Abstract
Lipin proteins (lipin 1, 2, and 3) regulate glycerolipid homeostasis by acting as phosphatidic acid phosphohydrolase (PAP) enzymes in the TG synthesis pathway and by regulating DNA-bound transcription factors to control gene transcription. Hepatic PAP activity could contribute to hepatic fat accumulation in response to physiological and pathophysiological stimuli. To examine the role of lipin 1 in regulating hepatic lipid metabolism, we generated mice that are deficient in lipin-1-encoded PAP activity in a liver-specific manner (Alb-Lpin1−/− mice). This allele of lipin 1 was still able to transcriptionally regulate the expression of its target genes encoding fatty acid oxidation enzymes, and the expression of these genes was not affected in Alb-Lpin1−/− mouse liver. Hepatic PAP activity was significantly reduced in mice with liver-specific lipin 1 deficiency. However, hepatocytes from Alb-Lpin1−/− mice had normal rates of TG synthesis, and steady-state hepatic TG levels were unaffected under fed and fasted conditions. Furthermore, Alb-Lpin1−/− mice were not protected from intrahepatic accumulation of diacylglyerol and TG after chronic feeding of a diet rich in fat and fructose. Collectively, these data demonstrate that marked deficits in hepatic PAP activity do not impair TG synthesis and accumulation under acute or chronic conditions of lipid overload.
Keywords: diacylglycerol, fatty acid metabolism, insulin signaling, phospholipids, lipid, triglyceride
The accumulation of lipid within the liver parenchyma (hepatic steatosis) is common in several hepatic disease states and may be linked to other abnormalities in liver metabolism and function. Indeed, in a subset of individuals, hepatic steatosis can progress to liver injury, dysfunction, and failure. Intrahepatic lipid accumulation is also usually highly correlated with systemic insulin resistance, hyperglycemia, dyslipidemias, and risk of cardiovascular disease. Hepatic steatosis is extremely prevalent in obese individuals, and with the epidemic of obesity, the occurrence of nonalcoholic fatty liver disease has risen dramatically, becoming the most common cause of liver disease in the United States (1, 2).
The primary storage form of lipid is TG, which, in the liver, is predominantly synthesized via the sequential acylation and dephosphorylation of glycerol-3-phosphate. In higher organisms, three genes (Lpin1, Lpin2, and Lpin3) encode canonical enzymes that catalyze the Mg2+-dependent dephosphorylation of phosphatidic acid (PA) to form diacylglycerol (DAG) at the endoplasmic reticulum (3, 4). The phosphatidic acid phosphohydrolase (PAP) reaction is not only the penultimate step in TG synthesis, but also a key metabolic branch point. Alterations in PA and DAG concentrations have been linked to regulation of important intracellular signaling cascades including protein kinase C (5, 6), protein kinase A (7), ERK MAPK kinase (8), and the molecular target of rapamycin (9–11). The regulation of PA and DAG concentrations has potentially important implications for hepatic nutrient homeostasis under conditions of fasting and overnutrition. Indeed, acute RNA interference (RNAi)-mediated depletion of lipin 1 or lipin 2 in mice fed a high-fat diet attenuated hepatic steatosis and led to improvements in hepatic insulin sensitivity (12, 13).
Lipins are not integral membrane proteins, and lipin 1 can translocate into the nucleus and also interact with DNA-bound transcription factors to regulate their activity (14). Lipin 1 coactivates the PPARs (14–16) and hepatic nuclear receptor 4α (HNF4α) (17) nuclear receptors but represses nuclear factor of activated T-cells 4c (18) and the sterol-response element binding protein 1 [SREBP1; (19)]. As a general rule, the transcriptional regulatory effects of lipin 1 do not require intact PAP catalytic activity and are mechanistically separable. The outlier to this generalization is repression of SREBP1, which requires both PAP activity and nuclear localization of lipin 1 (19). Thus, lipin 1 is poised to regulate intermediary fat metabolism at multiple regulatory levels.
Mice constitutively lacking lipin 1 [fatty liver dystrophic (fld) mice] exhibit very low PAP activity in most tissues, but because lipin 2 is highly expressed in liver, significant hepatic PAP activity can be detected in fld mice (4, 20). Indeed, lipin 2 likely encodes a significant proportion of hepatic PAP activity (21, 22). However, hepatic lipin 1 expression is robustly induced by a variety of metabolic stimuli that signal the need for increased PAP activity including fasting (14, 23), glucocorticoids and cAMP signaling (14, 23), experimental ethanol administration (24), sterols (25), and liver regeneration (26). These findings are congruent with older data, generated before the cloning of lipins, that detected increased PAP activity in response to these stimuli (27–30). Because lipin 1 has the highest intrinsic PAP activity of the lipin family proteins (4), the induction of lipin 1 expression by these stimuli fits with this being an adaptive response to augment the hepatic capacity for TG synthesis in response to increased supply of fatty acids. In support of this, the fasting-induced increase in hepatic PAP activity is markedly blunted in fld mice (20).
The caveat to interpreting results from fld mice is the profound lipodystrophic phenotype (31, 32) and its effects on lipid availability and metabolism. The marked deficiency in adipose tissue mass makes it difficult to isolate the effects of hepatic lipin 1 on liver TG metabolism. Recently, mice expressing a conditional allele of lipin 1 have been developed (8). These mice express, in a tissue-specific manner, lipin 1 protein that is N-terminally truncated and lacks PAP activity but retains its ability to coactivate PPARα (7). Herein, we show that mice expressing this allele in a liver-specific manner exhibit significant reductions in hepatic PAP activity but have normal rates of TG synthesis, and steady-state hepatic TG levels are unaffected. The ability of the truncated protein encoded by this allele to activate expression of fatty acid oxidation enzymes was intact. Collectively, these data demonstrate that marked deficits in hepatic PAP activity do not impair TG synthesis and accumulation under acute or chronic conditions of lipid overload.
METHODS
Mouse studies
Animal studies were approved by the institutional Animal Use and Care Committees of Washington University School of Medicine and fulfilled National Institutes of Health requirements for humane care. Generation of mice with liver-specific lipin 1 deficiency (Alb-Lpin1−/− mice) has been previously described (7, 33). Fasting studies were timed so that mice were euthanized at 0800, which was 2 h after the end of the dark cycle. Food removal was timed accordingly to achieve the desired duration of fasting.
To induce hepatic steatosis, mice were placed on a diet enriched with fat (40% kcal, mainly trans-fat; trans-oleic and trans-linoleic acids), cholesterol (2% weight [wt.]), and fructose (22% wt.) (HTF-C diet) (D09100301, Research Diets Inc.), which has been demonstrated to cause hepatic injury and inflammation (34). Mice were euthanized and tissues were harvested at the end of week 10 of the study after a 4 h fast. Liver, gonadal, and subcutaneous fat tissue samples were frozen in liquid nitrogen and stored at −80°C.
