SMSr_loxP_rev, 5′-TCT CAC TTC CTC CCT AGT TCC-3′). For both mouse lines the progeny were mated with Flp recombinase-expressing mice (33) to remove the neomycin resistance cassette and bred with C57BL/6 mice to reach a genetic background of at least 87.5% C57BL/6. For the conditional SMSrdelEx6 mouse line, the initially generated “floxed” animals were bred with mice expressing the Cre recombinase under control of the phosphoglycerate kinase (pgk) promoter (34) for ubiquitous deletion of exon 6. Animals were kept under standard conditions with a 12 h dark to light cycle with food and water ad libitum. All housing and experimental conditions were in accordance to instructions of local and state authorities. Prior to dissection, animals were anesthetized with Florene® (AbbVie, Germany) and transcardially perfused with PBS.
Southern blot hybridization
The correct insertion of the respective targeting vector into the SMSr locus was verified by Southern blot hybridization as described previously (35), using genomic DNA from the liver of wild-type heterozygous (+/SMSrD348E or +/SMSrdelEx6) and homozygous (SMSrD348E or SMSrdelEx6) mice. For the SMSrD348E mice, DNA was digested with BclI and a 5′ external probe binding downstream of exon 1 and upstream of the 5′HR was used. For analysis of SMSrdelEx6 mice, we used an internal probe binding in the 3′HR and external XmnI restriction sites (upstream of the 5′HR and downstream of the 3′HR).
CPE-synthase assay
Brain and liver samples from 10-week-old mice were homogenized in buffer R [20 mM HEPES-KOH (pH 7.2), 15 mM KCl, 5 mM NaCl, 250 mM sucrose] including freshly added PMSF (200 × stock 34.8 mg/ml in isopropanol) and protease inhibitor cocktail (1,000 × stock in DMSO containing 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 5 mg/ml antipain, 157 mg/ml benzamidine) using a Precellys® homogenizer (Peqlab Biotechnology, Germany). Microsomal fractions were obtained by centrifugation at 960 g for 10 min followed by centrifugation of the supernatant at 100,000 g for 1 h. The microsomal pellet was resuspended in buffer R, pressed 20 times through a 27 gauge needle, aliquoted, frozen in liquid nitrogen, and stored at −70°C. Protein concentration was determined using the BCA assay kit (Sigma-Aldrich, USA).
Aliquots of microsomes were normalized to protein content and resuspended in 400 μl end volume of ice-cold buffer R plus 0.1% v/v of 1,000 × protease inhibitor cocktail and kept on ice in 12 ml Pyrex tubes with lids. N-ethylmaleimide was added to this suspension to a final concentration of 0.5 mM from a 100 mM stock solution in ethanol. Then, C6 nitrobenzoxadiazole-ceramide (NBD-Cer) [6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl) sphingosine; 2 mM stock in absolute ethanol; Molecular Probes] was added to a final concentration of 2.5 μM. Samples were kept on ice for 5–10 min to allow incorporation of the NBD-Cer probe into the membrane. After this, samples were shifted to 37°C with continuous shaking for 2 h. The reaction was stopped by addition of 1 ml methanol and 0.5 ml chloroform, and vigorous vortexing. After this, lipids were extracted and dried essentially as described by Ternes et al. (23).
For analysis, the dried lipid film was redissolved in a suitable volume of chloroform:methanol (2:1 v/v) and applied to a TLC plate (NANO-ADAMANT #821150; Macherey-Nagel) using the CAMAG Linomat 5 (022.7808) operated at a dosage speed of 120 nl/s from a 100 μl syringe. TLC development was performed using the CAMAG ADC2 TLC developer, using the protocol essentially as described by Ternes et al. (23), which comprised two consecutive 1D-runs. The excess NBD-Cer was eluted in a first run (acetone). Then the NBD-SM and NBD-CPE formed were separated using a second run in the same dimension, with basic eluent [chloroform:methanol:ammonia (aq) (50:25:6 v/v/v)].
After thoroughly drying the TLC plate, NBD-lipid species were visualized by scanning on a Typhoon FLA 9500 imager (laser 473 nm, filter setting BPB1, PMT 290 V) at a resolution of 50 μm pixel size. Images were contrast-adjusted using the ImageQuant software and then exported to Tiff format for publication.
Construction of adenovirus expressing phosphatidylethanolamine N-methyltransferase
To construct recombinant adenovirus, a cDNA encoding human phosphatidylethanolamine N-methyltransferase (PEMT) was subcloned into a pADTrack-CMV shuttle vector, linearized with PmeI, and inserted into an adenovirus using pADEasy-1 system for homologous recombination in E. coli. This adenovirus (Ad.PEMT) expressed both PEMT and green fluorescent protein (GFP). An adenovirus expressing only GFP (Ad.GFP) was used as a control. Male C57BL/6 (backcrossed more than seven generations) Pemt+/+ and Pemt−/− mice were given free access to standard chow (#5001, LabDiet) (36). At 8 weeks of age, a single dose of Ad.GFP or Ad.PEMT (1.0 × 109 plaque-forming units) was injected into the tail vein. Seven days after injection, mice were fasted and anesthetized with metofane. Tissues were dissected immediately and stored at −80°C. All procedures were approved by the University of Alberta’s Institutional Animal Care Committee in accordance with guidelines of the Canadian Council on Animal Care. PEMT activity assay was performed as described previously (37).
Generation of custom-made SMSr-specific antibodies and immunoblotting
For the generation of primary antibodies targeting the SMSr protein, rabbits were injected with the C-terminal peptide of the mouse SMSr protein (CWPFSKPAIMKRLIG) by Pineda Antibody Service (Berlin, Germany). Antibodies were purified from the resulting serum by column affinity chromatography with the C-terminal peptide.
For SMSr expression pattern analysis, tissues from wild-type and SMSrdelEx6 mice were homogenized in 1 ml/g wet weight of modified RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1% NP-40, 0.25% Na-deoxycholate, 1 mM PMSF, and Complete® protease inhibitor cocktail (Roche Applied Sciences, Switzerland)] using a Precellys® homogenizer (Peqlab Biotechnology). Cell debris was removed by 10 min centrifugation at 4°C. Protein amounts were determined using a BCA assay kit. (Sigma-Aldrich). Depending on tissue, 25–50 μg of protein in urea buffer [40 mM Tris-HCl, 9 M urea, 1 mM EDTA, 5% SDS, 5% (v/v) β-mercaptoethanol, 0.01% (w/v) bromophenol blue (pH 6–8)] were separated by SDS-PAGE (12%) and blotted onto nitrocellulose membranes (Hybond-ECL, GE Healthcare, UK). Membranes were blocked for 1 h at room temperature in 5% milk powder in TBS-Tween. The newly generated SMSr polyclonal antibodies were used in a 1:1,000 dilution and incubated overnight at 4°C. The mouse anti-V5 antibody (Invitrogen, Germany) was used 1:1,000 overnight at 4°C. Secondary horseradish peroxidase-conjugated antibodies against rabbit and mouse (Dianova, Germany) were used 1:10,000 for 1 h at room temperature. For detection, Amersham ECL Prime Western blotting detection reagent (GE Healthcare) was used and membranes were developed using VersaDoc imaging system (Bio-Rad, USA).
β-Galactosidase staining
Tissues of adult mice were frozen on dry ice, embedded in Tissue-Tek OCT and prepared for cryostat sectioning. β-galactosidase staining was carried out according to Degen et al. (38). Stained sections were mounted on glass slides (Menzel, Germany), dried on a heating plate at 37°C, and mounted with Entellan (Merck Millipore Chemicals, Germany).
Mass spectrometric analysis of lipids
Tissues were homogenized in a small volume of water using a Precellys® homogenizer (Peqlab Biotechnology). Lipids were extracted according to Folch, Lees, and Stanley Sloane (39) with the standards (unnatural lipid species) added at the one-phase stage of extraction. After evaporation of the solvents, the lipids were dissolved in methanol/chloroform (2:1 v/v), the phosphate contents of the extracts were determined (40), and the samples were stored at −20°C. Glycerophospholipids and most sphingolipids were analyzed using direct infusion of the samples into the electrospray source of a Quattro Premier triple-quadrupole mass spectrometer (Waters Inc., USA) operated in the MS/MS mode. The analytes and the internal standards were detected based on class-specific precursor ion (PC, phosphatidylinositol, ceramide, hexosyl- and lactosyl-ceramides) or neutral loss [phosphatidylethanolamine (PE), phosphatidylserine] scanning as described previously (41). The data were analyzed with Microsoft Excel using the LIMSA add-on (42).
