Abstract
Cholesterol is essential to human health, and its levels are tightly regulated by a balance of synthesis, uptake, and efflux. Cholesterol synthesis requires the actions of more than twenty enzymes to reach the final product, through two alternate pathways. Here we describe a physical and functional interaction between the two terminal enzymes. 24-Dehydrocholesterol reductase (DHCR24) and 7-dehydrocholesterol reductase (DHCR7) coimmunoprecipitate, and when the DHCR24 gene is knocked down by siRNA, DHCR7 activity is also ablated. Conversely, overexpression of DHCR24 enhances DHCR7 activity, but only when a functional form of DHCR24 is used. DHCR7 is important for both cholesterol and vitamin D synthesis, and we have identified a novel layer of regulation, whereby its activity is controlled by DHCR24. This suggests the existence of a cholesterol “metabolon”, where enzymes from the same metabolic pathway interact with each other to provide a substrate channeling benefit. We predict that other enzymes in cholesterol synthesis may similarly interact, and this should be explored in future studies.
Keywords: 24-dehydrocholesterol reductase, 7-dehydrocholesterol reductase, desmosterol, 7-dehydrocholesterol
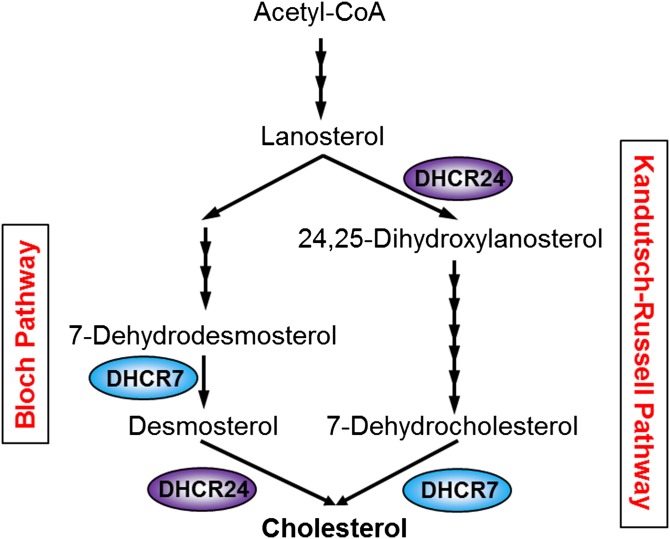
Cholesterol is a vital lipid in higher eukaryotes but is toxic in excess, and is thus tightly regulated to be kept within close margins by balancing cholesterol uptake, synthesis, and efflux. Cholesterol is synthesized via the mevalonate pathway (1); an energetically expensive multi-step process involving over 20 enzymes, starting with precursor acetyl-CoA, leading to the production of lanosterol as the first sterol (Fig. 1). Lanosterol then feeds into two pathways, the Bloch (2) and Kandutsch-Russell (3) pathways, which share the same enzymes to produce cholesterol from desmosterol or 7-dehydrocholesterol (7DHC), respectively, differing only in their entry and terminal enzymes involving 24-dehydrocholesterol reductase (DHCR24) and 7-dehydrocholesterol reductase (DHCR7). Activities of these terminal enzymes can affect membrane organization and dynamics, as the addition and position of the double bond in desmosterol and 7DHC confers very different properties (4). Moreover, 7DHC can be converted to vitamin D by UVB in the skin (5).
Fig. 1.
A simplified schematic of cholesterol synthesis. The mevalonate pathway begins with acetyl-CoA, undergoing many intermediary reactions to produce cholesterol via the Bloch pathway or the Kandutsch-Russell pathway.
Cholesterol is synthesized in the endoplasmic reticulum (ER), where most of the cholesterol biosynthetic enzymes reside, suggesting the possibility of a specialized metabolon or “cholestesome”, where cholesterol synthesis enzymes exist in an organized system in the ER membrane (1). There is evidence in the literature that the enzymes that synthesize ergosterol, the fungal equivalent of cholesterol, interact in yeast. Interacting enzymes include ERG27 and ERG7 (6, 7), although functional studies suggested that this was not the case for the mammalian orthologs, HSD17B7 and lanosterol synthase, respectively (8). Moreover, a systematic study examining the interaction between yeast ergosterol synthesis enzymes found that specific enzymes form a functional complex which the authors termed “ergosome” (7). The yeast ergosome is also supported by experimental evidence listed in the STRING database, and the high-confidence yeast protein-protein interaction dataset updated from (9), as described in (10), especially for post-lanosterol enzymes (supplementary Fig. 1). However, there is no experimental evidence that mammalian cholesterol synthesis proteins interact. Furthermore, because certain mammalian cholesterol biosynthetic enzymes are absent in yeast, such as the two terminal enzymes, DHCR24 (11) and DHCR7 (12), protein-protein interaction data for these enzymes are missing. Therefore, we investigated whether mammalian cholesterol synthetic enzymes interact in the ER membrane in an organized manner as the cholestesome, initially focusing our efforts on DHCR24.
DHCR24 is involved in a remarkable diversity of cellular functions (e.g., oxidative stress, neuroprotection, cell survival), and is implicated in many diseases including cardiovascular disease, hepatitis C, certain cancers, and neurodegenerative diseases [reviewed in (13)]. Deficient/defective DHCR24 results in a rare autosomal recessive developmental disorder called desmosterolosis, characterized by low cholesterol and high desmosterol levels (14). Thus, DHCR24 is an important enzyme in human health and disease. Here, we used DHCR24 as bait in a proteomic screen, and found that it interacts with DHCR7. Moreover, this interaction appears to have functional consequences on DHCR7 activity. Together, these results present the first case of mammalian cholesterol synthesis enzymes interacting, and indicate a novel mode of regulation of DHCR7.