Glucose tolerance test
The week prior to being euthanized, mice were fasted for 6 h, and tails were snipped to measure baseline blood glucose using a One-Touch Ultra glucometer (Life Scan Inc.). Following an overnight fast, a 10% d-glucose solution (1 g/kg) was administered via intraperitoneal injection and 30, 60, and 120 min after injection, and total area under the curve was calculated.
Protein isolation and Western blotting
Protein from frozen tissue was homogenized in 0.3–1 ml ice-cold homogenization buffer [25 mM HEPES, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, pH 8.0, supplemented with 1 mM activated Na3VO4, 1 mM phenylmethanesulfonyl fluoride, 5 mM sodium fluoride, and 1× Complete protease inhibitor cocktail tablet (Roche, Manneheim, Germany; cat. 04693116001)] using high-speed tissue disruption with the TissueLyser II (Qiagen, Valencia, CA). Tissue homogenates were subsequently solubilized by rotating at 4°C at 50 rpm for 1 h before being centrifuged (15,000 g for 15 min at 4°C) and collecting the supernatant. Protein from cultured cells were isolated by scraping adherent cells from culture plates, collecting them in the same homogenization buffer described above, and dispersing them via pipetting. Aliquots of the lysate from tissues and cultured cells were used to determine protein concentration by the bicinchoninic acid assay according to the manufacturer’s instructions (Pierce Biotechnology; no. 23227). The remaining lysates were stored at −80°C until further analyzed.
Lysates were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes. Blots were then rinsed with Tris-buffered saline plus Tween (TBST; 0.14 M NaCl, 0.02 M Tris base, pH 7.6, and 0.1% Tween), blocked with 5% BSA in TBST for 1 h at room temperature, washed 3 × 10 min at room temperature, and incubated with the relevant primary antibody 1:1,000 in 5% BSA overnight at 4°C. Blots were then washed 3 × 5 min with TBST, incubated with relevant secondary antibodies for 1 h at room temperature, washed again 3 × 10 min with TBST, and washed 2 × 10 min with TBS. Protein bands were visualized using the Odyssey Imaging System (LiCor Biosciences, Lincoln, NE). Lipin 1 (cat. sc-98450) antibody was obtained from Santa Cruz Biotechnology (Dallas, TX). Lipin 2 antibody has been previously described (21). Anti-α-Tubulin Clone B-5-1-2 (cat. T5168) was purchased from Sigma-Aldrich (St. Louis, MO). Goat anti-mouse IRDye 680 (cat. 926-32220), goat anti-rabbit IRDye 680 (cat. 926-68021), and goat anti-rabbit 800 (cat. 926-32211) secondary antibodies were obtained from LiCor Biosciences. Akt (cat. 9272), phospho-Akt Ser473 (cat. 9271), phospho-Akt Thr308 (cat. 9275), glycogen synthase kinase (GSK)-3β (cat. 9315), and phospho- GSK-3β Ser9 (cat. 9323) were obtained from Cell Signaling (Danvers, MA).
Quantitative RT-PCR analyses
For all analyses, total RNA from liver or hepatocytes was isolated in RNAzol (Invitrogen) reagent. RNA (500 ng) was subjected to reverse transcription and SYBR GREEN RT-PCR (Applied Biosystems) following the manufacturer’s instructions. Results were corrected to the expression of 36B4. Sequence of gene-specific primers is available upon request.
PAP activity
The method to assess PAP was modified from Martin et al. (35). Approximately 50 mg of liver tissue was homogenized in 1 ml of lysis buffer [0.01 M Tris-HCl (pH 7.3), 0.25 M Sucrose, 0.5% Tween-20, 1 mM DTT, 1× EDTA-free protease inhibitor (Roche, Manneheim, Germany; cat. 04693132001)] using a Tissue Master-125. Following homogenization, aliquots of the supernatant were used to determine protein concentration by the bicinchoninic acid assay. The [14C]PA used was prepared as follows: 15 µl 0.1 mCi/ml of the [14C]PA l-α-dioleoyl [oleoyl-1-14C] Na salt ([14C]PA) (American Radiolabeled Chemicals Inc., St. Louis, MO; cat. 1303) was added to 3 mM 3-sn-PA sodium salt (Sigma Aldrich; cat. P9511) and 2 mM l-α-phosphatidylcholine (Sigma Aldrich; cat. P3556) in 1 ml of chloroform, evaporated under a stream of N2, and resuspended in 1 ml of ice cold distilled deionized water. This mixture was sonicated 3 × 10 s and kept on ice in between sonications. To run the assay reaction, 5 µl of each sample was incubated with 40 µl of assay buffer [0.1 M Tris/maleate (pH 6.9), 10 mM MgCl2, 0.2% fatty acid-free BSA, 1 mM DTT, and 1× EDTA-free protease inhibitor with or without 12.5 mM N-ethylmaleimide (Sigma Aldrich; cat. E3876)], and 5 µl of [14C]PA was incubated at 37°C for 20 min. The reaction was stopped by adding 1 ml of a 19:1 (v/v) chloroform-methanol mixture containing 0.8% olive oil to each reaction and vortexing briefly. Then 500 mg of activated aluminum oxide was added to the tubes and capped securely. Three cycles of vortexing for 30 s and being left undisturbed for 10 min followed. Samples were spun down at 10,000 g for 5 min, and 350 µl of sample was added to scintillation fluid to count the 14C radioactivity.
Liver lipid content
For fasting studies, liver TG content was determined from samples prepared in the PAP Activity assay buffer, solubilizing the homogenate 1:1 (v/v) with 1% sodium deoxycholate, vortexing, incubating at 37°C for 5 min, and then using the Infinity TG Reagent (Thermo Fisher Scientific, Waltham, MA; cat: TR22421) as per the manufacturer’s instructions.
For HTF-C studies, liver lipids were quantified by using ESI/MS analysis as described (36, 37). In brief, lipids were extracted from mouse liver using a modified Bligh and Dyer technique in the presence of internal standards. ESI/MS analyses were performed utilizing a TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA), or an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA), equipped with an automated nanospray apparatus (Nanomate HD; Advion Bioscience Ltd., Ithaca, NY). TG molecular species were analyzed using TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer in the positive-ion mode in the presence of a small amount of LiOH. Diglyceride molecular species were analyzed by derivatization with dimethylglycine and direct infusion in the presence of 0.1% formic acid, and ionized in the positive ion mode. Due to the low abundance of PA, PA molecular species were analyzed by using LTQ Orbitrap mass spectrometer at resolving powers (at m/z 400 Th) of 60,000 in full-scan mode and with the lock-mass feature engaged. Quantitative analysis of PA molecular species was performed by ratiometric comparisons of the intensities of the high mass accuracy (<5 ppm) PA deprotonated ions with those of internal standard in the negative-ion mode.
Hepatocyte isolation and in vitro studies
Hepatocytes from WT and Alb-Lpin1−/− mice were isolated as previously described (38) and cultured in DMEM containing 5% FBS for 4 h, followed by a further 1 h incubation period in serum-free DMEM. Triacylglycerol synthesis rates were quantified by using [3H]glycerol in the presence of 0.5 mM BSA-conjugated oleic acid as previously described (39). Palmitate oxidation rates were assessed using [3H]palmitate as previously described (38, 40).