To quantify CPE (and SM), a volume of the extract containing 100 nmol of lipid phosphate was taken to dryness and dissolved in 1.5 ml of methanol. Then 0.5 ml of chloroform/methanol (9:1 v/v) containing 15:0-SM (1 nmol) and 17:0-CPE (0.25 nmol) standards and 0.5 ml of 0.3 M NaOH were added and the samples were incubated overnight at room temperature. The solution was neutralized by addition of 0.3 ml of 0.3 M HCl in water, and 3.5 ml of chloroform and 1.2 ml of water were added followed by thorough mixing. After centrifugation for 10 min at 3,000 rpm, the upper phase was removed and the lower chloroform phase was washed twice with 3.5 ml of theoretical Folch upper phase (39). After drying under a N2 stream, the lipids in the lower phase were dissolved in 0.2 ml of methanol, moved to conical MS vials, dried and dissolved in 40 μl of LC-MS quality methanol, and stored at −20°C.
The CPE and SM species were quantified using LC-MS. The chromatographic separation was carried out in the gradient mode using an Acquity Ultra Performance LC system equipped with an Acquity BEH-C18 1.0 × 150 mm column (Waters Inc.). The column temperature was 60°C and the flow rate 0.13 ml/min. Solvent A was acetonitrile/water (60:40 v/v) containing 10 mM ammonium formate and 1% NH4OH, while solvent B consisted of isopropanol/acetonitrile (90:10 v/v) containing 10 mM ammonium formate and 1% NH4OH. The gradient started from 40% B, changed linearly to 70% B in 10 min, and then linearly to 100% B in 4 min. After 2 min at 100%, B changed to 40% in 3 min and was kept there for 3 min prior the next injection. To detect the CPE and SM, the column eluent was infused to the ESI source of the Quattro Premier mass spectrometer operated in the positive ion mode using selective reaction monitoring. The CPE species were detected based on neutral loss of m/z 141 and the SM species based on precursor ion scan/EIC of m/z 184. The peak areas of the SM and CPE species and standards were obtained from the selective reaction monitoring chromatograms with the QuanLynx software (Waters Inc.) and the concentrations of the molecular species and the classes were determined with Microsoft Excel.
Electron microscopy3
Wild-type and SMSrD348E mice were fixed by perfusion via the left ventricle with 2.5% glutaraldehyde in 0.1 M cacodylate buffer. After fixation, small pieces of tissue were incubated in 1% (w/v) osmium tetroxide. After rinsing in cacodylate buffer, the specimens were treated with 30% and 50% ethanol followed by incubation with 1% uranyl acetate for 6 min in 70% ethanol in the dark. After dehydration in a graded series of ethanol and propylene oxide, tissues were embedded in EPON® according to the manufacturer’s protocol. Ultrathin sections were obtained with a Reichert-Jung UltracutE® ultramicrotome, mounted on copper grids and treated with 1% uranyl acetate for 20 min and 3 min with lead citrate (0.4%). A Zeiss transmission electron microscope (EM 902A) was used for final investigation. Digital image acquisition was performed using a MegaViewII slow-scan-CCD camera connected to a personal computer running ITEM® 5.0 software (Soft Imaging Systems, Germany); images were stored as uncompressed TIFF files in 16 bits of gray and processed with Adobe® Photoshop® CS5.
Statistical analyses
Analyses and graphical presentation were done using Microsoft Excel and Graph Pad Prism 5 software (Graph Pad Software). Statistical significance was determined by two-tailed Student’s t-test. Differences were considered statistically significant when P < 0.05.
RESULTS
CPE levels in mouse tissues are exceedingly low
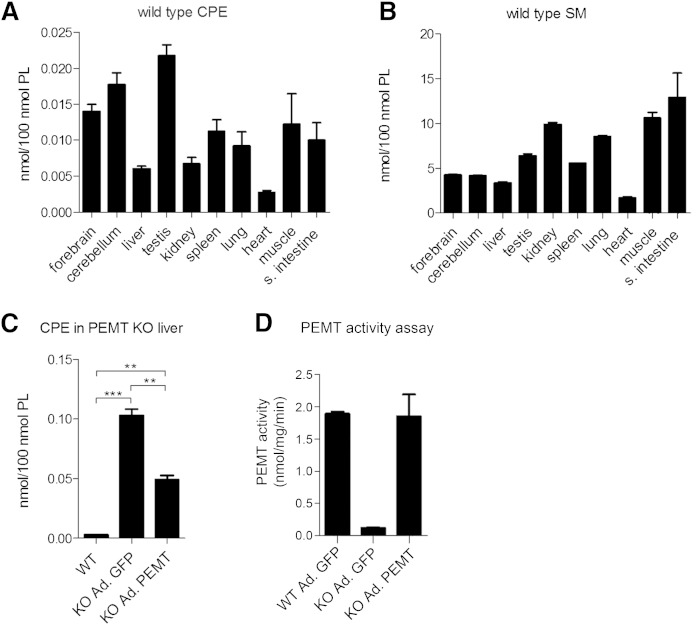
To support the functional analysis of SMSr in mice, we first quantified CPE levels in a variety of mouse tissues (i.e., forebrain, cerebellum, liver, testis, kidney, spleen, lung, heart, muscle, and small intestine). Because we were unable to detect CPE by direct infusion MS/MS, CPE levels were determined by LC-MS analysis and directly compared with SM levels. This revealed that in all tissues examined, the steady-state levels of CPE were exceedingly low, i.e., 300- to 1,500-fold lower than SM levels, depending on the tissue analyzed (Fig. 1A, B). The highest levels of CPE (∼0.020 mol% of total phospholipid) were observed in testis and brain, whereas CPE levels in heart and liver were particularly low (∼0.002–0.005 mol% of total phospholipid). These results are consistent with the low abundance of CPE in a variety of mammalian cell lines (24).
Fig. 1.
CPE and SM levels in different mouse tissues. A, B: LC-MS analysis of CPE and SM levels in forebrain, cerebellum, liver, testis, kidney, spleen, lung, heart, muscle, and small intestine (s. intestine) of wild-type mice (n = 4). C: LC-MS analysis of liver CPE content in wild-type (WT) mice, PEMT KO mice transduced with GFP (KO Ad. GFP), and PEMT KO mice transduced with functional PEMT (KO Ad. PEMT) (n = 2–3). D: PEMT activity assay from liver lysates of wild-type mice transduced with GFP (WT Ad. GFP), PEMT KO mice transduced with GFP (KO Ad. GFP), and PEMT KO mice transduced with functional PEMT (KO Ad. PEMT) (n = 3). Data are means ± SEM; **P < 0.005, ***P < 0.001.
CPE in liver is a putative substrate for PEMT-mediated SM production
SM biosynthesis in mammals mainly occurs through SM synthase-mediated headgroup transfer from PC to ceramide (9). However, an alternative pathway of SM production has been postulated in liver, which involves the step-wise methylation of CPE (21, 43). This reaction is analogous to the S-adenosylmethionine-dependent conversion of PE to PC catalyzed by the liver-specific enzyme PEMT (36, 37). To investigate whether PEMT contributes to the exceedingly low CPE content in liver, we analyzed the impact of PEMT removal on liver CPE levels. As shown in Fig. 1C, deletion of PEMT caused a 20-fold increase in liver CPE levels, i.e., from ∼0.005 to 0.10 mol% of total phospholipid. Adenovirus-mediated transduction of PEMT KO mice with a functional PEMT construct led to a 50% reduction in liver CPE levels (Fig. 1C, D). These results indicate that PEMT may utilize CPE as substrate in a methylation-dependent pathway of SM biosynthesis. As SM levels in wild-type and PEMT KO livers were indistinguishable and exceeded CPE levels at least 50-fold (data not shown), it appears that PEMT-mediated conversion of CPE to SM makes, at best, only a minor contribution to the total SM pool in liver.