MATERIALS AND METHODS
Materials
Chinese hamster ovary (CHO)-7 cells (15) were a generous gift from Drs. Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX). J774A.1 cells were from the Centre for Vascular Research (University of New South Wales). Dithiothreitol was from Astral Scientific. [2H6]desmosterol and [2H7]7DHC were from Avanti Polar Lipids (Alabaster, AL). FCS was from Bovogen (Vic, Australia). HRP-conjugated AffiniPure donkey anti-mouse and anti-rabbit IgG antibody was from Jackson ImmunoResearch. DMEM:Ham’s nutrient mixture F12 (1:1) (DMEM/F12), Dynabeads protein G magnetic beads, hygromycin B, Lipofectamine LTX transfection reagent, Lipofectamine RNAiMAX transfection reagent, newborn calf serum, and RPMI were from Life Technologies. Formic acid was from Merck. Immobilon Western chemiluminescent HRP substrate was from Millipore. Trypsin was from Promega. cOmplete ULTRA protease inhibitor cocktail tablets (EDTA-free) were from Roche. The 3-[(3-cholamidopropyl)dimethylammonio]1-propanesulfonate (CHAPS), 5α-cholestane, ammonium bicarbonate (NH4HCO3), compactin (mevastatin), iodoacetamide, methyl-β-cyclodextrin, mevalonate, N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane, primers, and protease inhibitor cocktail were from Sigma-Aldrich. Lipoprotein-deficient newborn calf serum (LPDS) and lipoprotein-deficient FCS were prepared from heat-inactivated newborn calf serum and FCS, respectively, as described (16, 17). [2H6]desmosterol and [2H7]7DHC were complexed to methyl-β-cyclodextrin as described (18).
Expression plasmids
A plasmid encoding the human DHCR24 sequence (NM_014762.3) containing a Flp recombinase target (FRT) recombination site (pcDNA5-DHCR24-V5/FRT) (19) was used to create the DHCR24 mutant, pcDNA5-DHCR24 Y471S-V5/FRT, using megaprimed site-directed mutagenesis (20). This was then used to generate the CHO-DHCR24 Y471S stable cell-line.
pcDNA5-DHCR7-myc/FRT contains the protein coding sequence of human DHCR7 (NM_001360.2) with a C-terminal myc epitope tag, and was cloned from cDNA from HeLa cells into pGEM T easy (Promega), and then subcloned into pcDNA5-FRT (Life Technologies) using restriction enzyme digestion.
Cell culture and treatments
CHO-7 cells were maintained and treated in 5% (v/v) LPDS/DMEM/F12. CHO-7 stable cell-lines were generated and maintained in 5% (v/v) LPDS/DMEM/F12 supplemented with 150 μg/ml hygromycin B. J774A.1 cells were maintained in 10% (v/v) FCS/RPMI and treated in 10% (v/v) lipoprotein-deficient FCS/RPMI. Statin treatment, employed to minimize basal cell cholesterol status, comprised 5 μM compactin and 50 μM mevalonate.
Stable cell-line generation
The Flp-In-CHO cell-line (for stable-cell generation) was generated in CHO-7 cells (21). CHO-7 cells overexpressing human DHCR24 [CHO-DHCR24-V5 (19)], or the pcDNA5/FRT empty vector (EV) [CHO-EV (21)], were previously generated in-house. The CHO-DHCR24 Y471S-V5 stable cell line was generated for this study using the Flp-In system, as described in (21). Protein expression was screened by Western blotting.
Plasmid and siRNA transfection
For plasmid DNA transfections, CHO-EV and CHO-DHCR24-V5 cells were seeded in 10 cm dishes and transfected with 5 μg DNA (pcDNA5-DHCR7-myc/FRT) for at least 4 h using 20 μl Lipofectamine LTX transfection reagent, according to the manufacturer’s instructions. After transfection, the cells were refreshed with medium, or statin treated overnight before harvesting for immunoprecipitation.
For siRNA transfections, cells were seeded in 6-well plates and transfected with 25 nM siRNA using 5 μl Lipofectamine RNAiMAX transfection reagent, according to the manufacturer’s instructions. After 24 h, cells were refreshed with medium overnight before treating/labeling/harvesting. Hamster-specific siRNA targeting DHCR24 [target sequence: GAGAGCCACGTGTGAAGCA; hamster versus human mRNA knockdown characterized previously (22)] and DHCR7 (target sequence: GAAGCCAGGAGTAAAGAAC) were designed by Sigma-Aldrich.
Quantitative real-time PCR
To measure mRNA levels, cells were seeded in a 12-well plate in triplicate wells per condition. The cells were transfected/treated accordingly, and harvested for total RNA using TRI reagent (Sigma), according to the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed into cDNA with Superscript III First Strand cDNA synthesis kit (Life Technologies). Quantitative real-time PCR was performed using a Rotor-Gene 3000 and analyzed using Rotor-Gene 6.1 (Corbett). SensiMix SYBR No-Rox (Bioline) was used as the amplification system using primers to amplify DHCR24 (23), DHCR7 (24), and the housekeeping control, porphobilinogen deaminase (PBGD) (25, 26). Changes in gene expression levels were normalized to PBGD for each sample by the ΔΔCt method.
Immunoprecipitation
Cells were washed with ice-cold PBS and harvested in either CHAPS buffer [0.5% (w/v) CHAPS, 150 mM NaCl, 2 mM EDTA, 25 mM HEPES (pH 7.4), and 5% (v/v) glycerol, supplemented with Roche’s protease inhibitor (one tablet per 10 ml buffer)] for LC-MS/MS experiments, or RIPA buffer [20 mM Tris-HCl (pH 7.4), 0.1% (w/v) SDS, 1% (v/v) Nonidet P-40, 0.5% sodium deoxycholate, 150 mM NaCl, 5 mM EDTA, and 1 mM sodium orthovanadate, supplemented with 2% (v/v) protease inhibitor cocktail (Sigma)] for Western blot detection (i.e., Fig. 2). The cell lysates were passed through a 22 gauge needle 20 times and centrifuged for 15 min at 20,000 g at 4°C. The cell lysates (supernatant) were adjusted to the same amount of protein in 1 ml buffer per sample. Dynabeads were rotated with the antibody for 1 h at room temperature, washed, and rotated with protein lysates overnight at 4°C. For the LC-MS/MS experiment, 3 mg protein lysate and 10 μg anti-V5 antibody were used per sample, and 1 mg protein lysate and 5 μg anti-myc antibody were used for the Western blotting experiment. The nonspecific proteins were then removed by a series of washes in their respective buffers (1 h wash, 30 min wash, and a 15 min wash). After the final wash, all supernatant was removed from the samples, and the pellets were resuspended in 50 μl immunoprecipitation loading buffer [2 vol of CHAPS or RIPA buffer, 2 vol 10% (w/v) SDS, and 1 vol fresh 5× Laemmli loading buffer (final concentration: 50 mM Tris-HCl (pH 6.8), 2% (w/v) SDS, 5% (v/v) glycerol, 0.04% (w/v) bromophenol blue, and 1% (v/v) β-mercaptoethanol)]. The samples were boiled at 95°C for 10 min with occasional vortexing before subjecting the supernatant to 10% (w/v) SDS-PAGE. The gels were then transferred for Western blotting or processed for LC-MS/MS analysis.