Lipin 1 adenoviral constructs
A mouse lipin 1 cDNA encoding a protein lacking the first 115 amino acids (Δ115-lipin 1) has previously been described (7). To construct an adenoviral expression vector, the truncated lipin 1 cDNA was subcloned into the AdTrack-cytomegalovirus shuttle vector and recombined using the AdEasy-1 system. The WT lipin 1 and control green fluorescent protein (GFP) adenovirus constructs have been previously described (14).
Transfection studies and luciferase reporter assays
HepG2 cells were maintained in DMEM-10% fetal calf serum. Transient transfections with luciferase reporter constructs were performed by calcium-phosphate coprecipitation. SV40-driven renilla luciferase expression construct was also included in each well. For all vectors, promoterless reporters or empty vector controls were included so that equal amounts of DNA were transfected into each well. Luciferase activity was quantified 48 h after transfection by using a luminometer and the Stop and Glo® dual luciferase kit (Promega). Assays were performed in duplicate. To control for transfection efficiency, firefly luciferase activity was corrected to renilla luciferase activity.
The 924 amino acid (WT lipin 1) and Δ115-lipin 1 proteins were fused to an N-terminal triple hemagglutinin tag were overexpressed using a pCDNA3.1 vector (7). The HNF4α (pMT-HNF4α) has been described (41, 42). Acadm.TKLuc (43, 44) was the gift of Daniel Kelly.
Statistical analysis
Statistical comparisons were made using ANOVA or t-test. All data are presented as the means ± SE, with a statistically significant difference defined as a P value <0.05.
RESULTS
Time course of fasting-mediated changes in lipin expression and PAP activity in liver
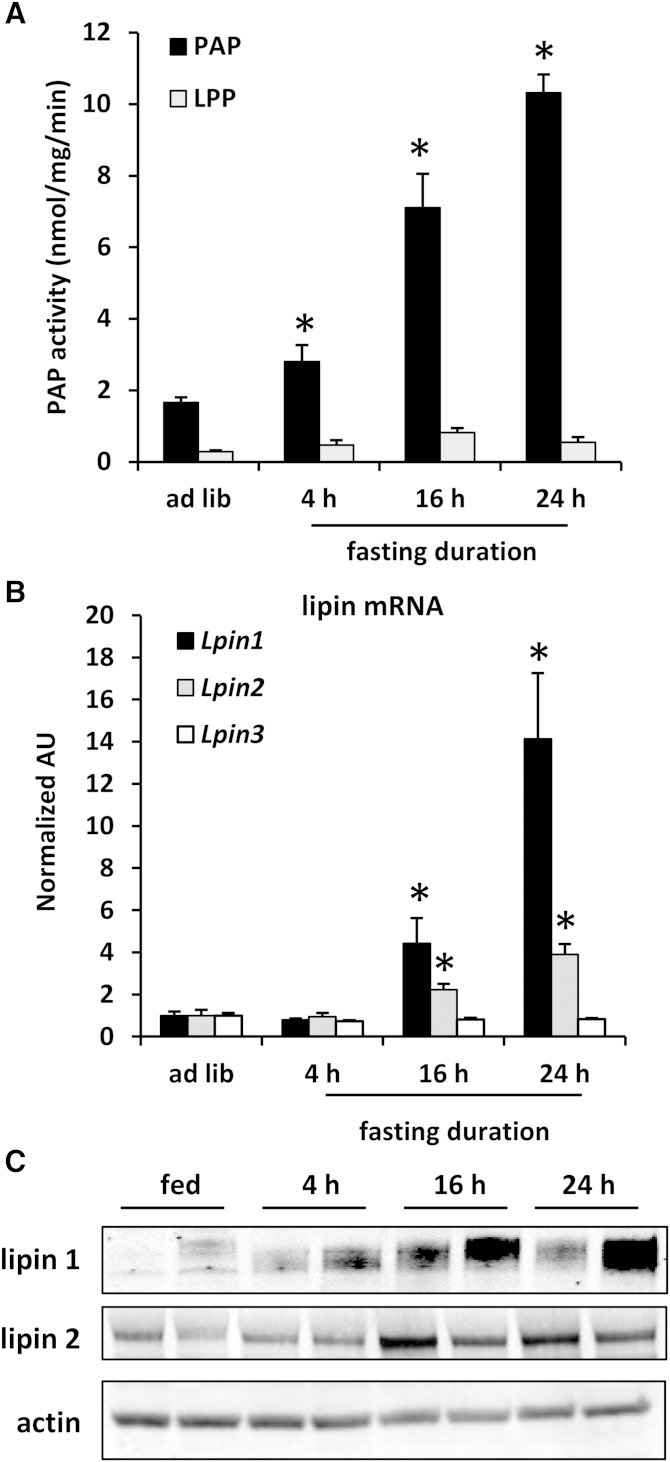
C57BL6/J mice were fasted for varying times, and PAP activity and lipin expression were characterized. Hepatic PAP activity was significantly elevated at 4, 16, and 24 h after food removal (Fig. 1A). In contrast, N-ethylmaleimide-insensitive dephosphorylation of PA (lipid phosphate phosphatase) was not affected by fasting. Lpin1 and Lpin2 mRNA expression was significantly increased at the 16 and 24 h time points (Fig. 1B). Lpin3 mRNA expression was unaffected by fasting. Lipin 1 protein abundance was increased by 4, 16, or 24 h of fasting (Fig. 1C). Lipin 2 protein abundance was increased only by 16 and 24 h of fasting. These data indicate that the time course of the induction in hepatic PAP activity is well correlated with the expression of lipin 1 and lipin 2.
Fig. 1.
Fasting activates lipin expression and activity in mouse liver. Hepatic PAP activity (A); Lpin1, Lpin2, and Lpin3 mRNA levels (B); and lipin 1 and lipin 2 protein abundance (C) in WT C57BL6/J mice is shown. Mice (n = 6) were either given ad libitum access to standard mouse chow or fasted for the indicated times. * P < 0.05 versus ad libitum fed. LPP, lipid phosphate phosphatase.