Mouse SMSr is a CPE synthase
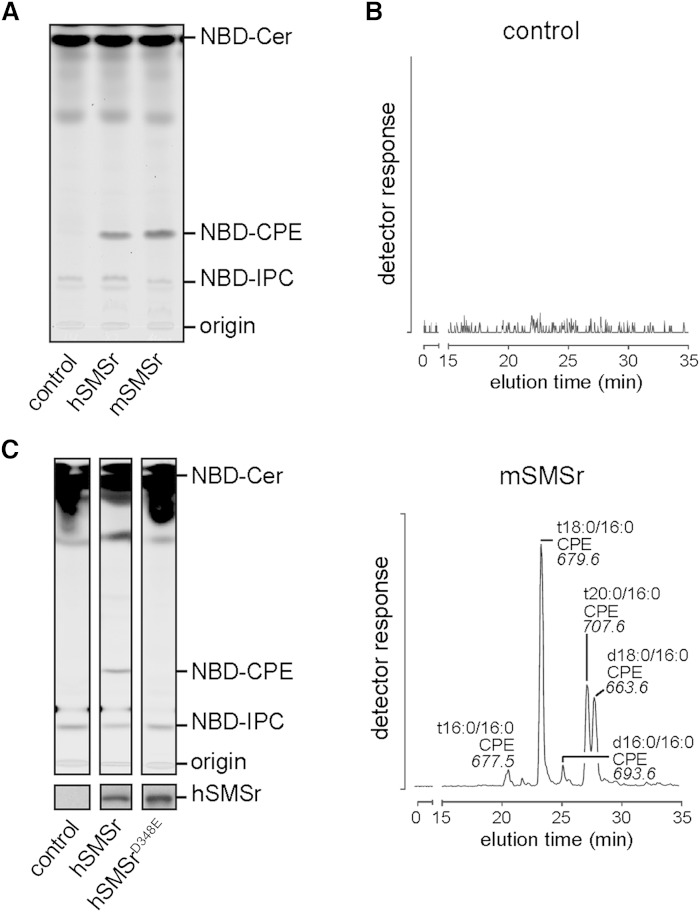
Mouse SMSr shares a common domain structure with other SMSr homologs, which includes six transmembrane helices, an N-terminal SAM domain, and a conserved active site comprising a catalytic triad of two histidine residues at positions 301 and 344, and an aspartate residue at position 348 (7, 9). We previously showed that human and Drosophila SMSr exhibit CPE synthase activity (24). To prove this also for mouse SMSr (mSMSr), a V5-tagged version of the protein was expressed in budding yeast, an organism lacking endogenous CPE synthase activity. As shown in Fig. 2A, lysates of yeast cells expressing V5 epitope-tagged mSMSr supported conversion of fluorescent C6-NBD-Cer into NBD-CPE. In addition, expression of mSMSr-V5 in a yeast strain producing ceramides that structurally resemble those found in mammals allowed the detection of several species of CPE by LC-MS/MS (Fig. 2B). In line with our previous findings (24, 26), substitution of the highly conserved aspartate residue at position 348 for a glutamate completely abolished SMSr-mediated CPE production (Fig. 2C). Together, this indicates that mSMSr is a true CPE-synthase and that the SMSrD348E point mutation leads to catalytic inactivation of the enzyme.
Fig. 2.
Mouse SMSr catalyzes CPE production. A: TLC separation of reaction products obtained when NBD-Cer was incubated with lysates of yeast strains expressing hSMSr or mSMSr. Part of the TLC (control and hSMSr) was already published (24), but is illustrated here for immediate comparison. B: Different molecular species of CPE detected by LC-MS/MS analysis (neutral loss of 141) after alkaline hydrolysis of glycerolipids in SMSr-expressing Conzelmann yeast strain in which endogenous ceramide synthases were substituted for mouse ceramide synthase 5; see (24) for further details. C: TLC separation of reaction products obtained when NBD-Cer was incubated with lysates of yeast strains expressing hSMSr or hSMSrD348E proving missing catalytic activity of the point mutated protein. Immunoblot analysis (below TLC) confirms that equal amounts of protein were used. hSMSr, human SMSr; mSMSr, mouse SMSr.
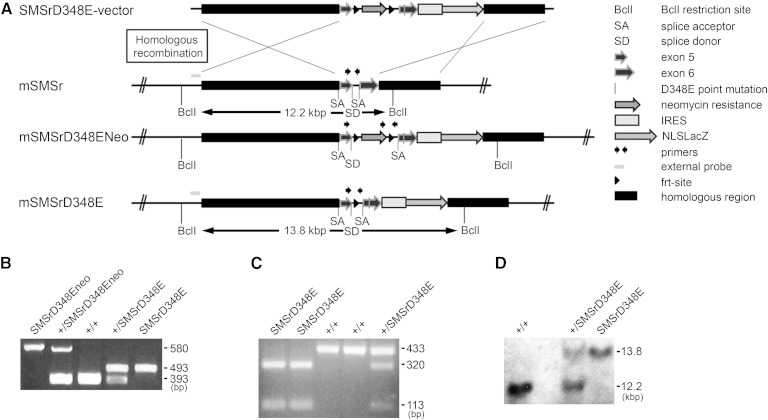
Generation of SMSrD348E and SMSrdelEx6 mice
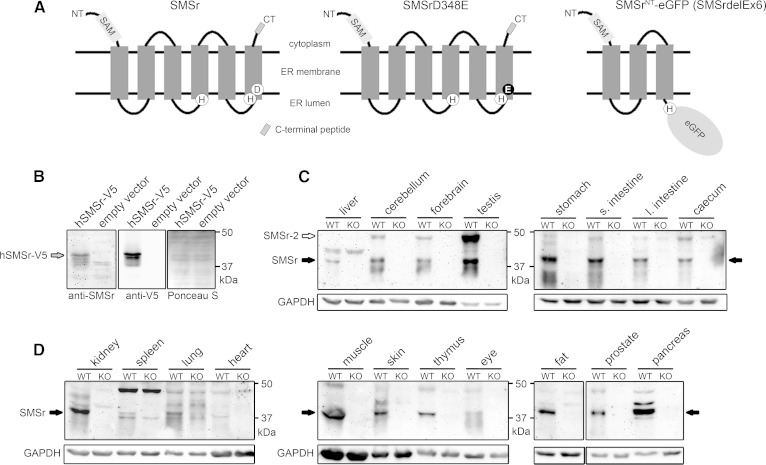
The mouse smsr gene is located on chromosome 14 and includes six exons. To study the biological functions of SMSr in mice, two transgenic mouse lines were generated expressing the SMSrD348E point mutation (SMSrD348E; Fig. 3) or a transcript lacking exon 6 of the smsr gene, including sequences for histidine residue 344 and aspartate residue 348 (SMSrdelEx6; supplementary Fig. 1). The SMSrD348E mice express a LacZ reporter protein and the SMSrdelEx6 mice express a SMSrNT-eGFP fusion protein (Fig. 3A; supplementary Figs. 1A, 5A) under transcriptional control of the endogenous SMSr promoter. Thus both should represent the expression of SMSr. For inactivation of the mouse smsr gene, we used the nonconditional SMSrD348E or the conditional SMSrdelEx6 vector (for details see Materials and Methods).
Fig. 3.
Generation of SMSrD348E mice. A: Scheme of homologous recombination for generation of SMSrD348E mice. B: Genotyping PCR for wild-type (+/+), heterozygous (+/SMSrD348E), and homozygous (SMSrD348E) mice; characteristic DNA fragments 393 bp (wild-type), 493 bp (D348E), 580 bp (Neo; mice with neomycin selection cassette). C: The sequence exchange leading to the SMSrD348E point mutation additionally produced an MfeI restriction site. This was validated by MfeI digestion of a 433 bp PCR amplicon generated from the D348E allele. The fragments of 320 bp and 113 bp after MfeI treatment confirm the identity of the point mutation. D: Southern blot analysis of SMSrD348E mice. Expected fragment sizes: BclI restriction, wild-type 12.2 kbp; D348E 13.8 kbp.
SMSrD348E and SMSrdelEx6 mice are viable and fertile
Heterozygous animals without the neomycin selection cassette were bred to generate homozygous SMSrD348E or a SMSrdelEx6 mice, respectively. Homozygous mice of both mouse lines were viable and fertile. The different genotypes were confirmed by PCR genotyping (Fig. 3B; supplementary Fig. 1B) and Southern blot hybridization (Fig. 3D; supplementary Fig. 1C). In the case of the SMSrD348E mice, an additional MfeI digestion confirmed the D348E point mutation (Fig. 3C). Offspring from matings with heterozygous mice were born at the expected Mendelian ratio for both mouse lines, indicating a nonessential role of SMSr during embryogenesis.