Fig. 2.
Overexpressed DHCR7 coimmunoprecipitates DHCR24. CHO-7 cells stably overexpressing EV or DHCR24-V5 were transfected with DHCR7-myc, and DHCR7-myc was immunoprecipitated with an anti-myc antibody. Whole cell lysates and pellets were subjected to SDS-PAGE and Western blotting with V5 (for DHCR24), myc (for DHCR7), α-tubulin, and 5×His (for both DHCR24 and DHCR7) antibodies. Western blots are representative of at least two separate experiments. IP, immunoprecipitation; WB, Western blot.
LC-MS/MS
After electrophoresis, gels were stained with EZ-Run protein gel staining solution (Thermo Fisher Scientific) according to the manufacturer’s instructions. Gel bands were excised, destained, reduced, and alkylated following the procedure described by Shevchenko et al. (27). For each gel slice, 40 ng of trypsin in 120 μl of 0.1 M NH4HCO3 was used for 16 h at 37°C. The digest solutions were transferred to new microfuge tubes and the gel slices treated with the following solutions sequentially for 30 min each: 80 μl of 0.1% (v/v) formic acid in 67% (v/v) acetonitrile, and 80 μl of 100% acetonitrile. Peptide solutions were then dried (Savant SPD1010, Thermo Fisher Scientific) before resuspending in 20 μl of 0.1% (v/v) formic acid. Proteolytic peptide samples were separated by nano-LC using an UltiMate 3000 HPLC and autosampler system (Dionex, Amsterdam, The Netherlands), and ionized using positive ion mode electrospray following experimental procedures described previously (28). Single-stage MS and MS/MS were performed using an LTQ Orbitrap Velos Pro (Thermo Electron, Bremen, Germany) hybrid linear ion trap and Orbitrap mass spectrometer. Survey scans m/z 350–2,000 were acquired in the Orbitrap (resolution = 30,000 at m/z 400, with an initial accumulation target value of 1,000,000 ions in the linear ion trap; lock mass applied to polycyclodimethylsiloxane background ions of exact m/z 445.1200 and 429.0887). Up to the five most abundant ions from an inclusion list (discussed below), followed by up to the ten most abundant ions (>5,000 counts) with charge states of >2 were sequentially isolated and fragmented via collision-induced dissociation with an activation q = 0.25, an activation time of 30 ms, normalized collision energy of 30%, and at a target value of 10,000 ions. Fragment ions were mass analyzed in the linear ion trap.
To ensure that peptides associated with cholesterol synthesis proteins were preferentially targeted for fragmentation during LC-MS/MS, MS/MS inclusion lists were generated with the aid of Skyline (version 0.7.0.2494, University of Washington). Amino acid sequences for the proteins listed in supplementary Table 1 were imported into Skyline, and m/z values for doubly charged theoretical proteotypic peptide ions associated with these proteins were generated using the following parameters: enzyme, trypsin (0 missed cleavages); and structural modifications, carbamidomethyl cysteine. Exported m/z values were incorporated into inclusion lists using the mixed targeted and untargeted LC-MS/MS experiments described above.
Peak lists derived from LC-MS/MS were submitted to the database search program Mascot (version 2.3, Matrix Science). The following search parameters were employed: instrument type was ESI-TRAP; precursor ion and peptide fragment mass tolerances were ±5 ppm and ±0.4 Da, respectively; variable modifications included were acrylamide (C), carbamidomethyl (C), and oxidation (M); enzyme specificity was trypsin with up to two missed cleavages; and rodentia taxonomies in the NCBI protein database (29) were searched. Peptide identifications were considered to be high confidence if they were statistically significant (P < 0.05) according to the Mascot expect metric.
DHCR24 protein expression
CHO-EV, CHO-DHCR24, and CHO-DHCR24 Y471S cells were harvested in 10% (w/v) SDS supplemented with 2% (v/v) protease inhibitor cocktail (Sigma). Equal amounts of protein were mixed with Laemmli loading buffer, boiled for 5 min, and subjected to SDS-PAGE and Western blotting.
Western blotting
Membranes were incubated with the following antibodies: anti-V5 (1:5,000; Life Technologies), anti-myc (1:5,000; Abcam), anti-α-tubulin (1:200,000; Sigma), and anti-5×His-HRP (1:10,000; Qiagen). Membranes were then visualized with the enhanced chemiluminescent detection system using the ImageQuant LAS 500 (GE Healthcare).