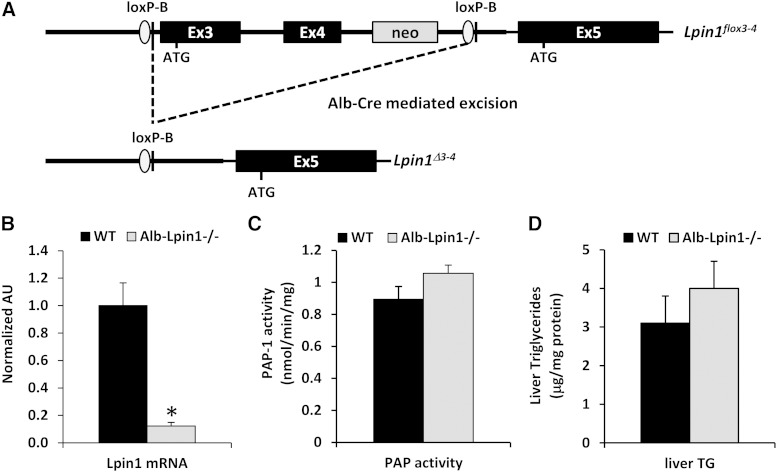
Generation of mice with loss of lipin 1 in liver
To generate mice with liver-specific lipin 1 deficiency, mice harboring a Lpin1 allele with LoxP sites flanking exons 3 and 4 were crossed with mice expressing Cre recombinase under control of the albumin promoter (Alb-Lpin1−/− mice; Fig. 2A). As expected, this led to a marked reduction in Lpin1 mRNA expression in Alb-Lpin1−/− mice compared with littermate controls as early as postnatal day 8 (P8; Fig. 2B). However, similar to our previous work with fld mice (45), loss of lipin 1 did not reduce hepatic PAP activity at this point in development (Fig. 2C). Loss of lipin 1 specifically in liver also did not affect liver TG content at P8 (Fig. 2D). This is consistent with our previous work indicating that loss of lipin 1 in adipose tissue leads to postnatal steatosis (7) and suggests that this aspect of the phenotype of fld mice is due to lipin 1 deficiency in extrahepatic tissues.
Fig. 2.
Generation of mice with liver-specific lipin 1 deficiency. A: The schematic depicts the targeting strategy used to generate an allele of Lpin1 that can be conditionally recombined. Lpin1 mRNA expression (B), PAP activity (C), and TG content (D) were determined in liver isolates from 8-day-old WT and Alb-Lpin1−/− mice (n = 5). * P < 0.05 versus WT mice.
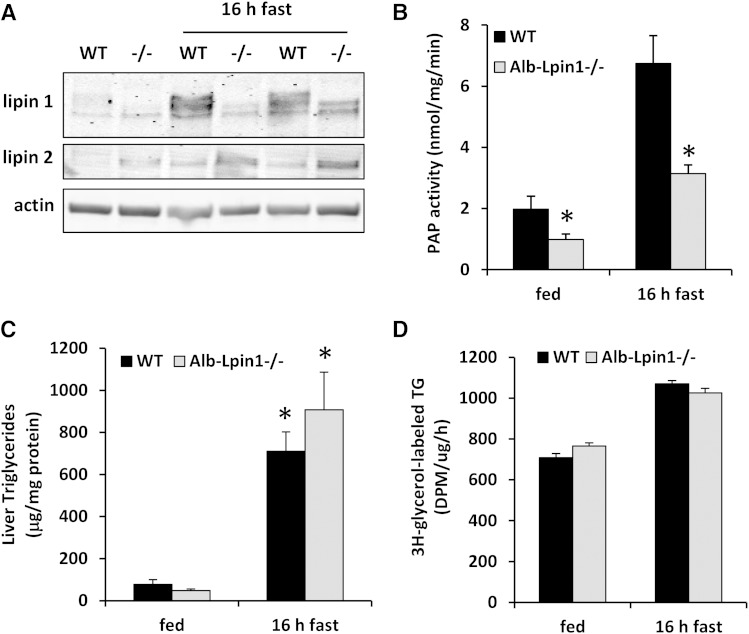
The fasting-induced increase in PAP activity is attenuated in Alb-Lpin1−/− mice without affecting TG content
At baseline, Western blotting detected no full-length protein in Alb-Lpin1−/− mice, but a truncated lipin 1 protein was observed upon prolonged exposure (Fig. 3A). We have shown that this truncated protein is the result of an alternative start codon in the first intact exon and encodes a protein lacking the first 115 amino acids (Δ115-lipin 1) (7). When WT and Alb-Lpin1−/− mice were fasted 16 h, the expression of WT and Δ115-lipin 1 protein was markedly induced (Fig. 3A). Lipin 2 protein abundance was moderately induced by fasting in liver and was increased in Alb-Lpin1−/− mice compared with WT comparators (Fig. 3A).
Fig. 3.
Effects of fasting on liver PAP activity and TG content in Alb-Lpin1−/− mice. A: Representative Western blots for lipin 1 and lipin 2 in livers of WT and Alb-Lpin1−/− (−/−) mice under fed and fasting conditions are shown. B: PAP activity in livers of WT and Alb-Lpin1−/− mice was quantified under fed and fasting conditions (n = 6). * P < 0.05 versus WT under same conditions. C: TG content of livers of WT and Alb-Lpin1−/− mice was determined under fed and fasting conditions (n = 6). * P < 0.05 versus fed mice. D: Rates of TG synthesis were quantified in hepatocytes isolated from WT and Alb-Lpin1−/− mice (n = 3). * P < 0.05 versus WT mice.
We have previously shown that the Δ115 allele of lipin 1 is intrinsically deficient in PAP activity (7). Consistent with this, we detected a significant decrease in hepatic PAP activity at baseline, and the fasting-induced increase in hepatic PAP activity was attenuated in Alb-Lpin1−/− mice (Fig. 3B). Despite this, hepatic TG content was not affected by lipin 1 deficiency under fed or fasted conditions (Fig. 3C), suggesting that hepatic PAP activity is not highly limiting for TG synthesis even under fasted conditions. Indeed, for hepatocytes isolated from mice that were either fed or fasted prior to isolation, we found no difference in rates of TG synthesis between the WT control and the Alb-Lpin1−/− mice (Fig. 3D). Loss of lipin-1-mediated PAP activity did not affect the plasma concentrations of glucose, TG, or NEFAs (Table 1).
TABLE 1.