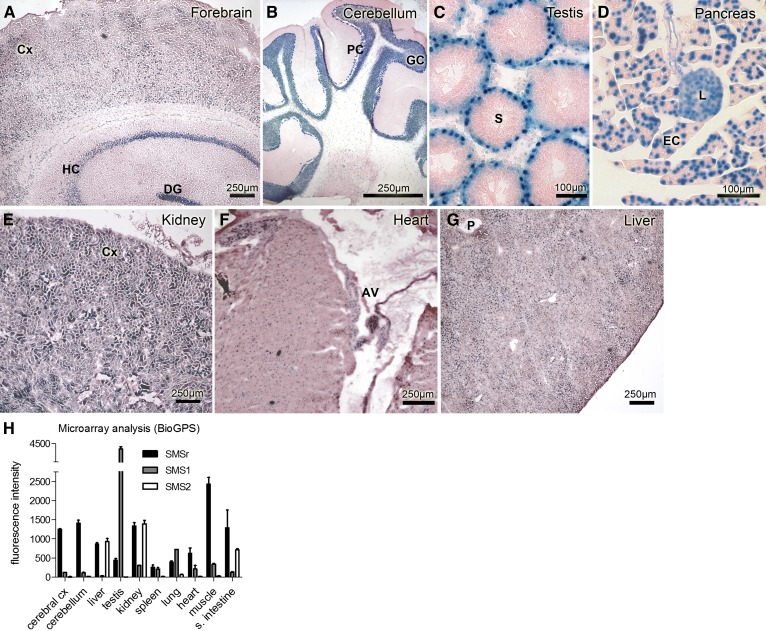
Mouse smsr is a widely expressed gene
SMSrD348E mice express the β-galactosidase reporter protein with a nuclear localization signal under the control of the endogenous smsr promoter (Fig. 3A). As there are essentially no data available regarding the tissue- and cell type-specific expression of SMSr, we analyzed the β-galactosidase expression in different tissues of the SMSrD348E mice (Fig. 4), using tissues from wild-type mice as controls (data not shown). We detected β-galactosidase staining in brain neuronal areas such as the forebrain cortex, the dentate gyrus, and the hippocampal regions CA1, CA2, CA3, and CA4, as well as the cerebellar and the granular cell layer (Fig. 4A, B). In testis, we detected staining signals adjacent to the basal epithelium of the seminiferous tubules indicating nuclear staining of spermatogonia (Fig. 4C). Furthermore, we detected β-galactosidase expression throughout the pancreas, with a prominent staining of the exocrine portion and a somewhat weaker staining of the endocrine β-islets (Fig. 4D). In kidney, we found smsr promoter activity predominantly in the cortex (Fig. 4E), whereas the medulla and papilla were devoid of β-galactosidase staining (data not shown). Cardiomyocytes of the myocard showed weak nuclear staining, but the smsr promoter activity in the atrioventricular node was more pronounced (Fig. 4F). In liver, β-galactosidase staining was observed throughout the whole tissue, but the staining was not homogenous, e.g., with stronger signals in the regions surrounding portal fields (Fig. 4G; data not shown). The results suggest a broad tissue distribution of smsr transcripts in mice. Moreover, microarray experiments revealed that smsr transcripts are relatively evenly expressed in mouse tissues, especially in comparison to sms1 and sms2 transcripts (Fig. 4H).
Fig. 4.
Mouse smsr is a ubiquitously expressed gene. β-galactosidase staining of cryo-sections from various SMSrD348E mouse tissues (A–G) correlates with smsr promoter activity. Neuronal regions of allo- and neocortical brain areas (A) and cerebellum (B) show prominent β-galactosidase staining mainly of nuclei and perinuclear areas of neurons. In testis, spermatogonia in the basal epithelium of the seminiferous tubules show similar nuclear β-galactosidase staining (C). In pancreas, β-galactosidase expression is displayed throughout the whole tissue, with a prominent staining of the exocrine portion and a somewhat weaker staining of the endocrine β-islets (D). In kidney, prominent β-galactosidase staining is displayed in the tubular system of the cortex (E). In heart, nuclei of all cardiomyocytes are β-galactosidase positive, especially those of the atrioventricular node (F). In liver, the distribution of β-galactosidase activity is diffuse with areas of more and others of less intensive staining that cannot be clearly associated to special regions of the hepatic lobules (G). AV, atrioventricular node; Cx, cortex; DG, dentate gyrus; EC, exocrine cells; GC, granular cell layer; HC, hippocampus; L, islets of Langerhans; P, portal field; PC, Purkinje cell layer; S, seminiferous tubule. H: Transcript expression profiles from microarray analysis of SMSr (Samd8), SMS2 (Sgms2), and SMS1 (Sgms1), modified according to BioGPS (47), data set Mouse MOE430 Gene Atlas (48). Microarray data were generated using an Affymetrix Mouse Genome 430 2.0 Array (GEO platform accession number GPL1261).
Tissue-specific distribution of SMSr protein
To complement our analysis of smsr expression in mice, we raised polyclonal antibodies against a peptide corresponding to the C terminus of SMSr, which is conserved between the mouse and human protein (CDGPIPDLSSDQYQY; Fig. 5A). The antibodies cross-reacted with a protein migrating at approximately 42 kDa in lysates of yeast cells expressing V5-tagged human SMSr; this protein was absent in control cells (Fig. 5B). To determine the tissue-specific expression of SMSr in mice, total protein extracts of 19 different tissues from wild-type and SMSrdelEx6 mice were subjected to immunoblot analysis. As the SMSrNT-eGFP fusion protein produced in SMSrdelEx6 mice lacks the C terminus of native SMSr, tissue samples from these mice served as control for the specificity of the anti-SMSr antibodies. In tissues from wild-type mice, a protein migrating at approximately 42 kDa was found to cross-react with the anti-SMSr antibodies. This protein was consistently absent from tissues of SMSrdelEx6 mice (Fig. 5C, D) and its estimated molecular mass is close to that of the 415-residue protein encoded by the mouse smsr gene. Therefore, we here refer to the 42 kDa protein as SMSr. In line with the β-galactosidase expression and microarray data, SMSr shows a broad tissue distribution, with the strongest expression in testis, brain, kidney, and pancreas. SMSr was also expressed in embryonic fibroblasts and bone marrow-derived macrophages (data not shown). An additional immuno-reactive protein, at approximately 48 kDa, was found in a subset of tissues from wild-type mice, but not in those from SMSrdelEx6 mice. This protein, which we here refer to as SMSr-2, was particularly abundant in testis (Fig. 5C). In line with these findings, database analysis revealed two distinct mouse smsr transcripts, one coding for the 415-residue protein (SMSr) and the other one coding for a 478-residue protein (Ensembl gene identification: ENSMUSG00000021770). Expression of the 478-residue protein, which likely corresponds to SMSr-2, is predicted to result from alternative splicing, which creates an alternative start codon in exon 1. The predominance of SMSr-2 over SMSr in testis might therefore be a consequence of the fact that alternative splicing is particularly prevalent in this tissue (44).
Fig. 5.
Tissue-specific expression of SMSr protein in mice. A: Predicted membrane topology of wild-type SMSr, SMSrD348E, and SMSrNT-eGFP fusion protein produced in SMSrdelEx6 mice. The eGFP reporter replaces the endogenous C terminus in the SMSrNT-eGFP (SMSrdelEx6) fusion protein and is predicted to be oriented toward the ER lumen. Lack of the C terminus allowed validating specificity of newly generated antibodies targeting the C terminus in immunoblot analysis (B–D). Sizes: Wild-type and SMSrD348E, 48.2 kDa (415 aa); SMSrNT-eGFP (SMSrdelEx6), 63.5 kDa (559 aa). B: Immunoblot analysis with lysates from yeast cells overexpressing an hSMSr-V5 vector confirmed specificity of the generated antibodies. The 49.6 kDa SMSr-V5 fusion protein was recognized by the SMSr antibodies, as well as by an antibody targeting the V5-tag. Ponceau S staining was used as loading control. C, D: Immunoblot analysis of wild-type tissues. Lysates from SMSrdelEx6 mice served as negative control and further confirmed specificity of the antibodies. Besides the 48.2 kDa SMSr protein, the antibodies recognize the predicted 54.7 kDa SMSr-2 isoform. Both isoforms did not migrate to positions expected for their theoretical mass. Using different protein standards, we show that SMSr and the corresponding SMSr-V5 are detected at about 42 kDa, whereas SMSr-2 was detected at about 48 kDa. GAPDH was used as loading control. Note: 25 μg of protein were applied for forebrain, cerebellum, and testis, 50 μg for liver and other tissues. (Data are representative of at least three independent experiments; s., small; l., large). hSMSr, human SMSr.