Lipid extraction and GC-MS
Cells were metabolically labeled with 1 μg/ml [2H6]desmosterol or [2H7]7DHC for 4 h. Cells were harvested, and protein levels were normalized before adding 0.2 μg 5α-cholestane as an internal standard for GC-MS. Lysates were saponified in 1 ml ethanol, 75% (w/v) KOH, 20 μl 20 mM EDTA, and 1 μl 20 mM butylated hydroxytoluene at 70°C for 1 h. Nonsaponifiable lipids were extracted in 2.5 ml hexane, and dried using nitrogen gas. Lipids were derivatized in BSTFA at 60°C for 1 h and then analyzed using a Thermo Trace gas chromatograph coupled with a Thermo DSQIII mass spectrometer and Thermo Triplus autosampler (Thermo Fisher Scientific), modified from a previous protocol (22). Samples (1.5 μl) were injected via a heated (300°C) splitless inlet into a Trace TR-50MS GC column (60 m × 0.25 mm, 0.25 μm film thickness) (Thermo Fisher Scientific). The column oven was initially held at 70°C for 0.7 min, then heated to 250°C at 20°C min−1, then to 270°C at 3°C min−1, and then to 305°C at 1.5°C min−1 (and held for 6 min in scan mode, or 3 min in selective ion monitoring mode). Helium was used as a carrier gas at a constant flow (1.2 ml min−1, with vacuum compensation on). MS conditions were: electron energy 70 eV, ion source temperature 200°C, transfer line temperature 305°C. The emission current was set to 130 μA and the detector gain to 6.0 × 105. Samples were analyzed either in scan mode (60–520 Da, 2.994 scans s−1) to obtain mass spectra for peak identification, or in selective ion monitoring mode to monitor the following sterols: 5α-cholestane, [2H6]cholesterol, [2H6]desmosterol, [2H7]cholesterol, and [2H7]7DHC (more detail in supplementary Fig. 2). Unlabeled cholesterol and desmosterol were not monitored, although they were also detected, as selected ions were also lowly present in unlabeled cholesterol and desmosterol, which were present in high abundance. Thermo Xcalibur software (version 2.1.0.1140) was used to acquire and process the data. Chromatographic peaks were integrated using the following parameters: baseline window 40–200, area noise factor 5, peak noise factor 10, multiplet threshold 10, and noise method set to “Incos noise”. Quantification of DHCR24 and DHCR7 activity was based on chromatographic peak areas of the confirmation ion measured for [2H6]cholesterol and [2H7]cholesterol relative to [2H6]desmosterol and [2H7]7DHC, respectively. Data are presented as mean + SEM. Statistical differences were determined by the Student’s paired t-test (two-tailed), where P values of ≤ 0.05 (*) and ≤ 0.01 (**) were considered statistically significant.
RESULTS
DHCR24 coimmunoprecipitates endogenous DHCR7
Interacting partner information for DHCR24 is currently unknown. To examine the potential interacting partner(s) of DHCR24, we employed CHO-7 cells stably overexpressing V5 epitope-tagged human DHCR24 (19) or the EV (21) as a negative control cell-line. An advantage of using the Flp-In stable system is that it expresses one copy per cell at a specific location (FRT site), which makes it more physiological and therefore less susceptible to immunoprecipitating false positive interacting partners. Cells were untreated, or statin treated to lower cellular cholesterol status, which therefore upregulates cholesterol synthesis gene/protein expression, and DHCR24 protein was immunoprecipitated with the anti-V5 antibody. Immunoprecipitated proteins were fractionated by SDS-PAGE and trypsin digested before analysis by LC-MS/MS. This was performed using a targeted approach in which peptides of masses potentially associated with hamster cholesterol synthesis proteins were preferentially selected for MS/MS (see LC-MS/MS in the Materials and Methods). Immunoprecipitation of DHCR24 resulted in the coimmunoprecipitation of a number of proteins, with immunoprecipitated cholesterol synthesis proteins listed in Table 1 (more information available in supplementary Table 2). Of note was DHCR7, which was pulled down in all instances, with and without statin treatment, indicating that DHCR7 is very likely an interacting partner of DHCR24.
TABLE 1.
Cholesterol synthesis proteins identified by LC-MS/MS after DHCR24 immunoprecipitation
| Accession ID | Name | Name Description | Experiment | Found? | Mascot Score | Protein Coverage (%) | Peptides |
| XP_003495787.1 | DHCR24 | 24-Dehydrocholesterol reductase | IP1a | Y | 82.75 | 5.39 | 3 |
| IP2a | Y | 196.93 | 8.14 | 6 | |||
| IP3 | Y | 51.96 | 3.10 | 3 | |||
| IP3b | Y | 63.57 | 3.10 | 3 | |||
| IP4b | Y | 698.73 | 17.05 | 10 | |||
| ERE81054 | DHCR7 | 7-Dehydrocholesterol reductase | IP1a | Y | 39.25 | 2.32 | 1 |
| IP2a | Y** | 93.06 | 7.01 | 2 | |||
| IP3 | Y | 59.04 | 2.32 | 1 | |||
| IP3b | Y* | 25.40 | 2.32 | 1 | |||
| IP4b | Y* | 29.28 | 1.81 | 1 | |||
| ERE90221 | SQS | Squalene synthase | IP1a | N | — | — | — |
| IP2a | Y | 48.40 | 2.99 | 1 | |||
| IP3 | N | — | — | — | |||
| IP3b | Y | 34.29 | 2.99 | 1 | |||
| IP4b | Y | 59.54 | 2.13 | 1 | |||
| EGW05732 | Thiolase 2 | Acetyl-CoA acetyltransferase, cytosolic | IP1a | Y | 105.71 | 6.80 | 2 |
| IP2a | Y | 133.91 | 14.11 | 4 | |||
| IP3 | N | — | — | — | |||
| IP3b | N | — | — | — | |||
| IP4b | N | — | — | — | |||
| EGW03272 | HMGCR | 3-Hydroxy-3-methylglutaryl-CoA reductase | IP1a | N | — | — | — |
| IP2a | N | — | — | — | |||
| IP3 | N | — | — | — | |||
| IP3b | N* | — | — | — | |||
| IP4b | Y** | 417.98 | 6.59 | 4 | |||
| EGV91499 | SQLE | Squalene monooxygenase | IP1a | N | — | — | — |
| IP2a | N | — | — | — | |||
| IP3 | N | — | — | — | |||
| IP3b | N | — | — | — | |||
| IP4b | Y** | 116.61 | 14.44 | 2 | |||
| ERE72972 | LBR | Lamin-B receptor | IP1a | N | — | — | — |
| IP2a | Y | 204.02 | 5.49 | 2 | |||
| IP3 | N | — | — | — | |||
| IP3b | N | — | — | — | |||
| IP4b | N | — | — | — | |||
| ERE73196 | HSD17B7 | 3-Keto-steroid reductase-like protein | IP1a | N | — | — | — |
| IP2a | N | — | — | — | |||
| IP3 | N | — | — | — | |||
| IP3b | N | — | — | — | |||
| IP4b | Y | 38.33 | 4.20 | 1 | |||
| EGW03873 | NSDHL | Sterol-4-α-carboxylate-3-dehydrogenase, decarboxylating | IP1a | N | — | — | — |
| IP2a | N | — | — | — | |||
| IP3 | N | — | — | — | |||
| IP3b | N | — | — | — | |||
| IP4b | Y | 41.75 | 6.25 | 1 |
The Mascot scores, protein coverage, and number of peptide matches are all derived from peptides identified with Mascot Expect values <0.05, except for underlined score. For more information, see supplementary Table 2. Y, protein found; Y*, protein inferred from a peptide identified with a Mascot Expect value >0.05, as MS/MS spectra derived from this peptide match those obtained in other experiments where the same peptide was identified with a higher Mascot Ion Score; Y**, protein found in EV as well, but with fewer peptide hits in EV; N, protein not found; N*, protein found with more peptide hits in EV (data omitted); ID, identification.