Plasma parameters in fed and fasted mice
| WT Fed | Alb-Lpin1−/− Fed | WT Fasted | Alb-Lpin1−/− Fasted | |
| Glucose (mg/dl) | 107.0 ± 3.0 | 123.5 ± 13.7 | 72.3 ± 2.4 | 81.3 ± 5.8 |
| TG (mg/dl) | 57.1 ± 14.0 | 33.6 ± 3.5 | 70.1 ± 11.6 | 67.4 ± 11.5 |
| NEFA (mM) | 0.23 ± 0.09 | 0.23 ± 0.4 | 1.92 ± 0.30 | 2.37 ± 0.52 |
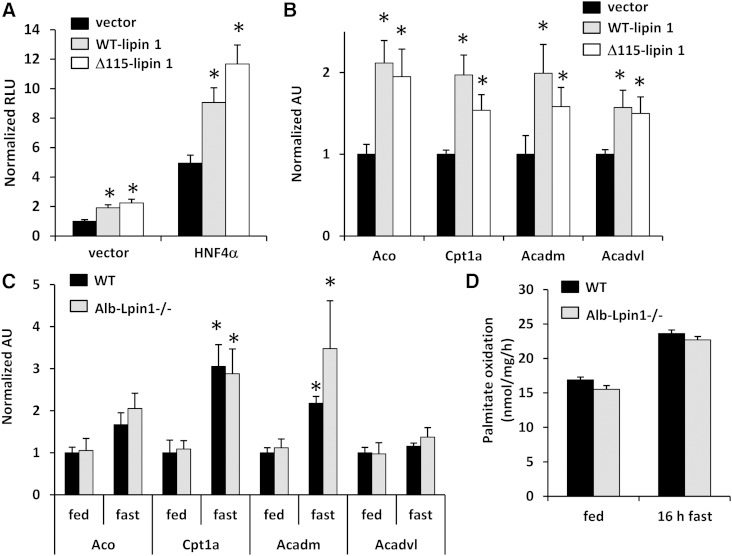
Lipin 1 Δ115 protein retains its transcriptional regulatory function in hepatocytes
We have recently shown that the truncated lipin 1 protein (Δ115-lipin 1) expressed in these mice retained its transcriptional regulatory function in promoter-luciferase reporter-based assays responsive to PPARα (7). Overexpression of the Δ115-lipin 1 protein was also sufficient to activate the liver-enriched transcription factor HNF4α on the Acadm promoter-luciferase reporter (Fig. 4A), which we have shown to be a target of lipin 1 (17). Consistent with this, adenovirus-mediated overexpression of lipin 1 WT or Δ115 in WT hepatocytes induced the expression of several genes encoding fatty acid oxidation enzymes known to be targets of HNF4α and PPARα (Fig. 4B). Given these findings with the truncation-producing allele, we next assessed the expression of several genes known to be under the control of lipin 1 in liver of mice under fed and fasted conditions. The hepatic expression of several genes encoding fatty acid oxidation enzymes, which was significantly induced by fasting, was not different in liver of Alb-Lpin1−/− mice compared with WT control mice (Fig. 4C). Consistent with this, rates of palmitate oxidation in hepatocytes isolated from Alb-Lpin1−/− mice were not different compared with control hepatocytes, even when mice were fasted prior to hepatocyte isolation to boost rates of oxidation (Fig. 4D). These findings indicate that liver-specific expression of lipin 1 Δ115 protein leads to reduced PAP activity, but does not affect the expression of target genes encoding fatty acid oxidation enzymes.
Fig. 4.
Lipin 1 Δ115 protein retains the ability to regulate gene transcription. A: Lipin 1 WT and Δ115 proteins enhance the activity of HNF4α on the HNF4-responsive luciferase reporter driven by the nuclear receptor response element from the promoter of the Acadm gene. * P < 0.05 versus vector control. B: The expression of known lipin 1 target genes was determined in hepatocytes from WT mice infected with adenovirus to overexpress lipin 1 WT or Δ115 truncation (n = 6). * P < 0.05 versus GFP control group. C: Expression of the indicated genes in livers of WT and Alb-Lpin1−/− mice fed ad libitum or fasted 16 h (n = 6) is shown. * P < 0.05 versus fed mice. D: Rates of palmitate oxidation were measured in hepatocytes isolated from mice fed ad libitum or fasted 16 h prior to being euthanized (n = 3).
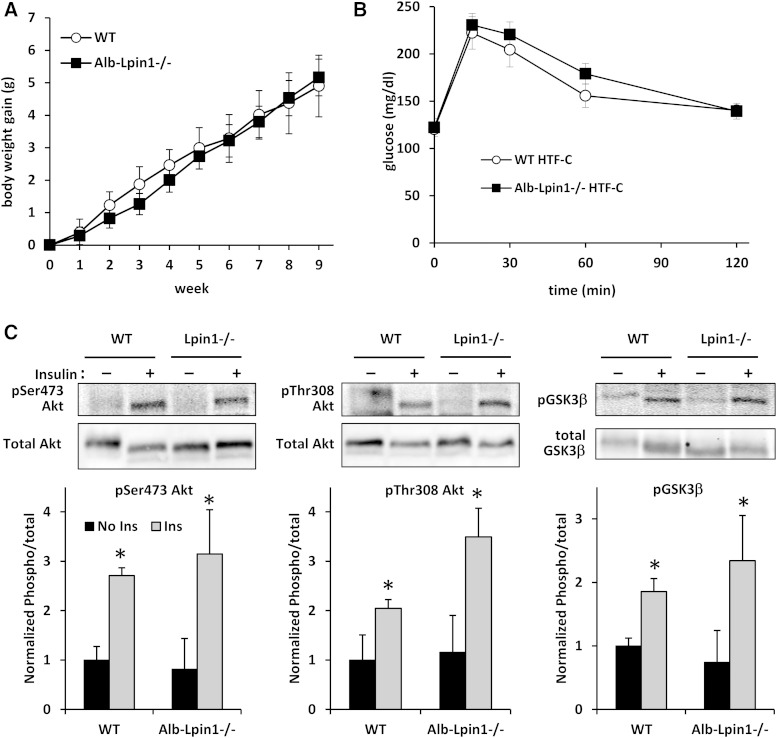
Liver-specific loss of lipin-1-mediated PAP activity does not affect glucose tolerance or insulin sensitivity in HTF-C diet-fed mice
Previous work has suggested that inhibition of hepatic lipin 1 expression in high fat diet fed mice improved insulin sensitivity and reduced hepatic steatosis (12). Therefore, we assessed the effects of lipin-1-mediated PAP activity on a model of chronic lipid oversupply. A high-fat/fructose/cholesterol (HTF-C) diet, which is known to increase hepatic TG storage, was administered to WT and Alb-Lpin1−/− mice. Weight gain on the HTF-C diet was not affected by genotype (Fig. 5A). Plasma insulin concentration was also unaffected by genotype, nor were the plasma concentrations of TG, NEFA, or cholesterol altered (Table 2). In contrast to the previous report, liver-specific loss of lipin-1-mediated PAP activity did not affect glucose tolerance (Fig. 5B). We also found that insulin-stimulated phosphorylation of insulin signaling proteins was not affected by loss of lipin-1-mediated PAP activity (Fig. 5C).
Fig. 5.
Loss of lipin-1-mediated PAP activity does not affect weight gain or glucose tolerance on HTF-C chow. A: Body weight gain of WT and Alb-Lpin1−/− mice on HTF-C diet for 10 weeks is shown. B: Glucose tolerance was quantified in WT and Alb-Lpin1−/− mice fed HTF-C diet for 10 weeks. C: Hepatic insulin signaling was measured by Western blotting for phosphorylation of proteins in that signaling cascade. The quantification of the normalized (WT saline = 1.0) ratio of phosphorylated to total protein abundance is shown in the graphs inset (n = 4). * P < 0.05 versus no insulin mice of the same genotype.
TABLE 2.