SMSr is the principal CPE synthase in brain
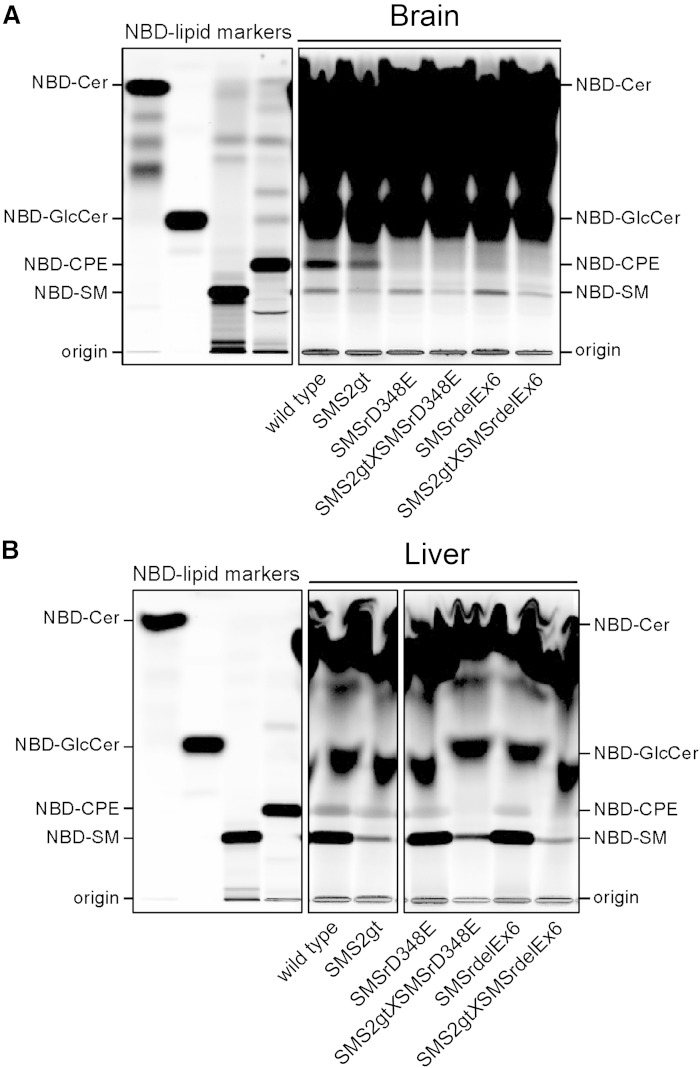
We next analyzed the impact of catalytic inactivation of SMSr on CPE synthase activity in mouse brain and liver, i.e., tissues in which this activity was first described (21). Because SMS2 is known to have dual activity as CPE synthase (23), we crossed the SMSr mutant mice with SMS2gt mice (27) and bred double mutants to determine the relative contribution of each enzyme to CPE biosynthesis. CPE-synthase activity was determined by incubating liver and brain microsomal preparations with NBD-Cer and analyzing the reaction products by TLC. As shown in Fig. 6A, incubation of NBD-Cer with brain microsomes from wild-type mice resulted in the formation of NBD-CPE, NBD-SM, and NBD-glucosylceramide (NBD-GlcCer). Formation of NBD-CPE was essentially abolished when NBD-Cer was incubated with microsomes from SMSrD348E or SMSrdelEx6 mice, while the production of NBD-SM and NBD-GlcCer was not affected. In brain microsomes from SMS2gt mice, on the other hand, the formation of NBD-SM was strongly diminished while NBD-CPE production was largely unaffected. These results indicate that SMSr operates as the principal CPE synthase in brain. In liver microsomes of wild-type mice, NBD-Cer was also converted into NBD-CPE, NBD-SM, and NBD-GlcCer (Fig. 6B). Formation of NBD-SM was unaffected by catalytic inactivation of SMSr, but almost completely abolished in liver microsomes from SMS2gt mice. Formation of NBD-CPE, on the other hand, was only slightly reduced in liver microsomes from SMSrD348E, SMSrdelEx6, and SMS2gt mice. However, NBD-CPE production was completely wiped out in liver microsomes from the double mutants (Fig. 6B). From this we conclude that in mouse liver, both SMSr and SMS2 contribute significantly to CPE biosynthesis.
Fig. 6.
Impact of SMSr and SMS2 inactivation on CPE and SMS activity in mouse liver and brain. A, B: TLC separation of reaction products formed after incubation of liver and brain microsomal preparations with NBD-Cer. NBD-CPE production in brain is completely abrogated in SMSrD348E and SMSrdelEx6 single mutants, but not in SMS2gt microsomes (A). In contrast, in the liver, production of NBD-CPE is only reduced in single mutants and not detectable in SMS2gt×SMSrD348E and SMS2gt×SMSrdelEx6 double mutants (B). (Data are representative of at least two independent experiments).
Impact of SMSr inactivation on tissue CPE levels
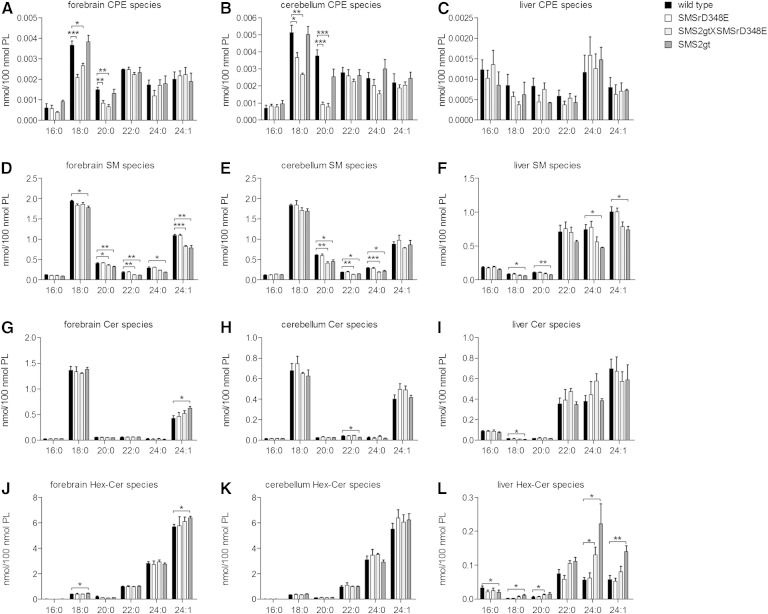
We next analyzed the impact of disrupting SMSr catalytic activity on CPE levels in various mouse tissues, including brain and liver. To this end, we used LC-MS to quantify individual CPE species in tissues from wild-type, SMSrD348E, SMS2gt, and SMSrD348E×SMS2gt double mutant mice. Forebrain and cerebellum of SMSrD348E and the double mutant mice displayed a 40–60% reduction in short-chain CPE species (C18:0, C20:0) relative to wild-type mice (Fig. 7A, B), consistent with SMSr being a major CPE synthase in brain (Fig. 6A). SMSr inactivation had, if any, only a minor impact on long-chain CPE levels (C22:0, C24:0, C24:1) and did not influence SM levels (Fig. 7D, E). In contrast, deletion of SMS2 did not affect CPE levels in these tissues, but caused a minor (up to 20%) reduction in SM levels, in line with its impact on brain-associated SM synthase activity (Fig. 6A).
Fig. 7.
LC-MS analysis of CPE, SM, ceramide, and hexoyslceramide species in forebrain, cerebellum, and liver of wild-type, SMSrD348E, SMS2gt×SMSrD348E, and SMS2gt mice. A–C: Analysis of main CPE species. D, E: Analysis of main SM species. F–H: Analysis of main ceramide species. I–K: Analysis of main hexosylceramide (Hex-Cer) species. Data are means ± SEM; *P < 0.05, **P < 0.005, ***P < 0.001; n = 3–4.