Experiments where only a selection of bands were subjected to LC-MS/MS analysis.
Experimental conditions that were statin treated overnight.
DHCR7 coimmunoprecipitates DHCR24
To confirm the interaction between DHCR24 and DHCR7, we transiently expressed human DHCR7 containing a myc epitope tag (DHCR7-myc) in CHO-EV and CHO-DHCR24-V5 cells, immunoprecipitated DHCR7 with an anti-myc antibody, and determined whether DHCR24 was coimmunoprecipitated. As both overexpressed DHCR24 and DHCR7 contain 6×His tags, we employed an anti-5×His antibody to simultaneously detect pulled down DHCR24 and DHCR7 protein. Because the immunoprecipitation for the proteomic screen (Table 1) was performed using a relatively low stringency CHAPS buffer, we used a more stringent RIPA buffer for the immunoprecipitation and wash steps, and found that DHCR7 immunoprecipitation also pulled down DHCR24, indicating that these proteins interact in cells (Fig. 2).
Gene silencing of DHCR24 decreases DHCR7 activity
Next, we examined whether the interaction between DHCR24 and DHCR7 has functional consequences. We employed specific siRNA to knock down hamster DHCR24 and DHCR7 in CHO-7 cells, and determined their effects on DHCR24 and DHCR7 activity. DHCR24 and DHCR7 siRNA decreased their respective target mRNA by approximately 93% and 88%, while having little or no effect on nontargeted mRNA (Fig. 3A).
Fig. 3.
DHCR24 knockdown decreases DHCR7 activity. CHO-7 cells were transfected with control, hamster-specific DHCR24, or DHCR7 siRNA (25 nM) for 24 h. A: The cells were washed with PBS and refed fresh media overnight. Total RNA was harvested and reverse transcribed to cDNA, and gene expression levels of DHCR24 and DHCR7 were quantified using quantitative real-time PCR, and normalized to the housekeeping gene, PBGD. Data are the mean + SEM from three separate experiments, each performed in triplicate cultures and presented relative to the control siRNA condition which has been set to 1. B: The cells were washed with PBS, refed fresh media overnight, and then labeled with 1 μg/ml [2H6]desmosterol or [2H7]7DHC for 4 h. Lipid extracts were derivatized in BSTFA before separation by GC-MS. Chromatogram peaks of [2H6]cholesterol ([2H6]-C), [2H6]desmosterol ([2H6]-Desm), [2H7]cholesterol ([2H7]-C), and [2H7]7DHC. The “c” denotes unlabeled cholesterol. Quantification of DHCR24 and DHCR7 activity was based on chromatographic peak areas of the confirmation ion measured for [2H6]cholesterol and [2H7]cholesterol relative to [2H6]desmosterol and [2H7]7DHC, respectively. Data are normalized to the control siRNA-transfected condition, which has been set to 1, and are the mean + SEM from three separate experiments. *P ≤ 0.05, **P ≤ 0.01 compared with the control siRNA-transfected condition.
To measure the specific activity of DHCR24 and DHCR7, we labeled CHO-7 cells with deuterated-substrates, [2H6]desmosterol for DHCR24 and [2H7]7DHC for DHCR7 (supplementary Fig. 2), and measured their conversion to deuterated-cholesterol by GC-MS, using the [2H6]cholesterol/[2H6]desmosterol ratio as a measure for specific DHCR24 activity [as in our previous study (22)], and the [2H7]cholesterol/[2H7]7DHC ratio as a measure for specific DHCR7 activity. As expected, knockdown of DHCR24 and DHCR7 decreased their respective enzymatic activity relative to the control siRNA-transfected condition, indicating that knockdown of mRNA translates to functional consequences downstream (Fig. 3B). Knockdown of DHCR7 had no effect on DHCR24 activity. Knockdown of DHCR24, however, decreased DHCR7 activity by ∼60%. Because knockdown of DHCR24 has no effect on DHCR7 mRNA (Fig. 3A), this implies that this phenomenon is occurring posttranscriptionally. Thus, the interaction between the two terminal steps of cholesterol synthesis appears to have functional consequences.
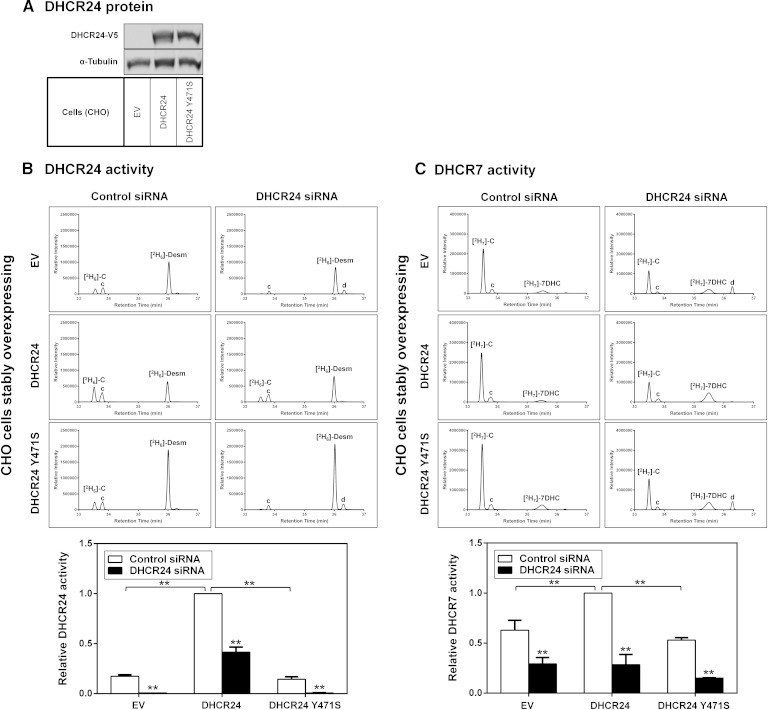
Overexpression of functional DHCR24 increases DHCR7 activity
As knocking down DHCR24 decreased DHCR7 activity (Fig. 3B), we next examined whether the converse is true, i.e., whether overexpressing DHCR24 increases DHCR7 activity. Thus, we compared DHCR7 activity in the CHO-EV and CHO-DHCR24 cell-lines. We also included a CHO cell-line stably overexpressing DHCR24 Y471S, a desmosterolosis mutant of DHCR24 that results in a complete loss of DHCR24 activity (11), to determine whether DHCR24 function plays a role in DHCR7 activity. As a positive control, DHCR24 activity was examined under these conditions, displaying an increase in activity in CHO-DHCR24 cells compared with CHO-EV and CHO-DHCR24 Y471S cells, even though CHO-DHCR24 and CHO-DHCR24 Y471S cells express similar levels of DHCR24 protein (Fig. 4A). As expected, DHCR24 knockdown decreased DHCR24 activity almost completely in CHO-EV and CHO-DHCR24 Y471S compared with CHO-DHCR24 cells (Fig. 4B), as the DHCR24 siRNA is targeted to hamster (endogenous) and not overexpressed human DHCR24 (22). As shown in Fig. 3B, knockdown of DHCR24 decreased DHCR7 activity in all cell-lines (Fig. 4C). Significantly, DHCR7 activity was ∼40% higher in cells overexpressing DHCR24 compared with CHO-EV (Fig. 4C). But when mutant DHCR24 was overexpressed, DHCR7 activity was comparable to CHO-EV, again consistent with the contention that DHCR24 function is important for DHCR7 activity.