Plasma parameters in HTF-C diet-fed mice
| WT | Alb-Lpin1−/− | |
| TG (mg/dl) | 67.0 ± 9.3 | 62.7 ± 3.3 |
| NEFA (mM) | 0.31 ± 0.07 | 0.44 ± 0.08 |
| Cholesterol (mg/dl) | 168.1 ± 9.0 | 132.9 ± 1 |
| Insulin (pg/ml) | 1,040.3 ± 155.5 | 1,192.5 ± 240.1 |
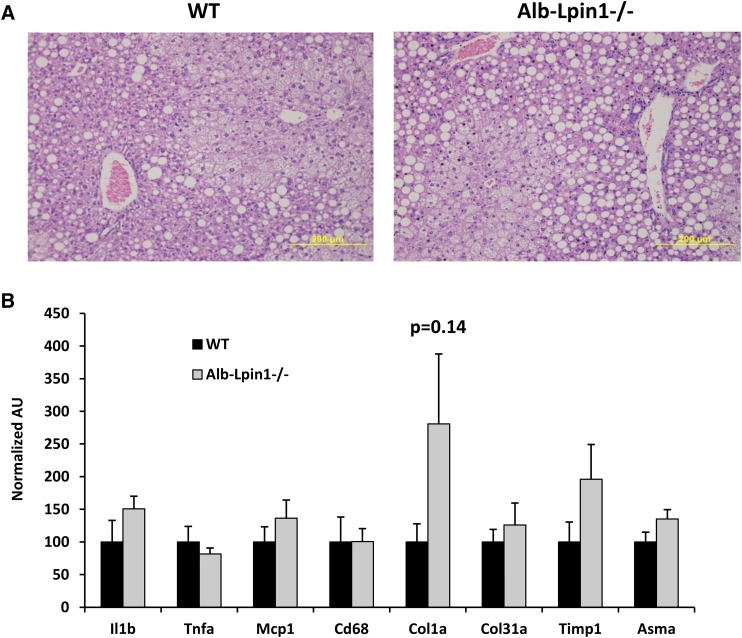
Histologic analyses demonstrated that HTF-C diet caused widespread lipid droplet accumulation and evidence of liver injury including hepatocyte ballooning and macrophage infiltration (Fig. 6A). These end points were not affected by genotype of the mice. We also assessed the hepatic expression of markers of liver injury, inflammation, and stellate cell activation. However, we found no evidence for an effect of lipin-1-mediated PAP activity deficiency on these parameters (Fig. 6B).
Fig. 6.
Liver injury caused by HTF-C chow is not affected by lipin 1 deficiency. A: Representative images of hematoxylin and eosin stained sections of liver from WT and Alb-Lpin1−/− mice on HTF-C diet for 10 weeks are shown. B: The expression of the indicated genes known to be markers of liver injury, macrophage infiltration, and stellate cell activation in liver of WT and Alb-Lpin1−/− mice on HTF-C diet for 10 weeks was determined.
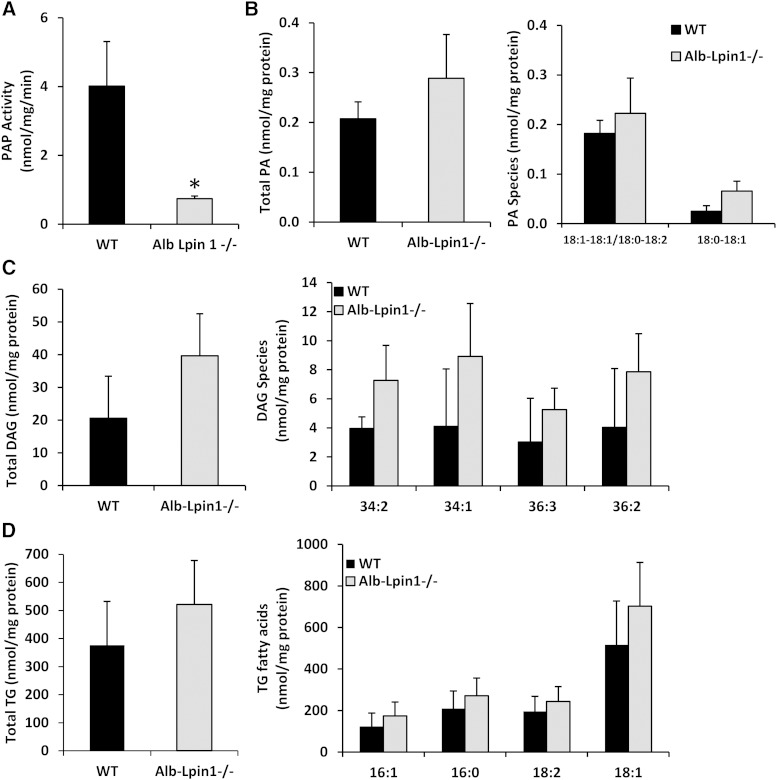
As expected, hepatic PAP activity was markedly reduced, compared with WT mice, in Alb-Lpin1−/− mice fed HTF-C diet (Fig. 7A). Loss of lipin-1-mediated PAP activity caused a strong trend toward accumulation of 18:0-18:1 PA species, but did not affect accumulation of other PA species or total PA content (Fig. 7B). Total DAG and TG content was not different in HTF-C-fed Alb-Lpin1−/− mice compared with WT controls (Fig. 7C, D). Additionally, there was no effect of genotype on specific DAG and TG species. Collectively, these findings indicate that chronic, liver-specific deficiency in lipin-1-mediated PAP activity does not impair the capacity of the liver for TG synthesis or the development of hepatic steatosis.
Fig. 7.
Liver-specific lipin 1 deficiency does not affect glycerolipid accumulation on HTF-C diet. A: Hepatic PAP activity of WT and Alb-Lpin1−/− mice on HTF-C diet for 10 weeks is shown. * P < 0.05 versus WT mice. Hepatic PA (B), DAG (C), and TG (D) content of liver of WT and Alb-Lpin1−/− mice fed HTF-C diet for 10 weeks was determined by MS. The abundance of specific species of these glycerolipids is also shown.
DISCUSSION
Lipin proteins have emerged as important regulators of hepatic intermediary metabolism via their enzymatic effects on TG biosynthesis as well as via nuclear effects on transcriptional regulation of metabolic enzymes. In this work, we characterized the phenotype of mice with liver-specific expression of an allele of lipin 1 that lacked PAP activity. Despite a reduction in PAP activity of >50% with loss of lipin 1, rates of TG synthesis and steady-state PA, DAG, and TG levels were unchanged even under conditions of chronic lipid overload. In contrast, knockdown of DAG acyltransferase (DGAT) 2, which catalyzes the step immediately downstream of lipin 1, with targeted antisense oligonucleotides led to a 50% reduction in DGAT activity, suppressed TG synthesis, and reduced steady-state TG content (46, 47). Similarly, suppression of 50% of glycerol-3-phosphate acyltransferase activity, the first step in this pathway, is also sufficient to attenuate hepatic TG synthesis and steatosis (48). These data, taken with previous work (21, 49), suggest that PAP activity is far from limiting for regulating intrahepatic TG synthesis. However, we cannot exclude that chronic deficiency in lipin-1-mediated PAP activity leads to compensatory mechanisms for synthesizing these glycerolipids.