We were unable to detect significant differences between CPE levels in liver from wild-type, SMSrD348E, and double mutant mice (Fig. 7C), even though a combined inactivation of SMSr and SMS2 essentially abolished liver-associated CPE synthase activity (Fig. 6A). However, a reliable quantification of CPE species in liver is complicated by the fact that their levels are barely detectable, presumably due to metabolic conversion of CPE by liver-specific PEMT (Fig. 1C, D). Removal of SMS2 resulted in a significant decrease in liver SM levels (Fig. 7C), in line with SMS2 corresponding to the major SM synthase activity in this tissue (Fig. 6B).
Further analysis revealed only a minor decrease in the levels of short-chain CPE species in testis, kidney, spleen, and lung of SMSrD348E mutant mice (supplementary Fig. 2). In the small intestine, a significant decrease was only observed for the double mutant (supplementary Figs. 2, 6), indicating that both SMSr and SMS2 contribute to the total CPE levels in this tissue. In heart and muscle, on the other hand, we could not detect any significant difference in CPE levels between wild-type, SMSrD348E, and double mutant mice (supplementary Figs. 2, 6). Removal of SMS2 caused a significant reduction in SM levels in kidney (supplementary Fig. 3), a tissue where this enzyme is well-expressed (Fig. 4H). Quantitative MS/MS analysis of the various glycerophospholipid classes revealed no obvious changes between tissues of wild-type and mutant mice, except for a minor increase in the overall levels of PE and phosphatidylserine in the spleen and heart of the mutants (supplementary Fig. 8).
In sum, these results indicate that SMSr significantly contributes to the CPE pool in the brain, and only to a minor extent to the CPE content of the testis, kidney, spleen, and lung. Curiously, combined inactivation of SMSr and SMS2 did not eliminate CPE in any of the tissues examined. This implies that mammals are equipped with CPE synthase(s) other than SMSr and SMS2. The identity of these enzyme(s) remains to be established.
SMSr inactivation does not affect ceramide and hexosylceramide levels in mouse tissues
Acute depletion or catalytic inactivation of SMSr in human HeLa or Drosophila S2 cells causes a substantial rise in ER ceramide and hexosylceramide levels (24). Unexpectedly, none of the tissues of SMSrD348E mutant mice displayed any increase in ceramide or hexosylceramide content when compared with wild-type (Fig. 7; supplementary Fig. 7), not even in brain, where SMSr inactivation essentially abolished CPE synthase activity (Fig. 6A). In the kidney, spleen, lung, and heart of SMS2gt×SMSrD348E mutant mice, we detected a minor increase in ceramide and hexosylceramide levels (supplementary Fig. 7). For the kidney, this could be a direct consequence of a block in SMS2-mediated SM production, as the accumulation of ceramides and hexosylceramides in this tissue was accompanied by a significant drop in SM levels (supplementary Fig. 6). Besides the species with incorporated sphingosine (d18:1), we also investigated the sphinganine (d18:0)- and sphingadiene (d18:2)-based species of ceramide, hexosylceramide, and SM, as well as species with hydroxylated and odd-numbered fatty acyl-chains and free long-chain bases (d18:0, d18:1, and d18:2) in liver and brain. In all cases, we did not find any significant differences in the levels of these species between wild-type and SMSrD348E mutant mice (data not shown). From this, we conclude that SMSr does not seem to operate as a critical regulator of ceramide levels in mice.
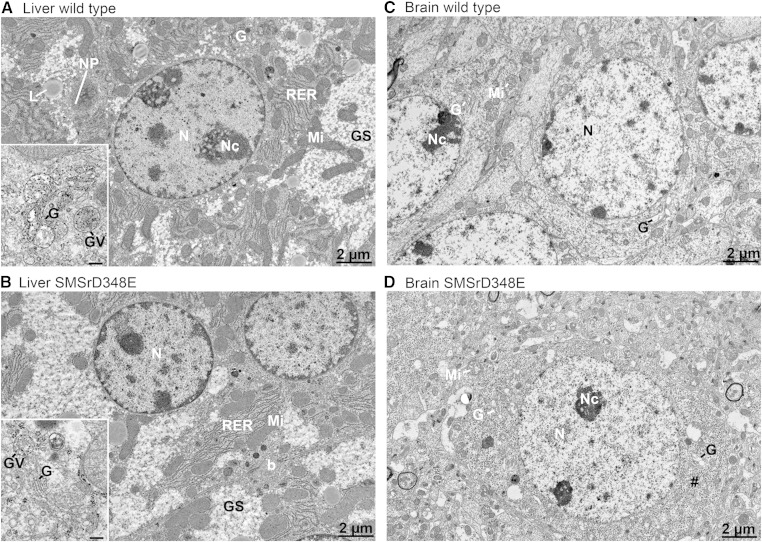
Disruption of SMSr catalytic activity has no impact on the structural integrity of mouse brain or liver cells
The rise in ER ceramides caused by an acute depletion of SMSr in cultured animal cells results in a fragmentation of ER exit sites, a structural collapse of the Golgi complex, and induction of mitochondrial apoptosis, which can be reverted by blocking de novo ceramide synthesis (24, 26). This led us to analyze liver and brain cells from SMSrD348E mice for ultrastructural aberrations in their secretory apparatus. We did not observe any obvious deviations in Golgi or ER morphology in hepatocytes from SMSrD348E mice when compared with their wild-type littermates (Fig. 8A, B). In addition, the integrity of mitochondria was not altered and we could not detect any difference in appearance or number of lipid droplets or glycogen granules. We also found no significant changes in the serum activities of liver transaminases and creatine phosphokinase between wild-type and SMSrD348E mutant mice (supplementary Fig. 9), indicating that catalytic inactivation of SMSr does not compromise the overall integrity of liver cells. A systematic analysis of neuronal cells in different regions of the brain in SMSrD348E mice also did not reveal any significant alterations in cellular integrity relative to wild-type mice (Fig. 8C, D). It therefore appears that cells of SMSrD348E mutant mice in general are devoid of phenotypes associated with a deregulation of ER ceramides.
Fig. 8.
Electron micrographs of hepatocytes and hippocampal neurons of wild-type and SMSrD348E mice. A: Wild-type mice reveal the typical cellular equipment of liver cells with abundant rough ER (RER), numerous mitochondria (Mi), lipid droplets (L), and large glycogen stores (Gs). Higher magnification of a small Golgi-apparatus, which is typical for liver cells, shows big vesicles containing lipoprotein complexes (insert). B: There are no obvious morphological differences detectable in cell organelles of a SMSrD348E hepatocyte. Higher magnification of a comparable Golgi-complex does show the same appearance with large vesicles containing lipoproteins (insert). C: Hippocampal neurons of wild-type mice revealed the typical morphology with abundant free ribosomes, mitochondria, and some Golgi-complexes. D: No obvious alterations in the morphology of cell organelles were observed in comparable SMSrD348E neurons. For analysis, littermates were used (n = 4). N, nucleus; Nc, nucleolus; b, bile canaliculus; G, Golgi-apparatus, L, lipid droplet; Gs, glycogen stores; GV, Golgi vesicle; Mi, mitochondria; #, Nissl substance; NP, nuclear pores; scale bar insert, 200 nm.
DISCUSSION
SMSr is a highly conserved member of the SMS family that, along with its relative SMS2, catalyzes production of the SM analog CPE. Studies in cultured cells previously revealed a critical role of SMSr in ceramide homeostasis and as suppressor of ceramide-induced mitochondrial apoptosis. We now show that ubiquitous catalytic inactivation of SMSr in mice primarily disrupts CPE biosynthesis in the brain without having any obvious impact on steady state ceramide levels, cell integrity, or survival. Besides pointing at a discrepancy between the in vitro and in vivo function of SMSr, our study provides a first inventory of CPE levels and CPE-biosynthetic enzymes in mammals.
The present analyses of enzyme activities in the brain and liver of SMS mutant mice complements our previous heterologous expression studies (23, 24), and now firmly establishes SMSr as monofunctional CPE synthase and SMS2 as a bifunctional enzyme with both SM and CPE synthase activity. In line with our previous findings in cultured mammalian cells (24), both enzymes seem to produce only trace amounts of CPE in mouse tissues, with steady state levels ∼300- to 1,500-fold below those of SM. Metabolic labeling experiments in cultured HeLa cells indicated that CPE is not a short-lived metabolic intermediate (24). However, our present data suggest that in liver cells, CPE may be used as precursor in a methylation-dependent pathway of SM production by the liver-specific methyl-transferase, PEMT. In fact, this could explain why steady-state CPE levels in the liver are particularly low in comparison to other tissues, in spite of expressing both SMSr and SMS2. While PEMT-catalyzed methylation of CPE remains to be verified in enzyme assays, our results indicate that the contribution of the PEMT-dependent pathway to the total liver-associated SM pool is at best very small and its physiological relevance is unclear.