Fig. 4.
DHCR24 function affects DHCR7 activity. A: CHO-7 cells stably overexpressing EV, DHCR24, or DHCR24-Y471S were seeded, harvested, and subjected to SDS-PAGE and Western blotting with V5 (for DHCR24), and α-tubulin antibodies. Western blots are representative of at least two separate experiments. B, C: CHO-7 cells stably overexpressing EV, DHCR24, or DHCR24-Y471S were transfected with control or DHCR24 siRNA (25 nM) for 24 h. The cells were washed with PBS, refed fresh media overnight, and then labeled with 1 μg/ml [2H6]desmosterol (B) or [2H7]7DHC (C) for 4 h. Lipid extracts were derivatized in BSTFA before separation by GC-MS. Chromatogram peaks of [2H6]cholesterol ([2H6]-C), [2H6]desmosterol ([2H6]-Desm), [2H7]cholesterol ([2H7]-C), and [2H7]7DHC. The “c” and “d” denote unlabeled cholesterol and desmosterol, respectively. Quantification of DHCR24 and DHCR7 activity was based on chromatographic peak areas of the confirmation ion measured for [2H6]cholesterol and [2H7]cholesterol relative to [2H6]desmosterol and [2H7]7DHC, respectively. Data are normalized to the CHO-DHCR24-V5 control siRNA-transfected condition, which has been set to 1, and are mean + SEM from at least three separate experiments. **P ≤ 0.01 compared with the control siRNA transfected condition. **P ≤ 0.01 of CHO-EV versus CHO-DHCR24 cells; no statistical difference between CHO-EV and CHO-DHCR24 Y471S.
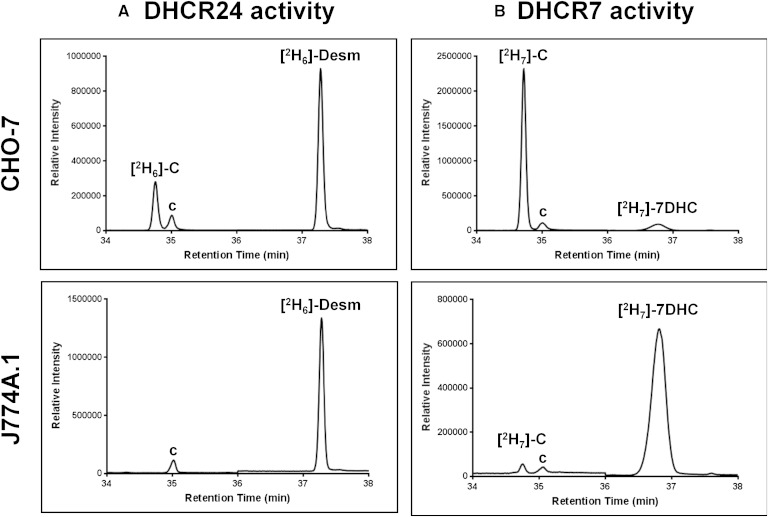
DHCR24 deficiency results in lowered DHCR7 activity
As proof of concept, we employed a cell-line deficient in DHCR24, the mouse macrophage-like cells, J774A.1 (30). As expected, no DHCR24 activity was detected in these cells (Fig. 5A). While J774A.1 cells were able to convert deuterated 7DHC to cholesterol, their DHCR7 activity was much lower compared with our model cell-line, CHO-7 (Fig. 5B).
Fig. 5.
DHCR24-deficient cells have lower basal DHCR7 activity. CHO-7 or J774A.1 cells were labeled with 1 μg/ml [2H6]desmosterol (A) or [2H7]7DHC (B) for 4 h. Lipid extracts were derivatized in BSTFA before separation by GC-MS. Chromatogram peaks of [2H6]cholesterol ([2H6]-C), [2H6]desmosterol ([2H6]-Desm), [2H7]cholesterol ([2H7]-C), and [2H7]7DHC. The “c” denotes unlabeled cholesterol.
DISCUSSION
In this study, we found that the two terminal enzymes of cholesterol synthesis interact physically (Table 1, Fig. 2) and functionally (Figs. 3, 4). Knockdown of DHCR24 decreased DHCR7 activity but did not completely impair its function, suggesting that DHCR24 aids DHCR7 function, but is not essential for it. This contention was supported by overexpression of DHCR24 further increasing DHCR7 activity, but only when functional DHCR24 was used.