The remaining PAP activity detected in the liver of Alb-Lpin1−/− mice is likely catalyzed by lipin 2, which is an important hepatic PAP enzyme and is the most abundant lipin family member in the liver under most conditions (21, 22). Although the intrinsic catalytic activity of lipin 2 is lower than lipin 1 (4), lipin 2 is subject to less regulation by phosphorylation (50). Lipin 2 protein abundance was increased in Alb-Lpin1−/− liver. We have previously described this phenomenon in fld mice (21), which is consistent with reciprocal regulation of lipin 1 and lipin 2 in liver and other cell types (51–53). We found Alb-Lpin1−/− mice retain sufficient hepatic PAP activity for robust TG synthesis under fasting and high-fat diet fed conditions. This is consistent with the finding that ethanol-induced hepatic steatosis in Alb-Lpin1−/− mice was not attenuated and actually exacerbated compared with WT control mice (33). In contrast, acute RNAi-mediated lipin 1 or lipin 2 depletion in liver of mice has been reported to attenuate hepatic steatosis and improve insulin sensitivity (12, 13). Again, it is possible that the lack of effect on these metabolic parameters in Alb-Lpin1−/− mice is due to chronic compensatory mechanisms that arise due to long-term lipin 1 deficiency or due to differences in dietary fat content or compositions used in the different studies.
Our previous work demonstrated that shRNA-mediated lipin 1 knockdown in liver blunted the increased expression of genes encoding fatty acid oxidation enzymes under fasting conditions (14). This was also observed in fld mouse liver after a shorter term fast (12 h) (49). These data, together with evidence for direct transcriptional regulation by lipin 1, suggest that lipin 1 is an inducible enhancer of hepatic fatty acid oxidation capacity via transcriptional mechanisms. The observation that Alb-Lpin1−/− mice exhibited no defect in fasting-induced expression of genes encoding fatty acid oxidation enzymes is consistent with our observation that the ability to regulate PPARα (7) and HNF4α (Fig. 4) is retained by the Δ115 allele.
In conclusion, we show that loss of lipin-1-mediated PAP activity fails to constrain TG synthesis or reduce steady-state DAG and TG content of the liver under acute or chronic conditions of high fatty acid availability. Our data further support the idea that the PAP activity and nuclear receptor transcriptional regulatory activity of lipin 1 can be functionally separated. Additional work will be needed to define the pathways and compensatory mechanisms that allow robust hepatic TG synthesis in the context of reduced PAP activity.
Acknowledgments
The authors thank Elizabeth M. Brunt for histopathological analyses.
Footnotes
Abbreviations:
- DAG
- diacylglycerol
- fld
- fatty liver dystrophic
- HNF4α
- hepatic nuclear receptor 4α
- PA
- phosphatidic acid
- PAP
- phosphatidic acid phosphohydrolase
- TBST
- Tris-buffered saline plus Tween
This work was supported by National Institutes of Health Grants (NIH) including R01 DK078187 (B.N.F.), R01 DK091176 (R.W.G.), P30 DK056341, P60 DK020579, and P30 DK052574. G.G.S. is supported by T32 HL007081 from the NIH. K.S.M. is an NIH Diabetes Research Postdoctoral Training Program fellow (T32 DK007296).
REFERENCES
- 1.Clark J. M., Diehl A. M. 2003. Nonalcoholic fatty liver disease: an underrecognized cause of cryptogenic cirrhosis. J. Am. Med. Assoc. 289: 3000–3004. [DOI] [PubMed] [Google Scholar]
- 2.Clark J. M., Brancati F. L., Diehl A. M. 2002. Nonalcoholic fatty liver disease. Gastroenterology. 122: 1649–1657. [DOI] [PubMed] [Google Scholar]
- 3.Han G. S., Wu W. I., Carman G. M. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457. [DOI] [PubMed] [Google Scholar]
- 5.Boni L. T., Rando R. R. 1985. The nature of protein kinase C activation by physically defined phospholipid vesicles and diacylglycerols. J. Biol. Chem. 260: 10819–10825. [PubMed] [Google Scholar]
- 6.Rando R. R., Young N. 1984. The stereospecific activation of protein kinase C. Biochem. Biophys. Res. Commun. 122: 818–823. [DOI] [PubMed] [Google Scholar]
- 7.Mitra M. S., Chen Z., Ren H., Harris T. E., Chambers K. T., Hall A. M., Nadra K., Klein S., Chrast R., Su X., et al. 2013. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc. Natl. Acad. Sci. USA. 110: 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadra K., de Preux Charles A. S., Medard J. J., Hendriks W. T., Han G. S., Gres S., Carman G. M., Saulnier-Blache J. S., Verheijen M. H., Chrast R. 2008. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22: 1647–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 294: 1942–1945. [DOI] [PubMed] [Google Scholar]
- 10.Yoon M. S., Sun Y., Arauz E., Jiang Y., Chen J. 2011. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J. Biol. Chem. 286: 29568–29574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Wendel A. A., Keogh M. R., Harris T. E., Chen J., Coleman R. A. 2012. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc. Natl. Acad. Sci. USA. 109: 1667–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu D., Oh K. J., Jo H. Y., Hedrick S., Kim Y. N., Hwang Y. J., Park T. S., Han J. S., Choi C. S., Montminy M., et al. 2009. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 9: 240–251. [DOI] [PubMed] [Google Scholar]
- 13.Ryu D., Seo W. Y., Yoon Y. S., Kim Y. N., Kim S. S., Kim H. J., Park T. S., Choi C. S., Koo S. H. 2011. Endoplasmic reticulum stress promotes LIPIN2-dependent hepatic insulin resistance. Diabetes. 60: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr, Kelly D. P. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4: 199–210. [DOI] [PubMed] [Google Scholar]
- 15.Kim H. E., Bae E., Jeong D. Y., Kim M. J., Jin W. J., Park S. W., Han G. S., Carman G. M., Koh E., Kim K. S. 2013. Lipin1 regulates PPARgamma transcriptional activity. Biochem. J. 453: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh Y. K., Lee M. Y., Kim J. W., Kim M., Moon J. S., Lee Y. J., Ahn Y. H., Kim K. S. 2008. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma 2. J. Biol. Chem. 283: 34896–34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z., Gropler M. C., Mitra M. S., Finck B. N. 2012. Complex interplay between the lipin 1 and the hepatocyte nuclear factor 4 alpha (HNF4alpha) pathways to regulate liver lipid metabolism. PLoS ONE. 7: e51320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H. B., Kumar A., Wang L., Liu G. H., Keller S. R., Lawrence J. C., Jr, Finck B. N., Harris T. E. 2010. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell. Biol. 30: 3126–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146: 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282: 277–286. [DOI] [PubMed] [Google Scholar]