All SMS enzymes utilize a lipid phosphate phosphatase-type reaction mechanism, involving a single lipid binding site and proceeding via transfer of the headgroup from PC or PE to a conserved histidine in the enzyme’s active site (9). When diacylglycerol is replaced by ceramide, the headgroup is transferred onto ceramide to generate SM or CPE, respectively. Why SMSr and SMS2 are intrinsically unable to produce bulk amounts of CPE is not known. The ability of SMS2 to produce large amounts of SM could be due to the enzyme having a higher affinity for PC than PE as headgroup donor in the transfer reaction or, alternatively, because of a high PC/PE ratio in the exoplasmic leaflet of the plasma membrane where its active site resides (7, 10). Another possibility is that catalytic activity of SMS2 and SMSr is under negative control by their product, CPE. In any case, bulk production of CPE in insects is catalyzed by an entirely different enzyme that belongs to an insect-specific branch of the CDP-alcohol transferase superfamily (25). This implies that the reaction mechanism used by SMS family members is unsuited for efficient production of CPE.
Remarkably, combined inactivation of SMSr and SMS2 significantly reduced, but did not eliminate, CPE levels in any of the mouse tissues examined. At present, we cannot rule out that residual CPE levels originate from the diet. Alternatively, enzymes other than SMSr and SMS2 might contribute to CPE production. For instance, enzymes that utilize ethanolamine in a headgroup transfer reaction with SM could help sustain the CPE pool. We recently noticed that Golgi-resident SM synthase SMS1 produces minute amounts of CPE, and that only a few substitutions of residues near the active site are needed to convert the enzyme into a monofunctional CPE synthase (M. Kol and J. C. M. Holthuis, unpublished observations). Therefore, our finding that combinatorial loss of SMSr and SMS2 has no impact on CPE levels in testis could be due to the relatively high expression of SMS1 in this tissue. Knowing the origin of the SMSr/SMS2-independent CPE pool should facilitate future studies on the biological role of this enigmatic sphingolipid in mammals.
We previously showed that acute disruption of SMSr catalytic activity in human HeLa or Drosophila S2 cells causes an accumulation of ER ceramides, a structural collapse of ER exit sites, Golgi fragmentation, and induction of a mitochondrial pathway of apoptosis (24, 26). Another study with cultured HeLa cells suggested that the accumulation of ceramides upon SMSr depletion is not due to an increased de novo synthesis (45). Nevertheless, the compromised structural integrity and viability of acutely SMSr-depleted cells could be suppressed by blocking de novo ceramide synthesis, stimulating ceramide export from the ER, or consuming excess ceramides by targeting SMS1 to the ER, indicating that these phenotypes are primarily due to a deregulation of ER ceramides (24, 26). Therefore, our present finding that catalytic inactivation of SMSr in mice does not significantly alter ceramide levels, cell survival, or integrity in any of the tissues examined came as a surprise. How can these two contradictory outcomes be reconciled?
A first point to consider is that the experiments with cultured cells focused on the short-term consequences of SMSr inactivation, i.e., within 3 days after siRNA-mediated depletion of the endogenous protein and its exchange for a siRNA-resistant catalytically inactive protein (SMSrD348E). This time span might be too short for some cells to adapt their metabolic activity toward normalizing ER ceramide levels before any perturbation of cell integrity and viability occurs. Because we analyzed adult mice, we cannot rule out that the SMSrD348E mutation causes a transient deregulation of ER ceramides in an early stage of development that individual embryonic cells are able to overcome. Ceramides serve as “metabolic hub” and cells are equipped with a multitude of enzymes to modulate their levels. This may explain why genetic ablation of SMS1 and SMS2 in mice causes only a modest increase in tissue ceramide levels [(13, 15, 27), and this study], even though these enzymes normally consume the bulk of newly synthesized ceramides (10). As tissues of SMSrD348E mutant mice did not contain any significantly elevated levels of sphingoid bases, hexosylceramides, or SM, it is unclear how embryonic cells with disrupted SMSr catalytic activity were able to overcome a deregulation of ER ceramides associated with a disruption in SMSr catalytic activity. One approach to investigate short-term effects of SMSr inactivation and explore compensatory mechanisms in vivo could be to crossbreed the conditional SMSrdelEx6 mice described in this study with mice expressing an inducible Cre recombinase (46). The presence of a functional SAM domain in SMSrD348E and SMSrdelEx6 mice might also explain why ceramide levels and cellular integrity are not altered, because this domain, similar to catalytic activity, proved to be essential in preventing ceramide accumulation and apoptosis in cultured HeLa cells (24, 26). Finally, we cannot rule out the existence of a protein with redundant function that copes with the consequences of SMSr inactivation. Loss of this protein in a genetically unstable cancer cell line such as HeLa would provide an alternative explanation for the discrepancy between data collected from cell lines and the mouse model.
In summary, our data indicate that SMSr does not qualify as an essential regulator of ceramide levels in mice kept under standardized conditions. While SMSr is a CPE synthase with homologs found throughout the animal kingdom, we find that SMSr-catalyzed CPE production is dispensable for mouse development, survival, and healthiness. This does not rule out a critical role of SMSr under stressed metabolic conditions. Therefore, challenging SMSrD348E mutant mice with inflammatory stimuli, altered diet conditions, or drugs that interfere with ceramide metabolism might shed further light on its in vivo function.
Supplementary Material
Acknowledgments
The authors thank Prof. Maarten Egmond (University of Utrecht) for synthesis and purification of SMSr immunogenic peptides, Brita Wilhelm (Bonn) and Susanne Lingrell (Helsinki) for technical assistance, as well as Christine Siegmund (Bonn) for blastocyst injection. Dorothea Schünke (Essen) is gratefully acknowledged for skillful preparation of the ultrathin sections. The authors thank Jana Koksch (Bonn) for excellent technical assistance in performing the AST, ALT, and CPK measurements in mouse serum.
Footnotes
Abbreviations:
- Ad.GFP
- adenovirus expressing only green fluorescent protein
- Ad.PEMT
- adenovirus expressing both phosphatidylethanolamine N-methyltransferase and green fluorescent protein
- CPE
- ceramide phosphoethanolamine
- DTA
- diphtheria toxin A
- eGFP
- (enhanced) green fluorescent protein
- ER
- endoplasmic reticulum
- ES
- embryonic stem
- Flp
- flippase
- frt
- Flp recognition target
- GFP
- green fluorescent protein
- HR
- homologous region
- lacZ
- β-galactosidase gene
- loxP
- locus of X-over P1
- mSMSr
- mouse sphingomyelin synthase-related protein
- NBD
- nitrobenzoxadiazole
- NBD-Cer
- nitrobenzoxadiazole-ceramide
- NBD-GlcCer
- nitrobenzoxadiazole glucosylceramide
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PEMT
- phosphatidylethanolamine N-methyltransferase
- SAM
- sterile α motif
- SMS
- sphingomyelin synthase
- SMSr
- sphingomyelin synthase-related protein
- UTR
- untranslated region
This study was supported by the German Research Foundation through the Collaborative Research Centre SFB-645, projects B2 to K.W. and Z4 to P.D. and SFB-944 project P14 to J.C.M.H. and by the grant Wi774/22-1 to E.W. and the European Union through the 7th FP Marie-Curie ITN “Sphingonet” to J.C.M.H., and the grant MOP5182 from the Canadian Institutes of Health Research to R.L.J. and D.E.V.
TEM was performed at the IMCES, University Clinics of Essen.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of nine figures.