We have uncovered a novel layer of regulation in cholesterol synthesis. Previously, virtually nothing was known about the posttranscriptional regulation of DHCR7, although it has recently been found that vitamin D inhibits DHCR7 activity in differentiated human keratinocytes through an unknown mechanism (31). Here, we have discovered that the activity levels of the alternate terminal enzyme, DHCR24, influences DHCR7 activity. Moreover, this is the first time that two mammalian cholesterol synthesis enzymes have been shown to interact, but we expect that many others also interact, probably in a coordinated fashion. Because DHCR7 and DHCR24 act sequentially in the Bloch pathway (Fig. 1), it is possible that their interaction provides a simple way for intermediates to be moved from one enzyme to the next. In this way, it may be expected that something of a “daisy chain” may be formed, where each enzyme interacts with both its preceding and subsequent enzymes.
This concept of enzymes from the same pathway interacting with their related enzymes (metabolon) has also been hypothesized for other metabolic pathways. For example, there is some experimental evidence for enzyme complexes forming in glycolysis, but the full details are yet to be elucidated [reviewed in (32)]. One of the evolutionary advantages suggested is similar to our hypothesis, the channeling of substrates. Physical interactions between five enzymes in the TCA cycle have also been observed in Bacillus subtilis (33), and again the authors suggested substrate channeling as a likely benefit.
Although evidence points toward the existence of metabolons, it has been suggested that these interactions are hard to detect using high-throughput assays because they only weakly or transiently interact under normal circumstances, and their assembly is regulated as required (34). This helps explain why we did not observe a stoichiometric interaction between DHCR24 and DHCR7 in our coimmunoprecipitation approach (apart from the fact that only a proportion of the cells would be transfected with the DHCR7 expression plasmid). We would expect only a subset of DHCR24 to be interacting with DHCR7 at any given time.
As DHCR24 is positioned not only at the end of the Bloch pathway but also at the gateway of the Kandutsch-Russell pathway, attuning its activity with the last step of this pathway (DHCR7) would ensure concerted control of cholesterol synthesis. Moreover, as 7DHC can be converted into cholesterol or vitamin D, DHCR7 represents a switch between these vital molecules in skin cells exposed to UVB, so it seems likely that this important branch point is regulated in some way.
In addition to DHCR24 acting at the start and end of each pathway, it is also able to act on any of the intermediates in the Bloch pathway to divert them into the Kandutsch-Russell pathway (35). This promiscuity may mean that it similarly interacts with other enzymes in the pathway, further promoting the metabolon concept. This possibility requires further investigation.
DHCR24 activity is regulated by a variety of factors, including side-chain oxysterols (19), progesterone (36), and phosphorylation (22). Based on our current work, these factors would also be expected to affect DHCR7 activity. Desmosterolosis resulting from DHCR24 deficiency is a very rare condition with only a handful of cases reported to date (13). We would predict that these patients would also have reduced DHCR7 activity, as we have observed in J774A.1 cells, which naturally lack DHCR24 (Fig. 5).
The precise mechanism by which DHCR24 affects DHCR7 activity remains to be determined. Genetic depletion of DHCR24 did not affect gene expression of DHCR7 (Fig. 3A), but we cannot exclude the possibility that protein expression may be affected, perhaps occurring through destabilization. We were unable to detect endogenous DHCR7 protein levels using several different commercial antibodies, and we are currently exploring different avenues to address this important question. There may also be a role of altered flux through the pathway. For example, desmosterol accumulates when DHCR24 is silenced, as can be seen in the CHO-EV and mutant DHCR24 cells in Fig. 4B. Desmosterol or other sterols could influence DHCR7 activity. Another possibility is that DHCR24 influences DHCR7 allosterically. Further work is required to elucidate the precise mechanism through which DHCR24 and DHCR7 interact, and the role this plays in the overall regulation of the cholesterol synthesis pathway. These studies will be assisted as more structural information for DHCR24 (37) and DHCR7 (38) becomes available.
Supplementary Material
Acknowledgments
The authors thank Dr. Eser Zerenturk for generating the CHO-DHCR24 Y471S cell-line, Anika Prabhu for cloning the pcDNA5-DHCR7-myc/FRT plasmid, and Dr. Martin Bucknall for technical advice on GC-MS. The authors also thank members of the Brown Laboratory for critically reviewing this manuscript.
Footnotes
Abbreviations:
- BSTFA
- N,O-bis(trimethylsilyl)trifluoroacetamide
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]1-propanesulfonate
- CHO
- Chinese hamster ovary
- 7DHC
- 7-dehydrocholesterol
- DHCR7
- 7-dehydrocholesterol reductase
- DHCR24
- 24-dehydrocholesterol reductase
- DMEM/F12
- DMEM:Ham’s nutrient mixture F12 (1:1)
- ER
- endoplasmic reticulum
- EV
- empty vector
- FRT
- Flp recombinase target
- LPDS
- lipoprotein-deficient newborn calf serum
- PBGD
- porphobilinogen deaminase
The Brown Laboratory is supported by a University of New South Wales Goldstar Award and a grant from the National Health and Medical Research Council (1060515).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and two tables.