- 21.Gropler M. C., Harris T. E., Hall A. M., Wolins N. E., Gross R. W., Han X., Chen Z., Finck B. N. 2009. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J. Biol. Chem. 284: 6763–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donkor J., Zhang P., Wong S., O’Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. 2009. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 284: 29968–29978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manmontri B., Sariahmetoglu M., Donkor J., Bou Khalil M., Sundaram M., Yao Z., Reue K., Lehner R., Brindley D. N. 2008. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 49: 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu M., Wang F., Li X., Rogers C. Q., Liang X., Finck B. N., Mitra M. S., Zhang R., Mitchell D. A., You M. 2012. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology. 55: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishimoto K., Nakamura H., Tachibana K., Yamasaki D., Ota A., Hirano K., Tanaka T., Hamakubo T., Sakai J., Kodama T., et al. 2009. Sterol-mediated regulation of human lipin 1 gene expression in hepatoblastoma cells. J. Biol. Chem. 284: 22195–22205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazit V., Weymann A., Hartman E., Finck B. N., Hruz P. W., Tzekov A., Rudnick D. A. 2010. Liver regeneration is impaired in lipodystrophic fatty liver dystrophy mice. Hepatology. 52: 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vavrecka M., Mitchell M. P., Hubscher G. 1969. The effect of starvation on the incorporation of palmitate into glycerides and phospholipids of rat liver homogenates. Biochem. J. 115: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau D. C., Roncari D. A. 1983. Effects of glucocorticoid hormones on lipid-synthetic enzymes from different adipose tissue regions and from liver. Can. J. Biochem. Cell Biol. 61: 1245–1250. [DOI] [PubMed] [Google Scholar]
- 29.Simpson K. J., Venkatesan S., Peters T. J., Martin A., Brindley D. N. 1989. Hepatic phosphatidate phosphohydrolase activity in acute and chronic alcohol-fed rats. Biochem. Soc. Trans. 17: 1115–1116. [DOI] [PubMed] [Google Scholar]
- 30.Sturton R. G., Butterwith S. C., Burditt S. L., Brindley D. N. 1981. Effects of starvation, corticotropin injection and ethanol feeding on the activity and amount of phosphatidate phosphohydrolase in rat liver. FEBS Lett. 126: 297–300. [DOI] [PubMed] [Google Scholar]
- 31.Reue K., Xu P., Wang X. P., Slavin B. G. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41: 1067–1076. [PubMed] [Google Scholar]
- 32.Péterfy M., Phan J., Xu P., Reue K. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- 33.Hu M., Yin H., Mitra M. S., Liang X., Ajmo J. M., Nadra K., Chrast R., Finck B. N., You M. 2013. Hepatic-specific lipin-1 deficiency exacerbates experimental alcohol-induced steatohepatitis in mice. Hepatology. 58: 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapper J. R., Hendricks M. D., Gu G., Wittmer C., Dolman C. S., Herich J., Athanacio J., Villescaz C., Ghosh S. S., Heilig J. S., et al. 2013. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am. J. Physiol. Gastrointest. Liver Physiol. 305: G483–G495. [DOI] [PubMed] [Google Scholar]
- 35.Martin A., Hales P., Brindley D. N. 1987. A rapid assay for measuring the activity and the Mg2+ and Ca2+ requirements of phosphatidate phosphohydrolase in cytosolic and microsomal fractions of rat liver. Biochem. J. 245: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han X., Gross R. W. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100. [DOI] [PubMed] [Google Scholar]
- 37.Han X., Yang K., Gross R. W. 2012. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 31: 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z., Gropler M. C., Norris J., Lawrence J. C., Jr, Harris T. E., Finck B. N. 2008. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler. Thromb. Vasc. Biol. 28: 1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z., Norris J. Y., Finck B. N. 2010. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) stimulates VLDL assembly through activation of cell death-inducing DFFA-like effector B (CideB). J. Biol. Chem. 285: 25996–26004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess S. C., Leone T. C., Wende A. R., Croce M. A., Chen Z., Sherry A. D., Malloy C. R., Finck B. N. 2006. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J. Biol. Chem. 281: 19000–19008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter M. E., Gulick T., Raisher B. D., Caira T., Ladias J. A., Moore D. D., Kelly D. P. 1993. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J. Biol. Chem. 268: 13805–13810. [PubMed] [Google Scholar]
- 42.Kressler D., Schreiber S. N., Knutti D., Kralli A. 2002. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 277: 13918–13925. [DOI] [PubMed] [Google Scholar]
- 43.Leone T. C., Cresci S., Carter M. E., Zhang Z., Lala D. S., Strauss A. W., Kelly D. P. 1995. The human medium chain acyl-CoA dehydrogenase gene promoter consists of a complex arrangement of nuclear receptor response elements and Sp1 binding sites. J. Biol. Chem. 270: 24622. [PubMed] [Google Scholar]
- 44.Pineda Torra I., Jamshidi Y., Flavell D. M., Fruchart J-C., Staels B. 2002. Characterization of the human PPAR{alpha} promoter: identification of a functional nuclear receptor response element. Mol. Endocrinol. 16: 1013–1028. [DOI] [PubMed] [Google Scholar]
- 45.Hall A. M., Brunt E. M., Chen Z., Viswakarma N., Reddy J. K., Wolins N. E., Finck B. N. 2010. Dynamic and differential regulation of proteins that coat lipid droplets in fatty liver dystrophic mice. J. Lipid Res. 51: 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., et al. 2005. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 42: 362–371. [DOI] [PubMed] [Google Scholar]
- 47.Choi C. S., Savage D. B., Kulkarni A., Yu X. X., Liu Z. X., Morino K., Kim S., Distefano A., Samuel V. T., Neschen S., et al. 2007. Suppression of diacylglycerol acyltransferase-2 (DGAT2), but not DGAT1, with antisense oligonucleotides reverses diet-induced hepatic steatosis and insulin resistance. J. Biol. Chem. 282: 22678–22688. [DOI] [PubMed] [Google Scholar]
- 48.Xu H., Wilcox D., Nguyen P., Voorbach M., Suhar T., Morgan S. J., An W. F., Ge L., Green J., Wu Z., et al. 2006. Hepatic knockdown of mitochondrial GPAT1 in ob/ob mice improves metabolic profile. Biochem. Biophys. Res. Commun. 349: 439–448. [DOI] [PubMed] [Google Scholar]
- 49.Kok B. P., Dyck J. R., Harris T. E., Brindley D. N. 2013. Differential regulation of the expressions of the PGC-1alpha splice variants, lipins, and PPARalpha in heart compared to liver. J. Lipid Res. 54: 1662–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eaton J. M., Takkellapati S., Lawrence R. T., McQueeney K. E., Boroda S., Mullins G. R., Sherwood S. G., Finck B. N., Villen J., Harris T. E. 2014. Lipin 2 binds phosphatidic acid by the electrostatic hydrogen bond switch mechanism independent of phosphorylation. J. Biol. Chem. 289: 18055–18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dwyer J. R., Donkor J., Zhang P., Csaki L. S., Vergnes L., Lee J. M., Dewald J., Brindley D. N., Atti E., Tetradis S., et al. 2012. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc. Natl. Acad. Sci. USA. 109: E2486–E2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimsey N., Han G. S., O’Hara L., Rochford J. J., Carman G. M., Siniossoglou S. 2008. Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J. Biol. Chem. 283: 29166–29174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Csaki L. S., Dwyer J. R., Li X., Nguyen M. H., Dewald J., Brindley D. N., Lusis A. J., Yoshinaga Y., de Jong P., Fong L., et al. 2014. Lipin-1 and lipin-3 together determine adiposity in vivo. Mol. Metab. 3: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]