REFERENCES
- 1.Holthuis J. C., Pomorski T., Raggers R. J., Sprong H., Van Meer G. 2001. The organizing potential of sphingolipids in intracellular membrane transport. Physiol. Rev. 81: 1689–1723. [DOI] [PubMed] [Google Scholar]
- 2.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 3.Breslow D. K., Weissman J. S. 2010. Membranes in balance: mechanisms of sphingolipid homeostasis. Mol. Cell. 40: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrill A. H. 2002. De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. Chem. 277: 25843–25846. [DOI] [PubMed] [Google Scholar]
- 5.Morad S. A. F., Cabot M. C. 2013. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer. 13: 51–65. [DOI] [PubMed] [Google Scholar]
- 6.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426: 803–809. [DOI] [PubMed] [Google Scholar]
- 7.Huitema K., van den Dikkenberg J., Brouwers J. F. H. M., Holthuis J. C. M. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279: 18688–18693. [DOI] [PubMed] [Google Scholar]
- 9.Tafesse F. G., Ternes P., Holthuis J. C. M. 2006. The multigenic sphingomyelin synthase family. J. Biol. Chem. 281: 29421–29425. [DOI] [PubMed] [Google Scholar]
- 10.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. M. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282: 17537–17547. [DOI] [PubMed] [Google Scholar]
- 11.Separovic D., Semaan L., Tarca A. L., Awad Maitah M. Y., Hanada K., Bielawski J., Villani M., Luberto C. 2008. Suppression of sphingomyelin synthase 1 by small interference RNA is associated with enhanced ceramide production and apoptosis after photodamage. Exp. Cell Res. 314: 1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyaji M., Jin Z-X., Yamaoka S., Amakawa R., Fukuhara S., Sato S. B., Kobayashi T., Domae N., Mimori T., Bloom E. T., et al. 2005. Role of membrane sphingomyelin and ceramide in platform formation for Fas-mediated apoptosis. J. Exp. Med. 202: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yano M., Watanabe K., Yamamoto T., Ikeda K., Senokuchi T., Lu M., Kadomatsu T., Tsukano H., Ikawa M., Okabe M., et al. 2011. Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice. J. Biol. Chem. 286: 3992–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano M., Yamamoto T., Nishimura N., Gotoh T., Watanabe K., Ikeda K., Garan Y., Taguchi R., Node K., Okazaki T., et al. 2013. Increased oxidative stress impairs adipose tissue function in sphingomyelin synthase 1 null mice. PLoS ONE. 8: e61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Fan Y., Liu J., Li Y., Huan C., Bui H. H., Kuo M-S., Park T-S., Cao G., Jiang X-C. 2012. Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 32: 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Zhang H., Liu J., Liang C-P., Li Y., Li Y., Teitelman G., Beyer T., Bui H. H., Peake D. A., et al. 2011. Reducing plasma membrane sphingomyelin increases insulin sensitivity. Mol. Cell. Biol. 31: 4205–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsutake S., Zama K., Yokota H., Yoshida T., Tanaka M., Mitsui M., Ikawa M., Okabe M., Tanaka Y., Yamashita T., et al. 2011. Dynamic modification of sphingomyelin in lipid microdomains controls development of obesity, fatty liver, and type 2 diabetes. J. Biol. Chem. 286: 28544–28555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowda S., Yeang C., Wadgaonkar S., Anjum F., Grinkina N., Cutaia M., Jiang X-C., Wadgaonkar R. 2011. Sphingomyelin synthase 2 (SMS2) deficiency attenuates LPS-induced lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 300: L430–L440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Huan C., Chakraborty M., Zhang H., Lu D., Kuo M-S., Cao G., Jiang X-C. 2009. Macrophage sphingomyelin synthase 2 deficiency decreases atherosclerosis in mice. Circ. Res. 105: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Dong J., Zhu X., Wang W., Yang Q. 2011. The effect of sphingomyelin synthase 2 (SMS2) deficiency on the expression of drug transporters in mouse brain. Biochem. Pharmacol. 82: 287–294. [DOI] [PubMed] [Google Scholar]
- 21.Malgat M., Maurice A., Baraud J. 1986. Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain. J. Lipid Res. 27: 251–260. [PubMed] [Google Scholar]
- 22.Malgat M., Maurice A., Baraud J. 1987. Sidedness of ceramide-phosphoethanolamine synthesis on rat liver and brain microsomal membranes. J. Lipid Res. 28: 138–143. [PubMed] [Google Scholar]
- 23.Ternes P., Brouwers J. F. H. M., van den Dikkenberg J., Holthuis J. C. M. 2009. Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J. Lipid Res. 50: 2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Brouwers J. F. H. M., Somerharju P., Rabouille C., Holthuis J. C. M. 2009. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J. Cell Biol. 185: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vacaru A. M., van den Dikkenberg J., Ternes P., Holthuis J. C. M. 2013. Ceramide phosphoethanolamine biosynthesis in Drosophila is mediated by a unique ethanolamine phosphotransferase in the Golgi lumen. J. Biol. Chem. 288: 11520–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tafesse F. G., Vacaru A. M., Bosma E. F., Hermansson M., Jain A., Hilderink A., Somerharju P., Holthuis J. C. M. 2014. Sphingomyelin synthase-related protein SMSr is a suppressor of ceramide-induced mitochondrial apoptosis. J. Cell Sci. 127: 445–454. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Zhang H., Li Z., Hailemariam T. K., Chakraborty M., Jiang K., Qiu D., Bui H. H., Peake D. A., Kuo M-S., et al. 2009. Sphingomyelin synthase 2 is one of the determinants for plasma and liver sphingomyelin levels in mice. Arterioscler. Thromb. Vasc. Biol. 29: 850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gierl M. S., Karoulias N., Wende H., Strehle M., Birchmeier C. 2006. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 20: 2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., Court D. L., Jenkins N. A., Copeland N. G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73: 56–65. [DOI] [PubMed] [Google Scholar]
- 30.Buchholz F., Angrand P. O., Stewart A. F. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16: 657–662. [DOI] [PubMed] [Google Scholar]
- 31.Magin T. M., McWhir J., Melton D. W. 1992. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 20: 3795–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy A., Gertsenstein M., Vintersten K., Behringer R. 2003. Manipulating the Mouse Embryo. 3rd edition. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25: 139–140. [DOI] [PubMed] [Google Scholar]
- 34.Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. 1998. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7: 105–112. [DOI] [PubMed] [Google Scholar]
- 35.Ginkel C., Hartmann D., vom Dorp K., Zlomuzica A., Farwanah H., Eckhardt M., Sandhoff R., Degen J., Rabionet M., Dere E., et al. 2012. Ablation of neuronal ceramide synthase 1 in mice decreases ganglioside levels and expression of myelin-associated glycoprotein in oligodendrocytes. J. Biol. Chem. 287: 41888–41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs R. L., Zhao Y., Koonen D. P. Y., Sletten T., Su B., Lingrell S., Cao G., Peake D. A., Kuo M-S., Proctor S. D., et al. 2010. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 285: 22403–22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridgway N. D., Vance D. E. 1992. Phosphatidylethanolamine N-methyltransferase from rat liver. Methods Enzymol. 209: 366–374. [DOI] [PubMed] [Google Scholar]
- 38.Degen J., Meier C., Van Der Giessen R. S., Söhl G., Petrasch-Parwez E., Urschel S., Dermietzel R., Schilling K., De Zeeuw C. I., Willecke K. 2004. Expression pattern of lacZ reporter gene representing connexin36 in transgenic mice. J. Comp. Neurol. 473: 511–525. [DOI] [PubMed] [Google Scholar]
- 39.Folch J., Lees M., Stanley Sloane G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 40.Bartlett E. M., Lewis D. H. 1970. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 36: 159–167. [DOI] [PubMed] [Google Scholar]
- 41.Kainu V., Hermansson M., Somerharju P. 2010. Introduction of phospholipids to cultured cells with cyclodextrin. J. Lipid Res. 51: 3533–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haimi P., Uphoff A., Hermansson M., Somerharju P. 2006. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 78: 8324–8331. [DOI] [PubMed] [Google Scholar]
- 43.Muehlenberg B. A., Sribney M., Duffe M. K. 1972. Occurrence and biosynthesis of ceramide phosphorylethanolamine in chicken and rat liver. Can. J. Biochem. 50: 166–173. [DOI] [PubMed] [Google Scholar]
- 44.Elliott D. J., Grellscheid S. N. 2006. Alternative RNA splicing regulation in the testis. Reproduction. 132: 811–819. [DOI] [PubMed] [Google Scholar]
- 45.Siow D. L., Wattenberg B. W. 2012. Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287: 40198–40204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feil S., Valtcheva N., Feil R. 2009. Inducible Cre mice. Methods Mol. Biol. 530: 343–363. [DOI] [PubMed] [Google Scholar]
- 47.Wu C., Macleod I., Su A. I. 2013. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41: D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lattin J. E., Schroder K., Su A. I., Walker J. R., Zhang J., Wiltshire T., Saijo K., Glass C. K., Hume D. A., Kellie S., et al. 2008. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.