REFERENCES
- 1.Sharpe L. J., Brown A. J. 2013. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288: 18707–18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloch K. 1965. The biological synthesis of cholesterol. Science. 150: 19–28. [DOI] [PubMed] [Google Scholar]
- 3.Kandutsch A. A., Russell A. E. 1960. Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J. Biol. Chem. 235: 2256–2261. [PubMed] [Google Scholar]
- 4.Shrivastava S., Paila Y. D., Dutta A., Chattopadhyay A. 2008. Differential effects of cholesterol and its immediate biosynthetic precursors on membrane organization. Biochemistry. 47: 5668–5677. [DOI] [PubMed] [Google Scholar]
- 5.Glossmann H. H. 2010. Origin of 7-dehydrocholesterol (provitamin D) in the skin. J. Invest. Dermatol. 130: 2139–2141. [DOI] [PubMed] [Google Scholar]
- 6.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dumpelfeld B., et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature. 440: 631–636. [DOI] [PubMed] [Google Scholar]
- 7.Mo C., Bard M. 2005. A systematic study of yeast sterol biosynthetic protein-protein interactions using the split-ubiquitin system. Biochim. Biophys. Acta. 1737: 152–160. [DOI] [PubMed] [Google Scholar]
- 8.Taramino S., Teske B., Oliaro-Bosso S., Bard M., Balliano G. 2010. Divergent interactions involving the oxidosqualene cyclase and the steroid-3-ketoreductase in the sterol biosynthetic pathway of mammals and yeasts. Biochim. Biophys. Acta. 1801: 1232–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertin N., Simonis N., Dupuy D., Cusick M. E., Han J. D., Fraser H. B., Roth F. P., Vidal M. 2007. Confirmation of organized modularity in the yeast interactome. PLoS Biol. 5: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erce M. A., Pang C. N., Hart-Smith G., Wilkins M. R. 2012. The methylproteome and the intracellular methylation network. Proteomics. 12: 564–586. [DOI] [PubMed] [Google Scholar]
- 11.Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., FitzPatrick D. R., Kelley R. I., Wanders R. J. 2001. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 69: 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moebius F. F., Fitzky B. U., Lee J. N., Paik Y. K., Glossmann H. 1998. Molecular cloning and expression of the human delta7-sterol reductase. Proc. Natl. Acad. Sci. USA. 95: 1899–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerenturk E. J., Sharpe L. J., Ikonen E., Brown A. J. 2013. Desmosterol and DHCR24: unexpected new directions for a terminal step in cholesterol synthesis. Prog. Lipid Res. 52: 666–680. [DOI] [PubMed] [Google Scholar]
- 14.FitzPatrick D. R., Keeling J. W., Evans M. J., Kan A. E., Bell J. E., Porteous M. E., Mills K., Winter R. M., Clayton P. T. 1998. Clinical phenotype of desmosterolosis. Am. J. Med. Genet. 75: 145–152. [PubMed] [Google Scholar]
- 15.Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. 1989. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 264: 15634–15641. [PubMed] [Google Scholar]
- 16.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260. [DOI] [PubMed] [Google Scholar]
- 17.Krycer J. R., Kristiana I., Brown A. J. 2009. Cholesterol homeostasis in two commonly used human prostate cancer cell-lines, LNCaP and PC-3. PLoS ONE. 4: e8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 10: 237–245. [DOI] [PubMed] [Google Scholar]
- 19.Zerenturk E. J., Kristiana I., Gill S., Brown A. J. 2012. The endogenous regulator 24(S),25-epoxycholesterol inhibits cholesterol synthesis at DHCR24 (seladin-1). Biochim. Biophys. Acta. 1821: 1269–1277. [DOI] [PubMed] [Google Scholar]
- 20.Tseng W. C., Lin J. W., Wei T. Y., Fang T. Y. 2008. A novel megaprimed and ligase-free, PCR-based, site-directed mutagenesis method. Anal. Biochem. 375: 376–378. [DOI] [PubMed] [Google Scholar]
- 21.Luu W., Sharpe L. J., Stevenson J., Brown A. J. 2012. Akt acutely activates the cholesterogenic transcription factor SREBP-2. Biochim. Biophys. Acta. 1823: 458–464. [DOI] [PubMed] [Google Scholar]
- 22.Luu W., Zerenturk E. J., Kristiana I., Bucknall M. P., Sharpe L. J., Brown A. J. 2014. Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis. J. Lipid Res. 55: 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe L. J., Wong J., Garner B., Halliday G. M., Brown A. J. 2012. Is seladin-1 really a selective Alzheimer’s disease indicator? J. Alzheimers Dis. 30: 35–39. [DOI] [PubMed] [Google Scholar]
- 24.Prabhu A. V., Sharpe L. J., Brown A. J. 2014. The sterol-based transcriptional control of human 7-dehydrocholesterol reductase (DHCR7): Evidence of a cooperative regulatory program in cholesterol synthesis. Biochim. Biophys. Acta. 1842: 1431–1439. [DOI] [PubMed] [Google Scholar]
- 25.Du X., Kristiana I., Wong J., Brown A. J. 2006. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol. Biol. Cell. 17: 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielar D., Dietmaier W., Langmann T., Aslanidis C., Probst M., Naruszewicz M., Schmitz G. 2001. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin. Chem. 47: 2089–2097. [PubMed] [Google Scholar]
- 27.Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- 28.Hart-Smith G., Raftery M. J. 2012. Detection and characterization of low abundance glycopeptides via higher-energy C-trap dissociation and orbitrap mass analysis. J. Am. Soc. Mass Spectrom. 23: 124–140. [DOI] [PubMed] [Google Scholar]
- 29.Pruitt K. D., Brown G. R., Hiatt S. M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C. M., Hart J., Landrum M. J., McGarvey K. M., et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42: D756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabas I., Feinmark S. J., Beatini N. 1989. The reactivity of desmosterol and other shellfish- and xanthomatosis-associated sterols in the macrophage sterol esterification reaction. J. Clin. Invest. 84: 1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou L., Porter T. D. 2014. Rapid suppression of 7-dehydrocholesterol reductase activity in keratinocytes by vitamin D. J. Steroid Biochem. Mol. Biol. In press. [DOI] [PubMed] [Google Scholar]
- 32.Menard L., Maughan D., Vigoreaux J. 2014. The structural and functional coordination of glycolytic enzymes in muscle: evidence of a metabolon? Biology (Basel). 3: 623–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer F. M., Gerwig J., Hammer E., Herzberg C., Commichau F. M., Volker U., Stulke J. 2011. Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab. Eng. 13: 18–27. [DOI] [PubMed] [Google Scholar]
- 34.Williamson M. P., Sutcliffe M. J. 2010. Protein-protein interactions. Biochem. Soc. Trans. 38: 875–878. [DOI] [PubMed] [Google Scholar]
- 35.Belič A., Pompon D., Monostory K., Kelly D., Kelly S., Rozman D. 2013. An algorithm for rapid computational construction of metabolic networks: a cholesterol biosynthesis example. Comput. Biol. Med. 43: 471–480. [DOI] [PubMed] [Google Scholar]
- 36.Jansen M., Wang W., Greco D., Bellenchi G. C., di Porzio U., Brown A. J., Ikonen E. 2013. What dictates the accumulation of desmosterol in the developing brain? FASEB J. 27: 865–870. [DOI] [PubMed] [Google Scholar]
- 37.Zerenturk E. J., Sharpe L. J., Brown A. J. 2014. DHCR24 associates strongly with the endoplasmic reticulum beyond predicted membrane domains: implications for the activities of this multi-functional enzyme. Biosci. Rep. 34: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Roberti R., Blobel G. 2015. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature. 517: 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.